Abstract
Currently, studies worldwide have comprehensively recognized the importance of Sphingomonadaceae bacteria and the mlrCABD gene cluster in microcystin (MC) degradation. However, knowledge about their degradation of nodularin (NOD) is still unclear. In this study, the degradation mechanism of NOD by Sphingopyxis sp. m6, an efficient MC degrader isolated from Lake Taihu, was investigated in several aspects, including degradation ability, degradation products, and potential application. The strain degraded NOD of 0.50 mg/L with a zero-order rate constant of 0.1656 mg/L/d and a half-life of 36 h. The average degradation rate of NOD was significantly influenced by the temperature, pH, and initial toxin concentrations. Moreover, four different biodegradation products, linear NOD, tetrapeptide H-Glu-Mdhb-MeAsp-Arg-OH, tripeptide H-Mdhb-MeAsp-Arg-OH, and dipeptide H-MeAsp-Arg-OH, were identified, of which the latter two are the first reported. Furthermore, the four mlr genes were upregulated during NOD degradation. The microcystinase MlrA encoded by the mlrA gene hydrolyzes the Arg-Adda bond to generate linear NOD as the first step of NOD biodegradation. Notably, recombinant MlrA showed higher degradation activity and stronger environmental adaptability than the wild strain, suggesting future applications in NOD pollution remediation. This research proposes a relatively complete NOD microbial degradation pathway, which lays a foundation for exploring the mechanisms of NOD degradation by MC-degrading bacteria.
Key Contribution:
A relatively complete pathway for NOD biodegradation by MC-degrading bacteria was elucidated. The role of microcystinase MlrA encoded by the mlrA gene in the degradation process was determined, and the application potential of recombinant enzymes was explored.
1. Introduction
Cyanotoxins such as nodularin (NOD) and microcystin (MC) released by cyanobacterial cells pose a severe threat to the environment and human health, as evidenced by the destruction of aquatic food webs [1,2], mortality of marine mammals and birds [3], massive fish kills [4], and human diseases and even death [5]. NOD is predominantly produced by Nodularia spumigena, an ancient organism of the Baltic Sea that grows uncontrollably in the summer months annually [6]. In recent years, however, the occurrence of NOD and its detrimental effects have been frequently reported around the world, including Australia [7], Europe [8], New Zealand [9], Spain [10], and Greece [11]. Besides exerting tumor-promoting activity by specifically inhibiting protein phosphatases 1 and 2A [12], Ohta et al. demonstrated that NOD penetrates more easily into hepatocytes and is also a tumor initiator [13]. Cyclic pentapeptide NOD is composed of D-MeAsp, L-Arg, Adda, D-Glu, and Mdhb, coincidentally similar to heptapeptide MC in structure and biological activity [14,15,16].
Removing cyanotoxins from water has become a top priority owing to their significant hazards [17]. Indigenous microbial degradation is accepted as the most important route to eliminate cyanotoxins from natural water owing to its low capital cost, safety, high efficiency, and environmental friendliness [18,19]. Dozens of MC-degrading bacteria strains have been isolated from natural water since 1994, and most of them belong to the Sphingomonadaceae family [20,21,22]. Bourne et al. first reported that MC biodegradation by Sphingomonas sp. ACM 3962 is primarily dependent on the enzymatic pathway encoded by gene cluster mlrBDAC [23,24]. Subsequently, the homologous mlr gene cluster was also detected in other isolated Sphingomonadaceae bacteria, such as Sphingopyxis sp. C-1 [25], X20 [26], YF1 [27], and USTB-05 [28], Sphingomonas sp. MD-1 [29] and B9 [30], and Novosphingobium sp. THN-1 [31], with genera Sphingopyxis and Sphingomonas accounting for the majority. The mlr gene cluster encodes three hydrolysis enzymes and one oligopeptide transporter protein. The first enzyme encoded by the mlrA gene is responsible for opening the highly stable cyclic structure of MC by cleaving the Arg-Adda bond. The resulting linear product is then degraded to a tetrapeptide by the second enzyme encoded by the mlrB gene. The enzyme MlrC encoded by the mlrC gene hydrolyzes the tetrapeptide to Adda as the final nontoxic product and also exerts cleavage activity on the linear product to degrade it directly to Adda. The transporter protein encoded by the mlrD gene is supposed to transport MC and its degradation products into bacterial cells [23,24]. Currently, novel studies on fundamental knowledge about the biodegradation of strains of the Sphingomonadaceae family have focused on MC, while NOD has received little attention [19,21,32].
To the best of our knowledge, views regarding the presence of microorganisms capable of degrading NOD in natural water date back to the 1990s [33]; however, the first native strain was not isolated and characterized until 2005 [34]. Until now, we have only found a few strains capable of degrading NOD [35,36], and few studies about the pathways, genes, and enzymes involved in NOD biodegradation have been conducted. Considering that NOD and MC are the most toxic and widely distributed, with similar structure and amino acid composition, the application of Sphingomonadaceae strains capable of degrading MC and/or NOD could facilitate understanding of biological attenuation for cyanotoxins, although this fact is rarely noticed. Therefore, there are gaps regarding whether it can degrade NOD, whether it shares a set of degradation genes with MC or mobilizes an entirely new degradation mechanism, and whether it has potential value in practical applications.
We recently found that Sphingopyxis sp. m6, an indigenous strain isolated from Taihu Lake with degradation activity against MC [37], was capable of degrading NOD. We evaluated the NOD degrading activity of this bacterium under different culturing environments. The possible pathway associated with NOD degradation was inferred from the discovery of several degradation products during the degradation process. Concomitantly, the expression dynamics of the MC degradation gene cluster carried by strain m6 were tracked throughout the degradation experiment. The correlation between the expression of the four mlr genes and NOD degradation rates was demonstrated. Finally, it was verified that MlrA encoded by the mlrA gene played an important hydrolytic role in NOD biodegradation, and the degradation efficiency and environmental adaptability of recombinant MlrA were investigated to preliminarily explore its application potential.
2. Results
2.1. Biodegradation Kinetics of NOD by Sphingopyxis sp. m6
After incubation of strain m6 with 0.50 mg/L NOD, the peak area value of NOD monitored by high performance liquid chromatography (HPLC) decreased over time and was almost zero at 72 h, which meant that the NOD concentration was reduced below the detection threshold for HPLC (Figure 1a–c). In contrast, almost no reduction in NOD was observed in the bacteria-free group, excluding physical, chemical, and other abiotic degradation. Figure S1 presents the standard curve of NOD quantified by HPLC.

Figure 1.
HPLC profiles for the microbial degradation of NOD by strain m6 at (a) 0 h, (b) 36 h, and (c) 72 h. (d) Biodegradation curve of NOD by strain m6. The error bars represent the standard deviation (n = 3).
The biodegradation kinetics of NOD showed that strain m6 decomposed NOD immediately without any lag period. The degradation rate initially increased and then decreased, reaching a maximum of 0.23 mg/L/d at 48 h, with an average degradation rate of 0.16 mg/L/d (Figure 1d). The kinetics model was set up based on the analysis of experimental dimensionless concentrations over the degradation time. Compared with first- or second-order reaction kinetics, the regression coefficient R2 value suggested that the zero-order reaction kinetics is better for degradation of NOD (R2 = 0.987). The rate constant for NOD biodegradation was computed utilizing the following equation:
where k is the rate constant, and are the initial concentration of NOD and the NOD concentration at time t, respectively. The degradation rate constant of NOD was calculated to be 0.1656 mg/L/d. The half-life (t1/2) of zero-order reaction kinetics can be estimated as follows:
Therefore, the t1/2 is approximately 36 h.
2.2. Factors Affecting Microbial Degradation of NOD
Single-factor experiments were performed under different incubation conditions. The results showed that the average degradation rate of NOD is strongly influenced by incubation temperatures, pH, and initial NOD concentrations. Under pH 7 and 0.50 mg/L NOD, bacterium m6 degraded NOD with an average rate of 0.14, 0.16, and 0.11 mg/L/d at 20, 30, and 37 °C, respectively (Figure 2a). The rate constant increased from 0.1344 mg/L/d to 0.1656 mg/L/d as the temperature rises from 20 °C to 30 °C. However, it decreased to 0.1152 mg/L/d when the temperature reached 37 °C. The optimal reaction temperature was 30 °C, whereas the toxin concentration barely decreased in the control and 40 °C groups. Figure 2b illustrated that at 30 °C, 0.50 mg/L NOD was removed, with the average rate of 0.08, 0.09, 0.16, 0.20, and 0.20 mg/L/d at pH 3, 5, 7, 9, and 11, respectively. The degradation of NOD was sensitive to the initial pH of the reaction system, with rate constants (0.1920–0.1928 mg/L/d) observed at pH 9 and 11 more than twice as high as under acidic environments. At 30 °C and pH 7, different initial concentrations of 1.00, 0.50, 0.10, 0.05 mg/L NOD were degraded at the rate of 0.22, 0.16, 0.10, and 0.03 mg/L/d, respectively (Figure 2c). Collectively, there was a lag period before degradation started only at pH 3 and 5. Strain m6 exhibited the highest degradation rate (0.22 mg/L/d) of NOD at 30 °C and pH 7 with toxin concentration of 1.00 mg/L.

Figure 2.
Effect of different incubation conditions on the NOD-degrading rate of stain m6. (a) Incubation temperatures (pH = 7, 0.50 mg/L NOD), (b) pH (30 °C, 0.50 mg/L NOD), and (c) NOD concentrations (30 °C, pH = 7). The error bars represent the standard deviation from triplicate experiments.
2.3. Identification of NOD Biodegradation Products
The total ion chromatogram (TIC) showed five substance peaks at different stages of degradation by strain m6 (Figure S2). The ten predominant fragment ions proved that the compound with m/z 825.4494 appearing at 8.359 min in TIC was NOD (Figures S2a and S3a,b; Table 1), as all amino acids were present. The MS spectrum of TIC at 7.836 min exhibited a major ion at m/z 692.3619 and a protonated molecular ion at m/z 843.4600 (Figures S2b and S3c). Based on all ions and bonds found in the fragmentation spectrum, except for the Arg-Adda bond, m/z 843.4600 with 18 Da more than the mass value of NOD was identified as hydrolyzed NOD [M + H2O + H]+ (Figure S3c,d; Table 1). The major ion at m/z 692.3619 was obtained by losing the amino NH2 group and the PhCH2CHOCH3 group (m/z 135.0805) from the linear product through random breakage. In the samples incubated with strain m6, two ions were present at 4.934 min of TIC (Figure S2d,e). Eight main fragment ions, 70.0646 [C4H8N]+, 158.0912 [Arg + OH − NH]+, 175.1186 [Arg-OH + 2H]+, 183.1146 [Mdhb-MeAsp + H − COOH]+, 201.0960 [Arg + OH + CO]+, 258.1558 [MeAsp-Arg + OH − COOH]+, 357.2228 [Mdhb-MeAsp-Arg + H − COOH]+, 401.2125 [M + OH + 2H]+ (Figure S4d; Table 1), provide evidence that one with m/z 401.2137 is a tripeptide H-Mdhb-MeAsp-Arg-OH (Figure S4c). The other molecular ion with m/z 304.1615 was fragmented into several ions, including m/z 70.0652 [C3H4NO]+, 158.0912 [Arg + OH − NH]+, 175.1195 [Arg-OH + 2H]+, and 304.1664 [M + OH + 2H]+ (Figure S5b; Table 1), demonstrating that this compound contained only MeAsp and Arg (Figure S5a). The last substance peak with m/z 530.2565 appeared in TIC at 4.789 min (Figures S2c and Figure S4a), revealing ten fragmentation products: 70.0655 [C4H8N]+, 158.0932 [Arg + OH − NH]+, 175.1186 [Arg-OH + 2H]+, 209.0922 [Glu-Mdhb + H − OH]+/[Mdhb-MeAsp + H − OH]+, 227.1009 [Glu-Mdhb + H]+/[Mdhb-MeAsp + H]+, 304.1632 [MeAsp-Arg-OH + 2H]+, 401.2285 [Mdhb-MeAsp-Arg-OH + 2H]+, 494.2389 [M + H − OH]+, and 512.2464 [M + H]+ (Figure S4b; Table 1), confirming that the substance was a tetrapeptide H-Glu-Mdhb-MeAsp-Arg-OH. No product was detected in the standard NOD samples.

Table 1.
Fragmentation ions of NOD and its biodegradation products.
The dynamics of the peak intensity for NOD and all degradation products detected by mass spectrometry are displayed in Figure S6. With the removal of NOD, degradation products progressively appeared. The signal intensity of linear NOD increased rapidly to a maximum at 72 h (Figure S6b), while the others required 108 h (Figure S6c–e). All products exhibited a downward trend after reaching the peak. Only the intensity value of the linear NOD dropped to almost zero (Figure S6b).
2.4. Expression of mlr Gene Cluster during NOD Biodegradation
After exposure to NOD, the expression dynamics of the four mlr genes in bacterium m6 were similar (Figure 3). Specifically, during the 144-h experimental period, the relative expression ratios of the mlr gene cluster experienced a rough three-step process of gradual increase, reaching the highest at 36 h and then decreasing to below the control level (Figure 3). At 36 h, the relative expression ratio of the mlrC gene was the largest (2.8-fold upregulation) (Figure 3c), while the mlrB gene had the minimum 2.4-fold upregulation (Figure 3b). After the peak, the expression level of the mlrD gene decreased most rapidly (Figure 3d). Figure 3 attempts to explain the kinetic relationships between the NOD degradation rate and the relative expression ratio of the four genes. For each, the rate of NOD degradation follows its expression trend, simultaneously reaching its maximum (Figure 3). There was a positive correlation between NOD degradation rate and the gene expression ratio of mlrA (R2 = 0.714, p < 0.01), mlrB (R2 = 0.769, p < 0.01), mlrC (R2 = 0.5723, p < 0.01), and mlrD (R2 = 0.5972, p < 0.01), respectively.

Figure 3.
Kinetic relationships between NOD degradation rate and mlr gene cluster expression. (a) mlrA gene, (b) mlrB gene, (c) mlrC gene, and (d) mlrD gene. Significant differences are indicated in comparison with the control by t-test. *, p < 0.05. The error bars represent the standard deviation of means for triplicate.
2.5. NOD Degradation and Its Product by Recombinant MlrA
NOD concentration was obviously decreased in both recombinant mlrA-HMBP-pET28a/BL21 and cell-free extracts (CEs) of the recombinant (Figure 4) while remaining essentially unchanged in the corresponding negative controls, suggesting that the mlrA gene-encoded enzyme (MlrA) may cause the loss of NOD. The absence of changes in the blank controls also ruled out self-catabolism. Nevertheless, divergent degradation rates of NOD were observed between intact recombinant cells and the CEs of the recombinant cells. A non-negligible phenomenon was that CEs from the recombinant sharply degraded more than 70% NOD within the first hour at a rate of 0.37 mg/L/h (Figure 4b). In contrast, intact cells only degraded 5% NOD in the first hour, which was significantly lower than its CEs at the same time point (p < 0.01). The average rate of intact recombinant was 0.12 mg/L/d (Figure 4a). Consequently, this series of tests indicated that recombinant MlrA readily degraded the majority of NOD in a short period of time, showing higher NOD removal efficiency than the wild strain and recombinant.

Figure 4.
NOD degradation curves of (a) intact recombinant mlrA-HMBP-pET28a/BL21 and (b) CEs from the recombinant. Bars represent the standard deviation of the means for triplicate.
The MS spectrum exhibited two major ion peaks at m/z 825.4562 and 843.4596. The monitored intensity of m/z 843.4596 consistently enhanced and showed no signs of attenuation, and its fragment ions were essentially identical to those previously described in Section 2.3. Hence, it was confirmed that MlrA is responsible for the first step in NOD bio-catalysis by strain m6, opening the Arg-Adda bond in the ring-like structure.
2.6. Environmental Adaptability Assay of Recombinant MlrA
As shown in Figure 5a, recombinant MlrA showed higher activity at 30, 37, and 40 °C, degrading more than 70% NOD in the first hour but 60% at 20 °C. However, there was no statistically significant difference in the degradation rates of recombinant MlrA (p > 0.05). Proteolytic activity was lower under solid acid (pH = 3) and strongly alkaline (pH = 11) conditions, with only approximately 30% NOD being degraded in the first hour at pH 3 compared to less than 30% at pH 11 (Figure 5b). The degradation curves for recombinant MlrA at pH 5, 7, and 9 largely overlapped (Figure 5b). Interestingly, no statistically significant differences in degradation rates were found between the temperature and pH groups (p > 0.05). In addition, enzyme activity was affected by the initial NOD concentration. The degradation rate in the first hour was proportional to the initial toxin concentration (Figure 5c). These features enhance the application potential of recombinant MlrA.
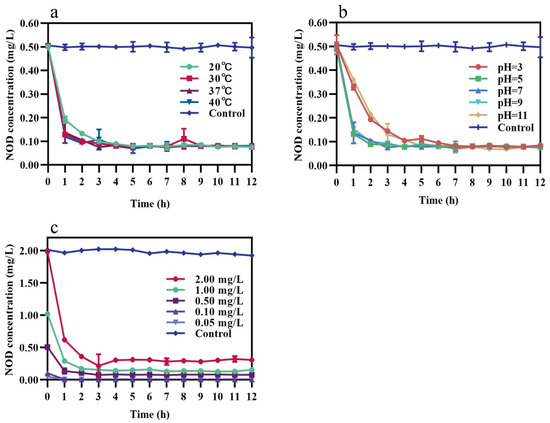
Figure 5.
Effect of different conditions on the NOD-degrading rate of recombinant MlrA. (a) Temperatures (pH = 7, 0.50 mg/L NOD), (b) pH (37 °C, 0.50 mg/L NOD), and (c) NOD initial concentrations (37 °C, pH = 7). The error bars represent the standard deviation from triplicate experiments.
3. Discussion
Globally, the frequency, intensity, and duration of harmful cyanobacterial blooms (HCBs) are increasing due to the eutrophication of water bodies [38,39]. Furthermore, cyanotoxins produced by HCBs have potent carcinogenic and even lethal effects on public health [40,41]. Among the numerous methods for removing cyanotoxins from natural water, biodegradation is the primary mechanism for eliminating toxins and is considered an eco-friendly and environmentally benign technology [42,43]. Studies on specific mechanisms for the degradation gene cluster mlr carried by Sphingomonadaceae bacteria, which account for a large proportion of MC-degrading bacteria, have advanced our understanding of the biological attenuation of MC in nature [20,21,44]. Nevertheless, the field of its bio-catalytic degradation mechanisms for NOD still needs to be addressed.
In the present study, Sphingopyxis sp. m6, an efficient MC degrader isolated from Lake Taihu [37], was also able to degrade NOD (Figure 1). Not all MC-degrading strains can decompose NOD, and only a few strains have been reported prior to this study; however, no further elaboration was given [34,35,45,46]. The lag phase is a significant factor affecting biodegradation kinetics [32]. Under average temperature and neutral environment, most reported functional bacteria have a lag period ranging from several hours to a few days before commencing degradation [32,47,48,49]. In contrast, for strain m6, there was no significant lag period for the degradation of either NOD or MC-LR at 30 °C and pH 7 (Figure 1d) [37]. This may be because strain m6 is an indigenous microorganism isolated from Lake Taihu, which has been chronically contaminated with blooms [50]. As indicated in some previous studies, the lag period can be shortened, and in some circumstances even eliminated, by pre-exposure or frequent exposure to cyanotoxins [51,52]. Therefore, the literature that integrates the history of algal toxin exposure into factors of biodegradation rate is rational [53]. The diversity of NOD concentrations used, reaction conditions, culture density of the studied strain, and the methods employed resulted in no comparable degradation rates between strain m6 and other previously reported bacteria [35,36,45]. However, at the same concentration, strain m6 did not respond consistently to NOD and MC-LR. This could be attributed to the higher relative specificity of the degradative enzymes for MC-LR, that is to say, the binding sites of the enzymes are more compatible with MC-LR.
Environmental conditions such as temperature or pH are related to cyanobacterial blooms and affect the biodegradation process of cyanobacterial toxins [16]. Many studies have shown that optimal biodegradation occurs at approximately 30 °C under neutral or weakly alkaline conditions [54,55,56]. Strain m6 showed the best degradation efficiency at 30 °C, but almost completely lost its degradation ability at 40 °C (Figure 2a). One reason for this is that higher temperatures have an adverse effect on the survival of organisms, although the probability of collisions between particles increases. The pH of natural water is usually above 8 during cyanobacterial blooms because of enhanced photosynthesis due to the consumption of carbon dioxide [57,58]. Interestingly, the average degradation rate of NOD by strain m6 was more rapid with an increase in pH, a phenomenon that is quite the opposite of its degradation of MC-LR (Figure 2b) [37]. A possible explanation could be that some sequences more adapted to high pH for regulating protease activity are mobilized only during the NOD degradation. Detailed studies are required to test this hypothesis. It is worth noting that under acidic conditions there was a lag period before strain m6 started to degrade NOD (Figure 2b), primarily because the synthesis and activation of degrading enzymes may be inhibited to some extent under unsuitable conditions. In addition, the average degradation rate correlated positively with the initial NOD concentration (Figure 2c), which is consistent with the results of previous studies [59,60].
The first study focusing on NOD biodegradation products was performed using Sphingomonas sp. B-9 cells. It found a degradation intermediate linear NOD and a final product Adda [45,61], similar to the degradation route discovered later by Sphingopyxis sp. USTB-05 [46]. In addition to two (linear NOD and Adda) belonging to previously reported compounds, Mazur-Marzec et al. found three novel degradation products in their study of NOD degradation by natural microbial communities in sediments: H-Glu-Mdhb-MeAsp-Arg-OH, H-Adda-Glu-Mdhb-MeAsp-OH, and H-Adda-Glu-Mdhb-MeAsp-Cit-OH [62]. Four degradation products were found in our study, including two new products: H-Mdhb-MeAsp-Arg-OH and H-MeAsp-Arg-OH (Figures S4c and S5a). The non-hydrolytic degradation pathways proposed by Edwards et al., such as demethylation of Mdhb, decarboxylation of MeAsp, and modification of Adda, were not observed during the degradation process in this study [63]. Figure 6 describes the specific degradation pathway of NOD by Sphingopyxis sp. m6. First, linear NOD was generated by breaking the Arg-Adda bond to open the cyclic pentapeptide. Next, the Adda-Glu bond was hydrolyzed to produce a tetrapeptide (H-Glu-Mdhb-MeAsp-Arg-OH). The tetrapeptide was further decomposed to a tripeptide (H-Mdhb-MeAsp-Arg-OH) by cleavage of the peptide bond at Glu-Mdhb. Finally, strain m6 successfully recognized and hydrolyzed the Mdhb-MeAsp bond to form a dipeptide (H-MeAsp-Arg-OH). These novel findings provided valuable supplements to previous studies on single strain and proposed a relatively complete NOD biodegradation pathway. Surprisingly, the entire degradation process did not reveal the presence of the traditional final product Adda and dimeric Adda that often occurs during NOD biodegradation. It should be noted that strain m6 has a metabolic pathway that degrades Adda so quickly that the signal intensity is below the detection threshold. Further degradation of Adda as a degradation intermediate has been reported [27,37].

Figure 6.
Proposed biodegradation pathway of NOD by strain m6.
Case studies of different Sphingomonadaceae strains have led to a consensus that the mlr gene cluster plays an essential role in the degradation of MC and its variants [20,21,44]. However, less information was available on NOD, even though it has a vital significance for understanding the environmental attenuation of NOD. In this study, detection of NOD biodegradation products, including linear NOD, implied that MlrA encoded by the mlrA gene acts similarly on NOD and MC by cleaving their ring structure at the Adda-Arg peptide bond. The results of the genetic analysis showed that exposure to NOD upregulates the expression of four mlr genes (Figure 3). The trend of mlr gene expression mostly coincided with that reported in other studies on MC degradation mechanisms, implying that this gene cluster is also involved in NOD degradation [37,64,65]. The positive correlation between NOD degradation rate and mlr gene cluster expression provides evidence to support a contention that biodegradation kinetics are regulated by the expression of this gene cluster, and the more considerable R2 value between mlrA gene expression and degradation rate indicates the importance of the mlrA gene in NOD biodegradation by strain m6.
This study further validated the role of the mlrA gene with the help of intact recombinant mlrA-HMBP-pET28a/BL21 and its CEs. The results definitively confirm that MlrA generates linear NOD as the first product by hydrolyzing the Arg-Adda bond in NOD. Combined with the previous study by Ding et al., it was suggested that the target substrates of MlrA in strain m6 are not limited to MC, which is consistent with the case of Sphingopyxis sp. USTB-05 [66]. It is noteworthy that this finding cannot be directly applied to all strains because the amino acid sequencing and structure of the specific protease encoded by the mlrA gene may differ in various bacteria. Bourne et al. found that MlrC recognizes and hydrolyzes the Adda-Glu bond of MC [24]. This finding was subsequently confirmed in many MC-degrading bacteria containing the mlr gene cluster [21,44]. Based on the fact that the Adda-Glu bond is also present in NOD and the upregulation expression of the mlrC gene during degradation, it is speculated that the mlrC gene regulates the hydrolysis reaction in the second step of NOD degradation pathway. However, the specific function of the mlrB gene needs further investigation due to the structural differences between NOD and MC. Degradation of intact recombinant indicated that recombinant MlrA does not exist in the host as an inclusion body (Figure 4a) [67]. Compared with the relatively moderate degradation ability of strain m6, the biodegradation curve of recombinant MlrA dropped sharply in the first hour (Figure 4b) and tended to be horizontal after 3 h, with trace amounts of toxin remaining even at 48 h (data not shown). This may be a consequence of the presence of other complex components in recombinant CEs preventing recombinant MlrA from binding to the substrate and performing catalysis when the NOD concentration decreases to a low level in a confined space [68]. Although the concentration of recombinant MlrA from CEs and the density of strain m6 could not be compared, it was concluded that recombinant MlrA could be activated within an extremely short period of time to initiate rapid degradation. The environmental adaptability of recombinant MlrA is discussed in this paper. Recombinant MlrA exhibited high catalytic activity even at high temperatures and extreme pH values (Figure 5). This finding paves the way for the subsequent introduction of recombinant enzymes into environmental applications, as corroborated by several studies on the potential applications of recombinant MlrA directly as a sustainable bioremediation product [68,69,70,71].
4. Conclusions
In this study, an indigenous bacterium, Sphingopyxis sp. m6, with strong MC degradation capability was able to degrade NOD. The biodegradation of NOD by strain m6 followed zero-order reaction kinetics. The average degradation rate of NOD was affected by the temperature, pH, and initial NOD concentrations. The MS analysis identified four products, of which two (H-Mdhb-MeAsp-Arg-OH and H-MeAsp-Arg-OH) have never been reported before. A relatively complete degradation pathway was proposed in this study; however, the Adda or Adda dimer was not found. The mlr gene cluster was highly transcribed during the catabolism of NOD by strain m6. It was further confirmed that MlrA encoded by the mlrA gene is responsible for the first step of the NOD biodegradation pathway by recognizing and hydrolyzing the Arg-Adda bond. Moreover, heterologous expression of recombinant MlrA significantly improved the biodegradation efficiency of NOD. The broad adaptability of recombinant MlrA to the environment provides preliminary evidence of its application potential.
5. Materials and Methods
5.1. Bacteria and Reagents
The previously obtained strains Sphingopyxis sp. m6, recombinant mlrA-HMBP-pET28a/BL21, and HMBP-pET28a/BL21 were investigated in this study [37]. Standard NOD (C41H60N8O10, ≥95% purity) purchased from APExBIO (Houston, USA) was dissolved in methanol at a concentration of 100 mg/L and stored at −20 °C. The mineral salt medium (MSM) was used as an essential component of the experimental systems. The real-time quantitative polymerase chain reaction (RT-qPCR) kit was supplied by TaKaRa (Kusatsu, Japan). Trifluoroacetic acid (Sinopharm Chemical Reagent Co., Ltd. Shanghai, China) and HPLC-grade methanol (TEDIA Co., Fairfield, OH, USA) were applied to the liquid phase. UPLC grade methanol (Merck KGaA, Darmstadt, Germany) and formic acid (Fisher Scientific, Shanghai, China) were used for mass spectrometry.
5.2. NOD Degradation Capability of Strain m6
Strain m6 was incubated in Luria-Bertani (LB) medium at 30 °C with shaking at 150 rpm, harvested using centrifugation (5000× g, 10 min, and 4 °C) during the logarithmic growth period (OD600 = 0.60 ± 0.05), washed three times, and then resuspended in 3 mL of sterile MSM containing 0.50 mg/L NOD. A certain amount (100 µL) of each sample was withdrawn every 12 h for six days. After centrifugation (12,000× g, 15 min, and 4 °C), the residual concentrations of NOD in the supernatants were measured using HPLC (Agilent 1260, Santa Clara, CA, USA). A bacteria-free group was used as a control.
5.3. NOD Degradation under Different Environmental Conditions
Three series of experiments were designed to simulate various environmental conditions. First, four different temperatures (20, 30, 37, and 40 °C) were tested with 0.50 mg/L NOD at pH 7. Second, five different pH values (pH = 3, 5, 7, 9, and 11) were executed with 0.50 mg/L NOD at 30 °C. Finally, four different initial NOD concentrations (0.05, 0.10, 0.50, and 1.00 mg/L) were conducted with pH 7 at 30 °C. The control groups contained 0.50 or 1.00 mg/L NOD without bacteria cells and were cultured at 30 °C and pH 7. Samples of 100 µL were taken every 12 h for six days and the concentrations of remaining NOD in supernatants were then detected by HPLC.
5.4. Quantification of NOD by HPLC
NOD was quantified by HPLC using a Zorbax Extend C18 column (2.1 × 50 nm, 1.8 µm, Agilent, Santa Clara, CA, USA) with a variable wavelength detector (VWD) at 238 nm. The mobile phase consisted of HPLC-grade methanol (B phase) and Milli-Q water containing 0.05% trifluoroacetic acid (A phase) (53:47, v/v) was set for chromatographic separation at a flow rate of 1.00 mL/min. The column temperature was maintained at 40 °C throughout the entire process, and the injection volume was 20 µL.
5.5. Identification of Degradation Products
Ultra-performance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS) (triple TOF 5600+, AB Sciex, Redwood, CA, USA) was used to identify NOD and its degradation products. First, 5 µL of Supernatant was injected into UPLC with an Acquity UPLC HSS T3 column (2.1 × 100 mm, 1.8 µm, Waters, Milford MA, USA). The mobile phase was a mixture of UPLC grade methanol (B phase) and Milli-Q water containing 0.1% formic acid (A phase). The percentage of B phase was started at 2% for 2 min, ramped linearly to 98% over 12 min, maintained at 98% from 14 min to 17 min, and then immediately dropped to 2% and held for 3 min at a constant flow rate of 0.30 mL/min for a total run time of 20 min. The MS system connected to the UPLC was equipped with a tandem quadrupole and atmospheric pressure chemical iosziaa-lion (APCI) source. The conditions were as follows: positive ionization mode, full scanning from m/z 100–1200, ionspray voltage floating of 5000 V, declustering potential of 80 V, and collision energy of 35 V.
5.6. Detection of mlr Gene Cluster Expression
To study whether degradation of NOD by strain m6 shares a standard set of degradation mechanisms with MC-LR, the five specific primers, qmlrAF/qmlrAR, qmlrBF/qmlrBR, qmlrCF/qmlrCR, qmlrDF/qmlrDR, and q16SF/q16SR, were designed based on the sequencing results of Ding et al. [37]. Total RNA from strain m6 in the degradation system was extracted with TRIzol (Life Technologies, Carlsbad, CA, USA) every 12 h until NOD disappeared entirely and was then reverse-transcribed to cDNA using a Reverse Transcription Kit (Takara Bio, Shiga, Japan). RT-qPCR was performed using SYBR Green Real-time PCR Master Mix (Toyobo, Osaka, Japan). The NOD-free control group was prepared in the same manner. The relative expression ratio was calculated by 2−ΔΔCt method, and the 16S gene was used as the reference gene [72].
5.7. NOD Degradation by Intact Recombinant and CEs of the Recombinant
The mlrA gene heterologous expression recombinant (mlrA-HMBP-pET28a/BL21) and control strain (HMBP-pET28a/BL21), which were previously constructed in our laboratory (specific information will be published in another article), were independently inoculated in LB medium containing 50 µg/mL kanamycin and incubated at 37 °C with a shaking rate of 200 rpm/min until OD600 reached 0.60 ± 0.05. Isopropyl-β-D-thiogalactopyranoside (IPTG) at a final concentration of 0.50 mM was added in order to induce the expression of MlrA. After 4 h, the organisms were collected by centrifugation at 5000× g for 10 min at 4 °C. The bacteria were washed three times with Milli-Q water before being resuspended in 1 mL Milli-Q water. The suspensions were then divided into two aliquots. One aliquot was used to test the NOD degradation capability of intact cells, and the other aliquot was sonicated in a 4 °C water bath for 20 min, alternating every working time of 3 s with 3 s intervals during this period to prevent high temperature from inactivating the target enzyme. The ultrasonic-treated suspension was centrifuged at 4 °C and 12,000× g for 30 min to discard debris, and the supernatants were collected as CEs of recombinant mlrA-HMBP-pET28a/BL21.
To test the NOD degradation activity of recombinant MlrA, the suspensions of intact recombinant and CEs of the recombinant were spiked into sterile water with an initial concentration of 0.50 mg/L NOD, respectively. The suspensions of HMBP-pET28a/BL21 and CEs from HMBP-pET28a/BL21 were used as negative controls. A reaction system containing only NOD was used as the blank control. All tubes were incubated at 37 °C and shaken at 200 rpm. Samples (100 µL) were removed periodically, and methanol (two-fold volume) was added to terminate the enzyme reaction. After centrifugation at 4 °C and 15,000× g for 15 min, the residual NOD concentration and degradation products in the supernatants were detected by HPLC and UPLC-MS/MS.
5.8. Activity Assay of Recombinant MlrA
To evaluate the application potential of recombinant MlrA, CEs from the recombinant were studied to test degradability at different temperatures (20, 30, 37, and 40 °C), pH (3, 5, 7, 9, and 11), and initial NOD concentrations (0.05, 0.10, 0.50, 1.00, and 2.00 mg/L). CEs from HMBP-pET28a/BL21 cells were used as controls.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/toxins13110813/s1, Figure S1: The standard curve of NOD quantified by HPLC (0.01–2.00 mg/L), Figure S2: Five substance peaks in the TIC during degradation. (a) NOD; (b) Linear NOD; (c) Tetrapeptide; (d) Tripeptide; (e) Dipeptide, Figure S3: UPLC-MS/MS profiles for NOD and linear NOD. (a) MS spectrum for NOD; (b) MS/MS spectrum for NOD; (c) MS spectrum for linear NOD; (d) MS/MS spectrum for linear NOD, Figure S4: UPLC-MS/MS profiles for degradation products. (a) MS spectrum for tetrapeptide; (b) MS/MS spectrum for tetrapeptide; (c) MS spectrum for tripeptide; (d) MS/MS spectrum for tripeptide, Figure S5: UPLC-MS/MS profile of dipeptide. (a) MS spectrum of dipeptide; (b) MS/MS spectrum of dipeptide, Figure S6: The dynamics of peak intensity of NOD and degradation products. (a) NOD; (b) linear NOD; (c) H-Glu-Mdhb-MeAsp-Arg-OH; (d) H-Mdhb-MeAsp-Arg-OH; (e) H-MeAsp-Arg-OH.
Author Contributions
Conceptualization, M.Y. and Y.P.; methodology, M.Y. and Q.D.; software, J.Z.; validation, M.Y., R.S. and L.Y.; formal analysis, Q.D.; investigation, M.Y.; resources, J.Z.; data curation, M.Y., Q.D., and R.S.; writing—original draft preparation, M.Y.; writing—review and editing, Y.P.; visualization, Q.D.; supervision, Y.P.; project administration, L.Y.; funding acquisition, Y.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Nature Science Foundation of China, grant number 81972997, and the Graduate Research and Innovation Projects of Jiangsu Province, grant number SJKY19_0140.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Su, R.C.; Meyers, C.M.; Warner, E.A.; Garcia, J.A.; Refsnider, J.M.; Lad, A.; Breidenbach, J.D.; Modyanov, N.; Malhotra, D.; Haller, S.T.; et al. Harmful Algal Bloom Toxicity in Lithobates catesbeiana Tadpoles. Toxins 2020, 12, 378. [Google Scholar] [CrossRef] [PubMed]
- Briland, R.D.; Stone, J.P.; Manubolu, M.; Lee, J.; Ludsin, S.A. Cyanobacterial blooms modify food web structure and interactions in western Lake Erie. Harmful Algae 2020, 92, 101586. [Google Scholar] [CrossRef] [PubMed]
- Akyol, C.; Ozbayram, E.G.; Accoroni, S.; Radini, S.; Eusebi, A.L.; Gorbi, S.; Vignaroli, C.; Bacchiocchi, S.; Campacci, D.; Gigli, F.; et al. Monitoring of cyanobacterial blooms and assessing polymer-enhanced microfiltration and ultrafiltration for microcystin removal in an Italian drinking water treatment plant. Environ. Pollut. 2021, 286, 117535. [Google Scholar] [CrossRef] [PubMed]
- Shahmohamadloo, R.S.; Ortiz Almirall, X.; Simmons, D.B.D.; Poirier, D.G.; Bhavsar, S.P.; Sibley, P.K. Fish tissue accumulation and proteomic response to microcystins is species-dependent. Chemosphere 2021, 287, 132028. [Google Scholar] [CrossRef] [PubMed]
- Refsnider, J.M.; Garcia, J.A.; Holliker, B.; Hulbert, A.C.; Nunez, A.; Streby, H.M. Effects of harmful algal blooms on stress levels and immune functioning in wetland-associated songbirds and reptiles. Sci. Total Environ. 2021, 788, 147790. [Google Scholar] [CrossRef]
- Sivonen, K.; Kononen, K.; Carmichael, W.W.; Dahlem, A.M.; Rinehart, K.L.; Kiviranta, J.; Niemela, S.I. Occurrence of the hepatotoxic cyanobacterium Nodularia spumigena in the Baltic Sea and structure of the toxin. Appl. Environ. Microbiol. 1989, 55, 1990–1995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Svircev, Z.; Lalic, D.; Bojadzija Savic, G.; Tokodi, N.; Drobac Backovic, D.; Chen, L.; Meriluoto, J.; Codd, G.A. Global geographical and historical overview of cyanotoxin distribution and cyanobacterial poisonings. Arch. Toxicol. 2019, 93, 2429–2481. [Google Scholar] [CrossRef]
- Karlson, B.; Andersen, P.; Arneborg, L.; Cembella, A.; Eikrem, W.; John, U.; West, J.J.; Klemm, K.; Kobos, J.; Lehtinen, S.; et al. Harmful algal blooms and their effects in coastal seas of Northern Europe. Harmful Algae 2021, 102, 101989. [Google Scholar] [CrossRef]
- Wood, S.A.; Kuhajek, J.M.; de Winton, M.; Phillips, N.R. Species composition and cyanotoxin production in periphyton mats from three lakes of varying trophic status. FEMS Microbiol. Ecol. 2012, 79, 312–326. [Google Scholar] [CrossRef] [Green Version]
- Beltran, E.; Ibanez, M.; Sancho, J.V.; Hernandez, F. Determination of six microcystins and nodularin in surface and drinking waters by on-line solid phase extraction-ultra high pressure liquid chromatography tandem mass spectrometry. J. Chromatogr. A 2012, 1266, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Zervou, S.K.; Moschandreou, K.; Paraskevopoulou, A.; Christophoridis, C.; Grigoriadou, E.; Kaloudis, T.; Triantis, T.M.; Tsiaoussi, V.; Hiskia, A. Cyanobacterial Toxins and Peptides in Lake Vegoritis, Greece. Toxins 2021, 13, 394. [Google Scholar] [CrossRef] [PubMed]
- Rinehart, K.L.; Harada, K.; Namikoshi, M.; Chen, C.; Harvis, C.A.; Munro, M.; Blunt, J.; Mulligan, P.E.; Beasley, V.R. Nodularin, microcystin, and the configuration of Adda. J. Am. Chem. Soc. 1988, 110, 8557–8558. [Google Scholar] [CrossRef]
- Ohta, T.; Sueoka, E.; Iida, N.; Komori, A.; Suganuma, M.; Nishiwaki, R.; Tatematsu, M.; Kim, S.J.; Carmichael, W.W.; Fujiki, H. Nodularin, a Potent Inhibitor of Protein Phosphatase-1 and Phosphatase-2a, Is a New Environmental Carcinogen in Male F344 Rat-Liver. Cancer Res. 1994, 54, 6402–6406. [Google Scholar]
- Van Apeldoorn, M.E.; van Egmond, H.P.; Speijers, G.J.; Bakker, G.J. Toxins of cyanobacteria. Mol. Nutr. Food Res. 2007, 51, 7–60. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Wang, L.; Wang, M.; Hu, T. Comprehensive insights into the occurrence and toxicological issues of nodularins. Mar. Pollut. Bull. 2021, 162, 111884. [Google Scholar] [CrossRef] [PubMed]
- Buratti, F.M.; Manganelli, M.; Vichi, S.; Stefanelli, M.; Scardala, S.; Testai, E.; Funari, E. Cyanotoxins: Producing organisms, occurrence, toxicity, mechanism of action and human health toxicological risk evaluation. Arch. Toxicol. 2017, 91, 1049–1130. [Google Scholar] [CrossRef]
- Chen, Y.; Shen, D.; Fang, D. Nodularins in poisoning. Clin. Chim. Acta 2013, 425, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Krausfeldt, L.E.; Steffen, M.M.; McKay, R.M.; Bullerjahn, G.S.; Boyer, G.L.; Wilhelm, S.W. Insight Into the Molecular Mechanisms for Microcystin Biodegradation in Lake Erie and Lake Taihu. Front. Microbiol. 2019, 10, 2741. [Google Scholar] [CrossRef]
- Massey, I.Y.; Yang, F. A Mini Review on Microcystins and Bacterial Degradation. Toxins 2020, 12, 268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ho, L.; Sawade, E.; Newcombe, G. Biological treatment options for cyanobacteria metabolite removal—A review. Water Res. 2012, 46, 1536–1548. [Google Scholar] [CrossRef]
- Li, J.; Li, R.; Li, J. Current research scenario for microcystins biodegradation—A review on fundamental knowledge, application prospects and challenges. Sci. Total Environ. 2017, 595, 615–632. [Google Scholar] [CrossRef]
- Jones, G.J.; Bourne, D.G.; Blakeley, R.L.; Doelle, H. Degradation of the cyanobacterial hepatotoxin microcystin by aquatic bacteria. Nat. Toxins 1994, 2, 228–235. [Google Scholar] [CrossRef] [PubMed]
- Bourne, D.G.; Jones, G.J.; Blakeley, R.L.; Jones, A.; Negri, A.P.; Riddles, P. Enzymatic pathway for the bacterial degradation of the cyanobacterial cyclic peptide toxin microcystin LR. Appl. Environ. Microbiol. 1996, 62, 4086–4094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bourne, D.G.; Riddles, P.; Jones, G.J.; Smith, W.; Blakeley, R.L. Characterisation of a gene cluster involved in bacterial degradation of the cyanobacterial toxin microcystin LR. Environ. Toxicol. 2001, 16, 523–534. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, K.; Maseda, H.; Okano, K.; Kurashima, T.; Kawauchi, Y.; Xue, Q.; Utsumi, M.; Zhang, Z.; Sugiura, N. Enzymatic pathway for biodegrading microcystin LR in Sphingopyxis sp. C-1. J. Biosci. Bioeng. 2012, 114, 630–634. [Google Scholar] [CrossRef]
- Qin, L.; Zhang, X.; Chen, X.; Wang, K.; Shen, Y.; Li, D. Isolation of a Novel Microcystin-Degrading Bacterium and the Evolutionary Origin of mlr Gene Cluster. Toxins 2019, 11, 269. [Google Scholar] [CrossRef] [Green Version]
- Yang, F.; Huang, F.; Feng, H.; Wei, J.; Massey, I.Y.; Liang, G.; Zhang, F.; Yin, L.; Kacew, S.; Zhang, X.; et al. A complete route for biodegradation of potentially carcinogenic cyanotoxin microcystin-LR in a novel indigenous bacterium. Water Res. 2020, 174, 115638. [Google Scholar] [CrossRef]
- Yan, H.; Wang, J.; Chen, J.; Wei, W.; Wang, H.; Wang, H. Characterization of the first step involved in enzymatic pathway for microcystin-RR biodegraded by Sphingopyxis sp. USTB-05. Chemosphere 2012, 87, 12–18. [Google Scholar] [CrossRef] [PubMed]
- Saitou, T.; Sugiura, N.; Itayama, T.; Inamori, Y.; Matsumura, M. Degradation characteristics of microcystins by isolated bacteria from Lake Kasumigaura. J. Water Supply Res. Technol. 2003, 52, 13–18. [Google Scholar] [CrossRef]
- Jin, H.; Nishizawa, T.; Guo, Y.; Nishizawa, A.; Park, H.D.; Kato, H.; Tsuji, K.; Harada, K.I. Complete Genome Sequence of a Microcystin-Degrading Bacterium, Sphingosinicella microcystinivorans Strain B-9. Microbiol. Resour. Announc. 2018, 7, e00898-18. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Y.; Shao, J.; Wu, X.; Xu, Y.; Li, R. Active and silent members in the mlr gene cluster of a microcystin-degrading bacterium isolated from Lake Taihu, China. FEMS Microbiol. Lett. 2011, 322, 108–114. [Google Scholar] [CrossRef] [Green Version]
- Kumar, P.; Hegde, K.; Brar, S.K.; Cledon, M.; Kermanshahi-Pour, A. Potential of biological approaches for cyanotoxin removal from drinking water: A review. Ecotoxicol. Environ. Saf. 2019, 172, 488–503. [Google Scholar] [CrossRef] [PubMed]
- Heresztyn, T.; Nicholson, B.C. Nodularin concentrations in Lakes Alexandrina and Albert, South Australia, during a bloom of the cyanobacterium (blue-green alga) Nodularia spumigena and degradation of the toxin. Environ. Toxic. Water 1997, 12, 273–282. [Google Scholar] [CrossRef]
- Rapala, J.; Berg, K.A.; Lyra, C.; Niemi, R.M.; Manz, W.; Suomalainen, S.; Paulin, L.; Lahti, K. Paucibacter toxinivorans gen. nov., sp. nov., a bacterium that degrades cyclic cyanobacterial hepatotoxins microcystins and nodularin. Int. J. Syst. Evol. Microbiol. 2005, 55, 1563–1568. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Idroos, E.S.; De Silva, B.G.D.N.K.; Manage, P.M. Biodegradation of microcystin analogues by Stenotrophomonas maltophilia isolated from Beira Lake Sri Lanka. J. Natl. Sci. Found. Sri Lanka 2017, 45, 91–99. [Google Scholar] [CrossRef] [Green Version]
- Toruńska-Sitarz, A.; Kotlarska, E.; Mazur-Marzec, H. Biodegradation of nodularin and other nonribosomal peptides by the Baltic bacteria. Int. Biodeterior. Biodegrad. 2018, 134, 48–57. [Google Scholar] [CrossRef]
- Ding, Q.; Liu, K.; Xu, K.; Sun, R.; Zhang, J.; Yin, L.; Pu, Y. Further Understanding of Degradation Pathways of Microcystin-LR by an Indigenous Sphingopyxis sp. in Environmentally Relevant Pollution Concentrations. Toxins 2018, 10, 536. [Google Scholar] [CrossRef] [Green Version]
- Tanvir, R.U.; Hu, Z.; Zhang, Y.; Lu, J. Cyanobacterial community succession and associated cyanotoxin production in hypereutrophic and eutrophic freshwaters. Environ. Pollut. 2021, 290, 118056. [Google Scholar] [CrossRef]
- Huisman, J.; Codd, G.A.; Paerl, H.W.; Ibelings, B.W.; Verspagen, J.M.H.; Visser, P.M. Cyanobacterial blooms. Nat. Rev. Microbiol. 2018, 16, 471–483. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Kang, X.; Chu, L.; Wang, Y.; Song, X.; Zhao, X.; Cao, X. Algicidal mechanism of Raoultella ornithinolytica against Microcystis aeruginosa: Antioxidant response, photosynthetic system damage and microcystin degradation. Environ. Pollut. 2021, 287, 117644. [Google Scholar] [CrossRef]
- Ibelings, B.W.; Chorus, I. Accumulation of cyanobacterial toxins in freshwater "seafood" and its consequences for public health: A review. Environ. Pollut. 2007, 150, 177–192. [Google Scholar] [CrossRef]
- Sun, R.; Sun, P.; Zhang, J.; Esquivel-Elizondo, S.; Wu, Y. Microorganisms-based methods for harmful algal blooms control: A review. Bioresour. Technol. 2018, 248, 12–20. [Google Scholar] [CrossRef]
- Yu, Y.; Zeng, Y.; Li, J.; Yang, C.; Zhang, X.; Luo, F.; Dai, X. An algicidal Streptomyces amritsarensis strain against Microcystis aeruginosa strongly inhibits microcystin synthesis simultaneously. Sci. Total Environ. 2019, 650, 34–43. [Google Scholar] [CrossRef]
- Massey, I.Y.; Zhang, X.; Yang, F. Importance of bacterial biodegradation and detoxification processes of microcystins for environmental health. J. Toxicol. Environ. Health B Crit. Rev. 2018, 21, 357–369. [Google Scholar] [CrossRef] [PubMed]
- Imanishi, S.; Kato, H.; Mizuno, M.; Tsuji, K.; Harada, K. Bacterial degradation of microcystins and nodularin. Chem. Res. Toxicol. 2005, 18, 591–598. [Google Scholar] [CrossRef]
- Feng, N.; Yang, F.; Yan, H.; Yin, C.; Liu, X.; Zhang, H.; Xu, Q.; Lv, L.; Wang, H. Pathway for Biodegrading Nodularin (NOD) by Sphingopyxis sp. USTB-05. Toxins 2016, 8, 116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Yang, X.; Yang, L.; Xiao, B.; Wu, X.; Wang, J.; Wan, H. An effective pathway for the removal of microcystin LR via anoxic biodegradation in lake sediments. Water Res. 2010, 44, 1884–1892. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Wang, C.; Tian, C.; Xiao, B.; Song, L. Evaluation of the potential of anoxic biodegradation of intracellular and dissolved microcystins in lake sediments. J. Hazard Mater. 2015, 286, 395–401. [Google Scholar] [CrossRef]
- Zhang, M.; Pan, G.; Yan, H. Microbial biodegradation of microcystin-RR by bacterium Sphingopyxis sp. USTB-05. J. Environ. Sci. 2010, 22, 168–175. [Google Scholar] [CrossRef]
- Tsuji, K.; Asakawa, M.; Anzai, Y.; Sumino, T.; Harada, K. Degradation of microcystins using immobilized microorganism isolated in an eutrophic lake. Chemosphere 2006, 65, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Holst, T.; Jørgensen, N.O.G.; Jørgensen, C.; Johansen, A. Degradation of microcystin in sediments at oxic and anoxic, denitrifying conditions. Water Res. 2003, 37, 4748–4760. [Google Scholar] [CrossRef]
- Smith, M.J.; Shaw, G.R.; Eaglesham, G.K.; Ho, L.; Brookes, J.D. Elucidating the factors influencing the biodegradation of cylindrospermopsin in drinking water sources. Environ. Toxicol. 2008, 23, 413–421. [Google Scholar] [CrossRef]
- Manage, P.M.; Edwards, C.; Singh, B.K.; Lawton, L.A. Isolation and identification of novel microcystin-degrading bacteria. Appl. Environ. Microbiol. 2009, 75, 6924–6928. [Google Scholar] [CrossRef] [Green Version]
- Dziga, D.; Maksylewicz, A.; Maroszek, M.; Budzynska, A.; Napiorkowska-Krzebietke, A.; Toporowska, M.; Grabowska, M.; Kozak, A.; Rosinska, J.; Meriluoto, J. The biodegradation of microcystins in temperate freshwater bodies with previous cyanobacterial history. Ecotoxicol. Environ. Saf. 2017, 145, 420–430. [Google Scholar] [CrossRef]
- Krishnan, A.; Zhang, Y.; Balaban, M.; Seo, Y.; Mou, X. Taxonomic and Genotypical Heterogeneity of Microcystin degrading Bacterioplankton in Western Lake Erie. Harmful Algae 2020, 98, 101895. [Google Scholar] [CrossRef]
- Ma, G.; Pei, H.; Hu, W.; Xu, X.; Ma, C.; Li, X. The removal of cyanobacteria and their metabolites through anoxic biodegradation in drinking water sludge. Bioresour. Technol. 2014, 165, 191–198. [Google Scholar] [CrossRef]
- Bullerjahn, G.S.; McKay, R.M.; Davis, T.W.; Baker, D.B.; Boyer, G.L.; D’Anglada, L.V.; Doucette, G.J.; Ho, J.C.; Irwin, E.G.; Kling, C.L.; et al. Global solutions to regional problems: Collecting global expertise to address the problem of harmful cyanobacterial blooms. A Lake Erie case study. Harmful Algae 2016, 54, 223–238. [Google Scholar] [CrossRef] [Green Version]
- Krausfeldt, L.E.; Farmer, A.T.; Castro Gonzalez, H.F.; Zepernick, B.N.; Campagna, S.R.; Wilhelm, S.W. Urea Is Both a Carbon and Nitrogen Source for Microcystis aeruginosa: Tracking (13)C Incorporation at Bloom pH Conditions. Front. Microbiol. 2019, 10, 1064. [Google Scholar] [CrossRef] [Green Version]
- Yang, F.; Guo, J.; Huang, F.; Massey, I.Y.; Huang, R.; Li, Y.; Wen, C.; Ding, P.; Zeng, W.; Liang, G. Removal of Microcystin-LR by a Novel Native Effective Bacterial Community Designated as YFMCD4 Isolated from Lake Taihu. Toxins 2018, 10, 363. [Google Scholar] [CrossRef] [Green Version]
- Yang, F.; Massey, I.Y.; Guo, J.; Yang, S.; Pu, Y.; Zeng, W.; Tan, H. Microcystin-LR degradation utilizing a novel effective indigenous bacterial community YFMCD1 from Lake Taihu. J. Toxicol. Environ. Health A 2018, 81, 184–193. [Google Scholar] [CrossRef]
- Kato, H.; Imanishi, S.Y.; Tsuji, K.; Harada, K.-i. Microbial degradation of cyanobacterial cyclic peptides. Water Res. 2007, 41, 1754–1762. [Google Scholar] [CrossRef]
- Mazur-Marzec, H.; Torunska, A.; Blonska, M.J.; Moskot, M.; Plinski, M.; Jakobkiewicz-Banecka, J.; Wegrzyn, G. Biodegradation of nodularin and effects of the toxin on bacterial isolates from the Gulf of Gdansk. Water Res. 2009, 43, 2801–2810. [Google Scholar] [CrossRef]
- Edwards, C.; Graham, D.; Fowler, N.; Lawton, L.A. Biodegradation of microcystins and nodularin in freshwaters. Chemosphere 2008, 73, 1315–1321. [Google Scholar] [CrossRef]
- Li, J.; Peng, L.; Li, J.; Qiao, Y. Divergent responses of functional gene expression to various nutrient conditions during microcystin-LR biodegradation by Novosphingobium sp. THN1 strain. Bioresour. Technol. 2014, 156, 335–341. [Google Scholar] [CrossRef]
- Wang, J.; Wang, C.; Li, Q.; Shen, M.; Bai, P.; Li, J.; Lin, Y.; Gan, N.; Li, T.; Zhao, J. Microcystin-LR Degradation and Gene Regulation of Microcystin-Degrading Novosphingobium sp. THN1 at Different Carbon Concentrations. Front. Microbiol. 2019, 10, 1750. [Google Scholar] [CrossRef] [Green Version]
- Xu, Q.; Ma, H.; Fan, J.; Yan, H.; Zhang, H.; Yin, C.; Liu, X.; Liu, Y.; Wang, H. Cloning and Expression of Genes for Biodegrading Nodularin by Sphingopyxis sp. USTB-05. Toxins 2019, 11, 549. [Google Scholar] [CrossRef] [Green Version]
- Wang, R.; Li, J.; Jiang, Y.; Lu, Z.; Li, R.; Li, J. Heterologous expression of mlrA gene originated from Novosphingobium sp. THN1 to degrade microcystin-RR and identify the first step involved in degradation pathway. Chemosphere 2017, 184, 159–167. [Google Scholar] [CrossRef]
- Liu, H.; Guo, X.; Liu, L.; Yan, M.; Li, J.; Hou, S.; Wan, J.; Feng, L. Simultaneous Microcystin Degradation and Microcystis aeruginosa Inhibition with the Single Enzyme Microcystinase A. Environ. Sci. Technol. 2020, 54, 8811–8820. [Google Scholar] [CrossRef]
- Dziga, D.; Tokodi, N.; Backovic, D.D.; Kokocinski, M.; Antosiak, A.; Puchalski, J.; Strzalka, W.; Madej, M.; Meriluoto, J.; Svircev, Z. The Effect of a Combined Hydrogen Peroxide-MlrA Treatment on the Phytoplankton Community and Microcystin Concentrations in a Mesocosm Experiment in Lake Ludos. Toxins 2019, 11, 725. [Google Scholar] [CrossRef] [Green Version]
- Dexter, J.; McCormick, A.J.; Fu, P.; Dziga, D. Microcystinase—A review of the natural occurrence, heterologous expression, and biotechnological application of MlrA. Water Res. 2021, 189, 116646. [Google Scholar] [CrossRef]
- Wu, X.; Wu, H.; Gu, X.; Zhang, R.; Sheng, Q.; Ye, J. Effect of the immobilized microcystin-LR-degrading enzyme MlrA on nodularin degradation and its immunotoxicity study. Environ. Pollut. 2020, 258, 113653. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).