Simple Summary
Melanoma accounts for the majority of skin-cancer related deaths. An early and correct diagnosis is essential for proper patient care. However, melanoma is known to mimic a great number of skin diseases, which makes the diagnosis difficult for both the clinician and the pathologist. Using a proper panel of immunohistochemical markers can greatly improve the diagnostic process. In addition to the diagnostic markers, there are some immunohistochemical markers, like BRAF, PD-L1, and Ki67, that need to be included in the final pathological report because of their predictive and prognostic roles.
Abstract
Background: With its incidence on the rise, a high mortality rate, and great costs associated with its treatment, melanoma represents an important challenge for healthcare systems, clinicians, and pathologists. Therefore, an emphasis should be placed on its early and correct diagnosis, as well as the appropriate assessment of prognostic and predictive factors. Immunohistochemistry (IHC) is an ancillary test that can provide invaluable information for diagnosing melanoma, especially in complex cases. Objective: The aim of this systematic review is to gather the available information regarding the use of IHC markers in the diagnosis, differential diagnosis, prognosis, staging, and treatment of melanoma in a format that is easy to access for clinicians and pathologists. Methods: A comprehensive search of the literature was conducted and resulted in one hundred and forty-seven studies being included in this systematic review. The results were grouped thematically by specific IHC markers. Results: The IHC markers specific to melanocytic differentiation, like S100, SOX10, and Melan-A/MART1, were consistent across studies as being positive in most cases of melanoma, with rare exceptions. HMB-45 and PRAME can provide additional information, especially for differential diagnoses between benign and malignant melanocytic lesions. MITF, Ki67, BRAF, and PD-L1 are associated with prognosis factors, like the Breslow thickness, tumour ulceration, type of inflammatory infiltrate, and response to treatment. Conclusions: IHC markers are an invaluable tool for the diagnosis and differential diagnosis of melanoma, especially in cases that lack the characteristic histopathological aspects. In addition, IHC provides prognostic factors and can help in predicting the tumour’s response to various treatments.
1. Introduction
Melanoma is defined as a malignant tumour of melanocytes and represents the most aggressive type of skin cancer [1]. The incidence of melanoma has increased over the last several decades, especially in fair-skinned populations. GLOBOCAN 2022 reported an incidence of 331,647 new cases [2]: 7012 cases more than the previous GLOBOCAN report from 2020 [3]. Melanoma also accounts for the majority of skin-cancer-related deaths worldwide, posing a significant public health challenge [2]. Moreover, melanoma also represents a challenge for healthcare systems as the costs of treating patients with melanoma has increased over the years [4]. Considering the aggressive behaviour of melanoma, it is critical to ensure an early and correct diagnosis, as the prognosis declines sharply between the radial and vertical growth phase [5].
Immunohistochemistry (IHC) is an ancillary test used widely in the pathological evaluations of various tissue samples, including skin cancer, in order to detect specific antigens [6]. In clinical practice, IHC is mainly used on formalin-fixed, paraffin-embedded tissues and provides additional, sometimes invaluable, information to the classic haematoxylin–eosin (H&E) examination [7]. This greatly facilitates the differentiation of melanoma from benign nevi and other non-melanocytic tumours [8]. Melanoma is known to mimic a broad spectrum of tumours, both epithelial and mesenchymal. Despite this, there are also the cases of amelanotic or hypomelanotic melanomas which, by definition, lack the presence of melanin pigment, an important clue regarding the tumour’s line of differentiation [9]. Other potential diagnostic pitfalls could be represented by the histopathological variants of melanoma, desmoplastic melanoma, nevoid melanoma, or malignant blue nevus [10]. The use of key immunohistochemical markers, such as S100, SOX10, Melan-A (MART1), HMB-45, and MITF, have demonstrated varying degrees of sensitivity and specificity for the diagnosis of melanoma, assessment of prognostic factors, and guiding of therapeutic decisions [11,12].
Despite its widespread use, the application of IHC in melanomas has some limitations. Technical issues occurring during the staining protocol, the variability of marker immunoreactivity, and interobserver variability when evaluating the slides could impact the diagnostic accuracy [13]. Someof these limitations could be resolved by the use of digital pathology, which has emerged as a transformative approach to the classic analysis of IHC slides. Integratinghigh-resolution scanning and imageanalysis intothe standard interpretation of histopathological slides could increase the consistency of the results across observers and reduce subjectivity [14].
Considering the expanding role of IHC in the diagnosis and therapeutic management of melanoma, a comprehensive evaluation of key immunohistochemical markers, with their strengths and limitations, is warranted. The aim of this systematic review is to synthesisethe existing evidence regarding the use of IHC markers in the diagnosis and treatment of melanoma.
2. Materials and Methods
2.1. Protocol and Registration
This systematic review was conducted in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines and checklist in Supplementary Materials. The review protocol was not prospectively registered in PROSPERO or any other systematic review registry.
2.2. Eligibility Criteria
The inclusion criteria were defined prior to data collection and were as follows:
- Original peer-reviewed research articles, clinical case series, or case reports focusing on the use of immunohistochemistry in melanoma;
- Studies that employed immunohistochemistry for the purpose of the diagnosis, differential diagnosis, prognosis, staging, or treatment of melanoma;
- The inclusion of primary cutaneous melanoma, metastatic melanoma, or histopathological variants such as nodular melanoma, lentigo maligna melanoma, superficial spreading melanoma, amelanotic melanoma, spitzoid melanoma, desmoplastic melanoma, or acral melanoma;
- Articles published in English between January 2000 and May 2025.
The exclusion criteria included the following:
- Animal or in vitro studies;
- Editorials, letters, reviews, or opinion pieces;
- Studies focusing on melanomas in special locations (e.g., mucosal, uveal, ocular);
- Studies not involving immunohistochemistry;
- Articles without the full text available;
- Duplicate publications or secondary analyses from the same dataset.
2.3. Information Sources and Search Strategy
A comprehensive search was conducted in two major databases, Pubmed and Scopus. The searches were completed on May 2025. The search strategies combined keywords and subject headings related to melanoma, immunohistochemistry, and relevant clinical applications. Boolean operators (AND, OR) were used to combine the search terms appropriately. Representative search terms included the following:
- “melanoma” AND “immunohistochemistry”.
- “melanoma” AND “S100” OR “SOX10” OR “HMB-45” OR “Melan-A” OR “MART1” OR “KI67” OR “MITF” OR “PRAME” OR “BRAF” OR “p16” OR “PD-L1”.
- “melanoma variants” AND “immunohistochemistry”.
Filters were used to exclude non-human melanomas, in vitro studies, articles not available in English and reviews.
2.4. Study Selection
All the identified articles were imported into the Zotero screening database for reference management. Duplicate records were removed. A title and abstract screening was conducted independently by A.I.P. and V.M.V., followed by a full-text screening, to assess eligibility. Discrepancies were resolved by consultation with a third reviewer (M.C.).
In total, 904 records were identified by the database search, of which, 156 were duplicates. After duplicate removal, 748 records were screened by their title and abstract, out of which, 439 were excluded. Thereafter, 309 reports were sought for retrieval, and 231 reports were retrieved in full and assessed for their eligibility in accordance with the mentioned criteria. Reports were removed for the following reasons: the study does not focus on IHC (55 reports);the study does not focus on melanoma (17 reports);the study type is a letter to the editor, commentary, or review (7 reports); the study is on cell cultures (4 reports); or the study is not in English (1 report). After the full-text assessment, 147 reports were included in the systematic review. The complete selection process is illustrated in the PRISMA 2020 flow diagram (Figure 1).
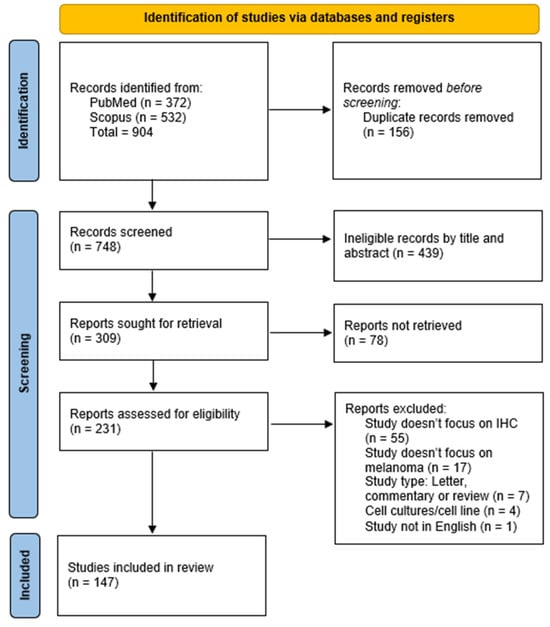
Figure 1.
PRISMA 2020 flowchart illustrating the study selection process.
Additional reports were included through the backward citation of the included articles.
2.5. Data Extraction
A standardised data extraction form was used to collect the following information from each included study:
- Melanoma subtype;
- IHC markers assessed;
- Diagnostic, differential diagnostic, staging, prognostic, or treatment applications.
2.6. Risk of Bias and Quality Assessment
No formal risk of bias tool was applied because of the heterogeneity of the studies included in terms of their design and reporting.
2.7. Data Synthesis
Due to the heterogeneity in the study designs, patient populations, and IHC markers assessed, a qualitative synthesis of findings was performed. The results were grouped thematically by specific IHC markers. No meta-analysis was conducted due to variability in the IHC markers included and the reporting standards.
3. Results
3.1. S100
S100, particularly the S100B isoform, is a nuclear marker that is broadly used in the diagnosis of primary cutaneous melanoma due to its high sensitivity for cells of melanocytic differentiation [15], with various studies reporting a sensitivity between 90% [11] and 100% for the diagnosis of primary cutaneous melanoma [16,17]. S100 is considered positive when staining is observed both in the cytoplasm and the nucleus (Figure 2).
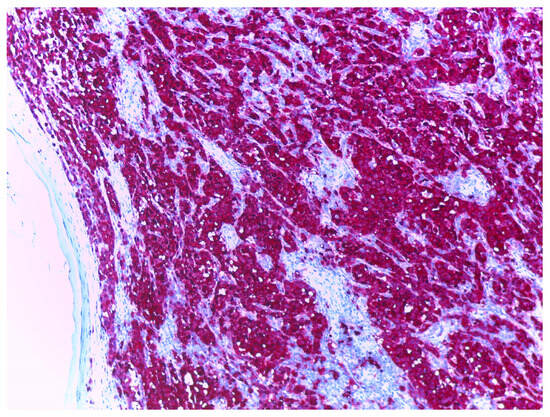
Figure 2.
Melanoma stained with S100, at 10× magnification. Nuclei and cytoplasm of melanoma tumour cells are highlighted in red, background stroma highlighted in blue.
In routine diagnostic histopathology, S100 is used among other markers of melanocytic differentiation, such as HMB-45 or Melan-A [15]. S100 positivity was reported in primary cutaneous melanomas [18,19,20], Spitzoid melanomas [21], amelanotic melanomas [22,23], nodular melanomas [24], and desmoplastic/spindle cell melanomas [25,26,27,28,29]. There are rare cases reported of S100-negative cutaneous melanomas [30], but more commonly encountered in the literature are the cases of desmoplastic melanoma that donot react with S100 [31]. Immunoreactivity is also maintained in metastases from cutaneous melanomas [32,33,34].
The tumour’s depth of invasion or the Breslow thickness is a critical prognostic marker and a part of the staging process of melanoma. It is measured in millimetres from the top of the granular layer of the epidermis or ulcer base, if present, to the deepest point of the tumour’s penetration and reported to the nearest 0.1 mm [35]. In most cases, the Breslow thickness is assessed on H&E evaluation. Using S100 when measuring the depth of invasion hasdemonstrated greater accuracy than H&E, as well as HMB-45 and Melan-A [36]. Evaluating the presence of lymph node metastases is another important prognostic factor thatis also used in staging melanomas. Lymph node metastases are usually assessed on H&E slides accurately. However, micrometastases are more challenging to detect. One study reported that only 30 to 45% percent of positive lymph nodes were correctly assessed on H&E slides [37], while in another study, 90% of positive lymph nodes were correctly identified with the use of S100 staining [38]. Drawbacks to the micrometastases evaluation by S100 staining include the nuclear expression of the marker, which hide nuclear details of tumoral cells and the reactivity of S100 with other cells from the lymph node, like dendritic cells [39]. The presence of lymphatic or blood vessel invasion is a negative prognostic marker, and its evaluation on H&E has demonstrated a sensitivity of 25% and a specificity of 100% [40]. Using dual staining with S100 and D2-40 demonstrated greater sensitivity(60%) in detecting the lymphatic or vascular invasion [40,41,42,43] factors correlated with a poor prognosis [44]. Other studies recommend pairing S100 with LYVE-1 for the detection of lymphatic or vascular invasion [45].
S100 is not a very specific marker for melanoma as it reacts with various other tumours, such as those derived from nerve sheath tumours, like neurofibromas, Schwannomas, and malignant peripheral nerve sheath tumours (MPNST) [28,46], as well as isolated spindle cells in cutaneous scars. Re-excision specimens occasionally contain rare, isolated, atypical spindle cells demonstrating S100 reactivity, raising the challenge of the differential diagnosis between desmoplastic melanoma and a cutaneous scar. Some authors consider these cells to be Schwann cells [47,48]. In addition, for the diagnosis of spindle cell or desmoplastic melanomas, there are studies that report p75 as being superior to S100 [49].
3.2. SOX10
SOX10 is a protein that is positively expressed in melanocytes, Schwann cells, myoepithelial cells, and the luminal cells of sweat glands and salivary glands [50]. In melanocytic lesions, SOX10 is expressed in benign melanocytic nevi [51], primary cutaneous melanomas [19], desmoplastic melanomas [52,53,54], both pure and mixed desmoplastic melanomas [55], and melanoma metastases [56]. SOX10 is highly sensitive and specific todesmoplastic melanoma. Challenges in the differential diagnosis of desmoplastic melanomas include spindle cell squamous cell carcinoma, atypical fibroxanthoma, MPNST, and sarcoma, which were included in a study that reported 100% of desmoplastic melanomas as being SOX10-positive, while all histological mimics were SOX10-negative. Only a few cases of MPNST demonstrated rare, isolated, positive cells [57]. However, others reported up to 49% of MPNSTs as being SOX10-positive, as well as diffuse expression in neurofibromas and Schwannomas [58,59]. There are also reported cases of SOX10-negative desmoplastic melanomas [31]. SOX10 is also expressed in breast carcinoma, which could pose another diagnostic challenge [51].
SOX10 is also expressed in histiocytes from dermal scars, which, in a manner similar to S100, could lead to diagnostic challenges, especially in cases of re-excision specimens. One study evaluated the morphology and immunoreactivity of histiocytes from dermal scars and reported that 71.3% had spindle cell morphology or angulated nuclei, while 86% presented with scattered SOX10-positive cells [60].
Measuring the Breslow thickness using SOX10 has demonstrated greater accuracy than H&E examination and no difference when compared with S100 assessment [36]. Regarding the status of lymph nodes, SOX10 has demonstrated greater accuracy than H&E, S100, Melan-A, and HMB45 in evaluating the presence of micrometastases, as it does not react with other background cells in the lymph node. One drawback of using SOX10 in lymph node sections is the staining of nodal nevi, which could raise a potential diagnostic challenge [61,62].
3.3. HMB-45
HMB-45 is a marker of melanocyte differentiation that is less sensitive, but more specific, than S100 for the diagnosis of primary cutaneous melanoma [15], with various studies reporting 56.3 to 77% of melanomas as being HMB-45-positive [11,63,64,65]. Desmoplastic melanomas are usually HMB-45-negative [25,26,27,52,66], with rare cases of positivity [63,67]. HMB-45 is also expressed in amelanotic melanomas (Figure 3) [22,68], including acral amelanotic melanomas [69], primary dermal melanomas [70], and melanoma metastases [63,71,72,73]. Despite being a marker of melanocytic differentiation, the specific pattern of expression allows it to be confidently used in the differential diagnosis between benign and malignant melanocytic lesions. The normal pattern shows HMB-45-positive cells at the junctional level and in the superficial layer of the dermis, while the deeper layers show weak positivity. This gradient is characteristically lost in melanomas, and HMB-45 becomes diffusely expressed [64]. Melanocytic lesions that show HMB-45 expressed in this gradient are more likely to be benign melanocytic lesions than melanomas [74]. HMB-45 is also expressed in blue nevi, Spitz nevi, congenital nevi, dysplastic nevi [64], and deep-penetrating nevi. Nevoid melanomas are usually completely negative in the dermal component [75]. In addition to melanocytic lesions, HMB-45 is also expressed in Schwannomas, angiomyolipomas, lymphangioleiomyomatosis, and clear cell “sugar”tumours [64].
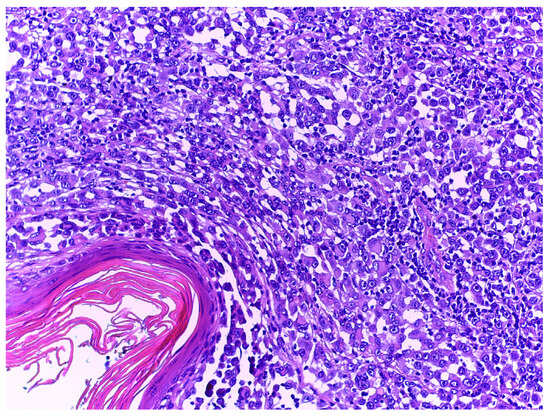
Figure 3.
Nodular amelanotic melanoma with H&E stain, at 20× magnification. Tumour cells demonstrate abundant cytoplasm with no melanotic pigment and irregular-shaped nuclei with evident nucleoli.
There are various results regarding the use of HMB-45 in evaluating the lymph node status. Some studies report that HMB-45 is not suitable as it also stains other cells from the lymph node [37], while others report 82% percent of micrometastasesbeing diagnosed with HMB-45 [38]. In addition to this, HMB-45 can differentiate between lymph node metastases and nodal nevi [62,76]. Measuring the Breslow thickness using HMB-45 stains demonstrated no difference in accuracy when compared with H&E [36,77].
3.4. Melan-A/MART1
Melan-A represents a melanocytic differentiation antigen encoded by the MART-1 gene and normally expressed in mature benign melanocytes. It is also expressed in benign melanocytic nevi and dysplastic nevi [63] with a diffuse and strong staining pattern [15]. Neurotized nevi demonstrated strong and diffuse positivity, in contrast to neurofibromas, whichare usually Melan-A-negative [78]. Primary cutaneous melanomas express Melan-A [79] in 73.3% to 83% of cases [11,63] and is also maintained in melanoma metastases in 71% of cases [63,80]. Desmoplastic melanomas are generally negative for Melan-A [57], with positivity reported in up to 27.3% of cases [67]. Desmoplastic nevi are more frequently positive for Melan-A [81]. Acral amelanotic melanomas also express Melan-A [82]. Some cases of metastatic lesions demonstrated a loss of Melan-Aafter treatment with vemurafenib [83].
Breslow thickness assessments on Melan-A stained slides demonstrated greater accuracy than H&E examination, without exceeding that of S100 [36]. One study reported 59.6% of melanoma biopsies being measured by Melan-A stain as having a greater depth of tumour invasion than was measured by H&E, which led to 33% cases of melanoma in situ to be re-classified as invasive melanoma [84]. Melan-A stain is also preferred in the assessment of the surgical margin during Mohs surgery [85]. It has demonstrated great sensitivity in frozen tissues, with good consistency, decreased background stain, and concordance with H&E margin evaluations on fixed tissue [86]. The lymph node status is accurately assessed by Melan-A stain, with 100% of negative lymph nodes and 98.2% of positive lymph nodes being correctly diagnosed. The percent increased to 100% when additional levels were cut [87], an accuracy level superior to both S100 and HMB-45 [39].
There are several limitations to the use of Melan-A in the primary and differential diagnosis of melanoma. Melan-A could potentially overestimate the number of melanocytes in sun-damaged skin [88]. Melan-A is also positive in “sugar” cell tumours of the lung, PEComa, and angiomyolipoma; however, these tumours usually present with a characteristic morphology that allows a differential diagnosis on H&E staining [89].
3.5. Ki67
Ki67 is a nuclear-DNA-bindingprotein used as both a diagnostic tool and a prognostic marker in melanoma. Ki67 expression is associated with melanoma progression. Benign melanocytic nevi typically exhibit a lower Ki67-labelling index than primary cutaneous melanomas, reflecting the increased proliferative activity [90,91]. The process of melanoma progression includes the transition from the radial growth phase to the vertical growth phase, a change marked by a shift in the behaviour of tumour cells. In the vertical growth phase, the cells are confined to the epidermis or the papillary dermis, and Ki67 is preferentially expressed in the epidermal component. Shifting to the vertical growth phase is characterised by a higher proliferation capacity and, therefore, a higher Ki67-labelling index [92]. In a similar manner, many studies report that melanoma metastases express a higher Ki67 index than primary tumours [90,93,94]. Also, desmoplastic melanomas express a higher proliferation index than desmoplastic nevi [66]. When assessing the Ki67 index in melanomas, it is recommended to perform a dual stain to ensure that Ki67 is positive in tumour cells (Figure 4).
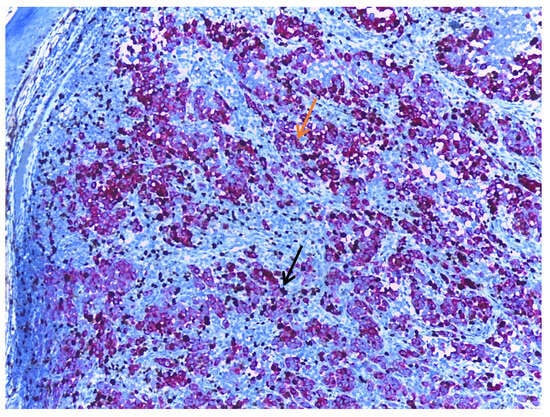
Figure 4.
Melanoma dual stained with Melan-A (orange arrow) and Ki67 (black arrow), at 10× magnification. When assessing the Ki67 index, it is recommended to use a dual stain, thus ensuring that the mitotic activity measured is in the tumour cells and not in the adjacent non-tumoral cells. The background stroma is stained blue.
From a prognostic perspective, higher Ki67 expression in melanoma has been associated with more-aggressive tumourbehaviour and a worse clinical outcome [90,93,95]. Studies have demonstrated that Ki67 expression is associated with the Breslow thickness [90,96]. When comparing the Ki67-labelling index between melanomas in situ and thin melanomas, no differences in proliferation activity were reported. However, metastasizing thin melanomas exhibit higher proliferation activity than non-metastasizing thin melanomas [97]. High Ki67 is also associated with the presence of ulceration, higher mitotic rates, the presence of vascular invasion, tumour necrosis, the Clark level [97], reduced overall survival [98], reduced melanoma-specific survival in acral melanomas [99], and reduced recurrence-free survival [100].
Ki67 assessment has several limitations. Interobserver variability with reduced reproducibility and differences in staining protocols can affect the consistency of results. Most laboratories report the percent of positive cells by eyeballing and using additional techniques, like the manual counting of printed images or automated counting by variousimage analysis software, with the latter greatly improving reproducibility [101].
3.6. MITF
MITF, or microphthalmia-associated transcription factor, is encoded by the MITF gene and expressed in melanocytes. In melanomas, MITF is expressed in 56% [65] to 100% of cases when excluding desmoplastic and spindle cells melanomas [102]. MITF seems to be sensitive and specific to epithelioid melanomas, but not for spindle cell and desmoplastic melanomas [103], with studies reporting 1–3% of desmoplastic melanomas as being MITF-positive [102,104]. MITF has exhibited equal or higher sensitivity and specificity than HMB-45 in the diagnosis of desmoplastic melanoma [105].In melanoma metastases, MITF expression is maintained in 23% [65] to 88% of cases [104]. As a marker of melanocytic differentiation, MITF is also expressed in benign melanocytic nevi, with studies reporting up to 83% of cases as positive [65], with similar results for Spitz nevi and dysplastic nevi [102].
MITF expression is associated with a good prognosis [98] and can be used in combination with D2-40 in order to detect lymphatic or vascular invasion [106]. Regarding differential diagnosis, MITF can be used to differentiate actinic keratosis with melanocyte hyperplasia from melanoma in situ with greater efficacy than SOX10 [107]. When evaluating melanocytes in sun-damaged skin, MITF melanocyte counting is more accurate than Melan-A, which tends to overestimate the melanocyte number. This derives from the nuclear expression of MITF compared with the cytoplasmic expression of Melan-A. Additionally, MITF can be used to evaluate the pagetoid spread [88].
3.7. PRAME
PRAME, or Preferentially-Expressed Antigen in Melanoma, is an IHC marker that is mainly used in the differential diagnosis of benign and malignant melanocytic lesions [108,109]. Positive staining is considered nuclear (Figure 5). Melanomas are traditionally diffusely positive for PRAME, which is defined as expression in >75% of tumour cells. Studies report various percentages of positivity for PRAME, from 59.1% up to 89.6% [64,110,111,112,113]. Its sensitivityhas been reported as 73.6% and its specificity as 96.5% [64]. Superficial spreading melanomas and nodular and lentigo maligna melanomas also express PRAME in 88.2–90.9% of cases [111]. Also, acral and subungual melanomas exhibit PRAME positivity [114,115]. Its sensitivity and specificity for subungual melanomas have been reported as 76.9% and 92.9%, respectively [116]. Spindle cell and desmoplastic melanomas are more frequently negative for PRAME staining, and in cases of mixed desmoplastic melanomas, staining is usually positive in the non-desmoplastic component [117]. There are rare cases of PRAME-negative melanomas, predominantly those with spindle-cell morphology [64,118]. PRAME is also expressed in melanomain situ cases [111] and can aid the early diagnosis of melanoma [119].
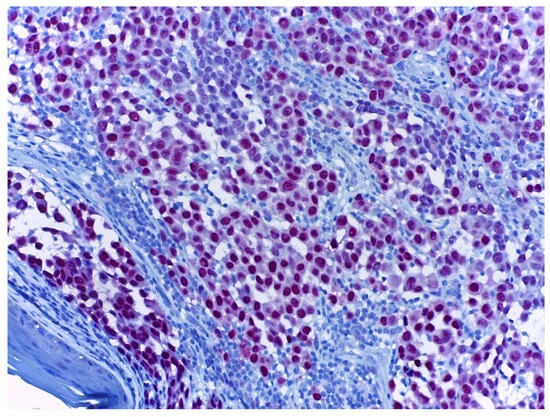
Figure 5.
Melanoma stained with PRAME (red), at 20× magnification. Melanoma tumour cells with red-stained nuclei demonstrate PRAME positivity.
When compared with S100 and SOX10, PRAME was demonstrated to be less sensitive but much more specific because of its limited expression in benign melanocytic lesions. Benign melanocytic nevi are usually negative for PRAME [64,120], with rare cases reported of scattered positive cells, especially at the junctional layer [121]. Spitz nevi, dysplastic nevi, and Reed nevi are also predominantly negative for PRAME, with rare cases of scattered positive cells [118]. In cases of melanoma developing on benign nevi, PRAME demonstrated positivity only in the melanoma component [122]. Metastases of primary cutaneous melanomas also exhibit PRAME expression [90]. Lymph node metastases exhibit PRAME in >50% of tumour cells, while nodal nevi can also exhibit PRAME positivity, usually in <50% of cells. In these cases, studies recommend pairing PRAME with p16 as the latter stains nodal nevi in >50% of cells [123]. One study reported a negative prognosis in correlation with PRAME staining [124].
Regarding differential diagnosis, PRAME can also differentiate between solar lentigo, non-lesional sun-damaged skin with melanocyte hyperplasia [125], melanocytic pseudonests in lichenoid reactions, and melanoma in situ [126]. PRAME results are in 90% concordance with FISH for melanoma and SNP-array [127].
This highly specific expression profile makes PRAME a valuable IHC stain in the histopathological examination of surgical margins [128], as normal adjacent or inflamed skin does not react with PRAME. This technique demonstrated equal, if not superior, results when compared with H&E assessment [117,129].
The limitations of PRAME use include positive expression in various other tumours, such as synovial sarcoma, myxoid liposarcoma, malignant peripheral nerve sheath tumours, breast cancer, renal cell carcinoma, and ovarian carcinoma [108].
3.8. BRAF
Around 50% of primary cutaneous melanomas show BRAF mutations, and out of these, 90% are a distinct mutant variant that forms by the replacement of valine by glutamic acid on exon 15 of the BRAF gene, named the BRAF V600E mutant variant [130]. The presence of the BRAF V600E mutant variant in melanoma is associated with several clinical characteristics. These mutations are more frequently encountered in melanomas that develop on the trunk, rather than the head and neck regions, or other sun-protected sites. This supports the theory of intermittent sun exposure in the pathophysiology of melanoma. Additionally, melanomas that develop on skin surfaces with signs of cumulative sun damage, like marked solar elastosis, show a low frequency of BRAF V600E mutations [131], but these cases show the less commonBRAF V600K mutation. This is characterised by the replacement of valine with lysine [132]. The BRAF V600K mutant melanoma is more frequently associated with older age, male gender [133], and scalp location [134] and have demonstrated more-aggressive behaviour than BRAF V600E, with an inferior response to treatment and shorter progression-free survival after a combination of BRAF and MEK inhibitors [135]. Melanomas harbouring BRAF V600E mutations tend to develop in younger patients, compared to the BRAF wild-type [136]. The BRAF V600E mutation is also associated with sentinel lymph node metastasis [137] but not with ulceration or the host immune response measured by TILs, as reported by some of the included studies. [138]. Spitzoid melanomas with BRAF mutations are also associated with worse clinical outcomes [139].
Other studies report that positive BRAF V600E expression is associated with the tumour thickness, the presence of ulceration, reduced overall survival [140], and non-brisk inflammation [141]. BRAF V600E mutations have also been reported in nodal nevi from patients with stage II melanoma. Out of these, 33% of patients also had positive lymph nodes in the same stations as the nodal nevi [76]. Metastatic melanomas usually present the same mutant variant as the primary tumour, with rare cases of discordance [142].
The presence of a BRAF mutation can confidently be assessed by IHC, having demonstrated comparable results with PCR and pyrosequencing [143]. One drawback to using IHC is the use of VE1 antibody that only recognises the BRAF V600E mutant variant [144,145].
BRAF V600E mutation acts as predictive marker for the response to specific treatments. Primary cutaneous melanomas that harbourthe mutation can respond to inhibitor treatments [146], like BRAF inhibitors, which include vemurafenib, dabrafenib, and encorafenib, and MEK inhibitors, like trametinib, binimetinib, and cobimetinib [147].
3.9. P16
P16 expression in primary cutaneous melanomas has exhibited variable behaviour, with some studies reporting all melanomas as being p16-negative [148], while others reported up to 72% positivity [149]. Because of this variable expression, p16 is more often used as an adjunct tool in challenging cases where histopathologic features do not clearly distinguish benign nevi from malignant melanoma. Benign nevi [112,150], as well as Spitz nevi [151,152] and congenital nevi, typically show moderate or strong staining for p16 [150,153]. In contrast, primary melanomas and metastatic lesions demonstrated a reductionor loss of p16 expression in a variety of cases, with exceptions as mentioned above [154]. Different expression patterns havealso been observed between the radial and vertical growth phases of melanoma [155], withstrong p16 expression in microinvasive dermal nests, marking the transition between the two phases [156]. Acral lentiginous melanomas retain p16 expression [157].
The loss of p16 expression is associated with a worse prognosis, reduced overall survival [158], vascular invasion, lymph node metastases, the recurrence of melanoma, metastases [159], and reduced inflammatory infiltrates in thetumoral microenvironment [160]. Regarding the Breslow thickness, the results are variable, with studies reporting an association between the loss of p16 and a greater depth of invasion [161], while others report no association [162]. In thin melanomas, p16 loss is associated with relapsing [163].
3.10. PD-L1
Programmed death-ligand 1 (PD-L1) is expressed on the surface of melanoma tumour cells and tumour-infiltrating immune cells. It binds to the PD-1 receptor on activated T-cells, leading to the suppression of the cytotoxic immune response. PD-L1 expression on tumour cells has been recognised as a negative prognostic marker as it correlates with the shorter 5-year survival rate, the Clark level, the presence of lymph node metastases [164], the mitotic rate, the Ki67 index, metastases, the presence of ulceration [165], and perineural invasion [166]. Other studies reported that PD-L1 expression in tumour cells is correlated with higher TIL infiltration in the tumour microenvironment [167]. Most cases of melanomas with PD-L1-positive expression develop on chronically sun-damaged skin [168]. Nodular-type melanomas are more frequently PD-L1-positive than superficial spreading, acral lentiginous, and lentigo malignamelanomas, even when adjusted for the tumour thickness [169]. Lymph node micrometastases express PD-L1 [170].
PD-L1 expression is an important marker in evaluating melanomas in the context of predicting their response to immune checkpoint inhibitors [171], like nivolumab or ipilimumab [172]. PD-L1 overexpression is associated with a better response to therapy [173]. PD-L1 expression is not the sole determinant of the immunotherapy response, and its level of expression is also affected by prior therapies and the presence of TILs. However, PD-L1 is still considered as a predictive marker in melanoma [174].
4. Conclusions
Melanoma represents a challenge from both the clinical and pathological point of view. With a high incidence that continues to increase, a high mortality rate, and enormous costs associated with treatment, this melanocytic malignancy is also a challenge for healthcare systems worldwide [2,4]. Thus, an emphasis must be placed on its early and correct diagnosis in order to improve patient’s prognosis. Melanoma is known as a great mimicker. From the clinician perspective, amelanotic or hypomelanotic types of melanomas could pose a significant challenge and delay a correct diagnosis and treatment. When evaluating the morphological characteristics, the list of possible lesions that could resemble melanoma and, thus, enter the differential diagnosis list is vast, especially when considering all the histologic variants of melanoma [9]. An especially difficult differential diagnosis for the pathologist is that between benign, atypical, and malignant melanocytic lesions, as the evolution, prognosis, and treatment of these entities differ considerably. Additionally, when confronted with a case of melanoma, it is not only important to correctly diagnose the case but also to include in the report all the prognostic and predictive factors [12]. This allows a clear view of the case, the prognosis of the patient, and the possible lines of treatment. Measuring the Breslow thickness; assessing vascular and perineural invasion; measuring and reporting surgical margins;and reporting the BRAF status, mitotic activity, presence or absence of ulceration, and the Clark level of invasion are all factors that need to be included in the final report of a melanoma diagnosis. All of these could potentially be assessed by classic H&E examination, but with greater interobserver variability, some factors, like perineural or vascular invasion, could be missed [175].
The potential differential diagnosis challenges and the assessment of prognostic and predictive factors could be greatly improved by using IHC staining. Using markers of melanocytic differentiation can establish the tumour’s cell line of differentiation, thus shortening the list of possible diagnoses. These are represented by S100, SOX10, and Melan-A/MART1 [37,176]. S100 is considered the most sensitive marker for melanoma, but its specificity is low, which limits its use as a singular marker. It is recommended, however, when paired with another IHC marker, usually HMB-45, which has demonstrated great specificity [176]. HMB-45 and PRAME are IHC markers to use when faced with a difficult diagnosis between benign and malignant melanocytic lesions. PRAME is expressed almost exclusively in malignant melanocytic lesions, while for HMB-45, the difference is made by its unique pattern of expression. The Ki67 proliferative index must be included in the final report, being a prognostic factor [94,177]. In the same manner, BRAF and PD-L1 must also be included in the report to establish the possible line of treatment, theirroles as predictive markers having been established in a great number of studies [168,178].
The list of IHC markers included in this review and their roles are summarised in Table 1.

Table 1.
The IHC markers included in the systematic reviews and the main findings for each.
Additional markers could be included in the IHC panel depending on the specific challenge the pathologist faces. MITF could provide important information when faced with the differential diagnosis of actinic keratosis with melanocyte hyperplasia and melanoma in situ, while p16 could help the diagnosis of a primary tumour and provides additional prognostic information [88,162].
The limitations of this systematic review include the heterogeneity of the study designs and methodologies, which make direct comparison difficult;a publication bias;the variability of IHC interpretation (manual vs. digital), which may affect the reproducibility and the advances in IHC technology over time, which may affect the relevance of the older studies included. In this context, performing a subgroup analysis based on methodological differences or more clearly defined inclusion criteria could ensure only the inclusion of studies reporting standardised scoring systems.
In conclusion, this systematic review proposes a comprehensive list of IHC markers for diagnosing melanoma and expands on their roles in the primary diagnosis, differential diagnosis, and assessment of prognostic and predictive factors. Future efforts may include focusing on studies with standardised scoring systems, which may also make it possible to conduct a meta-analysis.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/cancers17111769/s1, PRISMA 2020 checklist. Ref. [179] is cited in Supplementary Materials.
Author Contributions
Conceptualisation, V.-M.V., M.C., and A.-I.P.; research, V.-M.V., M.C., and A.-I.P.; image acquisition, M.C.; writing—original draft preparation, A.-I.P.; writing—review and editing, V.-M.V. and M.C.; All authors have read and agreed to the published version of the manuscript.
Funding
The authors acknowledge the support of the Foundation for Cellular and Molecular Medicine, Bucharest, in facilitating the publication of this review.
Data Availability Statement
No new data were created or analysed in this study. Data sharing is not applicable to this article.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Davis, L.E.; Shalin, S.C.; Tackett, A.J. Current State of Melanoma Diagnosis and Treatment. Cancer Biol. Ther. 2019, 20, 1366–1379. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global Cancer Statistics 2022: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Guy, G.P.; Machlin, S.R.; Ekwueme, D.U.; Yabroff, K.R. Prevalence and Costs of Skin Cancer Treatment in the U.S., 2002−2006 and 2007−2011. Am. J. Prev. Med. 2015, 48, 183–187. [Google Scholar] [CrossRef]
- Bellazzo, A.; Montico, B.; Guerrieri, R.; Colizzi, F.; Steffan, A.; Polesel, J.; Fratta, E. Unraveling the Role of Hypoxia-Inducible Factors in Cutaneous Melanoma: From Mechanisms to Therapeutic Opportunities. Cell Commun. Signal. 2025, 23, 177. [Google Scholar] [CrossRef]
- Lin, F.; Chen, Z. Standardization of Diagnostic Immunohistochemistry: Literature Review and Geisinger Experience. Arch. Pathol. Lab. Med. 2014, 138, 1564–1577. [Google Scholar] [CrossRef]
- Shi, S.R.; Key, M.E.; Kalra, K.L. Antigen Retrieval in Formalin-Fixed, Paraffin-Embedded Tissues: An Enhancement Method for Immunohistochemical Staining Based on Microwave Oven Heating of Tissue Sections. J. Histochem. Cytochem. 1991, 39, 741–748. [Google Scholar] [CrossRef]
- Chatterjee, D.; Bhattacharjee, R. Immunohistochemistry in Dermatopathology and Its Relevance in Clinical Practice. Indian Dermatol. Online J. 2018, 9, 234. [Google Scholar] [CrossRef]
- Ferringer, T. Immunohistochemistry in Dermatopathology. Arch. Pathol. Lab. Med. 2015, 139, 83–105. [Google Scholar] [CrossRef]
- Magro, C.M.; Crowson, A.N.; Mihm, M.C. Unusual Variants of Malignant Melanoma. Mod. Pathol. 2006, 19, S41–S70. [Google Scholar] [CrossRef]
- Viray, H.; Bradley, W.R.; Schalper, K.A.; Rimm, D.L.; Rothberg, B.E.G. Marginal and Joint Distributions of S100, HMB45 and Melan-A across a Large Series of Cutaneous Melanomas. Arch. Pathol. Lab. Med. 2013, 137, 1063–1073. [Google Scholar] [CrossRef] [PubMed]
- Pearlstein, M.V.; Zedek, D.C.; Ollila, D.W.; Treece, A.; Gulley, M.L.; Groben, P.A.; Thomas, N.E. Validation of the VE1 Immunostainfor the BRAF V600E Mutation in Melanoma. J. Cutan. Pathol. 2014, 41, 724–732. [Google Scholar] [CrossRef] [PubMed]
- Shakya, R.; Nguyen, T.H.; Waterhouse, N.; Khanna, R. Immune Contexture Analysis in Immuno-oncology: Applications and Challenges of Multiplex Fluorescent Immunohistochemistry. Clin. Transl. Immunol. 2020, 9, e1183. [Google Scholar] [CrossRef] [PubMed]
- Clarke, E.L.; Wade, R.G.; Magee, D.; Newton-Bishop, J.; Treanor, D. Image Analysis of Cutaneous Melanoma Histology: A Systematic Review and Meta-Analysis. Sci. Rep. 2023, 13, 4774. [Google Scholar] [CrossRef]
- Clarkson, K.S.; Sturdgess, I.C.; Molyneux, A.J. The Usefulness of Tyrosinase in the Immunohistochemical Assessment of Melanocytic Lesions: A Comparison of the Novel T311 Antibody (Anti-Tyrosinase) with S-100, HMB45, and A103 (Anti-Melan-A). J. Clin. Pathol. 2001, 54, 196–200. [Google Scholar] [CrossRef]
- Orchard, G. Evaluation of Melanocytic Neoplasms: Application of a Pan-Melanoma Antibody Cocktail. Br. J. Biomed. Sci. 2002, 59, 196–202. [Google Scholar] [CrossRef]
- Femel, J.; Hill, C.; Illa Bochaca, I.; Booth, J.L.; Asnaashari, T.G.; Steele, M.M.; Moshiri, A.S.; Do, H.; Zhong, J.; Osman, I.; et al. Quantitative Multiplex Immunohistochemistry Reveals Inter-Patient Lymphovascular and Immune Heterogeneity in Primary Cutaneous Melanoma. Front. Immunol. 2024, 15, 1328602. [Google Scholar] [CrossRef]
- Donati, M.; Nožička, J.; Kastnerova, L.; Hajkova, V.; Persichetti, P.; Michal, M.; Kazakov, D.V. Primary Cutaneous Amelanotic Melanoma and Gastrointestinal Stromal Tumor in Synchronous Evolution. Am. J. Dermatopathol. 2021, 43, 221–224. [Google Scholar] [CrossRef]
- Khan, S.; Al-Tariq, K.; Razi, S.; Haroon, A. Pyogenic Granuloma-like Amelanotic Melanoma of the Fingernail. Liaquat Natl. J. Prim. Care 2023, 5, 204–206. [Google Scholar] [CrossRef]
- Brown, G.T.; Cowen, E.W.; Lee, C.-C.R. Malignant Melanoma Masquerading as an Angiofibroma in a Patient with MEN-1. JAMA Dermatol. 2015, 151, 105–106. [Google Scholar] [CrossRef]
- Rekik, M.A.; Rouabeh, A.; Guermazi, Y.; Dahech, F.; Ellouz, Z.; Keskes, H. Spitzoid Melanoma of the Finger: A Case Report. J. Med Case Res. 2024, 18, 413. [Google Scholar] [CrossRef] [PubMed]
- Jia, J.; Wang, M.; Song, L.; Feng, Y. A Melanotic Malignant Melanoma Presenting as a Keloid: A Case Report. Medicine 2017, 96, e9047. [Google Scholar] [CrossRef] [PubMed]
- Rao, A.G.; Babu, V.A.; Koppada, D.; Haritha, M.; Chandana, P. Amelanotic Melanoma in the Vicinity of Acquired Melanocytic Nevi and Not Arising from Agminated Melanocytic Nevi: Masquerading as Pyogenic Granuloma. Indian J. Dermatol. 2016, 61, 122. [Google Scholar] [CrossRef] [PubMed]
- Alshaghel, M.M.; Almahairi, L.; Arian, R.; Alyousfi, M.S.; Majanni, W.; Alyousfi, R.; Etr, A. Amelanotic Nodular Melanoma Misdiagnosed as a Benign Skin Lesion: A Rare Case Report from Syria. Ann. Med. Surg. 2022, 74, 103316. [Google Scholar] [CrossRef]
- Weissinger, S.E.; Keil, P.; Silvers, D.N.; Klaus, B.M.; Möller, P.; Horst, B.A.; Lennerz, J.K. A Diagnostic Algorithm to Distinguish Desmoplastic from Spindle Cell Melanoma. Mod. Pathol. Off. J. U. S. Can. Acad. Pathol. Inc. 2014, 27, 524–534. [Google Scholar] [CrossRef]
- Marques, P.C.; Diniz, L.M.; Spelta, K.; Nogueira, P.S.E. Desmoplastic Melanoma: A Rare Variant with Challenging Diagnosis. An. Bras. Dermatol. 2019, 94, 82–85. [Google Scholar] [CrossRef]
- Andreevscaia, O.; Theate, I.; Goossens, C.; Vanhooteghem, O. Diagnostic Challenge of Desmoplastic Melanoma. Rare Tumors 2016, 8, 33–35. [Google Scholar] [CrossRef]
- Xu, X.; Chu, A.Y.; Pasha, T.L.; Elder, D.E.; Zhang, P.J. Immunoprofile of MITF, Tyrosinase, Melan-A, and MAGE-1 in HMB45-Negative Melanomas. Am. J. Surg. Pathol. 2002, 26, 82–87. [Google Scholar] [CrossRef]
- Javabal, P.; Subramanian, V. An Unusual Case of Desmoplastic Malignant Melanoma. J. Cutan. Aesthetic Surg. 2015, 8, 60–63. [Google Scholar] [CrossRef]
- Biernacka, A.; Linos, K.; Delong, P.; Suriawinata, A.; Padmanabhan, V.; Liu, X. A Case of S-100 Negative Melanoma: A Diagnostic Pitfall in the Workup of a Poorly Differentiated Metastatic Tumor of Unknown Origin. Cytojournal 2016, 13, 21. [Google Scholar] [CrossRef]
- Kooper-Johnson, S.; Mahalingam, M.; Loo, D.S. SOX-10 and S100 Negative Desmoplastic Melanoma: Apropos a Diagnostically Challenging Case. Am. J. Dermatopathol. 2020, 42, 697–699. [Google Scholar] [CrossRef] [PubMed]
- Chung, J.P.; Lee, S.J.; Lee, J.I.; Lee, K.S.; Lee, S.I. Duodenal Metastatic Amelanotic Melanoma. Gastrointest. Endosc. 2003, 58, 101. [Google Scholar] [PubMed]
- Heinig, J.; August, C.; Beckmann, V.; Konieczny, A. Endometrial Metastasis of Cutaneous Melanoma—A Case-Report Bearing Diagnostic Difficulties. Zentralbl. Gynakol. 2001, 123, 534–535. [Google Scholar] [CrossRef] [PubMed]
- Alves, M.G.O.; Chagas, L.R.; Carvalho, Y.R.; Cabral, L.A.G.; Coletta, R.D.; Almeida, J.D. Metastatic Melanoma of the Tongue: A Case Report with Immunohistochemical Profile. Gerodontology 2014, 31, 314–319. [Google Scholar] [CrossRef]
- Gershenwald, J.E.; Scolyer, R.A.; Hess, K.R.; Sondak, V.K.; Long, G.V.; Ross, M.I.; Lazar, A.J.; Faries, M.B.; Kirkwood, J.M.; McArthur, G.A.; et al. Melanoma Staging: Evidence-based Changes in the American Joint Committee on Cancer Eighth Edition Cancer Staging Manual. CA Cancer J. Clin. 2017, 67, 472–492. [Google Scholar] [CrossRef]
- Pop, A.M.; Monea, M.; Olah, P.; Moraru, R.; Cotoi, O.S. The Importance of Immunohistochemistry in the Evaluation of Tumor Depth of Primary Cutaneous Melanoma. Diagnostics 2023, 13, 1020. [Google Scholar] [CrossRef]
- Shidham, V.B.; Qi, D.; Rao, R.N.; Acker, S.M.; Chang, C.-C.; Kampalath, B.; Dawson, G.; Machhi, J.K.; Komorowski, R.A. Improved Immunohistochemical Evaluation of Micrometastases in Sentinel Lymph Nodes of Cutaneous Melanoma with ’MCW Melanoma Cocktail’--a Mixture of MonoclonalAntibodies to MART-1, Melan-A, and Tyrosinase. BMC Cancer 2003, 3, 15. [Google Scholar] [CrossRef]
- Abrahamsen, H.N.; Hamilton-Dutoit, S.J.; Larsen, J.; Steiniche, T. Sentinel Lymph Nodes in Malignant Melanoma: Extended Histopathologic Evaluation Improves Diagnostic Precision. Cancer 2004, 100, 1683–1691. [Google Scholar] [CrossRef]
- Shidham, V.B.; Qi, D.Y.; Acker, S.; Kampalath, B.; Chang, C.-C.; George, V.; Komorowski, R. Evaluation of Micrometastases in Sentinel Lymph Nodes of Cutaneous Melanoma: Higher Diagnostic Accuracy with Melan-A and MART-1 Compared With S-100 Protein and HMB-45. Am. J. Surg. Pathol. 2001, 25, 1039–1046. [Google Scholar] [CrossRef]
- Straker, R.J., 3rd; Taylor, L.A.; Neuwirth, M.G.; Sinnamon, A.J.; Shannon, A.B.; Abbott, J.; Miura, J.T.; Chu, E.Y.; Xu, X.; Karakousis, G.C. Optimizing Detection of Lymphatic Invasion in Primary Cutaneous Melanoma with the Use of D2-40 and a Paired Melanocytic Marker. Am. J. Dermatopathol. 2022, 44, 21–27. [Google Scholar] [CrossRef]
- Moy, A.P.; Duncan, L.M.; Kraft, S. Lymphatic Invasion and Angiotropism in Primary Cutaneous Melanoma. Lab. Invest. 2017, 97, 118–129. [Google Scholar] [CrossRef] [PubMed]
- Špirić, Z.; Erić, M.; Eri, Ž. Lymphatic Invasion and the Shields Index in Predicting Melanoma Metastases. J. Plast. Reconstr. Aesthetic Surg. 2017, 70, 1646–1652. [Google Scholar] [CrossRef] [PubMed]
- Petitt, M.; Allison, A.; Shimoni, T.; Uchida, T.; Raimer, S.; Kelly, B. Lymphatic Invasion Detected by D2-40/S-100 Dual Immunohistochemistry Does Not Predict Sentinel Lymph Node Status in Melanoma. J. Am. Acad. Dermatol. 2009, 61, 819–828. [Google Scholar] [CrossRef] [PubMed]
- Bayram, A.; Ozturk Sari, S.; Ozluk, Y.; Tas, F.; Buyukbabani, N. Multiple Combinations of Melanocytic and Vascular Endothelial Markers Enhance the Detection Rate of Lymphovascular Invasion in Cutaneous Melanoma. J. Cutan. Pathol. 2021, 48, 472–478. [Google Scholar] [CrossRef]
- Sahni, D.; Robson, A.; Orchard, G.; Szydlo, R.; Evans, A.V.; Russell-Jones, R. The Use of LYVE-1 Antibody for Detecting Lymphatic Involvement in Patients with Malignant Melanoma or Known Sentinel Node Status. J. Clin. Pathol. 2005, 58, 715–721. [Google Scholar] [CrossRef]
- Ireland, A.; Williams, B.; Rijhumal, A.; Mesbah Ardakani, N. Metastatic Melanoma to a Neurofibroma. Am. J. Dermatopathol. 2022, 44, 683–686. [Google Scholar] [CrossRef]
- Robson, A.; Allen, P.; Hollowood, K. S100 Expression in Cutaneous Scars: A Potential Diagnostic Pitfall in the Diagnosis of Desmoplastic Melanoma. Histopathology 2001, 38, 135–140. [Google Scholar] [CrossRef]
- Chorny, J.A.; Barr, R.J. S100-Positive Spindle Cells in Scars: A Diagnostic Pitfall in the Re-Excision of Desmoplastic Melanoma. Am. J. Dermatopathol. 2002, 24, 309–312. [Google Scholar] [CrossRef]
- Lazova, R.; Tantcheva-Poor, I.; Sigal, A.C. P75 Nerve Growth Factor Receptor Staining Is Superior to S100 in Identifying Spindle Cell and Desmoplastic Melanoma. J. Am. Acad. Dermatol. 2010, 63, 852–858. [Google Scholar] [CrossRef]
- Ordóñez, N.G. Value of SOX10 Immunostaining in Tumor Diagnosis. Adv. Anat. Pathol. 2013, 20, 275–283. [Google Scholar] [CrossRef]
- Mohamed, A.; Gonzalez, R.S.; Lawson, D.; Wang, J.; Cohen, C. SOX10 Expression in Malignant Melanoma, Carcinoma, and Normal Tissues. Appl. Immunohistochem. Mol. Morphol. 2013, 21, 506–510. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.A.; Zaheer, S.; Gulati, P.; Ahuja, S. Desmoplastic Melanoma of the Chest Wall: A Diagnostic Dilemma. Indian J. Surg. Oncol. 2024, 15, 164–167. [Google Scholar] [CrossRef] [PubMed]
- LaMonica, L.C.; Lang Houser, M.E.; Smith, E.H. Desmoplastic Melanoma Presenting as an Alopecic Patch in a Young Patient. JAAD Case Rep. 2023, 38, 96–99. [Google Scholar] [CrossRef] [PubMed]
- Kiuru, M.; McDermott, G.; Berger, M.; Halpern, A.C.; Busam, K.J. Desmoplastic Melanoma with Sarcomatoid Dedifferentiation. Am. J. Surg. Pathol. 2014, 38, 864–870. [Google Scholar] [CrossRef]
- Miller, D.D.; Emley, A.; Yang, S.; Richards, J.E.; Lee, J.E.; Deng, A.; Hoang, M.P.; Mahalingam, M. Mixed versus Pure Variants of Desmoplastic Melanoma: A Genetic and Immunohistochemical Appraisal. Mod. Pathol. Off. J. U. S. Can. Acad. Pathol. Inc. 2012, 25, 505–515. [Google Scholar] [CrossRef]
- Mikkelsen, J.; Hagen Wagenblast, A.L.; Behrendt, N.; Lock-Andersen, J. Melanoma in Situ with In-Transit Metastases. Open 2017, 11, 37–42. [Google Scholar] [CrossRef]
- Palla, B.; Su, A.; Binder, S.; Dry, S. SOX10 Expression Distinguishes Desmoplastic Melanoma from Its Histologic Mimics. Am. J. Dermatopathol. 2013, 35, 576–581. [Google Scholar] [CrossRef]
- Nonaka, D.; Chiriboga, L.; Rubin, B.P. Sox10: A Pan-Schwannian and Melanocytic Marker. Am. J. Surg. Pathol. 2008, 32, 1291–1298. [Google Scholar] [CrossRef]
- Moye, S.L.; Knowles, A.; Vandergriff, T.; Le, L.Q. Malignant Melanoma Developing in a Pre-Existing Cutaneous Neurofibroma from a Patient with Neurofibromatosis Type 1. JAAD Case Rep. 2024, 54, 46–49. [Google Scholar] [CrossRef]
- Behrens, E.L.; Boothe, W.; D’Silva, N.; Walterscheid, B.; Watkins, P.; Tarbox, M. SOX-10 Staining in Dermal Scars. J. Cutan. Pathol. 2019, 46, 579–585. [Google Scholar] [CrossRef]
- Ronchi, A.; Zito Marino, F.; Toni, G.; Pagliuca, F.; Russo, D.; Signoriello, G.; Moscarella, E.; Brancaccio, G.; Argenziano, G.; Franco, R.; et al. Diagnostic Performance of Melanocytic Markers for Immunocytochemical Evaluation of Lymph-Node Melanoma Metastases on Cytological Samples. J. Clin. Pathol. 2022, 75, 45–49. [Google Scholar] [CrossRef] [PubMed]
- Willis, B.C.; Johnson, G.; Wang, J.; Cohen, C. SOX10: A Useful Marker for Identifying Metastatic Melanoma in Sentinel Lymph Nodes. Appl. Immunohistochem. Mol. Morphol. 2015, 23, 109–112. [Google Scholar] [CrossRef]
- Orchard, G.E. Comparison of Immunohistochemical Labelling of Melanocyte Differentiation Antibodies Melan-A, Tyrosinase and HMB 45 with NKIC3 and S100 Protein in theEvaluation of Benign Naevi and Malignant Melanoma. Histochem. J. 2000, 32, 475–481. [Google Scholar] [CrossRef] [PubMed]
- Rasic, D.; Korsgaard, N.; Marcussen, N.; Precht Jensen, E.M. Diagnostic Utility of Combining PRAME and HMB-45 Stains in Primary Melanocytic Tumors. Ann. Diagn. Pathol. 2023, 67, 152211. [Google Scholar] [CrossRef] [PubMed]
- Nazarian, R.M.; Prieto, V.G.; Elder, D.E.; Duncan, L.M. Melanoma Biomarker Expression in Melanocytic Tumor Progression: A Tissue Microarray Study. J. Cutan. Pathol. 2010, 37 (Suppl. S1), 41–47. [Google Scholar] [CrossRef]
- Kucher, C.; Zhang, P.J.; Pasha, T.; Elenitsas, R.; Wu, H.; Ming, M.E.; Elder, D.E.; Xu, X. Expression of Melan-A and Ki-67 in Desmoplastic Melanoma and Desmoplastic Nevi. Am. J. Dermatopathol. 2004, 26, 452–457. [Google Scholar] [CrossRef]
- Chu, S.; Schrom, K.P.; Tripathi, R.; Conic, R.R.Z.; Ezaldein, H.H.; Scott, J.F.; Honda, K. Pure and Mixed Desmoplastic Melanomas: A Retrospective Clinicopathologic Comparison of 33 Cases. Am. J. Dermatopathol. 2021, 43, 776–780. [Google Scholar] [CrossRef]
- Mohammed Saeed, D.; Braniecki, M.; Groth, J.V. A Rare Case of Acral Amelanotic Melanoma, Nodular Type. Int. Wound J. 2019, 16, 1445–1449. [Google Scholar] [CrossRef]
- Zhang, J.; Yang, H.; Lin, J.; Zhang, F.; Shi, J.; Chen, R. Primary Acral Amelanotic Melanoma: A Rare Case Report. Mol. Clin. Oncol. 2020, 13, 1–4. [Google Scholar] [CrossRef]
- Cassarino, D.S.; Cabral, E.S.; Kartha, R.V.; Swetter, S.M. Primary Dermal Melanoma: Distinct Immunohistochemical Findings and Clinical Outcome Compared with Nodular and Metastatic Melanoma. Arch. Dermatol. 2008, 144, 49–56. [Google Scholar] [CrossRef]
- Gómez Portilla, A.; Cruz, A.; Juan, N.; Malo, P.; de Heredia, E.L.; Larrañaga, M. Isolated Rectus Abdominis Metastasis from Melanoma—An Extremely Rare Case. Int. J. Surg. Case Rep. 2016, 26, 121–123. [Google Scholar] [CrossRef] [PubMed]
- Amadeu, J.; Piazzetta, C.M.; Torres-Pereira, C.C.; Amenábar, J.M. Mandibular Metastasis of Cutaneous Melanoma. J. Oral Biol. Craniofacial Res. 2016, 6, 250–252. [Google Scholar] [CrossRef] [PubMed]
- Karateke, A.; Tuǧ, N.; Şahin, D. Metastatic Ovarian Malignant Melanoma with No Obvious Primary. J. Turk. Ger. Gynecol. Assoc. 2011, 12, 181–182. [Google Scholar] [CrossRef] [PubMed]
- Garola, R.; Singh, V. Utility of P16-Ki-67-HMB45 Score in Sorting Benign from Malignant Spitz Tumors. Pathol. Res. Pract. 2019, 215, 152550. [Google Scholar] [CrossRef]
- Ricci, C.; Dika, E.; Ambrosi, F.; Lambertini, M.; Veronesi, G.; Barbara, C. Cutaneous Melanomas: A Single Center Experience on the Usage of Immunohistochemistry Applied for the Diagnosis. Int. J. Mol. Sci. 2022, 23, 5911. [Google Scholar] [CrossRef]
- Siroy, A.E.; Aung, P.P.; Torres-Cabala, C.A.; Tetzlaff, M.T.; Nagarajan, P.; Milton, D.R.; Curry, J.L.; Ivan, D.; Prieto, V.G. Clinical Significance of BRAF V600E Mutational Status in Capsular Nevi of Sentinel Lymph Nodes in Patients with Primary Cutaneous Melanoma. Hum. Pathol. 2017, 59, 48–54. [Google Scholar] [CrossRef]
- Kamyab-Hesary, K.; Ghanadan, A.; Balighi, K.; Mousavinia, S.F.; Nasimi, M. Immunohistochemical Staining in the Assessment of Melanoma Tumor Thickness. Pathol. Oncol. Res. POR 2020, 26, 885–891. [Google Scholar] [CrossRef]
- Chen, Y.; Klonowski, P.W.; Lind, A.C.; Lu, D. Differentiating Neurotized Melanocytic Nevi from Neurofibromas Using Melan-A (MART-1) Immunohistochemical Stain. Arch. Pathol. Lab. Med. 2012, 136, 810–815. [Google Scholar] [CrossRef]
- Agostini, P.; Rivero, A.; Parra Martín, J.A.; Soares-de-Almeida, L. Pedunculated Polypoid Melanoma. A Case Report of a Rare Spindle-Cell Variant of Melanoma. Dermatol. Online J. 2015, 21, 13030. [Google Scholar] [CrossRef]
- Di Buono, G.; Maienza, E.; Rinaldi, G.; Buscemi, S.; Romano, G.; Agrusa, A. Malignant Metastatic Melanoma to the Gallbladder: Report of a Peculiar Case. Int. J. Surg. Case Rep. 2020, 77, S37–S39. [Google Scholar] [CrossRef]
- Sidiropoulos, M.; Sholl, L.M.; Obregon, R.; Guitart, J.; Gerami, P. Desmoplastic Nevus of Chronically Sun-Damaged Skin: An Entity to Be Distinguished from Desmoplastic Melanoma. Am. J. Dermatopathol. 2014, 36, 629–634. [Google Scholar] [CrossRef] [PubMed]
- Matusiak, L.; Bieniek, A.; Wozniak, Z.; Szepietowski, J.C. Amelanotic Malignant Melanoma in an Acral Location. Acta Dermatovenerol. Alp. Pannonica Adriat. 2008, 17, 72–74. [Google Scholar] [PubMed]
- Powell, M.R.; Sheehan, D.J.; Kleven, D.T. Altered Morphology and Immunohistochemical Characteristics in Metastatic Malignant Melanoma After Therapy with Vemurafenib. Am. J. Dermatopathol. 2016, 38, e137–e139. [Google Scholar] [CrossRef] [PubMed]
- Drabeni, M.; Lopez-Vilaró, L.; Barranco, C.; Trevisan, G.; Gallardo, F.; Pujol, R.M. Differences in Tumor Thickness between Hematoxylin and Eosin and Melan-A Immunohistochemically Stained Primary Cutaneous Melanomas. Am. J. Dermatopathol. 2013, 35, 56–63. [Google Scholar] [CrossRef]
- Ellison, P.M.; Zitelli, J.A.; Brodland, D.G. Mohs Micrographic Surgery for Melanoma: A Prospective Multicenter Study. J. Am. Acad. Dermatol. 2019, 81, 767–774. [Google Scholar] [CrossRef]
- Young, J.N.; Nguyen, T.A.; Freeman, S.C.; Hill, E.; Johnson, M.; Gharavi, N.; Bar, A.; Leitenberger, J. Permanent Section Margin Concordance after Mohs Micrographic Surgery with Immunohistochemistry for Invasive Melanoma and Melanoma in Situ: A RetrospectiveDual-Center Analysis. J. Am. Acad. Dermatol. 2023, 88, 1060–1065. [Google Scholar] [CrossRef]
- Gill, P.; Howell, J.; Naugler, C.; Daoud, M.S.A. Utility of Multistep Protocols in the Analysis of Sentinel Lymph Nodes in Cutaneous Melanoma: An Assessment of 194 Cases. Arch. Pathol. Lab. Med. 2019, 143, 1126–1130. [Google Scholar] [CrossRef]
- Nybakken, G.E.; Sargen, M.; Abraham, R.; Zhang, P.J.; Ming, M.; Xu, X. MITF Accurately Highlights Epidermal Melanocytes in Atypical Intraepidermal Melanocytic Proliferations. Am. J. Dermatopathol. 2013, 35, 25–29. [Google Scholar] [CrossRef]
- Huttenbach, Y.; Prieto, V.G.; Reed, J.A. Desmoplastic and Spindle Cell Melanomas Express Protein Markers of the Neural Crest but Not of Later Committed Stages of Schwann Cell Differentiation. J. Cutan. Pathol. 2002, 29, 562–568. [Google Scholar] [CrossRef]
- Parra, O.; Ma, W.; Li, Z.; Coffing, B.N.; Linos, K.; LeBlanc, R.E.; Momtahen, S.; Sriharan, A.; Cloutier, J.M.; Wells, W.A.; et al. PRAME Expression in Cutaneous Melanoma Does Not Correlate with Disease-Specific Survival. J. Cutan. Pathol. 2023, 50, 903–912. [Google Scholar] [CrossRef]
- Jian, J.; Guoying, W.; Jing, Z. Increased Expression of Sex Determining Region Y-Box 11 (SOX11) in Cutaneous Malignant Melanoma. J. Int. Med. Res. 2013, 41, 1221–1227. [Google Scholar] [CrossRef] [PubMed]
- Gimotty, P.A.; Van Belle, P.; Elder, D.E.; Murry, T.; Montone, K.T.; Xu, X.; Hotz, S.; Raines, S.; Ming, M.E.; Wahl, P.; et al. Biologic and Prognostic Significance of Dermal Ki67 Expression, Mitoses, and Tumorigenicity in Thin Invasive Cutaneous Melanoma. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2005, 23, 8048–8056. [Google Scholar] [CrossRef] [PubMed]
- Ladstein, R.G.; Bachmann, I.M.; Straume, O.; Akslen, L.A. Ki-67 Expression Is Superior to Mitotic Count and Novel Proliferation Markers PHH3, MCM4 and Mitosin as a Prognostic Factor in Thick Cutaneous Melanoma. BMC Cancer 2010, 10, 140. [Google Scholar] [CrossRef] [PubMed]
- Uguen, A.; Talagas, M.; Costa, S.; Duigou, S.; Bouvier, S.; De Braekeleer, M.; Marcorelles, P. A P16-Ki-67-HMB45 Immunohistochemistry Scoring System as an Ancillary Diagnostic Tool in the Diagnosis of Melanoma. Diagn. Pathol. 2015, 10, 195. [Google Scholar] [CrossRef]
- Nielsen, P.S.; Riber-Hansen, R.; Jensen, T.O.; Schmidt, H.; Steiniche, T. Proliferation Indices of Phosphohistone H3 and Ki67: Strong Prognostic Markers in a Consecutive Cohort with Stage I/II Melanoma. Mod. Pathol. Off. J. U. S. Can. Acad. Pathol. Inc. 2013, 26, 404–413. [Google Scholar] [CrossRef]
- Lawrence, N.F.; Hammond, M.R.; Frederick, D.T.; Su, Y.; Dias-Santagata, D.; Deng, A.; Selim, M.A.; Mahalingam, M.; Flaherty, K.T.; Hoang, M.P. Ki-67, P53, and P16 Expression, and G691S RET Polymorphism in Desmoplastic Melanoma (DM): A Clinicopathologic Analysis of Predictors of Outcome. J. Am. Acad. Dermatol. 2016, 75, 595–602. [Google Scholar] [CrossRef]
- Kaufmann, C.; Kempf, W.; Mangana, J.; Cheng, P.; Emberger, M.; Lang, R.; Kaiser, A.K.; Lattmann, E.; Levesque, M.; Dummer, R.; et al. The Role of Cyclin D1 and Ki-67 in the Development and Prognostication of Thin Melanoma. Histopathology 2020, 77, 460–470. [Google Scholar] [CrossRef]
- Jurmeister, P.; Bockmayr, M.; Treese, C.; Stein, U.; Lenze, D.; Jöhrens, K.; Friedling, F.; Dietel, M.; Klauschen, F.; Marsch, W.; et al. Immunohistochemical Analysis of Bcl-2, Nuclear S100A4, MITF and Ki67 for Risk Stratification of Early-Stage Melanoma—A Combined IHC Score for Melanoma Risk Stratification. J. Dtsch. Dermatol. Ges. 2019, 17, 800–808. [Google Scholar] [CrossRef]
- Du, Y.; Li, C.; Mao, L.; Wei, X.; Bai, X.; Chi, Z.; Cui, C.; Sheng, X.; Lian, B.; Tang, B.; et al. A Nomogram Incorporating Ki67 to Predict Survival of Acral Melanoma. J. Cancer Res. Clin. Oncol. 2023, 149, 13077–13085. [Google Scholar] [CrossRef]
- Väisänen, A.; Kuvaja, P.; Kallioinen, M.; Turpeenniemi-Hujanen, T. A Prognostic Index in Skin Melanoma through the Combination of Matrix Metalloproteinase-2, Ki67, and P53. Hum. Pathol. 2011, 42, 1103–1111. [Google Scholar] [CrossRef]
- Volynskaya, Z.; Mete, O.; Pakbaz, S.; Al-Ghamdi, D.; Asa, S.L. Ki67 Quantitative Interpretation: Insights Using Image Analysis. J. Pathol. Inform. 2019, 10, 8. [Google Scholar] [CrossRef] [PubMed]
- King, R.; Googe, P.B.; Weilbaecher, K.N.; Mihm, M.C., Jr.; Fisher, D.E. Microphthalmia Transcription Factor Expression in Cutaneous Benign, Malignant Melanocytic, and Nonmelanocytic Tumors. Am. J. Surg. Pathol. 2001, 25, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Granter, S.R.; Weilbaecher, K.N.; Quigley, C.; Fletcher, C.D.; Fisher, D.E. Microphthalmia Transcription Factor: Not a Sensitive or Specific Marker for the Diagnosis of Desmoplastic Melanoma and Spindle Cell (Non-Desmoplastic) Melanoma. Am. J. Dermatopathol. 2001, 23, 185–189. [Google Scholar] [CrossRef] [PubMed]
- Miettinen, M.; Fernandez, M.; Franssila, K.; Gatalica, Z.; Lasota, J.; Sarlomo-Rikala, M. Microphthalmia Transcription Factor in the Immunohistochemical Diagnosis of Metastatic Melanoma: Comparison with Four Other Melanoma Markers. Am. J. Surg. Pathol. 2001, 25, 205–211. [Google Scholar] [CrossRef]
- Koch, M.B.; Shih, I.-M.; Weiss, S.W.; Folpe, A.L. Microphthalmia Transcription Factor and Melanoma Cell Adhesion Molecule Expression Distinguish Desmoplastic/Spindle Cell Melanoma from Morphologic Mimics. Am. J. Surg. Pathol. 2001, 25, 58–64. [Google Scholar] [CrossRef]
- Feldmeyer, L.; Tetzlaff, M.; Fox, P.; Nagarajan, P.; Curry, J.; Ivan, D.; Cabala, C.A.T.; Prieto, V.G.; Aung, P.P. Prognostic Implication of Lymphovascular Invasion Detected by Double Immunostaining for D2-40 and MITF1 in Primary Cutaneous Melanoma. Am. J. Dermatopathol. 2016, 38, 484–491. [Google Scholar] [CrossRef]
- Buonaccorsi, J.N.; Prieto, V.G.; Torres-Cabala, C.; Suster, S.; Plaza, J.A. Diagnostic Utility and Comparative Immunohistochemical Analysis of MITF-1 and SOX10 to Distinguish Melanoma In Situ and Actinic Keratosis: A Clinicopathological and Immunohistochemical Study of 70 Cases. Am. J. Dermatopathol. 2014, 36, 124–130. [Google Scholar] [CrossRef]
- Parra, O.; Linos, K.; Li, Z.; Yan, S. PRAME Expression in Melanocytic Lesions of the Nail. J. Cutan. Pathol. 2022, 49, 610–617. [Google Scholar] [CrossRef]
- Koch, E.A.T.; Erdmann, M.; Berking, C.; Kiesewetter, F.; Kramer, R.; Schliep, S.; Heppt, M.V. Standardized Computer-Assisted Analysis of PRAME Immunoreactivity in Dysplastic Nevi and Superficial Spreading Melanomas. Int. J. Mol. Sci. 2023, 24, 6388. [Google Scholar] [CrossRef]
- Zengin, H.B.; Yildiz, B.; Pukhalskaya, T.; Smoller, B.R. FLI-1/Melan-A Dual Stain Is an Alternative to PRAME in Differentiating Metastatic Melanoma from Nodal Nevus: A Monocentric Retrospective Study. J. Cutan. Pathol. 2023, 50, 247–258. [Google Scholar] [CrossRef]
- Lezcano, C.; Jungbluth, A.A.; Busam, K.J. Immunohistochemistry for PRAME in Dermatopathology. Am. J. Dermatopathol. 2023, 45, 733–747. [Google Scholar] [CrossRef] [PubMed]
- Bahmad, H.F.; Oh, K.S.; Alexis, J. Potential Diagnostic Utility of PRAME and P16 Immunohistochemistry in Melanocytic Nevi and Malignant Melanoma. J. Cutan. Pathol. 2023, 50, 763–772. [Google Scholar] [CrossRef] [PubMed]
- Lo Bello, G.; Pini, G.M.; Giagnacovo, M.; Patriarca, C. PRAME Expression in 137 Primary Cutaneous Melanomas and Comparison with 38 Related Metastases. Pathol. Res. Pract. 2023, 251, 154915. [Google Scholar] [CrossRef] [PubMed]
- Miao, Q.-J.; Zang, J.; Shao, X.-B.; Sun, J.-F.; Chen, Y.-P.; Chen, H. Analysis of PRAME Immunocytochemistry in 109 Acral Malignant Melanoma in Situ. J. Clin. Pathol. 2024, 77, 417–420. [Google Scholar] [CrossRef]
- Kim, J.C.; Choi, J.W.; Kim, Y.C. Comparison of Melanocyte-Associated Immunohistochemical Markers in Acral Lentiginous Melanoma and Acral Benign Nevi. Am. J. Dermatopathol. 2023, 45, 748–752. [Google Scholar] [CrossRef]
- Kim, Y.J.; Jung, C.J.; Na, H.; Lee, W.J.; Chang, S.E.; Lee, M.W.; Park, C.-S.; Lim, Y.; Won, C.H. Cyclin D1 and PRAME Expression in Distinguishing Melanoma in Situ from Benign Melanocytic Proliferation of the Nail Unit. Diagn. Pathol. 2022, 17, 41. [Google Scholar] [CrossRef]
- Lezcano, C.; Jungbluth, A.A.; Nehal, K.S.; Hollmann, T.J.; Busam, K.J. PRAME Expression in Melanocytic Tumors. Am. J. Surg. Pathol. 2018, 42, 1456–1465. [Google Scholar] [CrossRef]
- Wakefield, C.; O’Keefe, L.; Heffron, C.C.B.B. Refining the Application of PRAME-a Useful Marker in High CSD and Acral Melanoma Subtypes. Virchows Arch. Int. J. Pathol. 2023, 483, 847–854. [Google Scholar] [CrossRef]
- Alomari, A.K.; Tharp, A.W.; Umphress, B.; Kowal, R.P. The Utility of PRAME Immunohistochemistry in the Evaluation of Challenging Melanocytic Tumors. J. Cutan. Pathol. 2021, 48, 1115–1123. [Google Scholar] [CrossRef]
- Rawson, R.V.; Shteinman, E.R.; Ansar, S.; Vergara, I.A.; Thompson, J.F.; Long, G.V.; Scolyer, R.A.; Wilmott, J.S. Diagnostic Utility of PRAME, P53 and 5-hmC Immunostaining for Distinguishing Melanomas from Naevi, Neurofibromas, Scars and Other Histological Mimics. Pathology 2022, 54, 863–873. [Google Scholar] [CrossRef]
- Hu, J.; Cai, X.; Lv, J.-J.; Wan, X.-C.; Zeng, X.-Y.; Feng, M.-L.; Dai, B.; Kong, Y.-Y. Preferentially Expressed Antigen in Melanoma Immunohistochemistry as an Adjunct for Differential Diagnosis in Acral Lentiginous Melanoma and Acral Nevi. Hum. Pathol. 2022, 120, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Ronchi, A.; Cazzato, G.; Ingravallo, G.; D’Abbronzo, G.; Argenziano, G.; Moscarella, E.; Brancaccio, G.; Franco, R. PRAME Is an Effective Tool for the Diagnosis of Nevus-Associated Cutaneous Melanoma. Cancers 2024, 16, 278. [Google Scholar] [CrossRef] [PubMed]
- See, S.H.C.; Finkelman, B.S.; Yeldandi, A.V. The Diagnostic Utility of PRAME and P16 in Distinguishing Nodal Nevi from Nodal Metastatic Melanoma. Pathol. Res. Pract. 2020, 216, 153105. [Google Scholar] [CrossRef] [PubMed]
- Asato, M.A.; Moraes Neto, F.A.; Moraes, M.P.d.T.; Ocanha-Xavier, J.P.; Takita, L.C.; Fung, M.A.; Marques, M.E.A.; Xavier-Júnior, J.C.C. The Utility of PRAME and Ki-67 as Prognostic Markers for Cutaneous Melanoma. Am. J. Dermatopathol. 2025, 47, 9–16. [Google Scholar] [CrossRef]
- Olds, H.; Utz, S.; Abrams, J.; Terrano, D.; Mehregan, D. Use of PRAME Immunostaining to Distinguish Early Melanoma in Situ from Benign Pigmented Conditions. J. Cutan. Pathol. 2022, 49, 510–514. [Google Scholar] [CrossRef]
- Roy, S.F.; Panse, G.; McNiff, J.M. PRAME Immunohistochemistry Can Distinguish Melanocytic Pseudonests of Lichenoid Reactions from Melanoma in Situ. J. Cutan. Pathol. 2023, 50, 450–454. [Google Scholar] [CrossRef]
- Lezcano, C.; Jungbluth, A.A.; Busam, K.J. Comparison of Immunohistochemistry for PRAME With Cytogenetic Test Results in the Evaluation of Challenging Melanocytic Tumors. Am. J. Surg. Pathol. 2020, 44, 893–900. [Google Scholar] [CrossRef]
- Ferreira, L.Á.; Kim, E.H.J.; Stelini, R.F.; Velho, P.E.N.F.; de Moraes, A.M.; Buffo, T.; Cintra, M.L. The Expression of PRAME as an Aid for Diagnosis and Evaluation of Histologic Margins of Intraepidermal Cutaneous Melanoma in Xeroderma Pigmentosum Patients. Appl. Immunohistochem. Mol. Morphol. 2024, 32, 272–279. [Google Scholar] [CrossRef]
- Gradecki, S.E.; Valdes-Rodriguez, R.; Wick, M.R.; Gru, A.A. PRAME Immunohistochemistry as an Adjunct for Diagnosis and Histological Margin Assessment in Lentigo Maligna. Histopathology 2021, 78, 1000–1008. [Google Scholar] [CrossRef]
- Falkenius, J.; Johansson, H.; Tuominen, R.; Frostvik Stolt, M.; Hansson, J.; Egyhazi Brage, S. Presence of Immune Cells, Low Tumor Proliferation and Wild Type BRAF Mutation Status Is Associated with a Favourable Clinical Outcome in Stage III Cutaneous Melanoma. BMC Cancer 2017, 17, 584. [Google Scholar] [CrossRef]
- Bauer, J.; Büttner, P.; Murali, R.; Okamoto, I.; Kolaitis, N.A.; Landi, M.T.; Scolyer, R.A.; Bastian, B.C. BRAF Mutations in Cutaneous Melanoma Are Independently Associated with Age, Anatomic Site of the Primary Tumor, and the Degree of Solar Elastosis at the Primary Tumor Site. Pigment. Cell Melanoma Res. 2011, 24, 345–351. [Google Scholar] [CrossRef] [PubMed]
- Menzies, A.M.; Haydu, L.E.; Visintin, L.; Carlino, M.S.; Howle, J.R.; Thompson, J.F.; Kefford, R.F.; Scolyer, R.A.; Long, G.V. Distinguishing Clinicopathologic Features of Patients with V600E and V600K BRAF -Mutant Metastatic Melanoma. Clin. Cancer Res. 2012, 18, 3242–3249. [Google Scholar] [CrossRef] [PubMed]
- Ponti, G.; Tomasi, A.; Maiorana, A.; Ruini, C.; Maccaferri, M.; Cesinaro, A.M.; Depenni, R.; Manni, P.; Gelsomino, F.; Giusti, F.; et al. BRAFp.V600E, p.V600K, and p.V600R Mutations in Malignant Melanoma: Do They Also Differ in Immunohistochemical Assessment and Clinical Features? Appl. Immunohistochem. Mol. Morphol. 2016, 24, 30–34. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Goto, K.; Yoshikawa, S.; Takai, T.; Tachibana, K.; Honma, K.; Isei, T.; Kukita, Y.; Oishi, T. Clinicopathologic and Genetic Characterization of Invasive Melanoma with BRAF V600K Mutation: A Study of 16 Cases. J. Cutan. Pathol. 2023, 50, 739–747. [Google Scholar] [CrossRef]
- Pires Da Silva, I.; Wang, K.Y.X.; Wilmott, J.S.; Holst, J.; Carlino, M.S.; Park, J.J.; Quek, C.; Wongchenko, M.; Yan, Y.; Mann, G.; et al. Distinct Molecular Profiles and Immunotherapy Treatment Outcomes of V600E and V600K BRAF-Mutant Melanoma. Clin. Cancer Res. 2019, 25, 1272–1279. [Google Scholar] [CrossRef]
- Urvanegia, A.C.; Tavoloni Braga, J.C.; Shitara, D.; Fregnani, J.H.; Neves, J.I.; Pinto, C.A.; Marghoob, A.A.; Duprat, J.P.; Rezze, G.G. Reflectance Confocal Microscopy Features of BRAF V600E Mutated Thin Melanomas Detected by Immunohistochemistry. PLoS ONE 2017, 12, e0179745. [Google Scholar] [CrossRef]
- Manninen, A.A.; Gardberg, M.; Juteau, S.; Ilmonen, S.; Jukonen, J.; Andersson, N.; Carpén, O. BRAF Immunohistochemistry Predicts Sentinel Lymph Node Involvement in Intermediate Thickness Melanomas. PLoS ONE 2019, 14, e0216043. [Google Scholar] [CrossRef]
- de Moll, E.H.; Fu, Y.; Qian, Y.; Perkins, S.H.; Wieder, S.; Gnjatic, S.; Remark, R.; Bernardo, S.G.; Moskalenko, M.; Yao, J.; et al. Immune Biomarkers Are More Accurate in Prediction of Survival in Ulcerated than in Non-Ulcerated Primary Melanomas. Cancer Immunol. Immunother. 2015, 64, 1193–1203. [Google Scholar] [CrossRef]
- Moysset, I.; Castrejon, N.; Garcia-Herrera, A.; Castillo, P.; Marginet, M.; Teixido, C.; Podlipnik, S.; Albero-Gonzalez, R.; Montironi, C.; Navarro, J.; et al. Restrospective Reappraisal of the Prognostic Classification of Spitzoid Melanocytic Neoplasms after BRAF and NRAS Mutation Characterisation: A SingleInstitution Experience. Histopathology 2024, 84, 1154–1166. [Google Scholar] [CrossRef]
- Hugdahl, E.; Kalvenes, M.B.; Puntervoll, H.E.; Ladstein, R.G.; Akslen, L.A. BRAF-V600E Expression in Primary Nodular Melanoma Is Associated with Aggressive Tumour Features and Reduced Survival. Br. J. Cancer 2016, 114, 801–808. [Google Scholar] [CrossRef]
- Aksenenko, M.B.; Kirichenko, A.K.; Ruksha, T.G. Russian Study of Morphological Prognostic Factors Characterization in BRAF-Mutant Cutaneous Melanoma. Pathol. Res. Pract. 2015, 211, 521–527. [Google Scholar] [CrossRef] [PubMed]
- Hannan, E.J.; O’Leary, D.P.; MacNally, S.P.; Kay, E.W.; Farrell, M.A.; Morris, P.G.; Power, C.P.; Hill, A.D.K. The Significance of BRAF V600E Mutation Status Discordance between Primary Cutaneous Melanoma and Brain Metastases: The Implications for BRAF Inhibitor Therapy. Medicine 2017, 96, e8404. [Google Scholar] [CrossRef] [PubMed]
- Suh, M.S.; Choi, Y.D.; Lee, J.-B.; Lee, S.-C.; Won, Y.H.; Yun, S.J. Sensitivity and Usefulness of VE1 Immunohistochemical Staining in Acral Melanomas with BRAF Mutation. Ann. Dermatol. 2018, 30, 556–561. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, H.; Zebary, A.; Vassilaki, I.; Omholt, K.; Ghaderi, M.; Hansson, J. BRAFV600E Protein Expression in Primary Cutaneous Malignant Melanomas and Paired Metastases. JAMA Dermatol. 2015, 151, 410–416. [Google Scholar] [CrossRef]
- Vallée, A.; Denis-Musquer, M.; Herbreteau, G.; Théoleyre, S.; Bossard, C.; Denis, M.G. Prospective Evaluation of Two Screening Methods for Molecular Testing of Metastatic Melanoma: Diagnostic Performance of BRAF V600E Immunohistochemistry and of a NRAS-BRAF Fully Automated Real-Time PCR-Based Assay. PLoS ONE 2019, 14, e0221123. [Google Scholar] [CrossRef]
- Menzer, C.; Menzies, A.M.; Carlino, M.S.; Reijers, I.; Groen, E.J.; Eigentler, T.; De Groot, J.W.B.; Van Der Veldt, A.A.M.; Johnson, D.B.; Meiss, F.; et al. Targeted Therapy in Advanced Melanoma with Rare BRAF Mutations. J. Clin. Oncol. 2019, 37, 3142–3151. [Google Scholar] [CrossRef]
- Zhong, J.; Yan, W.; Wang, C.; Liu, W.; Lin, X.; Zou, Z.; Sun, W.; Chen, Y. BRAF Inhibitor Resistance in Melanoma: Mechanisms and Alternative Therapeutic Strategies. Curr. Treat. Options Oncol. 2022, 23, 1503–1521. [Google Scholar] [CrossRef]
- Demirkan, N.C.; Kesen, Z.; Akdag, B.; Larue, L.; Delmas, V. The Effect of the Sun on Expression of Beta-Catenin, P16 and Cyclin D1 Proteins in Melanocytic Lesions. Clin. Exp. Dermatol. 2007, 32, 733–739. [Google Scholar] [CrossRef]
- Stefanaki, C.; Stefanaki, K.; Antoniou, C.; Argyrakos, T.; Patereli, A.; Stratigos, A.; Katsambas, A. Cell Cycle and Apoptosis Regulators in Spitz Nevi: Comparison with Melanomas and Common Nevi. J. Am. Acad. Dermatol. 2007, 56, 815–824. [Google Scholar] [CrossRef]
- Al Dhaybi, R.; Agoumi, M.; Gagné, I.; McCuaig, C.; Powell, J.; Kokta, V. P16 Expression: A Marker of Differentiation between Childhood Malignant Melanomas and Spitz Nevi. J. Am. Acad. Dermatol. 2011, 65, 357–363. [Google Scholar] [CrossRef]
- Mason, A.; Wititsuwannakul, J.; Klump, V.R.; Lott, J.; Lazova, R. Expression of P16 Alone Does Not Differentiate between Spitz Nevi andSpitzoid Melanoma. J. Cutan. Pathol. 2012, 39, 1062–1074. [Google Scholar] [CrossRef] [PubMed]
- Blokhin, E.; Pulitzer, M.; Busam, K.J. Immunohistochemical Expression of P16 in Desmoplastic Melanoma. J. Cutan. Pathol. 2013, 40, 796–800. [Google Scholar] [CrossRef] [PubMed]
- Harms, P.W.; Hocker, T.L.; Zhao, L.; Chan, M.P.; Andea, A.A.; Wang, M.; Harms, K.L.; Wang, M.L.; Carskadon, S.; Palanisamy, N.; et al. Loss of P16 Expression and Copy Number Changes of CDKN2A in a Spectrum of Spitzoid Melanocytic Lesions. Hum. Pathol. 2016, 58, 152–160. [Google Scholar] [CrossRef] [PubMed]
- Mihic-Probst, D.; Mnich, C.D.; Oberholzer, P.A.; Seifert, B.; Sasse, B.; Moch, H.; Dummer, R. P16 Expression in Primary Malignant Melanoma Is Associated with Prognosis and Lymph Node Status. Int. J. Cancer 2006, 118, 2262–2268. [Google Scholar] [CrossRef]
- Strickler, A.G.; Schaefer, J.T.; Slingluff, C.L.; Wick, M.R. Immunolabeling for P16, WT1, and Fli-1 in the Assignment of Growth Phase for Cutaneous Melanomas. Am. J. Dermatopathol. 2014, 36, 718–722. [Google Scholar] [CrossRef]
- Roncati, L.; Piscioli, F.; Pusiol, T.; Maiorana, A. Microinvasive Radial Growth Phase of Cutaneous Melanoma: A Histopathological and Immunohistochemical Study with Diagnostic Implications. Acta Dermatovenerol. Croat. 2017, 25, 39–45. [Google Scholar]
- Hsieh, R.; Firmiano, A.; Sotto, M.N. Expression of P16 Protein in Acral Lentiginous Melanoma. Int. J. Dermatol. 2009, 48, 1303–1307. [Google Scholar] [CrossRef]
- Gkionis, I.G.; Tzardi, M.; Alegakis, A.; Datseri, G.; Moustou, E.; Saridakis, G.; Michelakis, D.; Kruger-Krasagakis, S.; Krasagakis, K.; DE Bree, E. Cyclin E Expression and P16 Loss Are Strong Prognostic Biomarkers in Primary Invasive Cutaneous Melanoma. Anticancer. Res. 2025, 45, 625–637. [Google Scholar] [CrossRef]
- Hilliard, N.J.; Krahl, D.; Sellheyer, K. P16 Expression Differentiates between Desmoplastic Spitz Nevus and Desmoplastic Melanoma. J. Cutan. Pathol. 2009, 36, 753–759. [Google Scholar] [CrossRef]
- Castaneda, C.A.; Castillo, M.; Torres-Cabala, C.; Bernabe, L.A.; Casavilca, S.; Villegas, V.; Sanchez, J.; de la Cruz, M.; Dunstan, J.; Cotrina, J.M.; et al. Relationship between Tumor-Associated Immune Infiltrate and P16 Staining over Clinicopathological Features in Acral Lentiginous Melanoma. Clin. Transl. Oncol. 2019, 21, 1127–1134. [Google Scholar] [CrossRef]
- Richmond-Sinclair, N.M.; Lee, E.; Cummings, M.C.; Williamson, R.; Muller, K.; Green, A.C.; Hayward, N.K.; Whiteman, D.C. Histologic and Epidemiologic Correlates of P-MAPK, Brn-2, pRb, P53, and P16 Immunostaining in Cutaneous Melanomas. Melanoma Res. 2008, 18, 336–345. [Google Scholar] [CrossRef] [PubMed]
- Fauri, J.A.C.; Ricardi, F.; Diehl, E.S.; Cartell, A.; Furian, R.; Bakos, L.; Edelweiss, M.I. P16 Protein Expression in Primary Cutaneous Melanoma with Positive and Negative Lymph Node Biopsies: Particular Aspects of a Study Performed at the Hospital de Clinicas de Porto Alegre, Brazil. Can. J. Plast. Surg. 2011, 19, 77–81. [Google Scholar] [CrossRef] [PubMed]
- Fearfield, L.A.; Larkin, J.M.G.; Rowe, A.; A’Hern, R.; Fisher, C.; Francis, N.; MacKie, R.; McCann, B.; Gore, M.E.; Bunker, C.B. Expression of P16, CD95, CD95L and Helix Pomatia Agglutinin in Relapsing and Nonrelapsing Very Thin Melanoma. Br. J. Dermatol. 2007, 156, 440–447. [Google Scholar] [CrossRef] [PubMed]
- Cobankent Aytekin, E.; Unal, B.; Bassorgun, C.I.; Ozkan, O. Clinicopathologic Evaluation of CD80, CD86, and PD-L1 Expressions with Immunohistochemical Methods in Malignant Melanoma Patients. Turk. Patoloji Derg. 2024, 40, 16–26. [Google Scholar] [CrossRef]
- Kraft, S.; Fernandez-Figueras, M.-T.; Richarz, N.A.; Flaherty, K.T.; Hoang, M.P. PDL1 Expression in Desmoplastic Melanoma Is Associated with Tumor Aggressiveness and Progression. J. Am. Acad. Dermatol. 2017, 77, 534–542. [Google Scholar] [CrossRef]
- Frydenlund, N.; Leone, D.; Yang, S.; Hoang, M.P.; Deng, A.; Hernandez-Perez, M.; Singh, R.; Biswas, A.; Yaar, R.; Mahalingam, M. Tumoral PD-L1 Expression in Desmoplastic Melanoma Is Associated with Depth of Invasion, Tumor-Infiltrating CD8 Cytotoxic Lymphocytes and the Mixed Cytomorphological Variant. Mod. Pathol. 2017, 30, 357–369. [Google Scholar] [CrossRef]
- Škuciová, V.; Drahošová, S.; Výbohová, D.; Cígerová, V.; Adamkov, M. The Relationships between PD-L1 Expression, CD8+ TILs and Clinico-Histomorphological Parameters in Malignant Melanomas. Pathol. Res. Pract. 2020, 216, 153071. [Google Scholar] [CrossRef]
- Kaunitz, G.J.; Cottrell, T.R.; Lilo, M.; Muthappan, V.; Esandrio, J.; Berry, S.; Xu, H.; Ogurtsova, A.; Anders, R.A.; Fischer, A.H.; et al. Melanoma Subtypes Demonstrate Distinct PD-L1 Expression Profiles. Lab. Investig. J. Tech. Methods Pathol. 2017, 97, 1063–1071. [Google Scholar] [CrossRef]
- Giavina-Bianchi, M.; Giavina-Bianchi, P.; Sotto, M.N.; Rodig, S.; Mihm, M.J.; Festa Neto, C.; Duncan, L.M.; Kalil, J. Nodular Primary Cutaneous Melanoma Is Associated with PD-L1 Expression. Eur. J. Dermatol. 2020, 30, 352–357. [Google Scholar] [CrossRef]
- Tarhini, A.A.; Zahoor, H.; Yearley, J.H.; Gibson, C.; Rahman, Z.; Dubner, R.; Rao, U.N.M.; Sander, C.; Kirkwood, J.M. Tumor Associated PD-L1 Expression Pattern in Microscopically Tumor Positive Sentinel Lymph Nodes in Patients with Melanoma. J. Transl. Med. 2015, 13, 319. [Google Scholar] [CrossRef]
- Pinczewski, J.; Obeng, R.C.; Slingluff, C.L.J.; Engelhard, V.H. Phospho-β-Catenin Expression in Primary and Metastatic Melanomas and in Tumor-Free Visceral Tissues, and Associations with Expression of PD-L1 and PD-L2. Pathol. Res. Pract. 2021, 224, 153527. [Google Scholar] [CrossRef] [PubMed]
- Santos-Briz, A.; Cañueto, J.; Carmen, S.D.; Barrios, B.; Yuste, M.; Bellido, L.; Ludeña, M.D.; Román, C. Value of PD-L1, PD-1, and CTLA-4 Expression in the Clinical Practice as Predictors of Response to Nivolumab and Ipilimumab in Monotherapy in Patientswith Advanced Stage Melanoma. Am. J. Dermatopathol. 2021, 43, 423–428. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.; Ahn, S.; Yoo, K.H.; Kim, J.H.; Choi, S.-H.; Jang, K.-T.; Lee, J. Treatment Outcome of PD-1 Immune Checkpoint Inhibitor in Asian Metastatic Melanoma Patients: Correlative Analysis with PD-L1 Immunohistochemistry. Invest. New Drugs 2016, 34, 677–684. [Google Scholar] [CrossRef] [PubMed]
- Daud, A.I.; Wolchok, J.D.; Robert, C.; Hwu, W.-J.; Weber, J.S.; Ribas, A.; Hodi, F.S.; Joshua, A.M.; Kefford, R.; Hersey, P.; et al. Programmed Death-Ligand 1 Expression and Response to the Anti–Programmed Death 1 Antibody Pembrolizumab in Melanoma. J. Clin. Oncol. 2016, 34, 4102–4109. [Google Scholar] [CrossRef]
- Garbe, C.; Amaral, T.; Peris, K.; Hauschild, A.; Arenberger, P.; Basset-Seguin, N.; Bastholt, L.; Bataille, V.; Del Marmol, V.; Dréno, B.; et al. European Consensus-Based Interdisciplinary Guideline for Melanoma. Part 1: Diagnostics: Update 2022. Eur. J. Cancer 2022, 170, 236–255. [Google Scholar] [CrossRef]
- Prieto, V.G.; Shea, C.R. Use of Immunohistochemistry in Melanocytic Lesions. J. Cutan. Pathol. 2008, 35 (Suppl. S2), 1–10. [Google Scholar] [CrossRef]
- Cazzato, G.; Cascardi, E.; Colagrande, A.; Belsito, V.; Lospalluti, L.; Foti, C.; Arezzo, F.; Dellino, M.; Casatta, N.; Lupo, C.; et al. PRAME Immunoexpression in 275 Cutaneous Melanocytic Lesions: A Double Institutional Experience. Diagnostics 2022, 12, 2197. [Google Scholar] [CrossRef]
- Boursault, L.; Haddad, V.; Vergier, B.; Cappellen, D.; Verdon, S.; Bellocq, J.-P.; Jouary, T.; Merlio, J.-P. Tumor Homogeneity between Primary and Metastatic Sites for BRAF Status in Metastatic Melanoma Determined by Immunohistochemical and Molecular Testing. PLoS ONE 2013, 8, e70826. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).