Canopy-Level Spectral Variation and Classification of Diverse Crop Species with Fine Spatial Resolution Imaging Spectroscopy
Abstract
1. Introduction
2. Materials and Methods
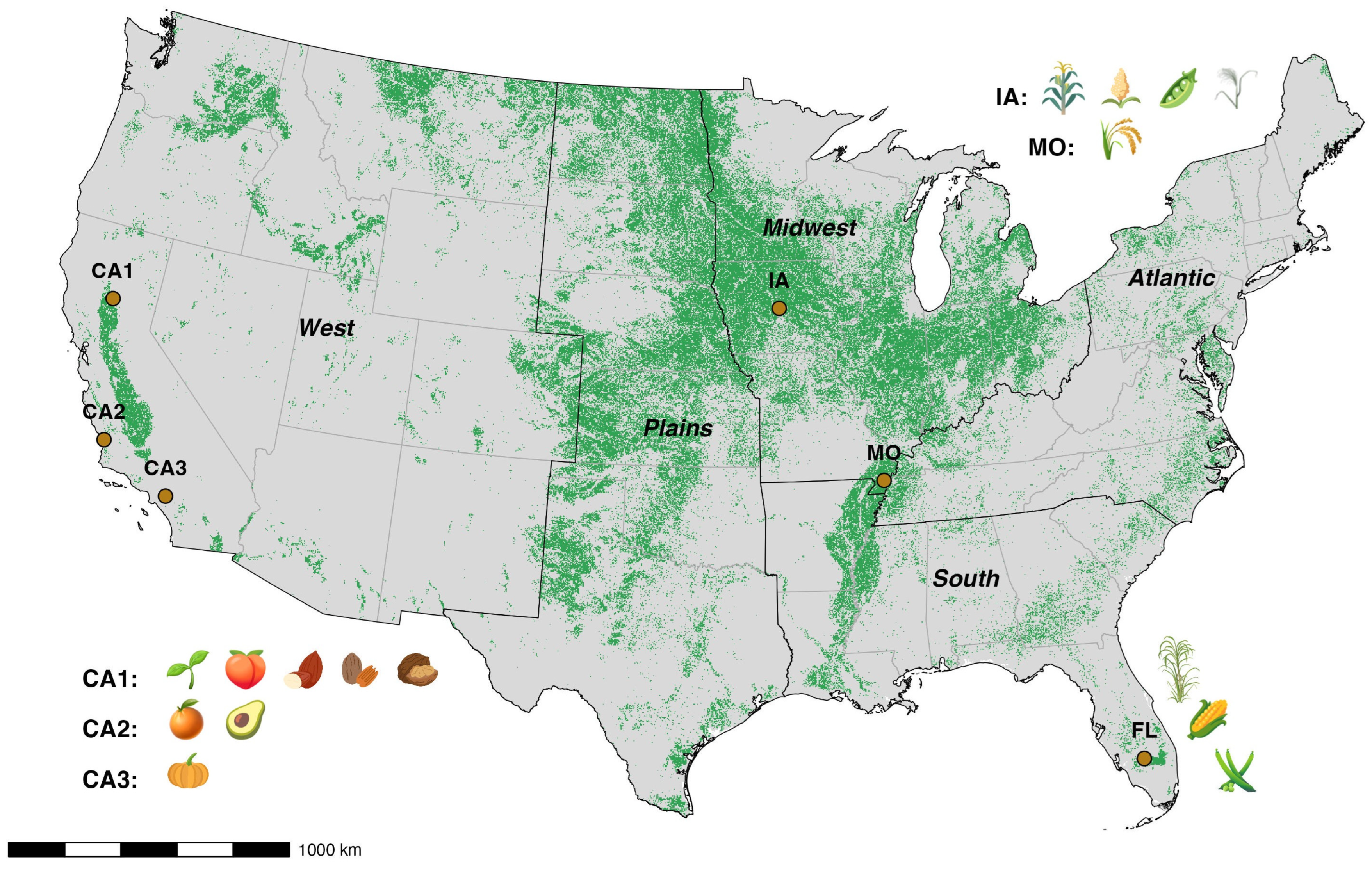
2.1. Study Sites
2.2. Data Collection and Processing
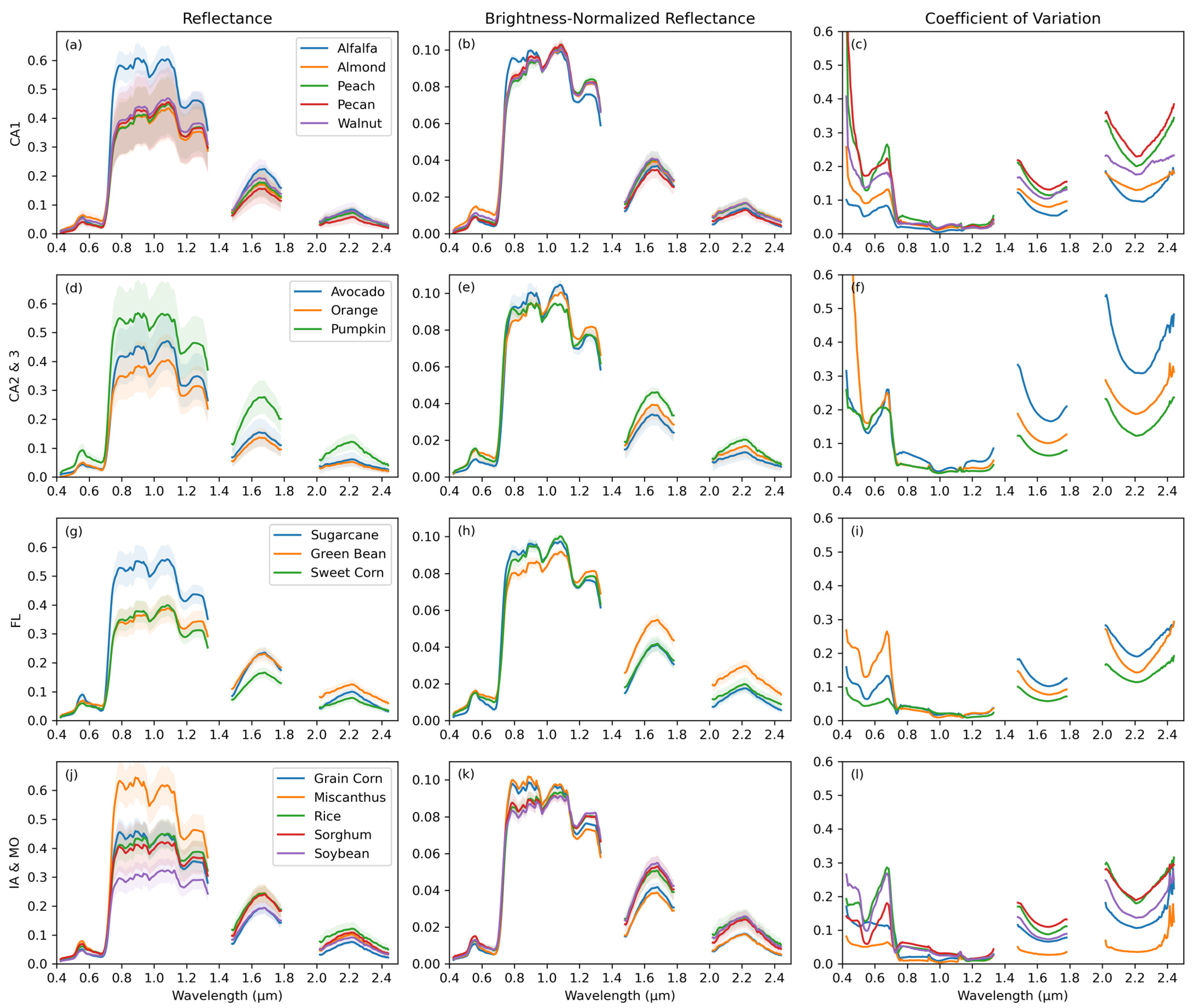
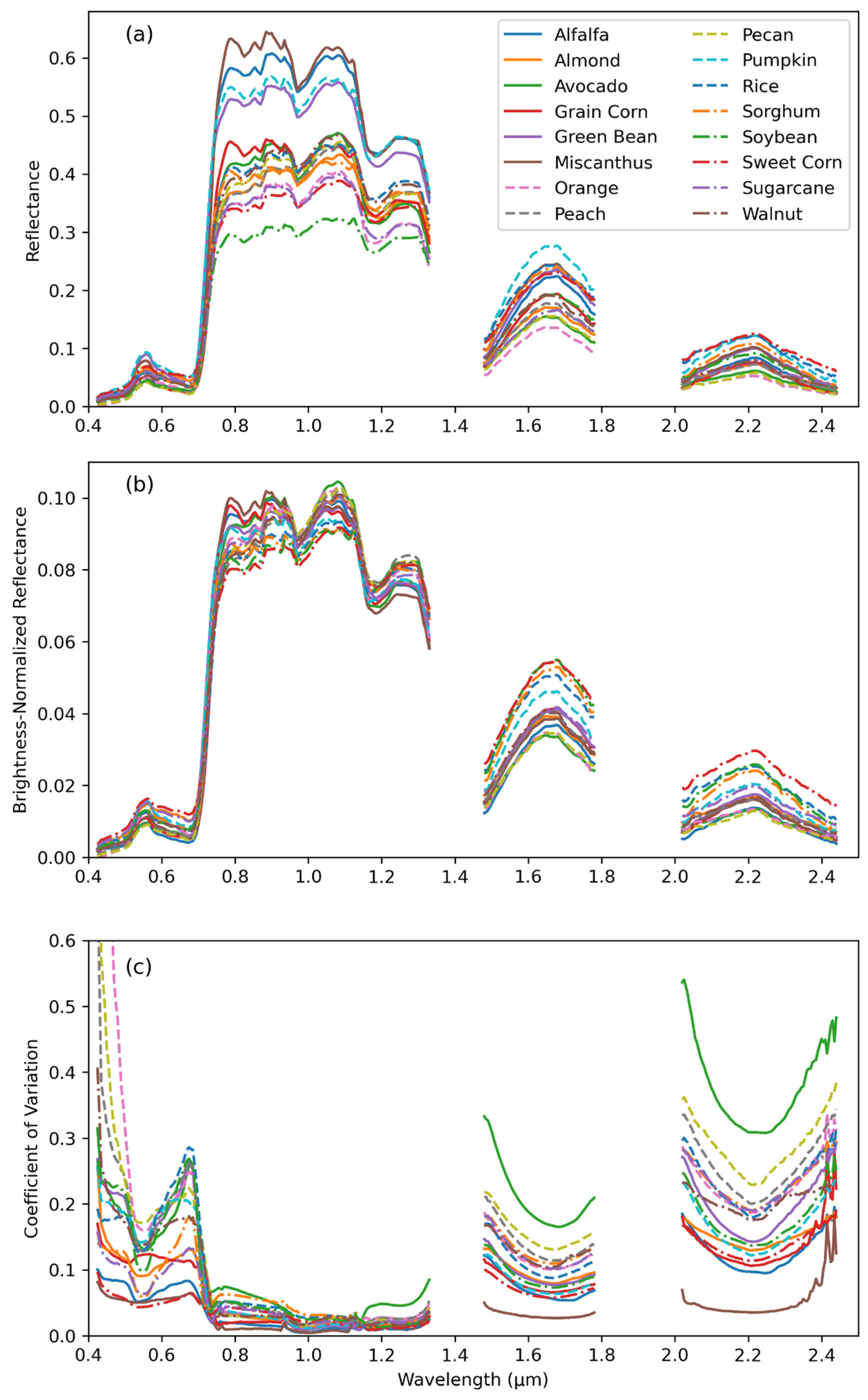
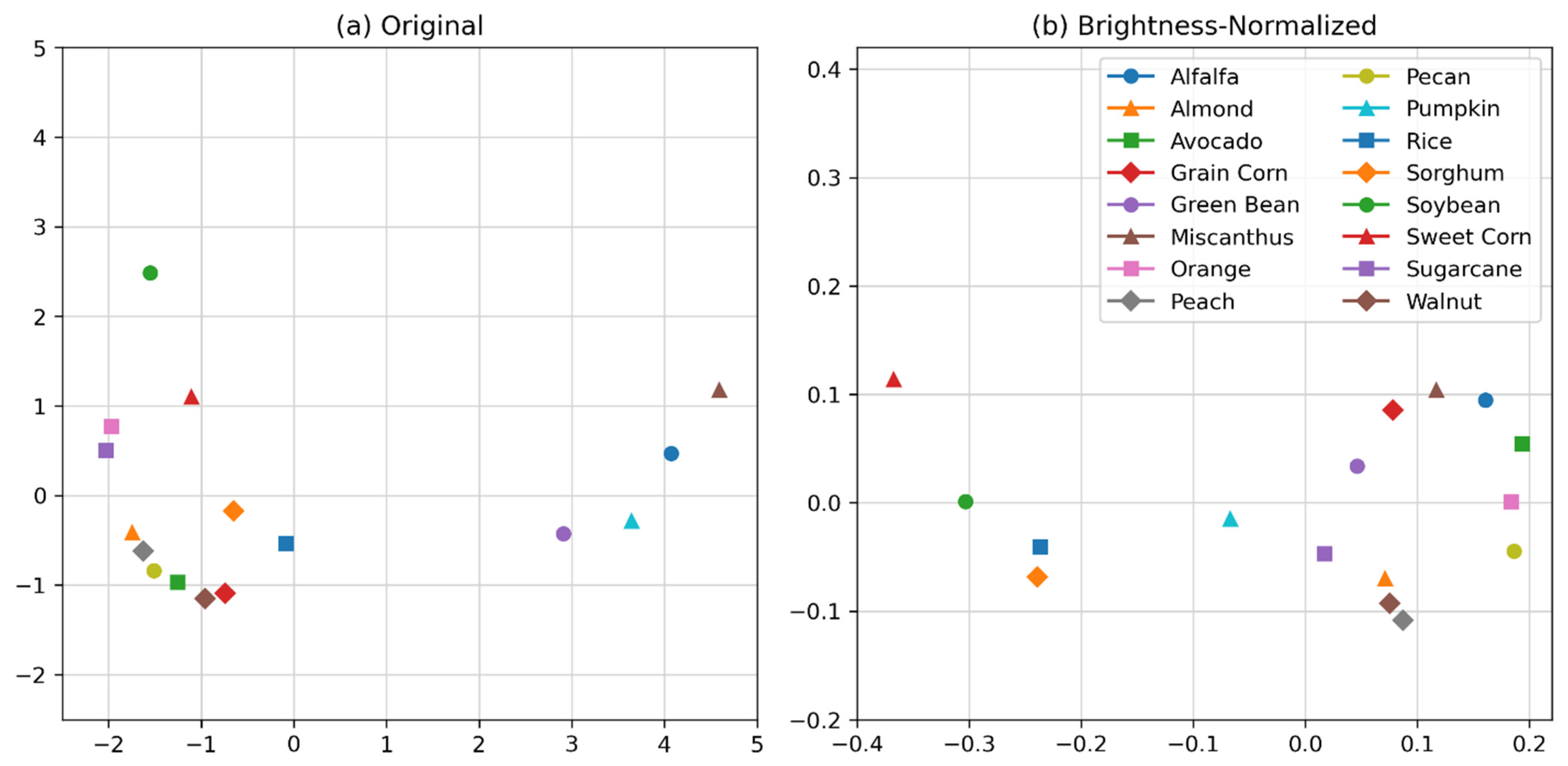
2.3. Spectral Variation
2.4. Classification Strategies
2.4.1. Training and Test Data
2.4.2. Full Spectrum vs. Selected Bands
2.4.3. Accuracy Assessment
3. Results
3.1. Crop Spectra and Spectral Variation
3.2. Classification Results
3.2.1. Training Sample Size and Cost Parameter
3.2.2. Different Classification Strategies
3.2.3. Pooled Classification
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
Appendix B
| Site | Farm Name | Lat | Lon | GAO Dates |
|---|---|---|---|---|
| FL | Rouge River Farm | 26.69 | −81.17 | 28 March–10 April |
| IA | Iowa State University Farm | 42.00 | −93.70 | 9 July |
| MO | University of Missouri Farm | 36.41 | −89.42 | 10 July |
| CA1 | Chico State University Farm | 39.68 | −121.82 | 4 September |
| CA2 | Cal Poly San Luis Obispo Farm | 35.30 | −120.67 | 3 September |
| CA3 | Cal Poly Pomona Farm | 34.04 | −117.82 | 6 September |
| Band | Wavelength | Band | Wavelength | Band | Wavelength | Band | Wavelength |
|---|---|---|---|---|---|---|---|
| 17 | 427.4 | 81 | 747.9 | 138 | 1033.3 | 269 | 1689.3 |
| 21 | 447.4 | 84 | 762.9 | 140 | 1043.3 | 271 | 1699.3 |
| 30 | 492.5 | 86 | 772.9 | 146 | 1073.4 | 283 | 1759.4 |
| 32 | 502.5 | 88 | 783.0 | 151 | 1098.4 | 342 | 2054.9 |
| 36 | 522.6 | 91 | 798.0 | 160 | 1143.5 | 344 | 2064.9 |
| 38 | 532.6 | 94 | 813.0 | 162 | 1153.5 | 352 | 2104.9 |
| 43 | 557.6 | 96 | 823.0 | 168 | 1183.6 | 358 | 2135.0 |
| 45 | 567.6 | 98 | 833.0 | 175 | 1218.6 | 364 | 2165.0 |
| 49 | 587.7 | 100 | 843.0 | 180 | 1243.6 | 366 | 2175.0 |
| 52 | 602.7 | 102 | 853.0 | 186 | 1273.7 | 372 | 2205.1 |
| 54 | 612.7 | 105 | 868.1 | 194 | 1313.7 | 382 | 2255.1 |
| 56 | 622.7 | 108 | 883.1 | 223 | 1459.0 | 390 | 2295.2 |
| 59 | 637.7 | 111 | 898.1 | 225 | 1469.0 | 395 | 2320.3 |
| 61 | 647.7 | 113 | 908.1 | 235 | 1519.1 | 400 | 2345.3 |
| 63 | 657.8 | 115 | 918.2 | 237 | 1529.1 | 402 | 2355.3 |
| 65 | 667.8 | 117 | 928.2 | 241 | 1549.1 | 411 | 2400.4 |
| 68 | 682.8 | 119 | 938.2 | 245 | 1569.1 | 417 | 2430.4 |
| 72 | 702.8 | 122 | 953.2 | 253 | 1609.2 | ||
| 75 | 717.8 | 125 | 968.2 | 255 | 1619.2 | ||
| 79 | 737.9 | 131 | 998.3 | 261 | 1649.2 |
| Alfalfa | Almond | Avocado | Grain Corn | Green Bean | Miscanthus | Orange | Peach | Pecan | Pumpkin | Rice | Sorghum | Soybean | Sweet Corn | Sugarcane | Walnut | |
| Alfalfa | 0 | 0.084 | 0.042 | 0.044 | 0.062 | 0.040 | 0.051 | 0.089 | 0.067 | 0.101 | 0.159 | 0.156 | 0.181 | 0.213 | 0.085 | 0.081 |
| Almond | 1.83 | 0 | 0.079 | 0.080 | 0.057 | 0.092 | 0.056 | 0.048 | 0.063 | 0.075 | 0.114 | 0.115 | 0.134 | 0.158 | 0.037 | 0.028 |
| Avocado | 1.61 | 0.40 | 0 | 0.065 | 0.080 | 0.057 | 0.039 | 0.081 | 0.054 | 0.116 | 0.164 | 0.167 | 0.190 | 0.217 | 0.083 | 0.075 |
| Grain Corn | 1.46 | 0.49 | 0.33 | 0 | 0.055 | 0.031 | 0.075 | 0.083 | 0.084 | 0.075 | 0.125 | 0.121 | 0.148 | 0.180 | 0.067 | 0.073 |
| Green Bean | 0.51 | 1.43 | 1.28 | 1.10 | 0 | 0.063 | 0.066 | 0.083 | 0.087 | 0.048 | 0.117 | 0.110 | 0.134 | 0.165 | 0.049 | 0.064 |
| Miscanthus | 0.35 | 2.08 | 1.86 | 1.69 | 0.72 | 0 | 0.075 | 0.102 | 0.093 | 0.089 | 0.145 | 0.143 | 0.170 | 0.199 | 0.080 | 0.090 |
| Orange | 2.18 | 0.45 | 0.60 | 0.79 | 1.82 | 2.44 | 0 | 0.068 | 0.039 | 0.105 | 0.157 | 0.158 | 0.179 | 0.207 | 0.074 | 0.058 |
| Peach | 1.76 | 0.23 | 0.37 | 0.45 | 1.39 | 2.01 | 0.55 | 0 | 0.049 | 0.098 | 0.115 | 0.120 | 0.136 | 0.164 | 0.060 | 0.025 |
| Pecan | 1.68 | 0.30 | 0.25 | 0.42 | 1.34 | 1.95 | 0.54 | 0.21 | 0 | 0.121 | 0.158 | 0.160 | 0.179 | 0.208 | 0.084 | 0.053 |
| Pumpkin | 0.61 | 1.73 | 1.61 | 1.41 | 0.40 | 0.64 | 2.13 | 1.68 | 1.67 | 0 | 0.080 | 0.070 | 0.095 | 0.126 | 0.057 | 0.080 |
| Rice | 1.54 | 0.71 | 0.81 | 0.61 | 1.10 | 1.72 | 1.11 | 0.66 | 0.80 | 1.26 | 0 | 0.033 | 0.039 | 0.059 | 0.092 | 0.109 |
| Sorghum | 1.70 | 0.55 | 0.72 | 0.53 | 1.26 | 1.90 | 0.91 | 0.54 | 0.69 | 1.45 | 0.28 | 0 | 0.031 | 0.069 | 0.095 | 0.111 |
| Soybean | 2.67 | 0.92 | 1.21 | 1.25 | 2.25 | 2.89 | 0.76 | 0.99 | 1.13 | 2.48 | 1.27 | 1.04 | 0 | 0.051 | 0.117 | 0.129 |
| Sweet Corn | 2.17 | 0.68 | 0.98 | 0.91 | 1.72 | 2.36 | 0.88 | 0.73 | 0.92 | 1.90 | 0.66 | 0.50 | 0.69 | 0 | 0.138 | 0.158 |
| Sugarcane | 2.15 | 0.37 | 0.63 | 0.73 | 1.76 | 2.39 | 0.29 | 0.48 | 0.58 | 2.04 | 0.92 | 0.71 | 0.61 | 0.61 | 0 | 0.044 |
| Walnut | 1.51 | 0.36 | 0.39 | 0.34 | 1.12 | 1.76 | 0.77 | 0.28 | 0.32 | 1.41 | 0.51 | 0.47 | 1.21 | 0.81 | 0.70 | 0 |
References
- THE 17 GOALS|Sustainable Development. Available online: https://sdgs.un.org/goals (accessed on 11 March 2024).
- Gomiero, T.; Pimentel, D.; Paoletti, M.G. Environmental Impact of Different Agricultural Management Practices: Conventional vs. Organic Agriculture. Crit. Rev. Plant Sci. 2011, 30, 95–124. [Google Scholar] [CrossRef]
- Weiss, M.; Jacob, F.; Duveiller, G. Remote Sensing for Agricultural Applications: A Meta-Review. Remote Sens. Environ. 2020, 236, 111402. [Google Scholar] [CrossRef]
- Howard, D.M.; Wylie, B.K.; Tieszen, L.L. Crop Classification Modelling Using Remote Sensing and Environmental Data in the Greater Platte River Basin, USA. Int. J. Remote Sens. 2012, 33, 6094–6108. [Google Scholar] [CrossRef]
- Kussul, N.; Lavreniuk, M.; Skakun, S.; Shelestov, A. Deep Learning Classification of Land Cover and Crop Types Using Remote Sensing Data. IEEE Geosci. Remote Sens. Lett. 2017, 14, 778–782. [Google Scholar] [CrossRef]
- Xie, H.; Tian, Y.Q.; Granillo, J.A.; Keller, G.R. Suitable Remote Sensing Method and Data for Mapping and Measuring Active Crop Fields. Int. J. Remote Sens. 2007, 28, 395–411. [Google Scholar] [CrossRef]
- Karthikeyan, L.; Chawla, I.; Mishra, A.K. A Review of Remote Sensing Applications in Agriculture for Food Security: Crop Growth and Yield, Irrigation, and Crop Losses. J. Hydrol. 2020, 586, 124905. [Google Scholar] [CrossRef]
- Townshend, J.; Justice, C.; Li, W.; Gurney, C.; McManus, J. Global Land Cover Classification by Remote Sensing: Present Capabilities and Future Possibilities. Remote Sens. Environ. 1991, 35, 243–255. [Google Scholar] [CrossRef]
- Wu, C.; Murray, A.T. Estimating Impervious Surface Distribution by Spectral Mixture Analysis. Remote Sens. Environ. 2003, 84, 493–505. [Google Scholar] [CrossRef]
- Gómez, C.; White, J.C.; Wulder, M.A. Optical Remotely Sensed Time Series Data for Land Cover Classification: A Review. ISPRS J. Photogramm. Remote Sens. 2016, 116, 55–72. [Google Scholar] [CrossRef]
- Yi, Z.; Jia, L.; Chen, Q. Crop Classification Using Multi-Temporal Sentinel-2 Data in the Shiyang River Basin of China. Remote Sens. 2020, 12, 4052. [Google Scholar] [CrossRef]
- Zhong, L.; Hu, L.; Zhou, H. Deep Learning Based Multi-Temporal Crop Classification. Remote Sens. Environ. 2019, 221, 430–443. [Google Scholar] [CrossRef]
- Rossi, C.; Kneubühler, M.; Schütz, M.; Schaepman, M.E.; Haller, R.M.; Risch, A.C. Remote Sensing of Spectral Diversity: A New Methodological Approach to Account for Spatio-Temporal Dissimilarities between Plant Communities. Ecol. Indic. 2021, 130, 108106. [Google Scholar] [CrossRef]
- Wang, R.; Gamon, J.A. Remote Sensing of Terrestrial Plant Biodiversity. Remote Sens. Environ. 2019, 231, 111218. [Google Scholar] [CrossRef]
- Asner, G.P. Biophysical and Biochemical Sources of Variability in Canopy Reflectance. Remote Sens. Environ. 1998, 64, 234–253. [Google Scholar] [CrossRef]
- Gamon, J.A.; Somers, B.; Malenovský, Z.; Middleton, E.M.; Rascher, U.; Schaepman, M.E. Assessing Vegetation Function with Imaging Spectroscopy. Surv. Geophys. 2019, 40, 489–513. [Google Scholar] [CrossRef]
- Ustin, S.L.; Gitelson, A.A.; Jacquemoud, S.; Schaepman, M.; Asner, G.P.; Gamon, J.A.; Zarco-Tejada, P. Retrieval of Foliar Information about Plant Pigment Systems from High Resolution Spectroscopy. Remote Sens. Environ. 2009, 113, S67–S77. [Google Scholar] [CrossRef]
- Green, R.O.; Eastwood, M.L.; Sarture, C.M.; Chrien, T.G.; Aronsson, M.; Chippendale, B.J.; Faust, J.A.; Pavri, B.E.; Chovit, C.J.; Solis, M.; et al. Imaging Spectroscopy and the Airborne Visible/Infrared Imaging Spectrometer (AVIRIS). Remote Sens. Environ. 1998, 65, 227–248. [Google Scholar] [CrossRef]
- Lu, B.; Dao, P.D.; Liu, J.; He, Y.; Shang, J. Recent Advances of Hyperspectral Imaging Technology and Applications in Agriculture. Remote Sens. 2020, 12, 2659. [Google Scholar] [CrossRef]
- Hank, T.B.; Berger, K.; Bach, H.; Clevers, J.G.P.W.; Gitelson, A.; Zarco-Tejada, P.; Mauser, W. Spaceborne Imaging Spectroscopy for Sustainable Agriculture: Contributions and Challenges. Surv. Geophys. 2019, 40, 515–551. [Google Scholar] [CrossRef]
- Agilandeeswari, L.; Prabukumar, M.; Radhesyam, V.; Phaneendra, K.L.N.B.; Farhan, A. Crop Classification for Agricultural Applications in Hyperspectral Remote Sensing Images. Appl. Sci. 2022, 12, 1670. [Google Scholar] [CrossRef]
- Aneece, I.; Thenkabail, P.S. New Generation Hyperspectral Sensors DESIS and PRISMA Provide Improved Agricultural Crop Classifications. Photogramm. Eng. Remote Sens. 2022, 88, 715–729. [Google Scholar] [CrossRef]
- Nidamanuri, R.R.; Zbell, B. Use of Field Reflectance Data for Crop Mapping Using Airborne Hyperspectral Image. ISPRS J. Photogramm. Remote Sens. 2011, 66, 683–691. [Google Scholar] [CrossRef]
- Wang, Z.; Zhao, Z.; Yin, C. Fine Crop Classification Based on UAV Hyperspectral Images and Random Forest. ISPRS Int. J. Geo-Inf. 2022, 11, 252. [Google Scholar] [CrossRef]
- Wei, L.; Yu, M.; Liang, Y.; Yuan, Z.; Huang, C.; Li, R.; Yu, Y. Precise Crop Classification Using Spectral-Spatial-Location Fusion Based on Conditional Random Fields for UAV-Borne Hyperspectral Remote Sensing Imagery. Remote Sens. 2019, 11, 2011. [Google Scholar] [CrossRef]
- Asner, G.P.; Knapp, D.E.; Boardman, J.; Green, R.O.; Kennedy-Bowdoin, T.; Eastwood, M.; Martin, R.E.; Anderson, C.; Field, C.B. Carnegie Airborne Observatory-2: Increasing Science Data Dimensionality via High-Fidelity Multi-Sensor Fusion. Remote Sens. Environ. 2012, 124, 454–465. [Google Scholar] [CrossRef]
- ARMS III Farm Production Regions Map. Available online: https://www.nass.usda.gov/Charts_and_Maps/Farm_Production_Expenditures/reg_map_c.php (accessed on 11 March 2024).
- Dai, J.; Jamalinia, E.; Vaughn, N.R.; Martin, R.E.; König, M.; Hondula, K.L.; Calhoun, J.; Heckler, J.; Asner, G.P. A General Methodology for the Quantification of Crop Canopy Nitrogen across Diverse Species Using Airborne Imaging Spectroscopy. Remote Sens. Environ. 2023, 298, 113836. [Google Scholar] [CrossRef]
- Asner, G.P.; Martin, R.E.; Anderson, C.B.; Knapp, D.E. Quantifying Forest Canopy Traits: Imaging Spectroscopy versus Field Survey. Remote Sens. Environ. 2015, 158, 15–27. [Google Scholar] [CrossRef]
- Seeley, M.M.; Martin, R.E.; Vaughn, N.R.; Thompson, D.R.; Dai, J.; Asner, G.P. Quantifying the Variation in Reflectance Spectra of Metrosideros Polymorpha Canopies across Environmental Gradients. Remote Sens. 2023, 15, 1614. [Google Scholar] [CrossRef]
- Feilhauer, H.; Asner, G.P.; Martin, R.E.; Schmidtlein, S. Brightness-Normalized Partial Least Squares Regression for Hyperspectral Data. J. Quant. Spectrosc. Radiat. Transf. 2010, 111, 1947–1957. [Google Scholar] [CrossRef]
- Wang, R.; Gamon, J.A.; Cavender-Bares, J.; Townsend, P.A.; Zygielbaum, A.I. The Spatial Sensitivity of the Spectral Diversity–Biodiversity Relationship: An Experimental Test in a Prairie Grassland. Ecol. Appl. 2018, 28, 541–556. [Google Scholar] [CrossRef] [PubMed]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-Learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Cortes, C.; Vapnik, V. Support-Vector Networks. Mach Learn 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Huang, C.; Davis, L.S.; Townshend, J.R.G. An Assessment of Support Vector Machines for Land Cover Classification. Int. J. Remote Sens. 2002, 23, 725–749. [Google Scholar] [CrossRef]
- Kumar, P.; Gupta, D.K.; Mishra, V.N.; Prasad, R. Comparison of Support Vector Machine, Artificial Neural Network, and Spectral Angle Mapper Algorithms for Crop Classification Using LISS IV Data. Int. J. Remote Sens. 2015, 36, 1604–1617. [Google Scholar] [CrossRef]
- Lin, Z.; Yan, L. A Support Vector Machine Classifier Based on a New Kernel Function Model for Hyperspectral Data. GIScience Remote Sens. 2016, 53, 85–101. [Google Scholar] [CrossRef]
- Hughes, G. On the Mean Accuracy of Statistical Pattern Recognizers. IEEE Trans. Inf. Theory 1968, 14, 55–63. [Google Scholar] [CrossRef]
- Thenkabail, P.S.; Mariotto, I.; Gumma, M.K.; Middleton, E.M.; Landis, D.R.; Huemmrich, K.F. Selection of Hyperspectral Narrowbands (HNBs) and Composition of Hyperspectral Twoband Vegetation Indices (HVIs) for Biophysical Characterization and Discrimination of Crop Types Using Field Reflectance and Hyperion/EO-1 Data. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2013, 6, 427–439. [Google Scholar] [CrossRef]
- Aneece, I.; Thenkabail, P. Accuracies Achieved in Classifying Five Leading World Crop Types and Their Growth Stages Using Optimal Earth Observing-1 Hyperion Hyperspectral Narrowbands on Google Earth Engine. Remote Sens. 2018, 10, 2027. [Google Scholar] [CrossRef]
- Aneece, I.; Thenkabail, P.S. Classifying Crop Types Using Two Generations of Hyperspectral Sensors (Hyperion and DESIS) with Machine Learning on the Cloud. Remote Sens. 2021, 13, 4704. [Google Scholar] [CrossRef]
- Czaplewski, R.L. Variance Approximations for Assessments of Classification Accuracy; U.S. Department of Agriculture, Forest Service, Rocky Mountain Forest and Range Experiment Station: Ft. Collins, CO, USA, 1994; p. RM-RP-316.
- Jamalinia, E.; Dai, J.; Vaughn, N.; Hondula, K.; König, M.; Heckler, J.; Asner, G. Application of Imaging Spectroscopy to Quantify Fractional Cover Over Agricultural Lands. In Proceedings of the IGARSS 2023—2023 IEEE International Geoscience and Remote Sensing Symposium, Pasadena, CA, USA, 16–21 July 2023; IEEE: Pasadena, CA, USA, 2023; pp. 681–684. [Google Scholar]
- Roth, K.L.; Roberts, D.A.; Dennison, P.E.; Alonzo, M.; Peterson, S.H.; Beland, M. Differentiating Plant Species within and across Diverse Ecosystems with Imaging Spectroscopy. Remote Sens. Environ. 2015, 167, 135–151. [Google Scholar] [CrossRef]
- Liu, N.; Townsend, P.A.; Naber, M.R.; Bethke, P.C.; Hills, W.B.; Wang, Y. Hyperspectral Imagery to Monitor Crop Nutrient Status within and across Growing Seasons. Remote Sens. Environ. 2021, 255, 112303. [Google Scholar] [CrossRef]
- Cawse-Nicholson, K.; Raiho, A.M.; Thompson, D.R.; Hulley, G.C.; Miller, C.E.; Miner, K.R.; Poulter, B.; Schimel, D.; Schneider, F.D.; Townsend, P.A.; et al. Intrinsic Dimensionality as a Metric for the Impact of Mission Design Parameters. J. Geophys. Res. Biogeosci. 2022, 127, e2022JG006876. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Vaughn, N.R.; Seeley, M.; Heckler, J.; Thompson, D.R.; Asner, G.P. Spectral Dimensionality of Imaging Spectroscopy Data over Diverse Landscapes and Spatial Resolutions. J. Appl. Remote Sens. 2022, 16, 044518. [Google Scholar] [CrossRef]

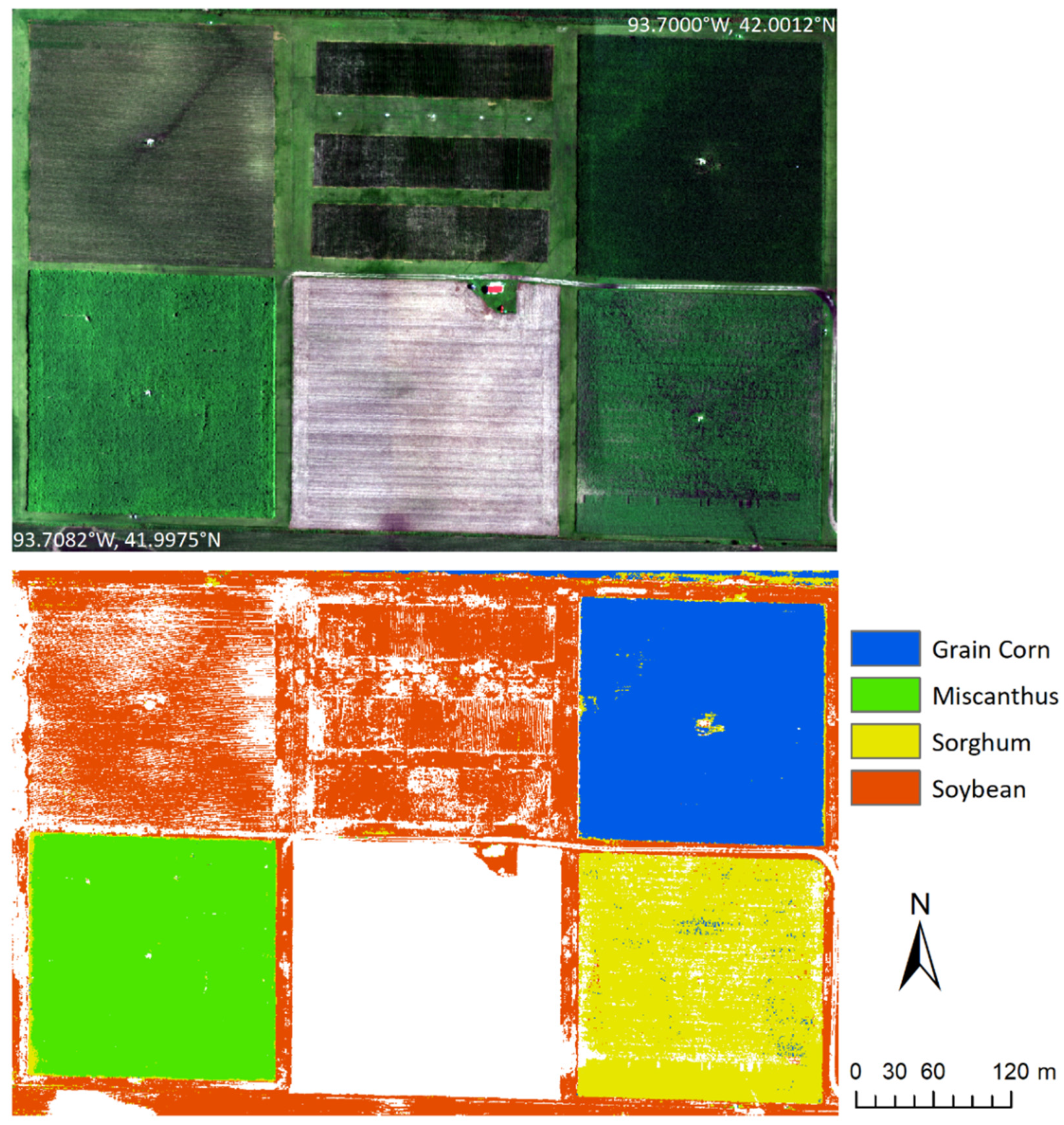
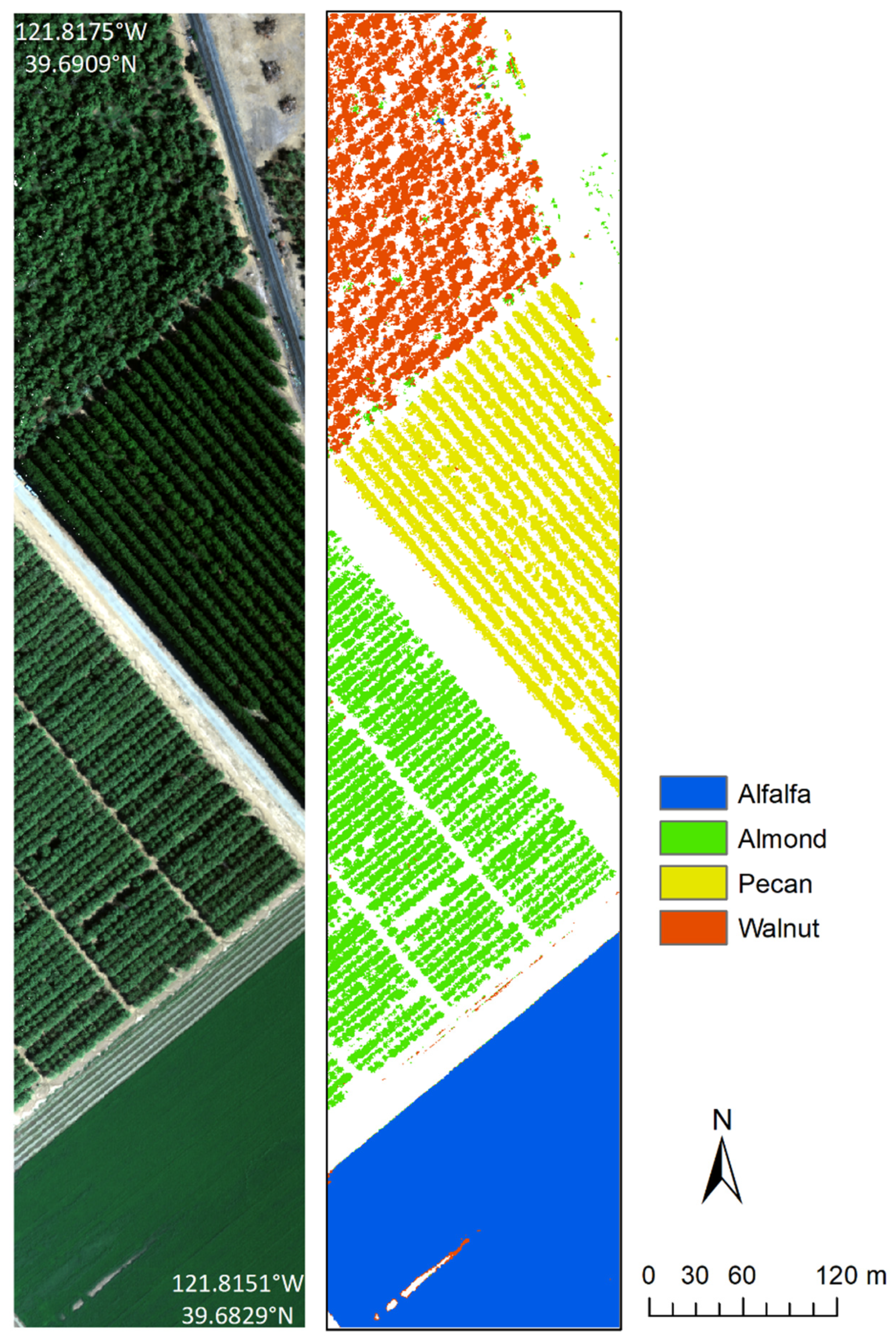
| Site | Crop | Polygons | Reflectance Spectra |
|---|---|---|---|
| CA1 | Alfalfa | 2 | 17,384 |
| Almond | 4 | 11,573 | |
| Peach | 2 | 6810 | |
| Pecan | 2 | 10,267 | |
| Walnut | 3 | 10,214 | |
| CA2 | Avocado | 2 | 8931 |
| Orange | 2 | 6361 | |
| Pumpkin | 2 | 9052 | |
| FL | Green Bean | 3 | 12,684 |
| Sugarcane | 4 | 11,433 | |
| Sweet Corn | 2 | 17,433 | |
| IA | Grain Corn | 2 | 13,624 |
| Miscanthus | 2 | 12,034 | |
| Sorghum | 2 | 20,253 | |
| Soybean | 2 | 17,281 | |
| MO | Rice | 2 | 13,224 |
| Spectra | C = 1 | C = 10 | C = 100 | C = 1000 |
|---|---|---|---|---|
| 50 | 0.39/0.39 | 0.80/0.76 | 0.97/0.95 | 1.00/0.97 |
| 100 | 0.54/0.53 | 0.85/0.85 | 0.98/0.97 | 1.00/0.99 |
| 200 | 0.64/0.64 | 0.91/0.91 | 0.99/0.99 | 1.00/1.00 |
| 500 | 0.75/0.75 | 0.96/0.95 | 1.00/0.99 | 1.00/1.00 |
| 1000 | 0.86/0.86 | 0.98/0.97 | 1.00/1.00 | 1.00/1.00 |
| 2000 | 0.91/0.91 | 0.99/0.99 | 1.00/1.00 | 1.00/1.00 |
| 3000 | 0.94/0.94 | 0.99/0.99 | 1.00/1.00 | 1.00/1.00 |
| Site | Species | Full Spectrum | 77 Bands | 33 Bands |
|---|---|---|---|---|
| CA1 | 5 | 0.96 | 0.96 | 0.94 |
| CA2 and CA3 | 3 | 0.99 | 0.99 | 0.99 |
| FL | 3 | 0.99 | 0.99 | 0.99 |
| IA and MO | 5 | 0.99 | 0.98 | 0.98 |
| Pooled | 16 | 0.97 | 0.97 | 0.97 |
| Alfalfa | Almond | Avocado | Grain Corn | Green Bean | Miscanthus | Orange | Peach | Pecan | Pumpkin | Rice | Sorghum | Soybean | Sweet Corn | Sugarcane | Walnut | |
| Alfalfa | 99.17 | 0.08 | 0.04 | 0.00 | 0.00 | 0.55 | 0.11 | 0.00 | 0.00 | 0.03 | ||||||
| Almond | 99.37 | 0.00 | 0.02 | 0.01 | 0.02 | 0.57 | ||||||||||
| Avocado | 0.02 | 0.20 | 96.32 | 0.00 | 0.03 | 1.21 | 0.21 | 0.01 | 0.18 | 0.14 | 0.34 | 1.33 | ||||
| Grain Corn | 0.03 | 0.00 | 0.00 | 98.28 | 0.25 | 0.00 | 0.02 | 0.15 | 1.26 | |||||||
| Green Bean | 0.02 | 0.01 | 98.78 | 0.01 | 0.01 | 0.02 | 0.01 | 0.36 | 0.04 | 0.07 | 0.49 | 0.15 | 0.03 | |||
| Miscanthus | 0.00 | 0.02 | 1.47 | 98.30 | 0.13 | 0.05 | 0.01 | 0.01 | 0.00 | |||||||
| Orange | 0.68 | 0.33 | 0.07 | 97.58 | 0.26 | 0.04 | 0.03 | 0.06 | 0.05 | 0.10 | 0.19 | 0.63 | ||||
| Peach | 0.01 | 0.48 | 0.01 | 97.53 | 0.24 | 0.01 | 0.02 | 0.00 | 0.28 | 0.10 | 1.32 | |||||
| Pecan | 0.68 | 0.18 | 0.05 | 2.18 | 96.15 | 0.06 | 0.01 | 0.10 | 0.05 | 0.55 | ||||||
| Pumpkin | 0.05 | 0.02 | 0.15 | 0.45 | 0.22 | 0.10 | 0.01 | 97.85 | 0.56 | 0.04 | 0.39 | 0.04 | 0.05 | 0.07 | ||
| Rice | 0.34 | 0.00 | 0.45 | 0.01 | 0.01 | 0.04 | 98.74 | 0.01 | 0.33 | 0.06 | 0.00 | |||||
| Sorghum | 0.20 | 1.65 | 0.00 | 0.00 | 0.03 | 0.05 | 97.45 | 0.61 | ||||||||
| Soybean | 0.02 | 0.27 | 0.01 | 0.01 | 0.02 | 0.62 | 98.96 | 0.08 | ||||||||
| Sweet Corn | 0.01 | 0.05 | 0.01 | 0.00 | 0.00 | 0.16 | 0.03 | 99.60 | 0.13 | |||||||
| Sugarcane | 0.03 | 0.20 | 0.10 | 0.26 | 0.01 | 0.42 | 98.98 | |||||||||
| Walnut | 0.03 | 1.88 | 0.24 | 0.01 | 0.06 | 0.01 | 0.10 | 3.01 | 0.52 | 0.05 | 0.36 | 0.00 | 0.53 | 0.46 | 0.08 | 92.66 |
| Crop | Site | PA | UA | Classified As | Classified |
|---|---|---|---|---|---|
| Alfalfa | CA1 | 99.3 | 99.2 | Rice | Rice |
| Almond | CA1 | 96.5 | 99.4 | Walnut | Walnut |
| Avocado | CA2 | 98.6 | 96.3 | Walnut | Peach |
| Grain Corn | IA | 96.1 | 98.3 | Sorghum | Sorghum |
| Green Bean | FL | 99.1 | 98.8 | Sweet Corn | Pumpkin |
| Miscanthus | IA | 99.5 | 98.3 | Grain Corn | Grain Corn |
| Orange | CA2 | 99.7 | 97.6 | Almond | Walnut |
| Peach | CA1 | 93.6 | 97.5 | Walnut | Walnut |
| Pecan | CA1 | 98.9 | 96.2 | Peach | Walnut |
| Pumpkin | CA3 | 99.3 | 97.8 | Rice | Green Bean |
| Rice | MO | 97.8 | 98.7 | Grain Corn | Pumpkin |
| Sorghum | IA | 97.9 | 97.5 | Grain Corn | Grain Corn |
| Soybean | IA | 98.0 | 99.0 | Sorghum | Sorghum |
| Sweet Corn | FL | 97.9 | 99.6 | Rice | Green Bean |
| Sugarcane | FL | 98.9 | 99.0 | Sweet Corn | Avocado |
| Walnut | CA1 | 95.3 | 92.7 | Peach | Avocado |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dai, J.; König, M.; Jamalinia, E.; Hondula, K.L.; Vaughn, N.R.; Heckler, J.; Asner, G.P. Canopy-Level Spectral Variation and Classification of Diverse Crop Species with Fine Spatial Resolution Imaging Spectroscopy. Remote Sens. 2024, 16, 1447. https://doi.org/10.3390/rs16081447
Dai J, König M, Jamalinia E, Hondula KL, Vaughn NR, Heckler J, Asner GP. Canopy-Level Spectral Variation and Classification of Diverse Crop Species with Fine Spatial Resolution Imaging Spectroscopy. Remote Sensing. 2024; 16(8):1447. https://doi.org/10.3390/rs16081447
Chicago/Turabian StyleDai, Jie, Marcel König, Elahe Jamalinia, Kelly L. Hondula, Nicholas R. Vaughn, Joseph Heckler, and Gregory P. Asner. 2024. "Canopy-Level Spectral Variation and Classification of Diverse Crop Species with Fine Spatial Resolution Imaging Spectroscopy" Remote Sensing 16, no. 8: 1447. https://doi.org/10.3390/rs16081447
APA StyleDai, J., König, M., Jamalinia, E., Hondula, K. L., Vaughn, N. R., Heckler, J., & Asner, G. P. (2024). Canopy-Level Spectral Variation and Classification of Diverse Crop Species with Fine Spatial Resolution Imaging Spectroscopy. Remote Sensing, 16(8), 1447. https://doi.org/10.3390/rs16081447







