Hsa_circ_0015278 Regulates FLT3-ITD AML Progression via Ferroptosis-Related Genes
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Acquisition and Processing
2.2. Identification of Differentially Expressed circRNAs (DECrics) and Differentially Expressed Genes (DEGs)
2.3. Establishment of the ceRNA Regulatory Network in AML
2.4. Functional Enrichment Analysis
2.5. PPI Network Construction and Hub Gene Screening
2.6. Survival and Prognosis Analysis
2.7. Immune Infiltration Analysis
2.8. Validation of Circrnas Relative Expression Level
2.9. Statistical Analysis
3. Results
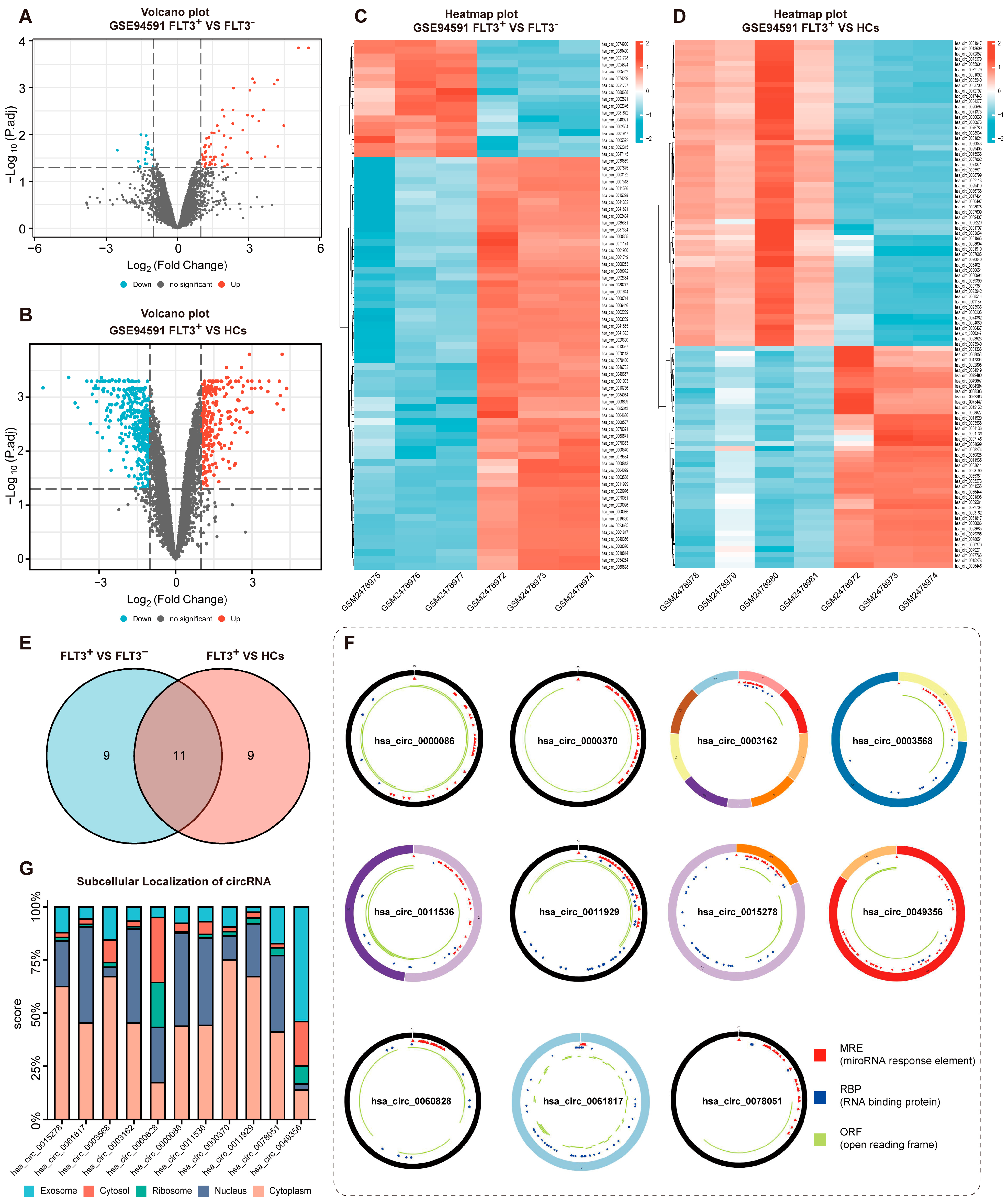
3.1. Identification of DECircs and DEGs in GEO Database
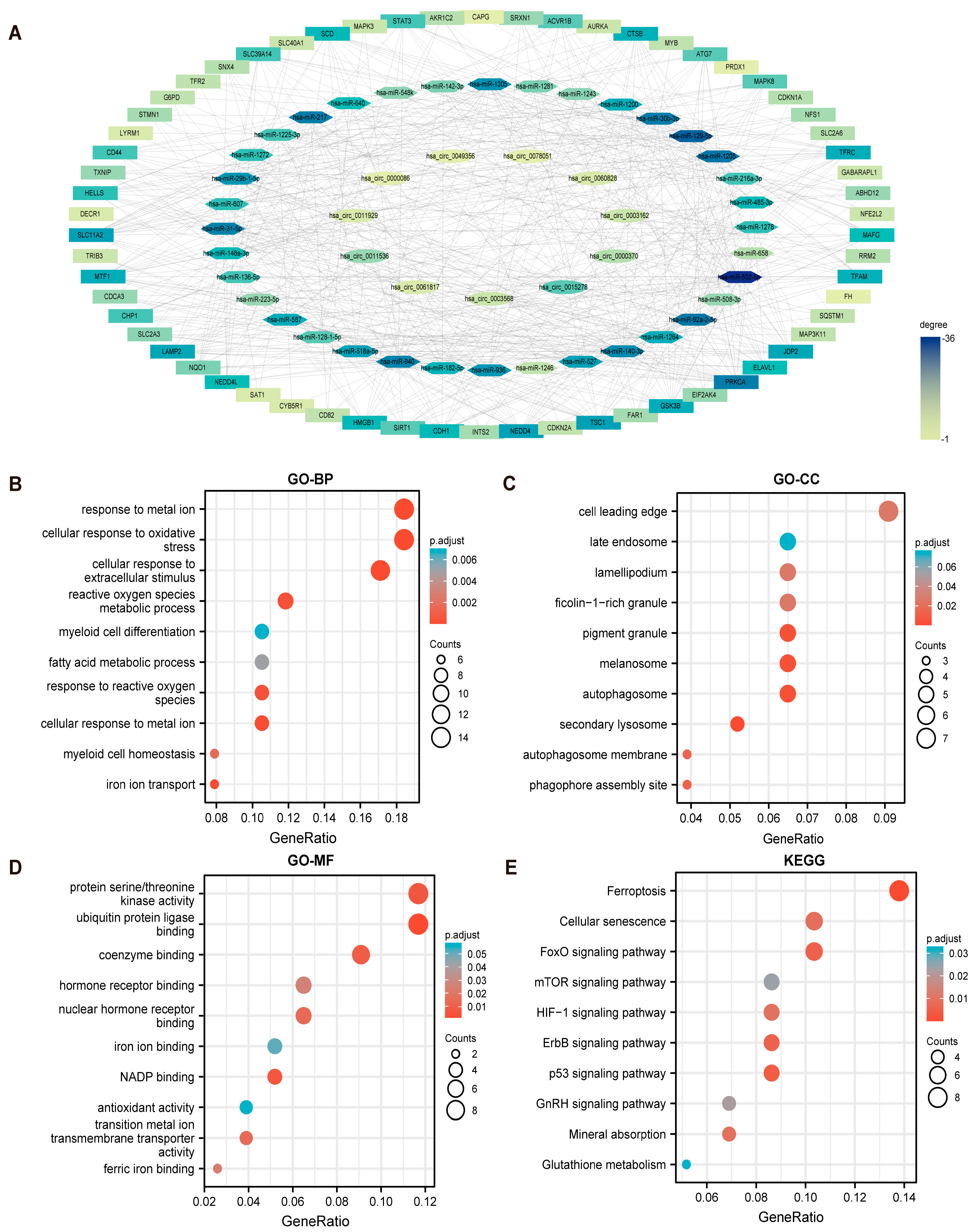
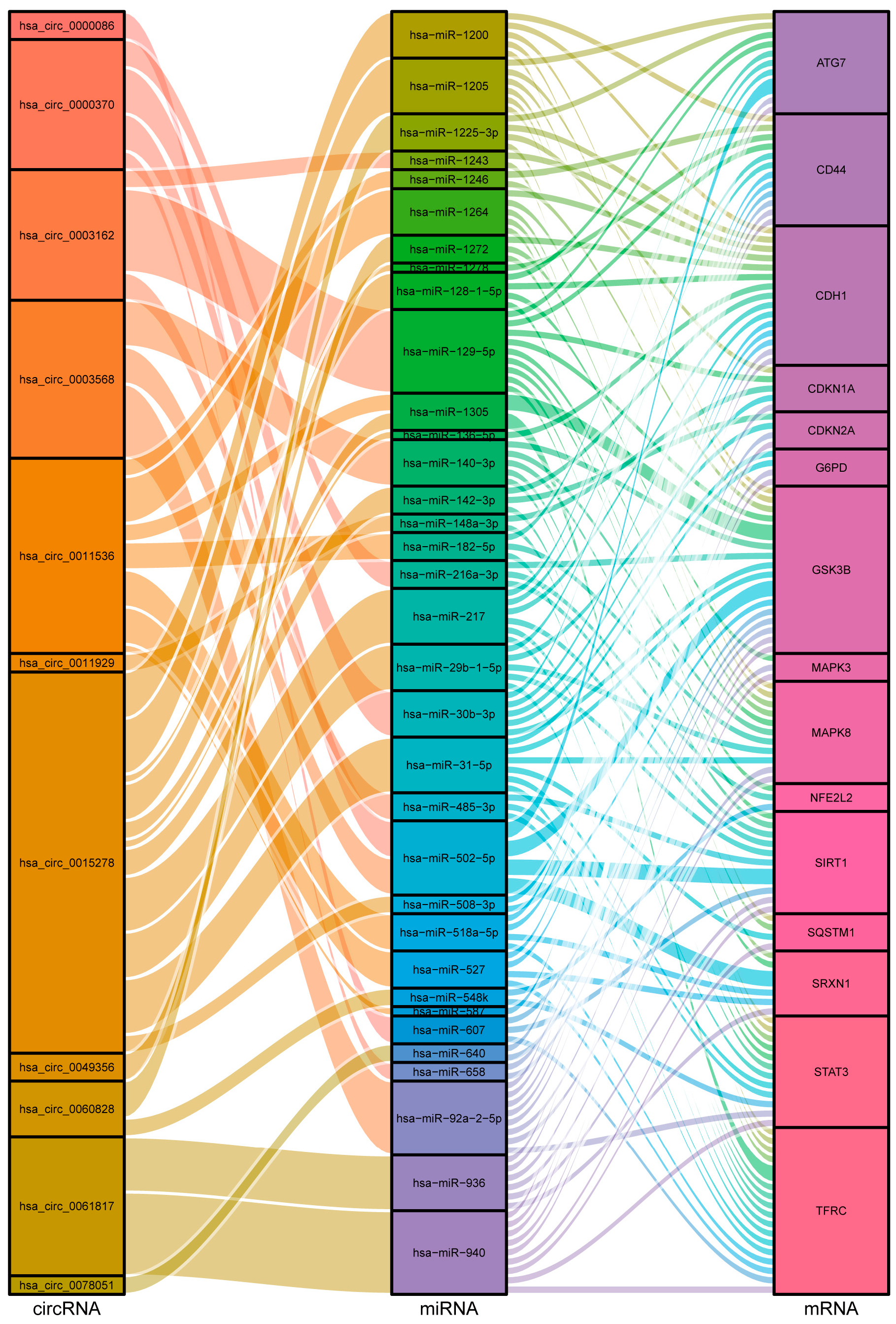
3.2. Construction of the circRNAs/ miRNAs/FerRGs Regulatory Network
3.3. GO and KEGG Functional Enrichment Analysis of FerRGs
3.4. Screening and Enrichment Analysis of Hub Genes
3.5. Validation of Expression and Prognosis of 15 Hub Genes
3.6. Validation of Relative Expression Levels and Structures of Candidate circRNAs
3.7. Construction of ceRNA Sub-Network and Correlation Analysis of Clinicopathological Parameters
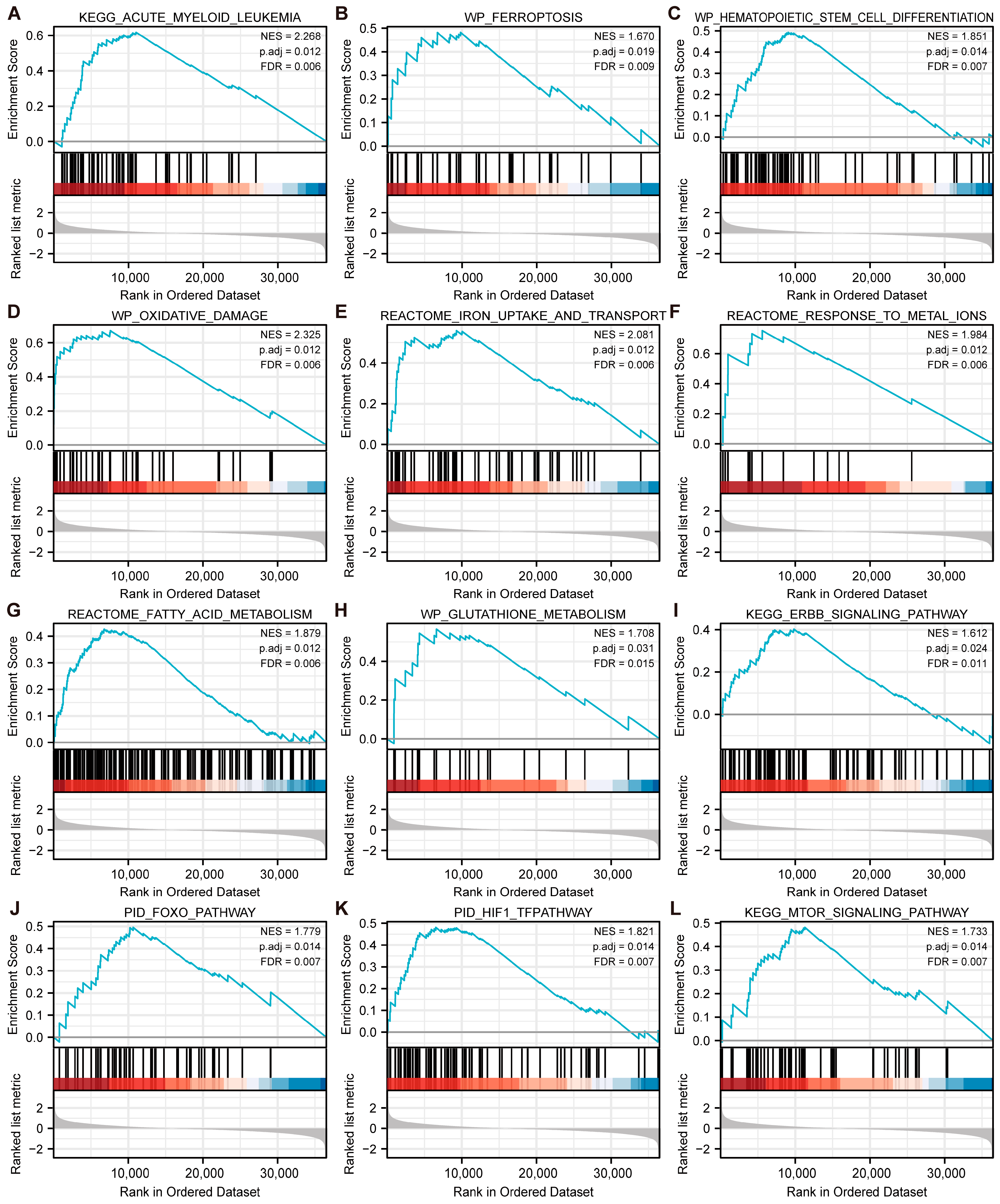
3.8. GSEA Enrichment Analysis of MAPK3 and CD44
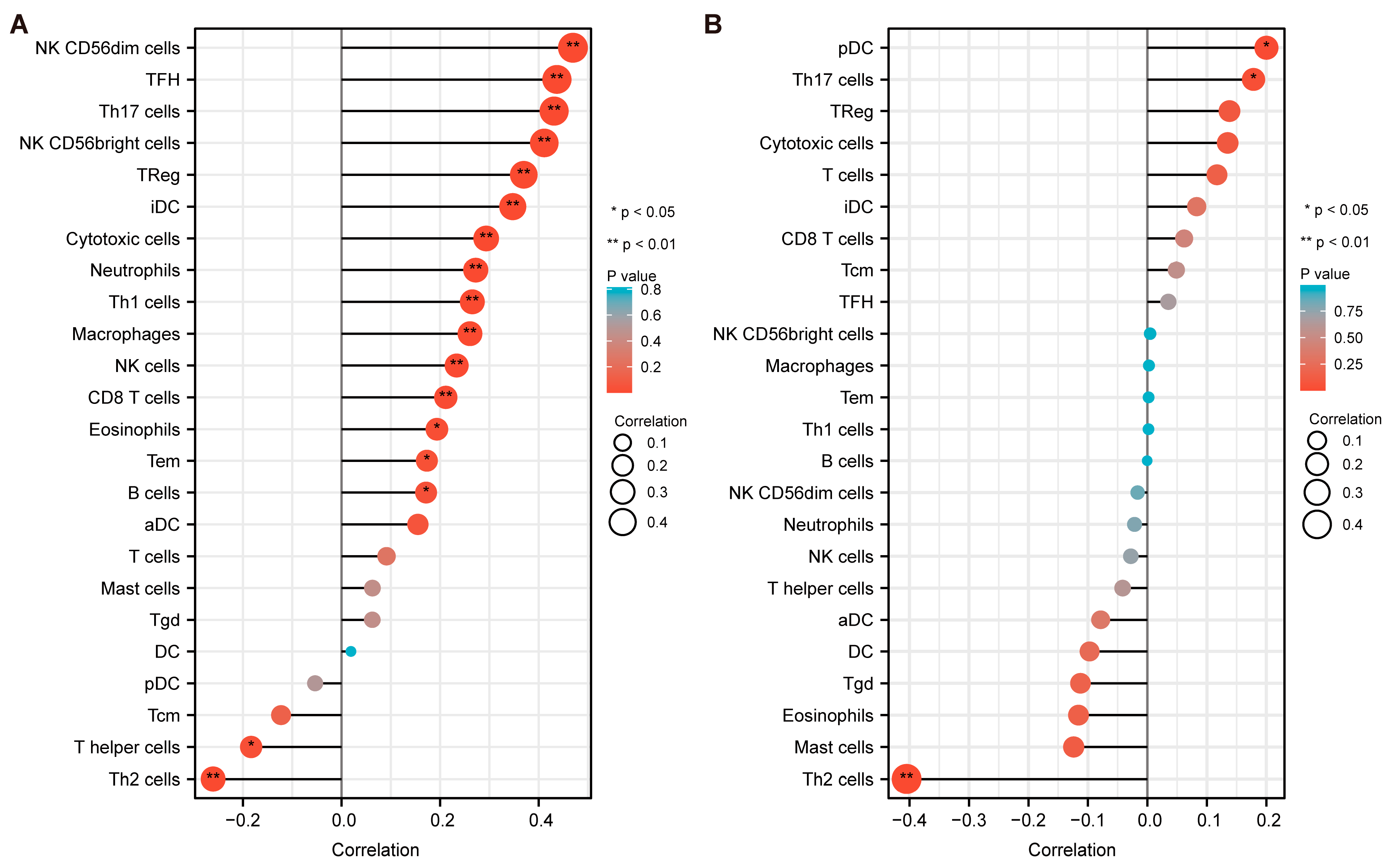
3.9. Immune Infiltration Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AML | Acute myeloid leukemia |
| FerRGs | ferroptosis-related genes |
| MREs | miRNA response elements |
| ceRNA | competitive endogenous RNA |
| DECircs | differentially expressed circRNAs |
| Ferr-Genes | ferroptosis genes |
| FDR | false discovery rate |
| DEGs | differentially expressed genes |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| CC | cellular component |
| MF | molecular function |
| BP | biological process |
| ssGSEA | single-sample Gene Set Enrichment Analysis |
| PPI | protein-protein interaction |
| STRING | Search Tool for the Retrieval of Interacting Genes |
| OS | overall survival |
| AUC | area under the curve |
References
- Estey, E.; Döhner, H. Acute myeloid leukaemia. Lancet 2006, 368, 1894–1907. [Google Scholar] [CrossRef]
- Zhou, J.; Zhou, L.Y.; Tang, X.; Zhang, J.; Zhai, L.L.; Yi, Y.Y.; Yi, J.; Lin, J.; Qian, J.; Deng, Z.Q. Circ-Foxo3 is positively associated with the Foxo3 gene and leads to better prognosis of acute myeloid leukemia patients. BMC Cancer 2019, 19, 930. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef]
- Kavanagh, S.; Murphy, T.; Law, A.; Yehudai, D.; Ho, J.M.; Chan, S.; Schimmer, A.D. Emerging therapies for acute myeloid leukemia: Translating biology into the clinic. JCI Insight 2017, 2, e95679. [Google Scholar] [CrossRef]
- Takahashi, S. Downstream molecular pathways of FLT3 in the pathogenesis of acute myeloid leukemia: Biology and therapeutic implications. J. Hematol. Oncol. 2011, 4, 13. [Google Scholar] [CrossRef] [PubMed]
- Levis, M.; Small, D. FLT3: ITDoes matter in leukemia. Leukemia 2003, 17, 1738–1752. [Google Scholar] [CrossRef] [PubMed]
- De Kouchkovsky, I.; Abdul-Hay, M. Acute myeloid leukemia: A comprehensive review and 2016 update. Blood Cancer J. 2016, 6, e441. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Li, Y. The interaction between ferroptosis and lipid metabolism in cancer. Signal Transduct. Target. Ther. 2020, 5, 108. [Google Scholar] [CrossRef] [PubMed]
- Mou, Y.; Wang, J.; Wu, J.; He, D.; Zhang, C.; Duan, C.; Li, B. Ferroptosis, a new form of cell death: Opportunities and challenges in cancer. J. Hematol. Oncol. 2019, 12, 34. [Google Scholar] [CrossRef]
- Lin, Z.; Song, J.; Gao, Y.; Huang, S.; Dou, R.; Zhong, P.; Huang, G.; Han, L.; Zheng, J.; Zhang, X.; et al. Hypoxia-induced HIF-1α/lncRNA-PMAN inhibits ferroptosis by promoting the cytoplasmic translocation of ELAVL1 in peritoneal dissemination from gastric cancer. Redox Biol. 2022, 52, 102312. [Google Scholar] [CrossRef]
- Sun, S.; Gao, T.; Pang, B.; Su, X.; Guo, C.; Zhang, R.; Pang, Q. RNA binding protein NKAP protects glioblastoma cells from ferroptosis by promoting SLC7A11 mRNA splicing in an m(6)A-dependent manner. Cell Death Dis. 2022, 13, 73. [Google Scholar] [CrossRef] [PubMed]
- Meng, C.; Zhan, J.; Chen, D.; Shao, G.; Zhang, H.; Gu, W.; Luo, J. The deubiquitinase USP11 regulates cell proliferation and ferroptotic cell death via stabilization of NRF2 USP11 deubiquitinates and stabilizes NRF2. Oncogene 2021, 40, 1706–1720. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Mo, Y.; Gong, Z.; Yang, X.; Yang, M.; Zhang, S.; Xiong, F.; Xiang, B.; Zhou, M.; Liao, Q.; et al. Circular RNAs in human cancer. Mol. Cancer 2017, 16, 25. [Google Scholar] [CrossRef] [PubMed]
- Papaioannou, D.; Volinia, S.; Nicolet, D.; Swierniak, M.; Petri, A.; Mrozek, K.; Bill, M.; Pepe, F.; Walker, C.J.; Walker, A.E.; et al. Clinical and functional significance of circular RNAs in cytogenetically normal AML. Blood Adv. 2020, 4, 239–251. [Google Scholar] [CrossRef]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef]
- Vo, J.N.; Cieslik, M.; Zhang, Y.; Shukla, S.; Xiao, L.; Zhang, Y.; Wu, Y.M.; Dhanasekaran, S.M.; Engelke, C.G.; Cao, X.; et al. The Landscape of Circular RNA in Cancer. Cell 2019, 176, 869–881.e13. [Google Scholar] [CrossRef]
- Hanniford, D.; Ulloa-Morales, A.; Karz, A.; Berzoti-Coelho, M.G.; Moubarak, R.S.; Sánchez-Sendra, B.; Kloetgen, A.; Davalos, V.; Imig, J.; Wu, P.; et al. Epigenetic Silencing of CDR1as Drives IGF2BP3-Mediated Melanoma Invasion and Metastasis. Cancer Cell 2020, 37, 55–70.e15. [Google Scholar] [CrossRef]
- Geng, Y.; Jiang, J.; Wu, C. Function and clinical significance of circRNAs in solid tumors. J. Hematol. Oncol. 2018, 11, 98. [Google Scholar] [CrossRef]
- Chen, L.L. The biogenesis and emerging roles of circular RNAs. Nat. Rev. Mol. Cell Biol. 2016, 17, 205–211. [Google Scholar] [CrossRef]
- Dong, Y.; He, D.; Peng, Z.; Peng, W.; Shi, W.; Wang, J.; Li, B.; Zhang, C.; Duan, C. Circular RNAs in cancer: An emerging key player. J. Hematol. Oncol. 2017, 10, 2. [Google Scholar] [CrossRef]
- Zheng, Q.; Bao, C.; Guo, W.; Li, S.; Chen, J.; Chen, B.; Luo, Y.; Lyu, D.; Li, Y.; Shi, G.; et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 2016, 7, 11215. [Google Scholar] [CrossRef]
- Kleaveland, B.; Shi, C.Y.; Stefano, J.; Bartel, D.P. A Network of Noncoding Regulatory RNAs Acts in the Mammalian Brain. Cell 2018, 174, 350–362.e17. [Google Scholar] [CrossRef]
- Yu, C.Y.; Li, T.C.; Wu, Y.Y.; Yeh, C.H.; Chiang, W.; Chuang, C.Y.; Kuo, H.C. The circular RNA circBIRC6 participates in the molecular circuitry controlling human pluripotency. Nat. Commun. 2017, 8, 1149. [Google Scholar] [CrossRef]
- Wu, D.M.; Wen, X.; Han, X.R.; Wang, S.; Wang, Y.J.; Shen, M.; Fan, S.H.; Zhang, Z.F.; Shan, Q.; Li, M.Q.; et al. Role of Circular RNA DLEU2 in Human Acute Myeloid Leukemia. Mol. Cell Biol. 2018, 38, e00259-18. [Google Scholar] [CrossRef]
- Fan, H.; Li, Y.; Liu, C.; Liu, Y.; Bai, J.; Li, W. Circular RNA-100290 promotes cell proliferation and inhibits apoptosis in acute myeloid leukemia cells via sponging miR-203. Biochem. Biophys. Res. Commun. 2018, 507, 178–184. [Google Scholar] [CrossRef]
- Wang, J.; Pan, J.; Huang, S.; Li, F.; Huang, J.; Li, X.; Ling, Q.; Ye, W.; Wang, Y.; Yu, W.; et al. Development and validation of a novel circular RNA as an independent prognostic factor in acute myeloid leukemia. BMC Med. 2021, 19, 28. [Google Scholar] [CrossRef]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets--update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef]
- Li, W.; Zhong, C.; Jiao, J.; Li, P.; Cui, B.; Ji, C.; Ma, D. Characterization of hsa_circ_0004277 as a New Biomarker for Acute Myeloid Leukemia via Circular RNA Profile and Bioinformatics Analysis. Int. J. Mol. Sci. 2017, 18, 597. [Google Scholar] [CrossRef]
- Tan, S.F.; Liu, X.; Fox, T.E.; Barth, B.M.; Sharma, A.; Turner, S.D.; Awwad, A.; Dewey, A.; Doi, K.; Spitzer, B.; et al. Acid ceramidase is upregulated in AML and represents a novel therapeutic target. Oncotarget 2016, 7, 83208–83222. [Google Scholar] [CrossRef]
- Zhou, N.; Yuan, X.; Du, Q.; Zhang, Z.; Shi, X.; Bao, J.; Ning, Y.; Peng, L. FerrDb V2: Update of the manually curated database of ferroptosis regulators and ferroptosis-disease associations. Nucleic Acids Res. 2022, gkac953. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Dudekula, D.B.; Panda, A.C.; Grammatikakis, I.; De, S.; Abdelmohsen, K.; Gorospe, M. CircInteractome: A web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016, 13, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Wang, Q.; Shen, J.; Yang, B.B.; Ding, X. Circbank: A comprehensive database for circRNA with standard nomenclature. RNA Biol. 2019, 16, 899–905. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Ji, P.; Zhao, F. CircAtlas: An integrated resource of one million highly accurate circular RNAs from 1070 vertebrate transcriptomes. Genome Biol. 2020, 21, 101. [Google Scholar] [CrossRef]
- Riffo-Campos, Á.L.; Riquelme, I.; Brebi-Mieville, P. Tools for Sequence-Based miRNA Target Prediction: What to Choose? Int. J. Mol. Sci. 2016, 17, 1987. [Google Scholar] [CrossRef]
- Li, J.H.; Liu, S.; Zhou, H.; Qu, L.H.; Yang, J.H. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014, 42, D92–D97. [Google Scholar] [CrossRef]
- Dweep, H.; Sticht, C.; Pandey, P.; Gretz, N. miRWalk--database: Prediction of possible miRNA binding sites by "walking" the genes of three genomes. J. Biomed. Inform. 2011, 44, 839–847. [Google Scholar] [CrossRef]
- Jia, A.; Xu, L.; Wang, Y. Venn diagrams in bioinformatics. Brief Bioinform. 2021, 22, bbab108. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed]
- Chin, C.H.; Chen, S.H.; Wu, H.H.; Ho, C.W.; Ko, M.T.; Lin, C.Y. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014, 8 (Suppl. 4), S11. [Google Scholar] [CrossRef]
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.C.; Muller, M. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011, 12, 77. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Pan, X.; Shen, H.B. lncLocator 2.0: A cell-line-specific subcellular localization predictor for long non-coding RNAs with interpretable deep learning. Bioinformatics 2021, 37, 2308–2316. [Google Scholar] [CrossRef] [PubMed]
- Cumbo, C.; Tarantini, F.; Anelli, L.; Zagaria, A.; Specchia, G.; Musto, P.; Albano, F. FLT3 mutational analysis in acute myeloid leukemia: Advantages and pitfalls with different approaches. Blood Rev. 2022, 54, 100928. [Google Scholar] [CrossRef]
- Chen, J.J.; Lei, P.; Zhou, M. hsa_circ_0121582 inhibits leukemia growth by dampening Wnt/β-catenin signaling. Clin. Transl. Oncol. 2020, 22, 2293–2302. [Google Scholar] [CrossRef]
- Long, J.; Jia, M.Y.; Fang, W.Y.; Chen, X.J.; Mu, L.L.; Wang, Z.Y.; Shen, Y.; Xiang, R.F.; Wang, L.N.; Wang, L.; et al. FLT3 inhibition upregulates HDAC8 via FOXO to inactivate p53 and promote maintenance of FLT3-ITD+ acute myeloid leukemia. Blood 2020, 135, 1472–1483. [Google Scholar] [CrossRef]
- Sykes, S.M.; Lane, S.W.; Bullinger, L.; Kalaitzidis, D.; Yusuf, R.; Saez, B.; Ferraro, F.; Mercier, F.; Singh, H.; Brumme, K.M.; et al. AKT/FOXO signaling enforces reversible differentiation blockade in myeloid leukemias. Cell 2011, 146, 697–708. [Google Scholar] [CrossRef]
- Liu, N.; Liang, Y.; Wei, T.; Zou, L.; Huang, X.; Kong, L.; Tang, M.; Zhang, T. The role of ferroptosis mediated by NRF2/ERK-regulated ferritinophagy in CdTe QDs-induced inflammation in macrophage. J. Hazard. Mater. 2022, 436, 129043. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Jiang, L.; Tavana, O.; Gu, W. The Deubiquitylase OTUB1 Mediates Ferroptosis via Stabilization of SLC7A11. Cancer Res. 2019, 79, 1913–1924. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Lu, Y.; Zhang, Y.; Wang, L. Construction of an Immune-Related Six-lncRNA Signature to Predict the Outcomes, Immune Cell Infiltration, and Immunotherapy Response in Patients with Hepatocellular Carcinoma. Front. Oncol. 2021, 11, 661758. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Sun, J.; Chen, H.; Ding, C.; Tang, Z.; Ruan, Y.; Liu, F.; Sun, Y. Establishment of an Immune Cell Infiltration Score to Help Predict the Prognosis and Chemotherapy Responsiveness of Gastric Cancer Patients. Front. Oncol. 2021, 11, 650673. [Google Scholar] [CrossRef]
- Sendker, S.; Reinhardt, D.; Niktoreh, N. Redirecting the Immune Microenvironment in Acute Myeloid Leukemia. Cancers 2021, 13, 1423. [Google Scholar] [CrossRef]
- Nahas, M.R.; Stroopinsky, D.; Rosenblatt, J.; Cole, L.; Pyzer, A.R.; Anastasiadou, E.; Sergeeva, A.; Ephraim, A.; Washington, A.; Orr, S.; et al. Hypomethylating agent alters the immune microenvironment in acute myeloid leukaemia (AML) and enhances the immunogenicity of a dendritic cell/AML vaccine. Br. J. Haematol. 2019, 185, 679–690. [Google Scholar] [CrossRef]
- Yan, H.; Qu, J.; Cao, W.; Liu, Y.; Zheng, G.; Zhang, E.; Cai, Z. Identification of prognostic genes in the acute myeloid leukemia immune microenvironment based on TCGA data analysis. Cancer Immunol. Immunother. 2019, 68, 1971–1978. [Google Scholar] [CrossRef]
- Misir, S.; Wu, N.; Yang, B.B. Specific expression and functions of circular RNAs. Cell Death Differ. 2022, 29, 481–491. [Google Scholar] [CrossRef]
- Ye, Y.; Wu, X.; Long, F.; Yue, W.; Wu, D.; Xie, Y. Circular RNA _0015278 inhibits the progression of non-small cell lung cancer through regulating the microRNA 1278/SOCS6 gene axis. Ann. Transl. Med. 2021, 9, 1255. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, J.; Feng, J.; Song, X.; Yang, Q.; Zhao, H.; Zhao, R.; He, X.; Tian, Y.; Wang, L.; Liu, Y. Hsa_circ_0015278 Regulates FLT3-ITD AML Progression via Ferroptosis-Related Genes. Cancers 2023, 15, 71. https://doi.org/10.3390/cancers15010071
Jiang J, Feng J, Song X, Yang Q, Zhao H, Zhao R, He X, Tian Y, Wang L, Liu Y. Hsa_circ_0015278 Regulates FLT3-ITD AML Progression via Ferroptosis-Related Genes. Cancers. 2023; 15(1):71. https://doi.org/10.3390/cancers15010071
Chicago/Turabian StyleJiang, Jiquan, Jing Feng, Xiangnan Song, Qing Yang, Hongbo Zhao, Rui Zhao, Xinrui He, Yaoyao Tian, Lianjie Wang, and Yanhong Liu. 2023. "Hsa_circ_0015278 Regulates FLT3-ITD AML Progression via Ferroptosis-Related Genes" Cancers 15, no. 1: 71. https://doi.org/10.3390/cancers15010071
APA StyleJiang, J., Feng, J., Song, X., Yang, Q., Zhao, H., Zhao, R., He, X., Tian, Y., Wang, L., & Liu, Y. (2023). Hsa_circ_0015278 Regulates FLT3-ITD AML Progression via Ferroptosis-Related Genes. Cancers, 15(1), 71. https://doi.org/10.3390/cancers15010071




