Simple Summary
Both genetic and epigenetic mechanisms contribute to the pathogenesis of breast cancer. Since Tahiliani et al. identified TET1 as the first methyl-cytosine dioxygenase in 2009, accumulating evidence has shown that aberrant 5mC oxidation and dysregulated TET family genes are associated with diseases, including breast cancer. In this review we provide an overview on the diagnosis and prognosis values of aberrant 5mC oxidation in breast cancer and emphasize the causes and consequences of such epigenetic alterations.
Abstract
DNA methylation (5-methylcytosine, 5mC) was once viewed as a stable epigenetic modification until Rao and colleagues identified Ten-eleven translocation 1 (TET1) as the first 5mC dioxygenase in 2009. TET family genes (including TET1, TET2, and TET3) encode proteins that can catalyze 5mC oxidation and consequently modulate DNA methylation, not only regulating embryonic development and cellular differentiation, but also playing critical roles in various physiological and pathophysiological processes. Soon after the discovery of TET family 5mC dioxygenases, aberrant 5mC oxidation and dysregulation of TET family genes have been reported in breast cancer as well as other malignancies. The impacts of aberrant 5mC oxidation and dysregulated TET family genes on the different aspects (so-called cancer hallmarks) of breast cancer have also been extensively investigated in the past decade. In this review, we summarize current understanding of the causes and consequences of aberrant 5mC oxidation in the pathogenesis of breast cancer. The challenges and future perspectives of this field are also discussed.
1. Introduction
Breast cancer is the most commonly diagnosed malignant tumor in women (2.3 million cases in 2020) and also one of the major causes of cancer-associated patient death (0.68 million deaths in 2020) worldwide [1]. The advances in the genetics and cell signaling studies have revolutionized our understanding of breast cancer pathogenesis and brought great progress in diagnosis and therapy. However, there is still a big gap in our understanding of the heterogeneity of breast cancer and their progressive metastasis and drug resistance, a process tightly associated with the plasticity of cancer genome in response to environmental changes.
Epigenetic regulation, which links the extracellular environments to the genome, plays an essential role in cell fate determination and homeostasis maintenance. Accumulating evidence indicates that the epigenetic alterations including DNA methylation changes are involved in the pathogenesis of breast cancer [2]. DNA methylation has been once regarded as a stable epigenetic modification until Tahiliani et al. identified TET1 as the first methyl-cytosine dioxygenase [3]. TET proteins catalyze 5mC oxidation and generate cytosine modifications such as 5hmC/5fC/5caC, leading to active or passive DNA demethylation [3,4,5]. Although TET1-MLL translocation was observed in acute myeloid leukemia (AML) as early as 2003 [6], the exact biochemical activity of TET proteins and their functional roles have been extensively studied since 2009 (Figure 1). Here we present the current knowledge on the dysregulated TET family genes and aberrant 5mC oxidation in breast cancer as well as its contributions to many aspects of cancer hallmarks.
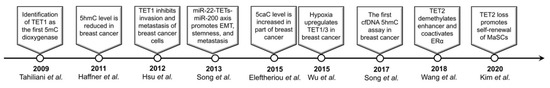
Figure 1.
Milestones of the research for TET/5mC oxidation in breast cancer.
2. Aberrant 5mC Oxidation in Breast Cancer
5hmC/5fC/5caC are derived from 5mC, while their abundances are much less than 5mC in cells [5]. Interestingly, these 5mC derivatives (i.e., 5hmC) display much more dynamic changes in response to the fluctuation of physiological or pathophysiological conditions [7]. In 2011, Haffner et al. first reported that breast cancer tissues as well as other tumor tissues showed much lower 5hmC levels compared with their adjacent normal tissues [8]. Of note, the 5hmC level in primary cultured cells (including mammary epithelial cells) declines quickly during the in vitro culture [9,10], suggesting that the TET/5hmC epigenetic pathway is dramatically altered by the culture environment. Alternatively, the 5mC oxidation may be sensitive to the status of cell proliferation as Bachman et al. found that the 5hmC level is refractory to cell mitosis except for embryonic stem cells [11].
Aberrant 5mC oxidation (mainly indicated by the level of 5hmC) has potential diagnostic, prognostic, and predictive values for breast cancer patients. For instance, Tsai et al. found that 5hmC has prognostic value only in ERα-negative breast cancer [12]. In their study, lower global 5hmC levels in tumor tissues were associated with poorer prognosis of patients with ER/PR-negative subtype. In contrast, Wu et al. observed an increase of 5hmC in breast cancer [13]. The reports regarding the prognostic value of 5hmC in breast cancer are contradictory, suggesting that the prognostic value of 5hmC might be highly dependent on the subtypes of breast cancer. The abundance of 5fC and 5caC is much lower than that of 5hmC. There is only one study reporting the 5caC levels in breast cancer [14]. Unlike 5hmC, the levels of 5caC are elevated in some breast cancers (27%) and surprisingly, the intensity of 5caC does not correlate with that of 5hmC. The authors proposed that the unexpected 5caC increase in some breast cancers may be due to thymine DNA glycosylase (TDG) loss-of-function.
In addition to the global changes of 5hmC/5fC/5caC levels, several labs have reported that the 5hmC levels on specific gene loci (i.e., LZTS1 and HLA-G) declined in breast cancer [15,16]. By comparing the genome-wide 5hmC and 5mC distribution in basal-like breast cancers (BLBCs), Collignon et al. found that 5hmC gain in TET1-high breast cancers compared with TET1-low breast cancer was mostly associated with 5mC loss [17], suggesting that TET1 participates in the maintenance of DNA hypomethylation in these regions.
With the rapid development of liquid biopsy, the plasma cell-free DNA (cfDNA) in peripheral blood samples has become an alternative source for 5hmC detection. Given that a small percentage of cfDNA may be derived from tumor cells in cancer patients, it is rational to detect them according to cancer cell-specific epigenetic information (i.e., 5mC and 5hmC). Like 5mC, 5hmC also has the potential to be an epigenetic marker in the cfDNA assay for breast cancer patients. Although the prognostic value of 5mC in cfDNA has been widely studied, the role of cfDNA 5hmC in breast cancer is largely unknown [18]. According to the progress of cfDNA 5hmC detection in other malignancies (i.e., lung cancer, liver cancer, colorectal cancer, and pancreatic cancer) [18,19,20,21,22], it is promising to identify the breast cancer-specific 5hmC signatures in the plasma cfDNA of breast cancer patients.
Taken together, the reduction of the global 5hmC level is likely an epigenetic hallmark of breast cancer as compared to healthy mammary gland tissue; however, the diagnostic, prognostic, and predictive values of the global 5hmC/5caC/5fC levels in breast cancer remain elusive. It still requires further intensive research to identify the subtype- and stage-specific 5hmC signatures at specific gene loci in breast cancer.
3. Dysregulation of TET Family Genes in Breast Cancer
3.1. Genetic Alterations of TET Family Genes in Breast Cancer
Among the three members of TET family genes (TET1, TET2, and TET3), TET1 chromosome translocation and TET2 mutations have been identified in hematopoietic malignancies such as AML and chronic myelomonocytic leukemia (CMML). Unlike the recurrent mutations in PI3KCA, TP53, NOTCH, and BRCA1/2, the genetic alterations of TET genes are rare in breast cancer. A recent GWAS analysis revealed a risk SNP associated with breast cancer is located within the TET2 gene [23]. However, it is unclear whether this SNP influences the expression or function of the TET2 gene.
Clonal hematopoiesis with indetermined potential (CHIP), a common age-associated genetic alteration, is associated with the increased risk of leukemia and cardiovascular diseases in elders [24,25]. Among the mutated genes, TET2 is the No.2 gene with the highest mutation rates in elders [26]. However, it is unclear whether CHIP has any contribution to the development and progression of breast cancer in elder populations. Moreover, it is also unknown whether breast cancer cells have a similar mechanism of clonal expansion and genetic selection. Given that luminal breast cancer mainly occurs in old population, it is of great interest to investigate the relationship between the hematopoietic TET2 mutations and the initiation and progression of breast cancer.
3.2. Aberrant Expression of TET Family Genes in Breast Cancer
As early as 2011, Yang et al. reported that all three members of TET family genes were lowly expressed in multiple types of tumor tissues (including breast cancer) as compared with their adjacent normal tissues [27]. Similarly, Ruan and colleagues found that, of the 140 pairs of sample tissue, 95 (68%) exhibited lower levels of TET1 mRNA in cancer tissues as compared with their normal-tissue control counterparts. Moreover, the TET1 expression level was inversely correlated with breast cancer cell invasion and tumor development. In 2015, Shao and colleagues also confirmed that reduced expression of TET family genes, as well as TDG, are associated with poor prognosis of patients with early breast cancer [28]. However, there are also a few opposite reports. For instance, Wu et al. found that TET1/3 expression levels were closely associated with tumor hypoxia, tumor malignancy, and poor prognosis in breast cancer patients [13]. In their study, they revealed that hypoxia conditions could upregulate TET1/3 expression via HIF1α and in turn increase the 5hmC level in tumor cells. Indeed, they found HREs (Hypoxia response elements) in the upstream promoter regions of TET1/3 genes. In 2019, Collignon et al. found that TET1 is repressed by NF-κb in BLBC and high TET1 is associated with low levels of immune and defense response markers [17]. Estrogen signaling plays critical roles in luminal breast cancer and endocrine therapy has been successfully applied to the therapy of luminal breast cancer. Interestingly, several groups identified that estrogen signaling can upregulate TET2 gene transcription through several distal enhancers [29,30].
Interestingly, epigenetic mechanisms also contribute to the aberrant expression of TET family genes during breast cancer pathogenesis. For instance, Sang et al. reported that TET1 was downregulated and hypermethylated in highly metastatic breast cancer cell lines. Tao and colleagues found that EZH2 inhibitors could reactivate TET1 expression in basal-like breast cancer cell lines, indicating that TET1 gene might be epigenetically repressed by H3K27me3 in basal-like breast cancer [31]. Another issue of gene transcription that may be tightly associated with epigenetic mechanisms is the activation of alternative promoter. Good et al. identified an isoform of TET1 driven by an alternative promoter in breast cancer, which yields a TET1 isoform without CXXC domain [32]. They found that the expression level of this TET1 isoform is high in triple-negative breast cancer (TNBC) and predicts poor prognosis. Similarly, an alternative promoter of the TET2 gene was also reported in different tissues and cell lines including breast cancer cells although the biological significance of the new isoform remains unclear [33]. In addition, many miRNAs contribute to fine-tuning the expression of TET mRNAs. In 2013, Song et al. identified TET genes as targets of miR-22 in mammary tissues [34]. They found that miR-22 binds to the 3′UTR of TET mRNAs and represses their mRNA stability and expression. Later, dozens of miRNA have been identified as regulators of TET genes in different tissues/cells. However, little of them have been proved in breast cancer cells and a systematic evaluation is required to evaluate to what extent these miRNAs contribute to the dysregulated TET family genes in breast cancers.
The levels of TET proteins are also precisely controlled at the post-translational layer. For instance, acetylation of TET2 as well as other TET members may be regulated by p300 and HDAC1/2 [35]. Phosphorylation of TET2 at S99, which is regulated by AMPK and PP2A, stabilize the TET2 protein [36,37,38]. Phosphorylation of TET2 at T1939 and T1964, which are regulated by the JAK2 pathway, influence the inflammatory signaling [39]. Autophagy is a strategy to control the recycling of macromolecules. As aforementioned, p53 could modulate the TET2 protein level through autophagy [40]. However, it requires further study to determine to what extent these post-translational modifications and corresponding signaling pathways contribute to the deregulated expression of TET family genes in breast cancer.
3.3. The Rewired Catalytic Activity of TET Proteins in Breast Cancer
Several co-factors, such as α-KG, oxygen, iron, and ascorbic acid, are required for TET proteins-catalyzed 5mC oxidation [7]. The changes in the intracellular concentrations of these co-factors influence the enzymatic activity of TET proteins. The IDH1/2 mutants, encoded by two recurrent mutated genes in glioma and AML, generate 2HD, so-called “oncometabolite”, which competes for α-KG and inhibits TET catalytic activity [41]. SDH and LDH mutants that influence the metabolism of succinate also inhibit TET activity [42]. In contrast, transient increase in α-KG concentration by glucose or glutamine shock increases 5hmC in vitro and in vivo [43]. Given that high glucose intake and consumption are features of most cancer cells, it will be of great interest to investigate whether the “Warburg effect” has any direct effect on 5mC oxidation catalyzed by TET proteins.
Ascorbic acid (also known as vitamin C) could recycle the iron for TET proteins-catalyzed 5mC oxidation [44]. Ascorbic acid has been applied to treat different cancer cells in a dish or mice and achieved exciting success. For instance, ascorbic acid treatment could reverse the leukemogenesis in mice by restoring the function of TET proteins [45]. A recent study has reported that ascorbic acid treatment could increase the 5hmC level and activate apoptosis gene expression such as TRAIL in breast cancer cells [46]. In theory, ascorbic acid treatment could increase 5hmC level and facilitate the maintenance of a healthy DNA methylome partially through modulating TET-catalyzed 5mC oxidation and DNA demethylation. Since ascorbic acid can be easily supplied as a nutrient in food (i.e., vegetables and fruits), more evidence is required to determine whether and how ascorbic acid could provide benefits in the prevention and therapy of breast cancer and to what extent such benefits are dependent on TET proteins-mediated 5mC oxidation.
Hypoxia is a common feature of solid tumor microenvironment due to the rapid growth of tumor cells and insufficient vascular blood supply. Since oxygen is essential for the oxidative reaction, low concentration of oxygen in tumor microenvironment could inhibit the catalytic activity of TET proteins, leading to a low 5hmC level in tumor cells [47]. This finding is completely opposite to the result of Wu et al. that hypoxia upregulates TET1/3 expression via HIF1α and thereby increases the 5hmC level [13], suggesting that TET has a complicated relationship with hypoxia. On one hand, TET1 gene is a target of HIF1α. There are a few HREs in the upstream regulatory region of TET1 gene and acute hypoxia can induce TET1 transcription. On the other hand, TET1 protein could synergize HIF1α to activate the expression of HIF1α target genes. However, hypoxia (if the O2 concentration is lower than the Km) itself may impair TET proteins-catalyzed 5mC oxidation. Given that sometimes these mechanisms may reach the opposite effect, it will be of importance to distinguish which mechanism plays the dominant role in certain conditions.
3.4. Aberrant Genomic Targeting of TET Proteins in Breast Cancer
Both TET1 and TET3 contain CXXC domain in their N-termini, which can facilitate their binding to CpG islands and thereby protect the CpG islands from DNA hypermethylation [48,49,50]. Although TET2 does not contain CXXC domain, CXXC4/IDAX and CXXC5/RINF, encoded by CXXC4 (a neighbor gene of TET2) and CXXC5, respectively, can bind to TET2 and facilitate its chromatin recruitment to CpG islands but also trigger the cleavage of TET2 by Caspase 3 [51]. In addition to the CXXC domain which prefers binding to DNA sequence with high-density GpG, transcriptional factors could interact with TET proteins and recruit them to specific gene loci for 5mC oxidation and DNA demethylation. In different tissues, various TFs have been identified to recruit TET2 protein to their unique genomic targets [52,53]. In breast cancer, FOXA1 which activates TET1 gene expression could also recruit TET1 to a subgroup of enhancer regions [54]. Therefore, the aberrant expression of such TFs or their mutations may change the action mode of TET proteins-mediated 5mC oxidation in breast cancer.
As a female prevalent cancer, breast cancer has tight connection with estrogen signaling. Activation of the nuclear receptor ERα and subsequent ERα-controlled transcriptome is the primary molecular mechanism through which estrogen exerts its physiological and pathophysiological functions. In fact, around 70% breast cancers belong to ERα+ breast cancer (so-called luminal A or B subtypes). It has been revealed that ERα cistrome and transcriptome were reprogrammed during the pathogenesis and progression of ERα+ breast cancer cells. A recent report found that TET2 functions as a co-activator of ERα and regulates the activity of enhancers containing estrogen response elements (EREs) through DNA demethylation [30]. Broome et al. proved the interaction between TET2 and ERα/GATA3 complex in breast cancer cells [55]. Co-localization of TET2 and ERα contributed to the maintenance of 5hmC at ERα-targeted enhancers, which regulated expression of ERα-targeted genes. Meanwhile, disrupting the TET2/ERα/GATA3 complex resulted in the dysregulation of cell cycle related gene expression. Our recent work also showed that TET2 loss led to DNA hypermethylation on a large proportion of enhancers including EREs in MCF7 cells [56]. Consistent to the notion that CpG methylation within EREs impairs ERα binding, our ERα ChIP-seq demonstrated that loss of TET2 impaired the ERα binding and gene transcription of a subgroup of E2-responsive genes in MCF7 cells. However, unlike several previous studies, we did not detect the binding between ERα and TET2 by co-IP experiment, indicating that other molecule (s) might be responsible for the recruitment of TET2 to these enhancers. Collectively, TET2-mediated 5mC oxidation is involved in the regulation of DNA methylation and transcriptional activity of certain enhancers including EREs. However, the detailed molecular mechanisms through which TET2 is recruited to these enhancers and how these different events are coordinated remain controversial and require further study.
4. Consequences of Dysregulated TET Family Genes and Aberrant 5mC Oxidation in Breast Cancer
Given the critical role of TET proteins-catalyzed 5mC oxidation in the regulation of DNA methylation dynamics, the dysregulation of TET family genes may participate in the development and progression of breast cancer. At the molecular level, aberrant 5mC oxidation could partially contribute to the well-known epigenetic hallmark of cancer (i.e., global DNA hypo-methylation but local DNA hypermethylation) [7]. In addition, TET proteins could also exert catalytic activity-independent function in the regulation of chromatin structure and gene transcription [7]. Here, we summarize the broad impact of dysregulated TET family genes and aberrant 5mC oxidation on multiple aspects (cancer hallmarks) of breast cancer (Figure 2).
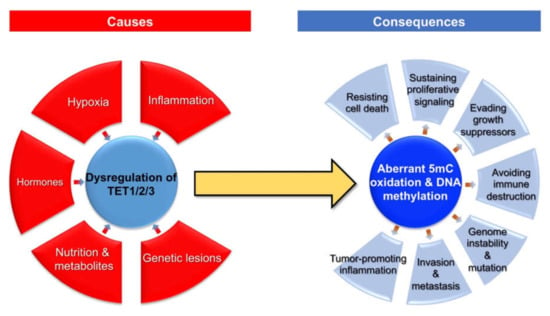
Figure 2.
Causes and consequences of aberrant 5mC oxidation in breast cancer.
4.1. Genomic Instability and Mutation
Chromosome abnormality and gene mutations are accumulated during the initiation and progression of breast cancer. DNA methylation plays essential role in the maintenance of genomic stability through epigenetic silencing of repeated elements [57]. In addition, the TDG-mediated base excision repair (BER) pathway as well as other DNA repair pathways (DNA deamination and glycosylation) have been proposed to work together with TET/5mC oxidation [4,58]. Cellular immunostaining and hMeDIP-seq analysis revealed that 5hmC signal was enriched in the regulatory and transcribed regions within the euchromatin regions but depleted from the heterochromatin regions [49,59,60]. The differential patterns of genomic 5hmC and 5mC distribution suggest that only the 5mC within the relatively open chromatin regions could undergo TET proteins-catalyzed oxidative reaction while the 5mC within the heterochromatin regions might be protected by certain mechanisms. Unexpectedly, there are several reports showing that the repeated regions (i.e., LINE1) might be activated by TET1/2 proteins [61,62,63]. However, a recent report showed that TET2 depletion leads to DNA hypomethylation in the intergenic repeated regions [64]. The authors speculate that this unexpected phenomenon might be caused by the redistribution of Dnmt3a/b from heterochromatin to euchromatin due to the loss of Tet2 competition in euchromatin.
4.2. Stemness
Loss of TETs results in DNA hyper-methylation and epigenetic silencing of miR-200 which leads to epithelial-mesenchymal transition (EMT) and increases the stemness of breast cancer cells [34]. Moreover, TET2 may suppress breast cancer stem cells (CSC) through the regulation of the miRNA200c/PKCζ axis [65]. The comparison of wild-type and Tet2−/− mice also revealed that loss of Tet2 enhances the self-renewal of mammary stem cells (MaSCs) while impairing the luminal lineage commitment [66]. However, there were also some opposite reports. Wu et al. revealed that increased TET1/3 levels in breast cancers are associated with TNF-α expression, which activates the p38-MAPK pathway and maintains breast tumor-initiating cells (BTIC) [13]. Slow-cycling cancer cells (SCCCs) were believed as the reservoir of chemoresistance during cancer progression. Puig and colleagues identified TET2 as a marker for the stem-like cell in tumors and showed that 5hmC was enriched in the persisting SCCCs [67]. Therefore, it remains a debate on whether TET family genes play a positive (oncogenic) or negative (tumor suppressive) role in the maintenance of cancer stem cells
4.3. Cell Proliferation
Hormone positive breast cancer cells showed addiction to E2/ER signaling. Ali and colleagues found that loss of TET2 impairs E2/ERα-stimulated cell growth [30], indicating an oncogenic role of TET2 in ERα+ breast cancer. In their study, TET2 acts as a co-activator to promote E2/ERα-induced gene transcription via DNA demethylation and MLL-dependent H3K4me1/2 generation at enhancers. Moreover, E2/ER promotes TET2 gene transcription in MCF7 cells due to the binding of ERα to 3 ERE upstream of the TET2 gene, suggesting a positive feedback between ERα mediated gene transcriptional activation and TET2-mediated enhancer DNA demethylation. Conversely, our work suggests a tumor-suppressive role of TET2 in ERα+ breast cancer [56]. In our work, TET2 depletion has no significant effect on the adherent cell growth while promotes the anchorage-independent growth of MCF7 cells. Further epigenomic and transcriptomic analyses revealed that TET2 depletion led to selective enhancer DNA methylation and subsequent impairment of TF binding and gene transcription. Interestingly, some apoptotic death genes were among these enhancers/genes, indicating that TET2 functions as a brake or negative feedback in E2/ERα signaling by maintaining the expression of those negative regulators.
4.4. Invasion and Metastasis
The first report on the functional role of TET genes in breast cancer was from Ruan and colleagues [68]. In 2012, they demonstrated a tumor-suppressive role of TET1 in breast cancer as knockdown of TET1 by shRNA promotes the invasion and metastasis of breast cancer cells. Mechanistically, they found that TET1 could promote the DNA demethylation on the promoters of TIMP2/3 genes, which encode the inhibitors of MMPs and inhibit extracellular matrix degradation [68]. Soon after, Sun et al. reported that TET1 activates HOXA9 gene via DNA demethylation, and the latter is involved in the regulation of migration and metastasis [69]. Pandolfi and colleagues found that TET proteins activate the expression of miRNA-200 via DNA demethylation of mir-200 promoter [34]. Low expression of TET genes leads to decreased miRNA-200a/b/c and enhanced epithelial–mesenchymal transition (EMT) of breast cancer cells, an important process not only in stemness but also in invasion and metastasis. TET2 could inhibit the migration and invasion of breast cancer cells through the demethylation of EpCAM and E-cadherin, again preceding their expression and activation [70].
4.5. Cell Death
The functional roles of TET family genes in cell death induced by chemotherapy and radiotherapy are controversial. A recent study showed that TET2 interacts with promyelocytic leukemia (PML), which plays a critical role in cell-cycle arrest, senescence, apoptosis, genome stability, and antiviral effects [71]. Loss of TET2 is associated with increased resistance of cancer cells to chemotherapy in vitro and in vivo [71]. Moreover, Sant et al. found that vitamin C, which activates TET family proteins, could induce apoptosis in several breast cancer cell lines through upregulating the expression of TRAIL [46,72]. However, another study in the neck and head tumor revealed that high 5hmC and increased TET2 expression are associated with dormancy and drug resistance to chemotherapy while knockdown of TET2 eliminates the dormant cancer cells [67]. As TET2 was downregulated by p53-mediated autophagy, p53 loss-induced TET2 increase facilitates the acquisition of chemoresistance in cancer cells [40]. Therefore, the molecular mechanism underlying this discrepancy is not fully understood and requires further in-depth research.
4.6. Inflammation and Tumor Immunity
The role of epigenetic regulation in inflammation and tumor immunity has attracted a lot of attention. Collignon et al. observed an anti-correlation between TET1 expression level and the extent of infiltration by the major types of leukocytes in BLBC, while such anti-correlation did not exist in other subtypes of breast cancers [17]. Together with the findings that TET1 expression is increased in some BLBC, these data indicate that TET1 may play an oncogenic role in BLBC and such oncogenic role is partially associated with decreased inflammatory response and anti-tumor response. Again, their findings also reinforce a subtype-specific role of TET1 in modulating the inflammation and cancer immunity in breast cancer.
Xiong and colleagues found that TET2 is essential for IFN-gamma induced chemokine gene expression in melanoma and colon cancer cells [73]. As one of the IFN-gamma induced genes, PD-L1 was also enhanced by TET2. Conversely, our recent work revealed that TET2 represses the expression of the PD-L1 gene in breast cancer either in the presence or absence of IFN-gamma treatment [74]. Mechanistically, we found that TET2-mediated PD-L1 repression was independent of DNA methylation, instead of the recruitment of HDAC1/2 by TET2 to the promoter of PD-L1 gene. The inhibitors of DNMTs, such as 5-Aza, could activate endogenous virus expression, which in turn activate type-I IFN response, converting the “cold” tumor to “hot” tumor. Given that TET2 knockout leads to intergenic DNA hypomethylation, it is possible that TET proteins also modulate the responsiveness of tumor cells to immunotherapy through similar mechanisms.
In addition to the well-studied cell-autonomous role of TET2 in tumor cells, our lab also revealed that the comprehensive knockout of Tet2 in mice leads to an impaired anti-tumor immunity due to the overactivation of granulocytic myeloid-derived suppressor cells (G-MDSCs) which subsequently decrease the number of cytotoxic CD8+ T cells [75]. Therefore, TET family genes regulate anti-tumor immunity of breast cancer from not only the aspect of cancer cells but also the non-tumor cells in tumor microenvironment especially the immune cells. Since targeting the immune checkpoint achieved great success in clinical cancer therapy including the TNBCs, it will be of great interest to investigate whether TETs are new targets to improve the anti-tumor immunity and immunotherapy efficiency of breast cancer.
5. Conclusions
As the initial step of DNA demethylation, TET proteins-catalyzed 5mC oxidation regulates the stability and plasticity of the epigenomes of mammary tissues. The dysregulation of TET family genes and aberrant 5mC oxidation are involved in breast cancer pathogenesis. Of note, the value of 5hmC/5fC/5caC and TET gene expression in the diagnosis and prognosis of breast cancer is largely dependent on the subtypes of breast cancer, suggesting a double-edged sword role of TET/5mC oxidation pathway in breast cancer. Therefore, caution should be taken when distinguishing the exact role of TET family genes in different subtypes, aspects, and stages of breast cancer before we transform this knowledge in the diagnosis and therapy of breast cancer.
Author Contributions
Conceptualization, H.W. and L.T.; critical review of literature, B.X., H.W. and L.T.; writing—original draft preparation, B.X. and L.T.; writing—review and editing, H.W. and L.T.; funding acquisition, H.W. and L.T.; supervision, H.W. and L.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (82073113, 31871291, 81672785, and 81301615), the National Key R&D Project of China (2016YFA0101800), the Shanghai Municipal Key Clinical Specialty (shslczdzk03603), and the innovative research team of high-level local university in Shanghai.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Łukasiewicz, S.; Czeczelewski, M.; Forma, A.; Baj, J.; Sitarz, R.; Stanisławek, A. Breast Cancer—Epidemiology, Risk Factors, Classification, Prognostic Markers, and Current Treatment Strategies—An Updated Review. Cancers 2021, 13, 4287. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Martinez, L.; Zhang, Y.; Nakata, Y.; Chan, H.L.; Morey, L. Epigenetic mechanisms in breast cancer therapy and resistance. Nat. Commun. 2021, 12, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Tahiliani, M.; Koh, K.P.; Shen, Y.; Pastor, W.A.; Bandukwala, H.; Brudno, Y.; Agarwal, S.; Iyer, L.M.; Liu, D.R.; Aravind, L.; et al. Conversion of 5-Methylcytosine to 5-Hydroxymethylcytosine in Mammalian DNA by MLL Partner TET1. Science 2009, 324, 930–935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, Y.-F.; Li, B.-Z.; Li, Z.; Liu, P.; Wang, Y.; Tang, Q.; Ding, J.; Jia, Y.; Chen, Z.; Li, L.; et al. Tet-Mediated Formation of 5-Carboxylcytosine and Its Excision by TDG in Mammalian DNA. Science 2011, 333, 1303–1307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ito, S.; Shen, L.; Dai, Q.; Wu, S.C.; Collins, L.B.; Swenberg, J.A.; He, C.; Zhang, Y. Tet Proteins Can Convert 5-Methylcytosine to 5-Formylcytosine and 5-Carboxylcytosine. Science 2011, 333, 1300–1303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lorsbach, R.B.; Moore, J.; Mathew, S.; Raimondi, S.C.; Mukatira, S.T.; Downing, J.R. TET1, a member of a novel protein family, is fused to MLL in acute myeloid leukemia containing the t(10;11)(q22;q23). Leukemia 2003, 17, 637–641. [Google Scholar] [CrossRef] [Green Version]
- Tan, L.; Shi, Y.G. Tet family proteins and 5-hydroxymethylcytosine in development and disease. Development 2012, 139, 1895–1902. [Google Scholar] [CrossRef] [Green Version]
- Haffner, M.C.; Chaux, A.; Meeker, A.K.; Esopi, D.M.; Gerber, J.; Pellakuru, L.G.; Toubaji, A.; Argani, P.; Iacobuzio-Donahue, C.; Nelson, W.G.; et al. Global 5-hydroxymethylcytosine content is significantly reduced in tissue stem/progenitor cell compartments and in human cancers. Oncotarget 2011, 2, 627–637. [Google Scholar] [CrossRef] [Green Version]
- Nestor, C.E.; Ottaviano, R.; Reddington, J.; Sproul, D.; Reinhardt, D.; Dunican, D.; Katz, E.; Dixon, J.M.; Harrison, D.J.; Meehan, R.R. Tissue type is a major modifier of the 5-hydroxymethylcytosine content of human genes. Genome Res. 2011, 22, 467–477. [Google Scholar] [CrossRef] [Green Version]
- Nestor, C.E.; Ottaviano, R.; Reinhardt, D.; Cruickshanks, H.A.; Mjoseng, H.K.; McPherson, R.C.; Lentini, A.; Thomson, J.P.; Dunican, D.S.; Pennings, S.; et al. Rapid reprogramming of epigenetic and transcriptional profiles in mammalian culture systems. Genome Biol. 2015, 16, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Bachman, M.; Uribe-Lewis, S.; Yang, X.; Williams, M.; Murrell, A.; Balasubramanian, S. 5-Hydroxymethylcytosine is a predominantly stable DNA modification. Nat. Chem. 2014, 6, 1049–1055. [Google Scholar] [CrossRef] [Green Version]
- Tsai, K.-W.; Li, G.-C.; Chen, C.-H.; Yeh, M.-H.; Huang, J.-S.; Tseng, H.-H.; Fu, T.-Y.; Liou, H.-H.; Pan, H.-W.; Huang, S.-F.; et al. Reduction of global 5-hydroxymethylcytosine is a poor prognostic factor in breast cancer patients, especially for an ER/PR-negative subtype. Breast Cancer Res. Treat. 2015, 153, 219–234. [Google Scholar] [CrossRef]
- Wu, M.Z.; Chen, S.F.; Nieh, S.; Benner, C.; Ger, L.P.; Jan, C.I.; Ma, L.; Chen, C.H.; Hishida, T.; Chang, H.T.; et al. Hypoxia Drives Breast Tumor Malignancy through a TET-TNFalpha-p38-MAPK Signaling Axis. Cancer Res. 2015, 75, 3912–3924. [Google Scholar] [CrossRef] [Green Version]
- Eleftheriou, M.M.; Pascual, A.J.A.; Wheldon, L.L.; Perry, C.C.; Abakir, A.H.; Arora, A.A.; Johnson, A.D.A.; Auer, D.D.; Ellis, I.O.; Madhusudan, S.S.; et al. 5-Carboxylcytosine levels are elevated in human breast cancers and gliomas. Clin. Epigenet. 2015, 7, 1–6. [Google Scholar] [CrossRef]
- Matthias, W.; Liou, W.; Pulverer, W.; Singer, C.F.; Rappaport-Fuerhauser, C.; Kandioler, D.; Egger, G.; Weinhäusel, A. Cytosine 5-Hydroxymethylation of the LZTS1 Gene Is Reduced in Breast Cancer. Transl. Oncol. 2013, 6, 715–721. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.; An, X.; Li, Z.; Zhang, S. Role of gene promoter methylation regulated by TETs and DNMTs in the overexpression of HLA-G in MCF-7 cells. Exp. Ther. Med. 2019, 17, 4709–4714. [Google Scholar] [CrossRef]
- Collignon, E.; Canale, A.; Al Wardi, C.; Bizet, M.; Calonne, E.; Dedeurwaerder, S.; Garaud, S.; Naveaux, C.; Barham, W.; Wilson, A.; et al. Immunity drives TET1 regulation in cancer through NF-kappaB. Sci. Adv. 2018, 4, eaap7309. [Google Scholar] [CrossRef] [Green Version]
- Song, C.-X.; Yin, S.; Ma, L.; Wheeler, A.; Chen, Y.; Zhang, Y.; Liu, B.; Xiong, J.; Zhang, W.; Hu, J.; et al. 5-Hydroxymethylcytosine signatures in cell-free DNA provide information about tumor types and stages. Cell Res. 2017, 27, 1231–1242. [Google Scholar] [CrossRef] [Green Version]
- Cai, J.; Chen, L.; Zhang, Z.; Zhang, X.; Lu, X.; Liu, W.; Shi, G.; Ge, Y.; Gao, P.; Yang, Y.; et al. Genome-wide mapping of 5-hydroxymethylcytosines in circulating cell-free DNA as a non-invasive approach for early detection of hepatocellular carcinoma. Gut 2019, 68, 2195–2205. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Han, X.; Gao, C.; Xing, Y.; Qi, Z.; Liu, R.; Wang, Y.; Zhang, X.; Yang, Y.-G.; Li, X.; et al. 5-Hydroxymethylome in Circulating Cell-free DNA as A Potential Biomarker for Non-small-cell Lung Cancer. Genom. Proteom. Bioinform. 2018, 16, 187–199. [Google Scholar] [CrossRef]
- Li, W.; Zhang, X.; Lu, X.; You, L.; Song, Y.; Luo, Z.; Zhang, J.; Nie, J.; Zheng, W.; Xu, D.; et al. 5-Hydroxymethylcytosine signatures in circulating cell-free DNA as diagnostic biomarkers for human cancers. Cell Res. 2017, 27, 1243–1257. [Google Scholar] [CrossRef] [Green Version]
- Zeng, C.; Stroup, E.K.; Zhang, Z.; Chiu, B.C.-H.; Zhang, W. Towards precision medicine: Advances in 5-hydroxymethylcytosine cancer biomarker discovery in liquid biopsy. Cancer Commun. 2019, 39, 12–19. [Google Scholar] [CrossRef] [Green Version]
- Li, N.; Rowley, S.M.; Thompson, E.R.; McInerny, S.; Devereux, L.; Amarasinghe, K.C.; Zethoven, M.; Lupat, R.; Goode, D.; Li, J.; et al. Evaluating the breast cancer predisposition role of rare variants in genes associated with low-penetrance breast cancer risk SNPs. Breast Cancer Res. 2018, 20, 3. [Google Scholar] [CrossRef] [Green Version]
- Jaiswal, S.; Fontanillas, P.; Flannick, J.; Manning, A.; Grauman, P.V.; Mar, B.; Lindsley, C.; Mermel, C.; Burtt, N.; Chavez, A.; et al. Age-Related Clonal Hematopoiesis Associated with Adverse Outcomes. N. Engl. J. Med. 2014, 371, 2488–2498. [Google Scholar] [CrossRef] [Green Version]
- Xie, M.; Lu, C.; Wang, J.; McLellan, M.D.; Johnson, K.J.; Wendl, M.C.; McMichael, J.F.; Schmidt, H.K.; Yellapantula, V.; Miller, C.A.; et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat. Med. 2014, 20, 1472–1478. [Google Scholar] [CrossRef]
- Busque, L.; Buscarlet, M.; Mollica, L.; Levine, R.L. Concise Review: Age-Related Clonal Hematopoiesis: Stem Cells Tempting the Devil. Stem Cells 2018, 36, 1287–1294. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Liu, Y.; Bai, F.; Zhang, J.-Y.; Ma, S.-H.; Liu, J.; Xu, Z.-D.; Zhu, H.-G.; Ling, Z.-Q.; Ye, D.; et al. Tumor development is associated with decrease of TET gene expression and 5-methylcytosine hydroxylation. Oncogene 2013, 32, 663–669. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Yu, S.-J.; Hong, Q.; Yang, Y.; Shao, Z.-M. Reduced Expression of TET1, TET2, TET3 and TDG mRNAs Are Associated with Poor Prognosis of Patients with Early Breast Cancer. PLoS ONE 2015, 10, e0133896. [Google Scholar] [CrossRef]
- Sklias, A.; Halaburkova, A.; Vanzan, L.; Jimenez, N.F.; Cuenin, C.; Bouaoun, L.; Cahais, V.; Ythier, V.; Sallé, A.; Renard, C.; et al. Epigenetic remodelling of enhancers in response to estrogen deprivation and re-stimulation. Nucleic Acids Res. 2021, 49, 9738–9754. [Google Scholar] [CrossRef]
- Wang, L.; Ozark, P.A.; Smith, E.R.; Zhao, Z.; Marshall, S.A.; Rendleman, E.J.; Piunti, A.; Ryan, C.; Whelan, A.L.; Helmin, K.A.; et al. TET2 coactivates gene expression through demethylation of enhancers. Sci. Adv. 2018, 4, eaau6986. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Li, C.; Mao, H.; Du, Z.; Chan, W.Y.; Murray, P.; Luo, B.; Chan, A.T.C.; Mok, T.; Chan, F.K.; et al. Epigenetic inactivation of the CpG demethylase TET1 as a DNA methylation feedback loop in human cancers. Sci. Rep. 2016, 6, 26591. [Google Scholar] [CrossRef] [PubMed]
- Good, C.R.; Madzo, J.; Patel, B.; Maegawa, S.; Engel, N.; Jelinek, J.; Issa, J.-P.J. A novel isoform of TET1 that lacks a CXXC domain is overexpressed in cancer. Nucleic Acids Res. 2017, 45, 8269–8281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lou, H.; Li, H.; Ho, K.J.; Cai, L.; Huang, A.S.; Shank, T.R.; Verneris, M.R.; Nickerson, M.L.; Dean, M.; Anderson, S.K. The Human TET2 Gene Contains Three Distinct Promoter Regions with Differing Tissue and Developmental Specificities. Front. Cell Dev. Biol. 2019, 7, 99. [Google Scholar] [CrossRef] [PubMed]
- Song, S.J.; Poliseno, L.; Song, M.S.; Ala, U.; Webster, K.; Ng, C.; Beringer, G.; Brikbak, N.J.; Yuan, X.; Cantley, L.C.; et al. MicroRNA-antagonism regulates breast cancer stemness and metastasis via TET-family-dependent chromatin remodeling. Cell 2013, 154, 311–324. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.W.; Wang, Z.; Xie, W.; Cai, Y.; Xia, L.; Easwaran, H.; Luo, J.; Yen, R.-W.C.; Li, Y.; Baylin, S.B. Acetylation Enhances TET2 Function in Protecting against Abnormal DNA Methylation during Oxidative Stress. Mol. Cell 2017, 65, 323–335. [Google Scholar] [CrossRef] [Green Version]
- Wu, D.; Hu, D.; Chen, H.; Shi, G.; Fetahu, I.; Wu, F.; Rabidou, K.; Fang, R.; Tan, L.; Xu, S.; et al. Glucose-regulated phosphorylation of TET2 by AMPK reveals a pathway linking diabetes to cancer. Nat. Cell Biol. 2018, 559, 637–641. [Google Scholar] [CrossRef]
- Chen, H.; Yu, D.; Fang, R.; Rabidou, K.; Wu, D.; Hu, D.; Jia, P.; Zhao, Z.; Wu, Z.; Peng, J.; et al. TET2 stabilization by 14-3-3 binding to the phosphorylated Serine 99 is deregulated by mutations in cancer. Cell Res. 2019, 29, 248–250. [Google Scholar] [CrossRef] [Green Version]
- Kundu, A.; Shelar, S.; Ghosh, A.P.; Ballestas, M.; Kirkman, R.; Nam, H.; Brinkley, G.; Karki, S.; Mobley, J.A.; Bae, S.; et al. 14-3-3 proteins protect AMPK-phosphorylated ten-eleven translocation-2 (TET2) from PP2A-mediated dephosphorylation. J. Biol. Chem. 2020, 295, 1754–1766. [Google Scholar] [CrossRef]
- Jeong, J.J.; Gu, X.; Nie, J.; Sundaravel, S.; Liu, H.; Kuo, W.L.; Bhagat, T.D.; Pradhan, K.; Cao, J.; Nischal, S.; et al. Cytokine-Regulated Phosphorylation and Activation of TET2 by JAK2 in Hematopoiesis. Cancer Discov. 2019, 9, 778–795. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Tan, P.; Guo, L.; Gong, J.; Ma, J.; Li, J.; Lee, M.; Fang, S.; Jing, J.; Johnson, G.; et al. p53-dependent autophagic degradation of TET2 modulates cancer therapeutic resistance. Oncogene 2019, 38, 1905–1919. [Google Scholar] [CrossRef]
- Xu, W.; Yang, H.; Liu, Y.; Yang, Y.; Wang, P.; Kim, S.-H.; Ito, S.; Yang, C.; Wang, P.; Xiao, M.-T.; et al. Oncometabolite 2-Hydroxyglutarate Is a Competitive Inhibitor of α-Ketoglutarate-Dependent Dioxygenases. Cancer Cell 2011, 19, 17–30. [Google Scholar] [CrossRef] [Green Version]
- Hoekstra, A.S.; De Graaff, M.A.; Bruijn, I.H.B.-D.; Ras, C.; Seifar, R.M.; Van Minderhout, I.; Cornelisse, C.J.; Hogendoorn, P.; Breuning, M.H.; Suijker, J.; et al. Inactivation of SDH and FH cause loss of 5hmC and increased H3K9me3 in paraganglioma/pheochromocytoma and smooth muscle tumors. Oncotarget 2015, 6, 38777–38788. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Lin, H.; Xu, H.; Zhang, L.; Cheng, L.; Wen, B.; Shou, J.; Guan, K.; Xiong, Y.; Ye, D. TET-catalyzed 5-methylcytosine hydroxylation is dynamically regulated by metabolites. Cell Res. 2014, 24, 1017–1020. [Google Scholar] [CrossRef] [Green Version]
- Yin, R.; Mao, S.-Q.; Zhao, B.; Chong, Z.; Yang, Y.; Zhao, C.; Zhang, D.; Huang, H.; Gao, J.; Li, Z.; et al. Ascorbic Acid Enhances Tet-Mediated 5-Methylcytosine Oxidation and Promotes DNA Demethylation in Mammals. J. Am. Chem. Soc. 2013, 135, 10396–10403. [Google Scholar] [CrossRef]
- Agathocleous, M.; Meacham, C.E.; Burgess, R.J.; Piskounova, E.; Zhao, Z.; Crane, G.; Cowin, B.L.; Bruner, E.; Murphy, M.M.; Chen, W.; et al. Ascorbate regulates haematopoietic stem cell function and leukaemogenesis. Nat. Cell Biol. 2017, 549, 476–481. [Google Scholar] [CrossRef]
- Sant, D.W.; Mustafi, S.; Gustafson, C.B.; Chen, J.; Slingerland, J.M.; Wang, G. Vitamin C promotes apoptosis in breast cancer cells by increasing TRAIL expression. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef]
- Thienpont, B.; Steinbacher, J.; Zhao, H.; D’Anna, F.; Kuchnio, A.; Ploumakis, A.; Ghesquière, B.; Van Dyck, L.; Boeckx, B.; Schoonjans, L.; et al. Tumour hypoxia causes DNA hypermethylation by reducing TET activity. Nat. Cell Biol. 2016, 537, 63–68. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, X.; Clark, E.; Mulcahey, M.; Huang, S.; Shi, Y.G. TET1 is a DNA-binding protein that modulates DNA methylation and gene transcription via hydroxylation of 5-methylcytosine. Cell Res. 2010, 20, 1390–1393. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Wu, F.; Tan, L.; Kong, L.; Xiong, L.; Deng, J.; Barbera, A.J.; Zheng, L.; Zhang, H.; Huang, S.; et al. Genome-wide Regulation of 5hmC, 5mC, and Gene Expression by Tet1 Hydroxylase in Mouse Embryonic Stem Cells. Mol. Cell 2011, 42, 451–464. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Xu, C.; Kato, A.; Tempel, W.; Abreu, J.G.; Bian, C.; Hu, Y.; Hu, D.; Zhao, B.; Cerovina, T.; et al. Tet3 CXXC Domain and Dioxygenase Activity Cooperatively Regulate Key Genes for Xenopus Eye and Neural Development. Cell 2012, 151, 1200–1213. [Google Scholar] [CrossRef] [Green Version]
- Ko, M.; An, J.; Bandukwala, H.S.; Chavez, L.; Äijö, T.; Pastor, W.A.; Segal, M.F.; Li, H.; Koh, K.P.; Lähdesmäki, H.; et al. Modulation of TET2 expression and 5-methylcytosine oxidation by the CXXC domain protein IDAX. Nat. Cell Biol. 2013, 497, 122–126. [Google Scholar] [CrossRef]
- Rasmussen, K.D.; Jia, G.; Johansen, J.V.; Pedersen, M.T.; Rapin, N.; Bagger, F.O.; Porse, B.T.; Bernard, O.A.; Christensen, J.; Helin, K. Loss of TET2 in hematopoietic cells leads to DNA hypermethylation of active enhancers and induction of leukemogenesis. Genes Dev. 2015, 29, 910–922. [Google Scholar] [CrossRef] [Green Version]
- Sardina, J.L.; Collombet, S.; Tian, T.; Gómez, A.; Di Stefano, B.; Berenguer, C.; Brumbaugh, J.; Stadhouders, R.; Segura-Morales, C.; Gut, M.; et al. Transcription Factors Drive Tet2-Mediated Enhancer Demethylation to Reprogram Cell Fate. Cell Stem Cell 2018, 23, 905–906. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.A.; Zhao, J.C.; Fong, K.-W.; Kim, J.; Li, S.; Song, C.-X.; Song, B.; Zheng, B.; He, C.; Yu, J. FOXA1 potentiates lineage-specific enhancer activation through modulating TET1 expression and function. Nucleic Acids Res. 2016, 44, 8153–8164. [Google Scholar] [CrossRef]
- Broome, R.; Chernukhin, I.; Jamieson, S.; Kishore, K.; Papachristou, E.K.; Mao, S.-Q.; Tejedo, C.G.; Mahtey, A.; Theodorou, V.; Groen, A.J.; et al. TET2 is a component of the estrogen receptor complex and controls 5mC to 5hmC conversion at estrogen receptor cis-regulatory regions. Cell Rep. 2021, 34, 108776. [Google Scholar] [CrossRef]
- Lyu, R.; Zhu, X.; Shen, Y.; Xiong, L.; Liu, L.; Liu, H.; Wu, F.; Argueta, C.; Tan, L. Tumour suppressor TET2 safeguards enhancers from aberrant DNA methylation and epigenetic reprogramming in ERalpha-positive breast cancer cells. Epigenetics 2021, 2021, 1–15. [Google Scholar] [CrossRef]
- Sheaffer, K.L.; Elliott, E.N.; Kaestner, K.H. DNA Hypomethylation Contributes to Genomic Instability and Intestinal Cancer Initiation. Cancer Prev. Res. 2016, 9, 534–546. [Google Scholar] [CrossRef] [Green Version]
- Guo, J.U.; Su, Y.; Zhong, C.; Ming, G.-L.; Song, H. Hydroxylation of 5-Methylcytosine by TET1 Promotes Active DNA Demethylation in the Adult Brain. Cell 2011, 145, 423–434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ficz, G.; Branco, M.; Seisenberger, S.; Santos, F.; Krueger, F.; Hore, T.; Marques, J.; Andrews, S.; Reik, W. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nat. Cell Biol. 2011, 473, 398–402. [Google Scholar] [CrossRef] [PubMed]
- Williams, K.; Christensen, J.; Pedersen, M.T.; Johansen, J.V.; Cloos, P.; Rappsilber, J.; Helin, K. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nat. Cell Biol. 2011, 473, 343–348. [Google Scholar] [CrossRef]
- Guallar, D.; Bi, X.; Pardavila, J.A.; Huang, X.; Saenz, C.; Shi, X.; Zhou, H.; Faiola, F.; Ding, J.; Haruehanroengra, P.; et al. RNA-dependent chromatin targeting of TET2 for endogenous retrovirus control in pluripotent stem cells. Nat. Genet. 2018, 50, 443–451. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Ludwig, A.K.; Hastert, F.D.; Rausch, C.; Lehmkuhl, A.; Hellmann, I.; Smets, M.; Leonhardt, H.; Cardoso, M.C. L1 retrotransposition is activated by Ten-eleven-translocation protein 1 and repressed by methyl-CpG binding proteins. Nucleus 2017, 8, 548–562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De La Rica, L.; Deniz, Ö.; Cheng, K.C.L.; Todd, C.; Cruz, C.; Houseley, J.; Branco, M.R. TET-dependent regulation of retrotransposable elements in mouse embryonic stem cells. Genome Biol. 2016, 17, 1–14. [Google Scholar] [CrossRef] [Green Version]
- López-Moyado, I.F.; Tsagaratou, A.; Yuita, H.; Seo, H.; Delatte, B.; Heinz, S.; Benner, C.; Rao, A. Paradoxical association of TET loss of function with genome-wide DNA hypomethylation. Proc. Natl. Acad. Sci. USA 2019, 116, 16933–16942. [Google Scholar] [CrossRef] [Green Version]
- Wu, M.J.; Kim, M.R.; Chen, Y.S.; Yang, J.Y.; Chang, C.J. Retinoic acid directs breast cancer cell state changes through regulation of TET2-PKCzeta pathway. Oncogene 2017, 36, 3193–3206. [Google Scholar] [CrossRef]
- Kim, M.R.; Wu, M.-J.; Zhang, Y.; Yang, J.-Y.; Chang, C.J. TET2 directs mammary luminal cell differentiation and endocrine response. Nat. Commun. 2020, 11, 1–13. [Google Scholar] [CrossRef]
- Puig, I.; Tenbaum, S.P.; Chicote, I.; Arqués, O.; Martínez-Quintanilla, J.; Cuesta-Borrás, E.; Ramírez, L.; Gonzalo, P.; Soto, A.; Aguilar, S.; et al. TET2 controls chemoresistant slow-cycling cancer cell survival and tumor recurrence. J. Clin. Investig. 2018, 128, 3887–3905. [Google Scholar] [CrossRef]
- Hsu, C.-H.; Peng, K.-L.; Kang, M.-L.; Chen, Y.-R.; Yang, Y.-C.; Tsai, C.-H.; Chu, C.-S.; Jeng, Y.-M.; Chen, Y.-T.; Lin, F.-M.; et al. TET1 Suppresses Cancer Invasion by Activating the Tissue Inhibitors of Metalloproteinases. Cell Rep. 2012, 2, 568–579. [Google Scholar] [CrossRef] [Green Version]
- Sun, M.; Song, C.X.; Huang, H.; Frankenberger, C.A.; Sankarasharma, D.; Gomes, S.; Chen, P.; Chen, J.; Chada, K.K.; He, C.; et al. HMGA2/TET1/HOXA9 signaling pathway regulates breast cancer growth and metastasis. Proc. Natl. Acad. Sci. USA 2013, 110, 9920–9925. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.-Y.; Luo, C.-W.; Lai, Y.-S.; Wu, C.-C.; Hung, W.-C. Lysine demethylase KDM2A inhibits TET2 to promote DNA methylation and silencing of tumor suppressor genes in breast cancer. Oncogenesis 2017, 6, e369. [Google Scholar] [CrossRef] [Green Version]
- Song, C.; Wang, L.; Wu, X.; Wang, K.; Xie, D.; Xiao, Q.; Li, S.; Jiang, K.; Liao, L.; Yates, J.R., 3rd; et al. PML Recruits TET2 to Regulate DNA Modification and Cell Proliferation in Response to Chemotherapeutic Agent. Cancer Res 2018, 78, 2475–2489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blaschke, K.; Ebata, K.; Karimi, M.M.; Zepeda-Martínez, J.A.; Goyal, P.; Mahapatra, S.; Tam, A.; Laird, D.J.; Hirst, M.; Rao, A.; et al. Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nat. Cell Biol. 2013, 500, 222–226. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.-P.; Lv, L.; Liu, Y.; Smith, M.D.; Li, W.-C.; Tan, X.-M.; Cheng, M.; Li, Z.; Bovino, M.; Aubé, J.; et al. Tumor suppressor TET2 promotes cancer immunity and immunotherapy efficacy. J. Clin. Investig. 2019, 129, 4316–4331. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Liu, L.; Wang, M.; Xu, B.; Lyu, R.; Shi, Y.; Tan, L. TET2 Inhibits PD-L1 Gene Expression in Breast Cancer Cells through Histone Deacetylation. Cancers 2021, 13, 2207. [Google Scholar] [CrossRef]
- Li, S.; Feng, J.; Wu, F.; Cai, J.; Zhang, X.; Wang, H.; Fetahu, I.S.; Iwanicki, I.; Ma, D.; Hu, T.; et al. TET2 promotes anti-tumor immunity by governing G-MDSCs and CD8(+) T-cell numbers. EMBO Rep. 2020, 21, e49425. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).