The authors would like to make a correction to their published paper [1].
In Figure 1, the ATAC-seq section appears empty, and should be changed for a complete Figure 1. The correct Figure 1 is as below:
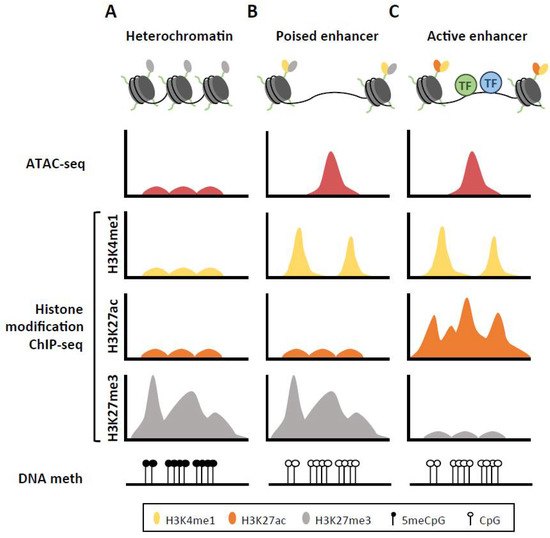
Figure 1.
Chromatin landscape for heterochromatin, poised and active enhancer regions. (A) The inactive DNA is tightly packed around histone proteins marked with H3K27me3 modification, in the form of heterochromatin. This structure prevents any interactions between transcription factors (TF) and the DNA sequence. (B) When the enhancer region is preactivated or poised, addition of H3K4me1 to the histone tails make the nucleosomes mobile, allowing for their displacement to form highly accessible DNA regions, which get frequently demethylated. (C) Upon activation of enhancer region, nucleosomes flanking this region acquire H3K27ac, losing the repressing H3K27me3 mark, which subsequently recruits the corresponding transcription factors.
The change does not affect the review results.
The rest of the manuscript does not need to be changed. The authors would like to apologize for any inconvenience caused. The manuscript will be updated, and the original will remain available on the article webpage.
Conflicts of Interest
The authors declare no conflict of interest.
Reference
- Ordoñez, R.; Martínez-Calle, N.; Agirre, X.; Prosper, F. DNA methylation of enhancer elements in myeloid neoplasms: Think outside the promoters? Cancers 2019, 11, 1424. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).