Abstract
Inactivation of phosphatase and tensin homolog (PTEN) is caused by multiple mechanisms, and loss of PTEN activity is related to the progression of various cancers. In gastric cancer (GC), the relationship between the loss of PTEN protein expression and various genetic alterations remains unclear. The effects of microsatellite instability (MSI), Epstein–Barr virus (EBV), HER2 overexpression, and PD-L1 expression on PTEN mutation have not been fully explored. We performed comprehensive cancer panel tests with a cohort of 322 tumor samples from patients with advanced GC. Immunohistochemistry for PTEN protein was performed in all cases, and the loss of protein expression was defined as a complete absence of nuclear staining. In total, 34 cases (10.6%) had pathogenic PTEN mutations, of which 19 (55.9%) showed PTEN protein loss. The most common PTEN variants associated with protein loss were p.R130 (n = 4) followed by p.R335, p.L265fs, and deletions (n = 2). All the ten nonsense mutations identified in the samples resulted in PTEN inactivation. In the remaining 288 GC cases with wild-type PTEN, protein loss was found in 35 cases (12.2%). Thus, PTEN mutations were significantly associated with PTEN protein loss (p = 5.232 × 10−10), high MSI (p = 3.936 × 10−8), and EBV-positivity (p = 0.0071). In conclusion, our results demonstrate that loss-of-function mutations in PTEN are a frequent genetic mechanism of PTEN inactivation in GC.
1. Introduction
Mechanisms of DNA damage repair are crucial to preserve the genomic integrity of the cell. In normal cells, DNA damage triggers the activation of DNA damage response (DDR) pathways to repair the damage, and these depend on the type of DNA damage. The phosphatase and tensin homolog (PTEN), a well-known tumor suppressor gene, is involved in double-strand break repair, and nucleotide excision repair, and regulates the DDR pathways by interacting with Chk1 and p53 [1,2]. In addition, PTEN is a dual-specificity protein and lipid phosphatase that regulates various cellular processes and signal pathways, such as the induction of apoptosis by inhibiting phosphatidylinositol 3-kinase (PI3K)/Akt pathway and control of cell adhesion, migration, and tumor invasion, by downregulating the activity of focal adhesion kinases (FAKs) [3].
Somatic PTEN mutation, a frequent event in endometrial cancer, is associated with microsatellite instability (MSI) status ranging from 25% to 83% [4], and PTEN inactivation is thought to be an early step in the development and progression of endometrial cancer [5]. PTEN inactivation is also frequently observed in glioblastoma, with hemizygous or homozygous deletions in over 90% of primary glioblastomas [6]. In gastric cancers (GCs), the frequency of PTEN mutation is relatively low (7–11%) and mostly found in advanced GCs [7,8]. PTEN inactivation is closely linked to disease progression in GC. A gradual decrease in PTEN expression has been reported during the course of GC development from normal gastric mucosae, atrophic gastritis, intestinal metaplasia, dysplasia, and early stage GC to advanced stage GC [9,10]. Low PTEN expression is also associated with lymph node metastasis, advanced stage, diffuse type histology, and poor prognosis [11,12,13]. Functional inactivation of the tumor suppressor protein PTEN has been detected in multiple cases of GC, and already shown to be closely linked to the development, progression, and prognosis of the disease [3].
PTEN inactivation is known to be caused by various mechanisms in GC, including gene mutations, loss of heterozygosity, promoter hypermethylation, miRNA-mediated regulation, and post-translational phosphorylation [3]. Previous studies on PTEN mutation in GC showed no consensus on the frequency and type of PTEN mutation, because they were based on a small number of GC cases in various stages, and used direct sequencing or polymerase chain reaction single strand conformation polymorphism (PCR-SSCP) to detect the PTEN mutations, which also detected nonpathogenic mutations [14,15,16,17,18]. The relationship between PTEN protein loss and genetic variants remains unclear, and their effect on MSI, EBV, and PD-L1 expression has not yet been studied. Therefore, it is necessary to understand the association between PTEN mutations and its expression and other biomarkers of immunotherapy in advanced GC using a large cohort and a new detection method that can cover non-hot-spot mutations and exclude nonpathogenic mutations.
In this study, we performed next generation sequencing (NGS) in 322 patients with advanced GC and evaluated the association of PTEN mutation with its MSI, EBV, and PD-L1 expressions to evaluate its clinical significance in GC.
2. Results
2.1. Clinicopathological Characteristics of Patients with PTEN-Mutated GC
Of the 322 GC cases with NGS data included in this study, 38 showed pathogenic PTEN alterations confirmed by COSMIC [19], Polyphen [20], and SIFT [21]. Among them, three cases with low sequencing depth and one with poor quality were excluded, and finally, 34 cases (10.6%) were confirmed to harbor pathogenic PTEN mutations.
In the 34 GC cases with PTEN mutations, the median age of the patients was 64 years (40–85 years), and 22 (64.7%) patients were male. All the samples used for NGS analysis were from the stomach, of which 20 (58.8%) were resection specimens. In the pathologic subtypes, 31 cases (91.2%) were tubular adenocarcinoma, two (5.9%) were papillary adenocarcinoma, and one (2.9%) was a signet ring cell carcinoma. Eight cases (23.5%) were MSI-high, two (5.9%) were EBV-positive, and 24 (70.6%) were PD-L1 positive (CPS ≥ 1) among the 33 available cases. Five (14.7%) of the 34 cases were HER2-positive. The clinicopathologic characteristics of PTEN-mutated GCs are summarized in Table 1.

Table 1.
Clinicopathologic characteristics of phosphatase and tensin homolog (PTEN)-mutated gastric cancers.
2.2. Clinicopathologic Characteristics of PTEN Wild-Type GC
In the GCs without PTEN alteration, the median age of the patients was 61 years (29–85 years), and 181 (62.8%) patients were male. All the samples used for NGS analysis were resected stomach specimens. Nine (3.1%) cases were MSI-high, 39 cases (13.5%) were HER2-positive, and seven (2.4%) cases were EBV-positive. PD-L1 positivity (CPS ≥ 1) was present in 182 (63.2%) out of the 287 available cases. Clinicopathologic characteristics of PTEN wild-type GCs are summarized in Table S1.
2.3. PTEN Mutations in Relationship with PTEN Protein Loss, MSI, EBV, and PD-L1 Expression
In the 34 PTEN-mutated GCs, we detected a total of 38 PTEN alterations of which four cases had two variants each. PTEN alterations were composed of 18 missense mutations, ten nonsense mutations, seven frameshift mutations, two deletions, and one nonframeshift deletion. The most frequent PTEN mutation was p.R130 (4/38, 10.5%), followed by p.L265fs (3/38, 7.9%). Among them, more than half of the cases showed PTEN protein loss (19/34, 55.9%), which was found in ten nonsense mutations, four missense mutations, three frameshift mutations, two deletions, and one non-frameshift deletion of PTEN, and one case with PTEN inactivation had two nonsense mutations. All cases with nonsense mutation showed PTEN protein loss. The most common PTEN mutation associated with protein loss was p.R130 (n = 4), followed by p.R335, p.L265fs, and deletion (n = 2). Representative cases of PTEN IHC and PTEN mutation confirmed by Integrative Genomics Viewer are shown in Figure 1. PTEN mutation was significantly associated with PTEN protein loss (p = 5.232 × 10−10), MSI-high (p = 3.936 × 10−8), and EBV-positivity (p = 0.0071) but was not associated with sex (p = 0.9805), age (p = 0.175), HER2 status (p = 0.79), and PD-L1 CPS (p = 0.111).
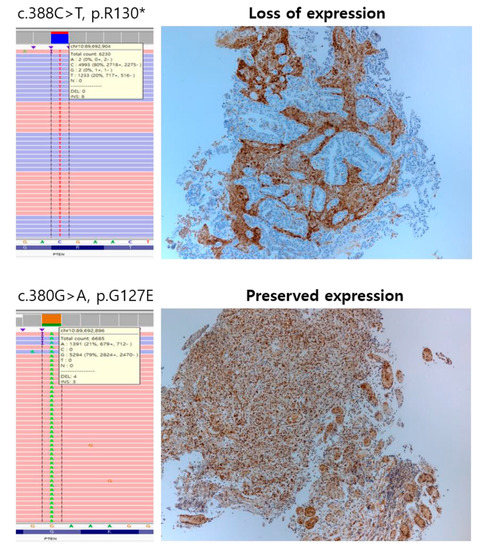
Figure 1.
Representative cases of Phosphatase and Tensin Homolog (PTEN) immunohistochemistry and PTEN mutation confirmed by Integrative Genomics Viewer.
Comparison of the clinicopathologic characteristics of PTEN-mutated and PTEN wild-type GCs are summarized in Table 2. PTEN inactivation was significantly associated with disease stages (p = 0.04), but did not predict overall survival (p = 0.083).

Table 2.
Comparison of clinicopathologic characteristics between phosphatase and tensin homolog (PTEN)-mutated gastric carcinomas and PTEN wild-type gastric carcinomas.
2.4. Co-Occurring Genetic Alterations Associated with PTEN Mutation
Co-occurring genetic alterations with PTEN mutation were detected in 26 genes. The most common co-occurring genetic alteration was TP53 mutation, found in 14 cases (41.2%), followed by PIK3CA mutation (10 cases; 29.4%), ERBB2 mutation (8 cases; 23.5%), and KRAS mutation (5 cases; 14.7%). Translocation was not found in any of the cases. The distribution and prevalence of co-occurring genetic alterations associated with PTEN, MSI status, EBV, HER2, and PD-L1 are summarized in Figure 2.
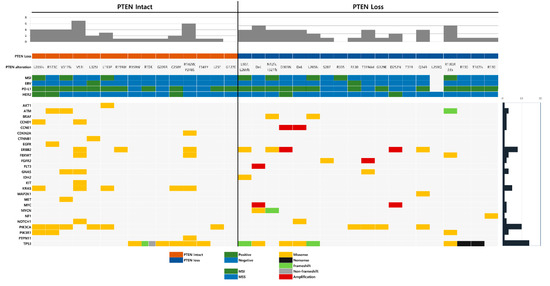
Figure 2.
The distribution and prevalence of co-occurring genetic alterations associated with phosphatase and tensin homolog (PTEN), microsatellite instability (MSI) status, Epstein–Barr virus (EBV), HER2, and PD-L1.
3. Discussion
We performed comprehensive cancer panel tests in 322 patients with advanced GC and found PTEN mutations in 34 cases (10.6%), of which 19 (55.9%) showed PTEN protein loss. Among the cases with PTEN inactivation, nonsense mutation was the most common type, and all cases with nonsense mutation showed PTEN loss. PTEN mutations were significantly associated with PTEN protein loss, MSI-high, and EBV-positivity.
The frequency of PTEN mutation varies ranging from 0% to 20%, except for synonymous mutations [14,15,16,17,18]. Previous studies were mostly based on a small number of cases and included various disease stages. Further, the PTEN mutations were detected using Sanger sequencing or PCR-SSCP, which also included nonpathogenic mutations. In the present study, using NGS test (covering all hot and non-hot spot mutations) in cases with advanced GCs, the frequency of PTEN mutation observed was similar to the frequency (7–11%) previously reported in GCs [7,8].
Several mechanisms of PTEN inactivation have been suggested previously, including gene mutations, loss of heterozygosity, promoter hypermethylation, miRNA-mediated regulation, and post-translational phosphorylation [3]. In the present study, we focused on gene mutation and showed a significant association of PTEN mutation with the loss of its expression (p = 5.232 × 10−10). A previous study on PTEN mutations in GC used IHC in cases with PTEN nonsense mutations and reported an association between PTEN mutation and its expression. Further, the study showed that nonsense mutation reduced PTEN expression [15]. It has been suggested that PTEN gene inactivation, mainly due to mutation and/or loss of heterozygosity, plays a pivotal role in tumor progression. In TCGA GC data, all but one case with truncating mutations of PTEN showed decreased mRNA expression [7,22]. The present study confirmed that mutation, especially nonsense mutation, is one of the mechanisms of PTEN inactivation in GC. Given that a nonsense mutation results in a truncated and usually nonfunctional protein, we proved this change by directly comparing the type of PTEN mutation and protein expression in GC.
We also found that PTEN mutation was significantly associated with MSI-high (p = 3.936 × 10−8), and EBV-positivity (p = 0.0071). Significant associations of PTEN mutation/inactivation and MSI-high were reported in colorectal cancers [23,24] and endometrial cancers [4]. The inactivation of PTEN has been reported in EBV-positive nasopharyngeal carcinoma through altered EBV-miR-BART1 expression [25] or promoter hypermethylation of PTEN in GC [26]. Several drugs are currently in clinical trials for the treatment of patients with PTEN-deficient cancers [27]. As EBV and MSI-high are predictive biomarkers for immunotherapy [28], either immunotherapy or combined therapy targeting PTEN would be a good option for this type of GC. A previous study investigated the association of DDR mutations (including PTEN) with MSI, PD-L1 expression, and tumor mutational burden (TMB) in GCs, and found that MSI-high, high PD-L1 expression, and TMB-high were significantly more frequent in DDR mutated cancers. In the study, PTEN was a prevalent DDR mutations found in MSI-H and TMB-high GCs (≥10 mt/MB) but not in GCs with high PD-L1 expression (CPS > 10) [29]. Another group reported that DDR mutated GCs had high TMB, but they did not assess the effect of PTEN on TMB [30]. As our study focused on PTEN alterations rather than DDR mutations, further studies are recommended to confirm the role and mechanism of failure of DNA repair in tumor immune dynamics in this era of cancer immunotherapy [31,32]. In previous studies, PTEN inactivation was significantly associated with tumor progression [3]; however, this difference was evident in the progression of early stage GC to advanced stage GC and the prognostic significance in patients with advanced GC is not evident [33,34]. In the present study with advanced stage GC, we also failed to find any significant correlations between PTEN inactivation and survival of the patients.
We analyzed co-occurring genetic alterations associated with PTEN mutations in GC and found alterations of TP53 (41.2%), PIK3CA (29.4%), ERBB2 (23.5%), and KRAS (14.7%) in all 34 PTEN-mutated cases. High frequency of coexistent mutations of PIK3CA and PTEN genes has been reported previously in endometrial carcinoma but not in GC [35]. These concomitant mutations can accelerate tumor progression through aberrant PI3K/AKT pathway. Although the relationship between PTEN and ERBB2 has not been studied in GC, the frequency of PTEN inactivation in ERBB2-amplified GC has been reported in 16.4–34.5% of cases [36,37,38]. In the present study, five PTEN mutations (11.4%) and five PTEN protein losses (11.4%) were found in 44 GCs with HER2 overexpression. In HER2-positive GCs, PTEN inactivation has been linked to trastuzumab resistance [37,38,39]. The clinical significance of concurrent KRAS and PTEN mutations is unknown in humans but has been reported in murine carcinogenesis [40,41,42].
Overall, our NGS results showed PTEN mutation in 34 cases (10.6%) out of 322 patients with advanced GC. More than half of the GCs (55.9%) with PTEN mutation showed PTEN inactivation, of which all nonsense mutation showed PTEN inactivation. Although this was a retrospective study and the number of PTEN-mutated cases analyzed was limited, we demonstrated for the first time that PTEN mutation is significantly associated with PTEN protein loss, MSI-high, and EBV-positivity. Given that MSI-high and EBV-positive GCs are biomarkers for immunotherapy and the repertoire of PTEN functions has recently been expanded with new potential implications for immunotherapy-based approaches [43], further studies are warranted.
4. Materials and Methods
4.1. Patients and Data Collection
The study population consisted of a total of 322 patients with primary advanced gastric carcinoma registered at the Samsung Medical Center (Seoul, Republic of Korea) from January 2017 to July 2019. The study was approved by the institutional review board (IRB 2020-01-051). The median age of the study cohort was 61 years (29–85), and 203 (63.0%) patients were male. We obtained all tumor samples from the stomach (308 resections and 14 endoscopic biopsies). Clinicopathological information, including age, sex, sample type, and the method of sample collection were obtained from electronic medical records. All but two patients did not receive chemotherapy or radiotherapy prior to obtaining the specimen. Formalin-fixed paraffin-embedded (FFPE) tissues of specimens were used for several tests including NGS cancer panel test, MSI test, and immunohistochemistry (IHC) to find molecular characteristics of the GC.
4.2. Cancer Panel Test with NGS
We performed NGS in all cases to detect copy number alterations (CNAs); single nucleotide variants (SNVs); small insertions and deletions (indels); and gene fusions using the Oncomine comprehensive cancer panel v1 (Thermo Fisher Scientific, Waltham, MA, USA), which examines 143 oncogenes and tumor suppressor genes. Our comprehensive cancer panel showed sensitivity of 99% for SNVs and 93% for indels at 10% allele frequency on the validation of analytical procedures using Acrometrix® Oncology Hotspot Control (Thermo Fisher Scientific) and 5-Fusion Multiplex FFPE RNA Reference Standard (Horizon Discovery, Waterbeach, UK). It also showed 100% sensitivity when the sample was diluted to 23% (CNAs) and 10% (fusions). For cancer panel tests, we used FFPE tissue samples, and the minimum tumor volume was 10%. After deparaffinization and nucleic acid extraction, targeted DNA and RNA amplification of the tumor was performed using the Ion AmpliSeq Library kit 2.0 (Thermo Fisher Scientific). Partial digestion of primer sequences, ligation of adapters to amplicons, and purification and quantitation of the libraries were performed using the Ion Xpress Barcode Adapter 1–96 kit (Thermo Fisher Scientific), Ion AmpliSeq Library Kit, and the Ion Library TaqMan Quantitation Kit (Thermo Fisher Scientific). Eight constructed libraries were loaded on an Ion 540 chip, and sequencing was performed using the Ion S5XL system. We used Ion Torrent software (Ion ReporterTM 5.2, Thermo Fisher Scientific) and Oncomine Knowledgebase for automated data analysis. We set the following criteria and evaluated the sequencing quality accordingly: mapped reads > 5,000,000, on-target rate > 90%, mean depth > 1200, and uniformity > 90%. A sequencing coverage of 250X, variant coverage of 25X, and variant allelic frequency of 5% were set as cutoffs to avoid false-positive and false-negative results. An average copy number ≥ 4 was interpreted as a gain (amplification) and <1 as a loss (deletion). For translocations, read counts ≥ 20 and total valid mapped reads ≥ 50,000 were interpreted as positive results.
4.3. Immunohistochemistry
To evaluate the correlation between genetic mutation and protein expression, we performed IHC with anti-PTEN antibody (138G6, CST, Danvers, MA, USA). Loss of nuclear and cytoplasmic staining in the tumor cells was considered as loss of PTEN expression. For HER2 IHC, we used anti-HER2 antibody (4B5, Ventana Mdical Systems, Tucson, AZ, USA), and scored HER2 status according to gastric criteria proposed by Ruschoff et al [44]. For EBV ISH, we used BOND-MAX with an EBV-encoded RNA probe (Leica, Newcastle, UK), and considered cases showing strong signals in most of the tumor cells as positive. For PD-L1 IHC, PD-L1 IHC 22C3 pharmDx kit (Agilent technologies, Carpinteria, CA, USA) was used with a Dako Autostainer Link 48 system (Agilent Technologies, Carpinteria, CA, USA). We measured the combined positive score (CPS) and the ratio of the number of all PD-L1 expressing cells to the number of viable tumor cells, as described previously [45].
4.4. MSI Test
MSI status was determined by multiplex PCR to amplify five quasimonomorphic mononucleotide repeat markers (BAT-25, BAT-26, NR-21, NR-24, and NR-27). Genomic DNA was isolated from the FFPE tissue blocks using a QIAamp DNA Mini Kit (Qiagen, Hilden, Germany). Sense primers were fluorescently end-labeled with FAM, HEX, or NED. Amplicons were analyzed on an ABI Prism 3130 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA). Allelic sizes were estimated with GeneMapper 4.1 (Applied Biosystems, Foster City, CA, USA), and tumors with allelic size variation in <2 and ≥2 microsatellites were classified as microsatellite stable and instable, respectively.
4.5. Statistical Analysis
The χ2 test, Fisher’s Exact test, and Kruskal–Wallis rank sum test were used to compare categorical variables between PTEN-mutated and PTEN wild-type GCs. The Wilcoxon rank sum test was used to compare a continuous variable between PTEN-mutated and PTEN wild-type GC. The p-value was two-sided, and p < 0.05 was considered significant using R software version 3.4.4 (https://www.r-project.org/foundation/).
5. Conclusions
In conclusion, PTEN loss-of-function mutation is an important genetic mechanism of PTEN inactivation.
Supplementary Materials
The following are available online at https://www.mdpi.com/2072-6694/12/7/1724/s1, Table S1: Clinicopathologic characteristics of phosphatase and tensin homolog (PTEN) wild-type gastric carcinomas.
Author Contributions
Conceptualization and methodology, K.-M.K. and B.K.; formal analysis and investigation, D.K., S.Y.K., and Y.J.H.; data curation, B.K.; visualization, Y.J.H.; writing—original draft preparation, B.K.; writing—review and editing, K.-M.K. and B.K.; supervision, project administration, and funding acquisition, K.-M.K. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Science and ICT (NRF-2017R1E1A1A01075005 and NRF-2017R1A2B4012436).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Pellegrino, B.; Mateo, J.; Serra, V.; Balmana, J. Controversies in oncology: Are genomic tests quantifying homologous recombination repair deficiency (HRD) useful for treatment decision making? ESMO Open 2019, 4, e000480. [Google Scholar] [CrossRef] [PubMed]
- Ming, M.; He, Y.Y. PTEN in DNA damage repair. Cancer Lett. 2012, 319, 125–129. [Google Scholar] [CrossRef]
- Xu, W.T.; Yang, Z.; Lu, N.H. Roles of PTEN (Phosphatase and Tensin Homolog) in gastric cancer development and progression. Asian Pac. J. Cancer Prev. APJCP 2014, 15, 17–24. [Google Scholar] [CrossRef]
- Bilbao, C.; Rodriguez, G.; Ramirez, R.; Falcon, O.; Leon, L.; Chirino, R.; Rivero, J.F.; Falcon, O., Jr.; Diaz-Chico, B.N.; Diaz-Chico, J.C.; et al. The relationship between microsatellite instability and PTEN gene mutations in endometrial cancer. Int. J. Cancer 2006, 119, 563–570. [Google Scholar] [CrossRef]
- Kanamori, Y.; Kigawa, J.; Itamochi, H.; Shimada, M.; Takahashi, M.; Kamazawa, S.; Sato, S.; Akeshima, R.; Terakawa, N. Correlation between loss of PTEN expression and Akt phosphorylation in endometrial carcinoma. Clin. Cancer Res. 2001, 7, 892–895. [Google Scholar] [PubMed]
- Verhaak, R.G.; Hoadley, K.A.; Purdom, E.; Wang, V.; Qi, Y.; Wilkerson, M.D.; Miller, C.R.; Ding, L.; Golub, T.; Mesirov, J.P.; et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010, 17, 98–110. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research, N. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014, 513, 202–209. [Google Scholar] [CrossRef] [PubMed]
- Cristescu, R.; Lee, J.; Nebozhyn, M.; Kim, K.M.; Ting, J.C.; Wong, S.S.; Liu, J.; Yue, Y.G.; Wang, J.; Yu, K.; et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat. Med. 2015, 21, 449–456. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Kuang, L.G.; Zheng, H.C.; Li, J.Y.; Wu, D.Y.; Zhang, S.M.; Xin, Y. PTEN encoding product: A marker for tumorigenesis and progression of gastric carcinoma. World J. Gastroenterol. 2003, 9, 35–39. [Google Scholar] [CrossRef]
- Zheng, H.C.; Chen, Y.; Kuang, L.G.; Yang, L.; Li, J.Y.; Wu, D.Y.; Zhang, S.M.; Xin, Y. Expression of PTEN-encoding product in different stages of carcinogenesis and progression of gastric carcinoma. Zhonghua Zhong Liu Za Zhi 2003, 25, 13–16. [Google Scholar]
- Bai, Z.G.; Ye, Y.J.; Shen, D.H.; Lu, Y.Y.; Zhang, Z.T.; Wang, S. PTEN expression and suppression of proliferation are associated with Cdx2 overexpression in gastric cancer cells. Int. J. Oncol. 2013, 42, 1682–1691. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Sun, H.; Song, L.; Gao, X.; Chang, W.; Qin, X. Immunohistochemical expression of mTOR negatively correlates with PTEN expression in gastric carcinoma. Oncol. Lett. 2012, 4, 1213–1218. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Cui, J.; Zhang, C.H.; Yang, D.J.; Chen, J.H.; Zan, W.H.; Li, B.; Li, Z.; He, Y.L. High-expression of DJ-1 and loss of PTEN associated with tumor metastasis and correlated with poor prognosis of gastric carcinoma. Int. J. Med Sci. 2013, 10, 1689–1697. [Google Scholar] [CrossRef] [PubMed]
- Sato, K.; Tamura, G.; Tsuchiya, T.; Endoh, Y.; Sakata, K.; Motoyama, T.; Usuba, O.; Kimura, W.; Terashima, M.; Nishizuka, S.; et al. Analysis of genetic and epigenetic alterations of the PTEN gene in gastric cancer. Virchows Arch. 2002, 440, 160–165. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.Y.; Huang, T.J.; Chen, F.M.; Hsieh, M.C.; Lin, S.R.; Hou, M.F.; Hsieh, J.S. Mutation analysis of the putative tumor suppressor gene PTEN/MMAC1 in advanced gastric carcinomas. Virchows Arch. 2003, 442, 437–443. [Google Scholar] [CrossRef]
- Wen, Y.G.; Wang, Q.; Zhou, C.Z.; Qiu, G.Q.; Peng, Z.H.; Tang, H.M. Mutation analysis of tumor suppressor gene PTEN in patients with gastric carcinomas and its impact on PI3K/AKT pathway. Oncol. Rep. 2010, 24, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Lima, E.M.; Araujo, J.J.; Harada, M.L.; Assumpcao, P.P.; Burbano, R.R.; Casartelli, C. Molecular study of the tumour suppressor gene PTEN in gastric adenocarcinoma in Brazil. Clin. Exp. Med. 2005, 5, 129–132. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.G.; Chen, Y.J.; Perng, L.I.; Wang, N.M.; Kao, M.C.; Yang, T.Y.; Chang, C.P.; Tsai, C.H. Mutation analysis of the PTEN/MMAC1 gene in cancers of the digestive tract. Eur. J. Cancer 1999, 35, 647–651. [Google Scholar] [CrossRef]
- Forbes, S.A.; Beare, D.; Boutselakis, H.; Bamford, S.; Bindal, N.; Tate, J.; Cole, C.G.; Ward, S.; Dawson, E.; Ponting, L.; et al. COSMIC: Somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017, 45, D777–D783. [Google Scholar] [CrossRef]
- Adzhubei, I.A.; Schmidt, S.; Peshkin, L.; Ramensky, V.E.; Gerasimova, A.; Bork, P.; Kondrashov, A.S.; Sunyaev, S.R. A method and server for predicting damaging missense mutations. Nat. Methods 2010, 7, 248–249. [Google Scholar] [CrossRef]
- Kumar, P.; Henikoff, S.; Ng, P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009, 4, 1073–1081. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.P.; Loukola, A.; Salovaara, R.; Nystrom-Lahti, M.; Peltomaki, P.; de la Chapelle, A.; Aaltonen, L.A.; Eng, C. PTEN mutational spectra, expression levels, and subcellular localization in microsatellite stable and unstable colorectal cancers. Am. J. Pathol. 2002, 161, 439–447. [Google Scholar] [CrossRef]
- Guanti, G.; Resta, N.; Simone, C.; Cariola, F.; Demma, I.; Fiorente, P.; Gentile, M. Involvement of PTEN mutations in the genetic pathways of colorectal cancerogenesis. Hum. Mol. Genet. 2000, 9, 283–287. [Google Scholar] [CrossRef] [PubMed]
- Cai, L.; Ye, Y.; Jiang, Q.; Chen, Y.; Lyu, X.; Li, J.; Wang, S.; Liu, T.; Cai, H.; Yao, K.; et al. Epstein-Barr virus-encoded microRNA BART1 induces tumour metastasis by regulating PTEN-dependent pathways in nasopharyngeal carcinoma. Nat. Commun. 2015, 6, 7353. [Google Scholar] [CrossRef]
- Hino, R.; Uozaki, H.; Murakami, N.; Ushiku, T.; Shinozaki, A.; Ishikawa, S.; Morikawa, T.; Nakaya, T.; Sakatani, T.; Takada, K.; et al. Activation of DNA methyltransferase 1 by EBV latent membrane protein 2A leads to promoter hypermethylation of PTEN gene in gastric carcinoma. Cancer Res. 2009, 69, 2766–2774. [Google Scholar] [CrossRef]
- Dillon, L.M.; Miller, T.W. Therapeutic targeting of cancers with loss of PTEN function. Curr. Drug Targets 2014, 15, 65–79. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.T.; Cristescu, R.; Bass, A.J.; Kim, K.M.; Odegaard, J.I.; Kim, K.; Liu, X.Q.; Sher, X.; Jung, H.; Lee, M.; et al. Comprehensive molecular characterization of clinical responses to PD-1 inhibition in metastatic gastric cancer. Nat. Med. 2018, 24, 1449–1458. [Google Scholar] [CrossRef] [PubMed]
- Cerniglia, M.; Xiu, J.; Grothey, A.; Pishvaian, M.J.; Hwang, J.J.; Marshall, J.; Vanderwalde, A.M.; Shields, A.F.; Lenz, H.-J.; Salem, M.E.; et al. Association of DNA damage response and repair genes (DDR) mutations and microsatellite instability (MSI), PD-L1 expression, tumor mutational burden (TMB) in gastroesophageal cancers. J. Clin. Oncol. 2019, 37, 60. [Google Scholar] [CrossRef]
- Cai, H.; Jing, C.; Chang, X.; Ding, D.; Han, T.; Yang, J.; Lu, Z.; Hu, X.; Liu, Z.; Wang, J.; et al. Mutational landscape of gastric cancer and clinical application of genomic profiling based on target next-generation sequencing. J. Transl. Med. 2019, 17, 189. [Google Scholar] [CrossRef] [PubMed]
- Mouw, K.W.; Goldberg, M.S.; Konstantinopoulos, P.A.; D’Andrea, A.D. DNA Damage and Repair Biomarkers of Immunotherapy Response. Cancer Discov. 2017, 7, 675–693. [Google Scholar] [CrossRef] [PubMed]
- Teo, M.Y.; Seier, K.; Ostrovnaya, I.; Regazzi, A.M.; Kania, B.E.; Moran, M.M.; Cipolla, C.K.; Bluth, M.J.; Chaim, J.; Al-Ahmadie, H.; et al. Alterations in DNA Damage Response and Repair Genes as Potential Marker of Clinical Benefit From PD-1/PD-L1 Blockade in Advanced Urothelial Cancers. J. Clin. Oncol. 2018, 36, 1685–1694. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Qin, X.; Fei, M.; Hou, W.; Greshock, J.; Bachman, K.E.; Kang, J.; Qin, C.Y. Loss and reduced expression of PTEN correlate with advanced-stage gastric carcinoma. Exp. Ther. Med. 2013, 5, 57–64. [Google Scholar] [CrossRef]
- Kang, Y.H.; Lee, H.S.; Kim, W.H. Promoter methylation and silencing of PTEN in gastric carcinoma. Lab. Investig. 2002, 82, 285–291. [Google Scholar] [CrossRef]
- Oda, K.; Stokoe, D.; Taketani, Y.; McCormick, F. High frequency of coexistent mutations of PIK3CA and PTEN genes in endometrial carcinoma. Cancer Res. 2005, 65, 10669–10673. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Hong, M.; Kim, S.T.; Park, S.H.; Kang, W.K.; Kim, K.M.; Lee, J. The impact of concomitant genomic alterations on treatment outcome for trastuzumab therapy in HER2-positive gastric cancer. Sci. Rep. 2015, 5, 9289. [Google Scholar] [CrossRef]
- Kim, C.; Lee, C.K.; Chon, H.J.; Kim, J.H.; Park, H.S.; Heo, S.J.; Kim, H.J.; Kim, T.S.; Kwon, W.S.; Chung, H.C.; et al. PTEN loss and level of HER2 amplification is associated with trastuzumab resistance and prognosis in HER2-positive gastric cancer. Oncotarget 2017, 8, 113494–113501. [Google Scholar] [CrossRef]
- Deguchi, Y.; Okabe, H.; Oshima, N.; Hisamori, S.; Minamiguchi, S.; Muto, M.; Sakai, Y. PTEN loss is associated with a poor response to trastuzumab in HER2-overexpressing gastroesophageal adenocarcinoma. Gastric Cancer 2017, 20, 416–427. [Google Scholar] [CrossRef] [PubMed]
- Linos, K.; Sheehan, C.E.; Ross, J.S. Correlation of HER2 and PTEN status with clinical outcome in esophageal (E), gastric (G), and gastroesophageal junction (GEJ) adenocarcinomas (ACs). J. Clin. Oncol. 2011, 29, 4066. [Google Scholar] [CrossRef]
- Lin, Y.K.; Fang, Z.; Jiang, T.Y.; Wan, Z.H.; Pan, Y.F.; Ma, Y.H.; Shi, Y.Y.; Tan, Y.X.; Dong, L.W.; Zhang, Y.J.; et al. Combination of Kras activation and PTEN deletion contributes to murine hepatopancreatic ductal malignancy. Cancer Lett. 2018, 421, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Davies, E.J.; Marsh Durban, V.; Meniel, V.; Williams, G.T.; Clarke, A.R. PTEN loss and KRAS activation leads to the formation of serrated adenomas and metastatic carcinoma in the mouse intestine. J. Pathol. 2014, 233, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Ma, T.; Brozick, J.; Babalola, K.; Budiu, R.; Tseng, G.; Vlad, A.M. Effects of Kras activation and Pten deletion alone or in combination on MUC1 biology and epithelial-to-mesenchymal transition in ovarian cancer. Oncogene 2016, 35, 5010–5020. [Google Scholar] [CrossRef] [PubMed]
- Piro, G.; Carbone, C.; Carbognin, L.; Pilotto, S.; Ciccarese, C.; Iacovelli, R.; Milella, M.; Bria, E.; Tortora, G. Revising PTEN in the Era of Immunotherapy: New Perspectives for an Old Story. Cancers 2019, 11, 1525. [Google Scholar] [CrossRef] [PubMed]
- Ruschoff, J.; Hanna, W.; Bilous, M.; Hofmann, M.; Osamura, R.Y.; Penault-Llorca, F.; van de Vijver, M.; Viale, G. HER2 testing in gastric cancer: A practical approach. Mod. Pathol. 2012, 25, 637–650. [Google Scholar] [CrossRef] [PubMed]
- Kulangara, K.; Hanks, D.A.; Waldroup, S.; Peltz, L.; Shah, S.; Roach, C.; Juco, J.W.; Emancipator, K.; Stanforth, D. Development of the combined positive score (CPS) for the evaluation of PD-L1 in solid tumors with the immunohistochemistry assay PD-L1 IHC 22C3 pharmDx. J. Clin. Oncol. 2017, 35, e14589. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).