1. Introduction
Forests provide crucial ecosystem services such as biomass production, air purification, and carbon storage. However, their efficiency and resilience is closely linked to tree species richness [
1]. Especially in the context of climate change and its potential impact on the distribution of forest compositions [
2,
3,
4], accurate mapping of tree species will remain a fundamental challenge for large scale forest management as well as small scale local forest inventories and biodiversity assessments [
5].
To meet the growing demand for spatially explicit data on the distribution of tree species, several classification approaches based on a variety of satellite data were proposed [
6,
7,
8,
9,
10]. Since these data have a relatively coarse spatial resolution, they are, however, not well suited for classifying tree species at single-tree-level in complexly structured species-rich forests.
Tree species classification approaches based on passive multispectral or hyperspectral imaging data and active LiDAR or RADAR scans recorded with airplanes or unmanned aerial vehicles (UAVs) reach higher resolution levels and thus perform better on single-tree-level [
11,
12,
13,
14,
15]. For a comprehensive overview of the latest advances in tree species classification methodologies based on airborne observations, see Fassnacht et al. [
5] and Parkan [
16]. Despite the general success of these approaches, they usually share the following insufficiencies: Most studies are based on the use of more than one data collection system which complicates data acquisition. As they are also often dependent on expensive sensor technology, the economic viability of the approaches is not always guaranteed [
5].
Although the collected data have a high spatial resolution, in most cases pixel-based-only classification approaches are chosen. Hence, information on spatial patterns in the data are ignored. Varying weather conditions and seasonal as well as diurnal variations in the solar inclination cause changing illumination characteristics. This again, results in variations in the signal patterns reaching the sensor during different data acquisition times. Unfortunately, pixel-based classification models trained on data of passive collection approaches (e.g., multi- and hyperspectral data) cannot handle these variations adequately as they can only make their classification decision on the spectral signal of one single pixel at a time. This results in pixel-based models not being transferable to unstudied regions or data collection times that differ from the acquisition time of the training data.
Another limitation results from the fact that the remote sensing data collection was often only carried out at one or very few specific dates (often even temporally separated from the collection of ground-based training and validation data). As the season-dependent phenological stage significantly influences the spectral characteristics of forests, models trained on these data are therefore not directly applicable to data collected at a different time of the year. A further critical aspect of recent classification approaches concerns the acquisition of field data obtained during ground level site inspections for training and validation of the classification models. Geolocating field data in forests is not trivial due to poor GPS reception. Also, linking single-tree data obtained at ground level to canopy level (as seen from a UAV/airplane/satellite) is complicated and time-consuming and thus makes co-locating ground-based and remote sensing data even more difficult.
To carry out a tree species classification independent of the prevailing illumination conditions, it is advantageous not to perform a pixel-based classification, but to use spatial patterns within the canopy data, the recognition of which is essentially feasible independent of the illumination situation. We also know that, to a large extent, species information is encoded within the leaf shape and branch structure of a tree, which is why forest managers mainly use these features to identify tree species. Since these characteristics are contained within the structural patterns of the canopy (provided the resolution in the remote sensing data is sufficiently high), they can be used to improve the accuracy of tree species classifications. Provided that the training data were collected appropriately, this approach also largely avoids the problems of existing pixel-based methods and allows the training and application of classification models independently of the phenological stage, time of day and year, tree vitality and prevailing atmospheric conditions.
Spatially zero-dimensional machine learning models that were used in recent studies (mainly random forests (RF) or support vector machines (SVM)) are not able to recognize the spatial-structural patterns of leaves and branches in two-dimensional image data. On the other hand, modern convolutional neural networks (CNN), which were developed within the research field of computer vision, are capable of classifying image data on the basis of their spatial-structural content with accuracies that exceed human capabilities [
17]. These models can also be used profitably in the classification process of plants in general. CNN-models were already trained on single-leaf data sets such as Flavia [
18] or ICL [
19] and were shown to achieve good performance [
20,
21,
22,
23]. However, these models are not transferable to RGB image data automatically gathered over forests as these are much more heterogeneous than the images from Flavia/ICL which were taken under laboratory conditions (white background, just one leave per image, no complex illumination and shadow conditions). Community based plant classification approaches on the basis of photos taken with smartphones such as Pl@ntNet [
24,
25] or Flora Incognita [
26] also showed to be very successful. However, these approaches can only be used for selectively identifying plants but they cannot be used directly to produce spatially explicit data on plant occurrence.
A few recent studies use the high spatial resolution of remote sensing data acquired by satellite [
27], airplane [
28,
29] or UAV [
30,
31] and apply CNNs in their classification process. Although all these studies show convincing results, they share the insufficiencies of previous studies: As their classification approach is dependent on multi- or hyperspectral image data, expensive hardware has to be employed during data acquisition. Also, the data dimensionality has to be decreased due to the Hughes phenomenon preliminary to the classification process [
32]. Furthermore, data was acquired only on a single or very few dates and thus the models were not able to learn the characteristics of different phenological stages and weather conditions. Due to the chosen flight altitudes, the resolution of the data was also not sufficient to make leaf and branch structures recognizable. The only recent UAV-based approach that does not depend on hyperspectral data and that takes into account different phenological stages and changing illumination conditions is that of Natesan et al. [
33]. Unfortunately, only two coniferous tree species are distinguished here and the validation results of 51% accuracy under strict temporal separation of training and test data set are in need of improvement.
Additionally, only very few studies have dealt with the examination of an optimal resolution in the input data for the classification task so far [
5]. This is problematic as a high image resolution is at the expense of the flight altitude and thus the area that can be covered within a single flight mission.
In summary, we identified three main problems associated with tree species classification in remote sensing: (1) Most studies are based on expensive sensor ensembles. (2) Very few studies have dealt with changes in the spectral signal due to variations in the diurnal and seasonal illumination conditions as well as due to phenological modifications. (3) The influence of the spatial resolution of the input data on the classification result has not yet been investigated thoroughly.
To overcome these shortcomings, the aim of this study was to investigate the feasibility of a tree species classification based on high-resolution photos (simple RGB data) obtained with a low-price ready-to-fly UAV. In order to take into account different illumination conditions and phenological stages, the photos were taken at different times of day and during different seasons throughout the entire vegetation period. In order to be able to exploit the spatial-structural properties of the image material, a CNN model was used for the classification procedure.
Two broadleaf and two coniferous tree species were investigated, namely Quercus robur and Fagus sylvatica as well as Larix decidua and Picea abies. In the following, only the common English terms beech, oak, larch and spruce will be used.
In the context of this work, the following two hypotheses were tested:
Hypotheses 1 (H1). It is possible to classify tree species on the basis of high-resolution RGB image data independent of phenological stage, time of day, season and prevailing atmospheric conditions.
Hypotheses 2 (H2). With decreasing spatial resolution below the detectability of leaf and branch structures, the quality of the classification result drops significantly.
Finally, the feasibility of our approach to create a gridded product for the distribution of tree species was tested on an exemplary test site within the study area.
The paper is structured as follows:
Section 2 describes the data acquisition and the applied methodology in detail. In
Section 3 the results are presented and discussed. A final conclusion is given in
Section 4.
2. Data and Methods
2.1. Study Area
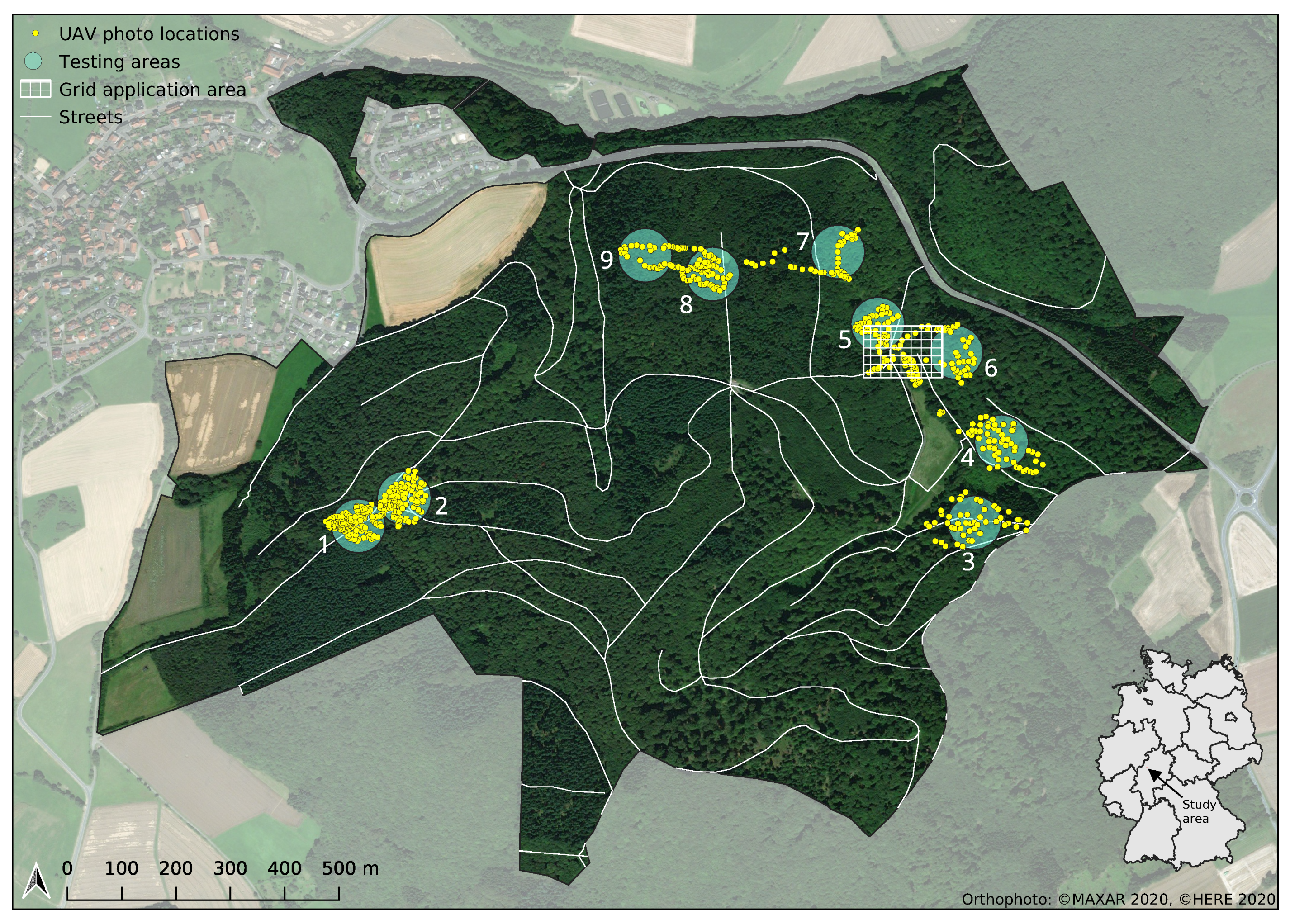
This study was conducted in the Marburg Open Forest, a teaching and research forest of the Philipps-University of Marburg, located in Central Germany (see
Figure 1). The forest is a mixed forest typical for this region. Besides occasional birches, ash trees and a few other rarely occurring tree species, it mainly consists of the leading tree species beech, oak, larch, spruce, fir, and Douglas fir. The forest is well mixed in terms of age, with average tree ages varying between 40 and 160 years. Although it is partly managed for the purpose of timber production, logging activities are compensated for by appropriate sustainability measures.
2.2. Data Collection
Image data were collected using a DJI Phantom 4 Pro UAV. A high-resolution RGB-camera with a 1” CMOS sensor and effectively 20 mega pixels with an edge length of 5472 × 3648 pixels is integrated into the vehicle. Although DJI has since expanded its consumer UAV series with additional models, the camera sensor with its 20 mega pixel resolution is still at the top of the technical range in the consumer drone market. The camera’s field of view is specified as 84
, which corresponds approximately to the 35 mm format. With an initial aperture of 2.8, the camera is also relatively light-sensitive [
34]. Images were taken vertically (top-down perspective) in auto-mode to automatically adjust to different illumination conditions and stored in JPEG format. No further image calibration was conducted.
To be able to perform automated flights close to the canopy, in a preliminary step grid flights were conducted at an altitude of 100
. Using the open source software OpenDroneMap [
35], a digital surface model (DSM) was derived from the image data obtained in these flights via the structure-from-motion approach. On the basis of the DSM, single tree locations were then manually identified and marked with waypoints. These waypoints were subsequently used for the preparation of automated flights close to the canopy level (10
distance). In order to account for different phenological stages as well as changing weather situations and thus altering illumination conditions, image data were gathered during multiple flights in different forest divisions during most part of the vegetation period of 2020.
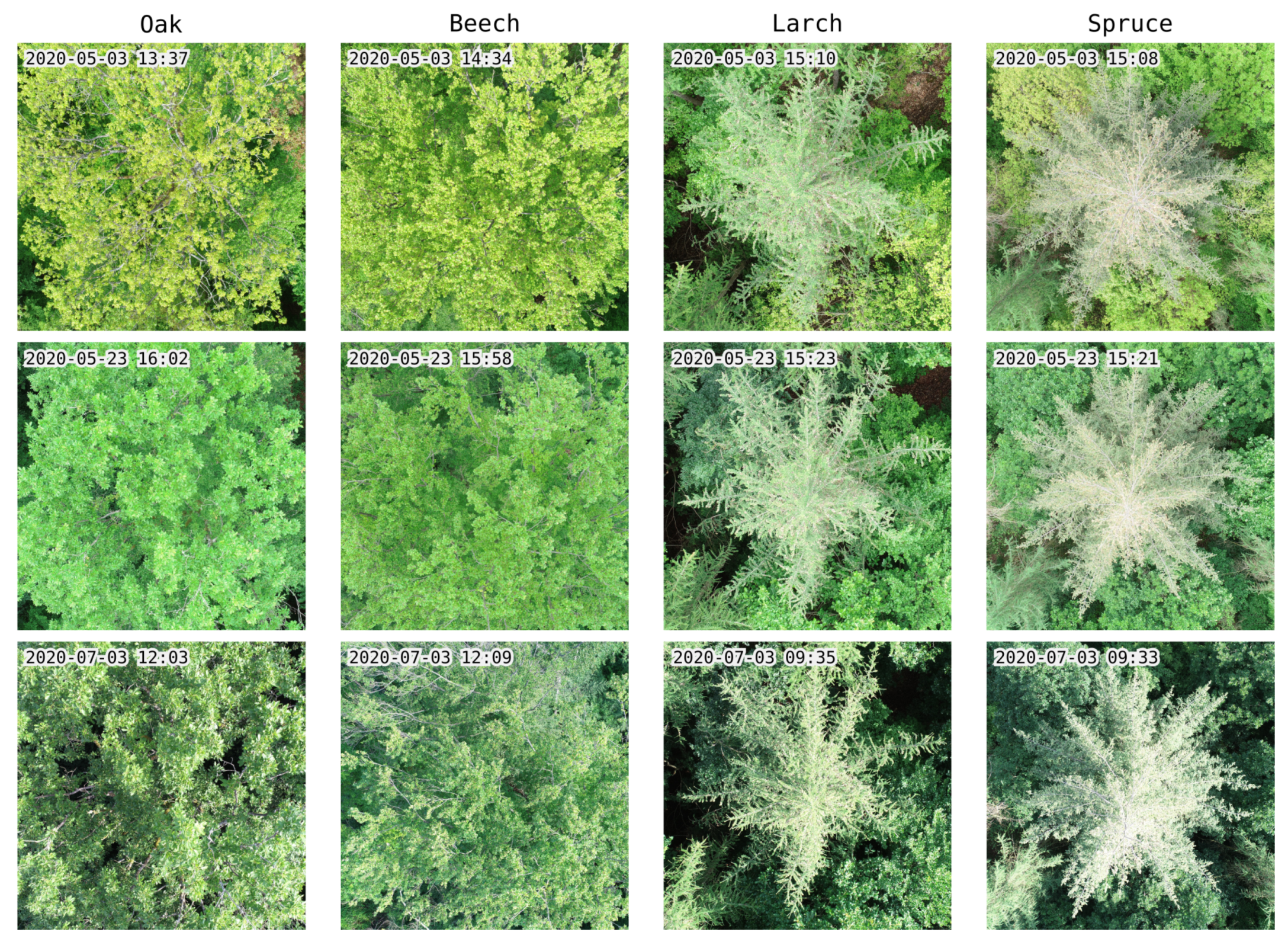
In total, 1556 photos were taken from 477 individual trees during six flight missions. Exemplary photos of each tree species assessed are shown in
Figure 2. For a detailed overview of all conducted flight missions see
Table 1 and
Figure 1.
2.3. Data Preparation
The performance of machine learning models at predicting space-time related properties is often strongly overestimated due to spatial and temporal auto-correlation in the data when using conventional cross-validation methods with simple random picks [
36]. Thus, in order to evaluate the true performance of the classification model and its generalizability, care was taken to ensure that the training/validation data set was strictly separated from the testing data set both in time and space, following the leave-location-and-time-out (LLTO) cross validation procedure as described in Meyer et al. [
36]. This means that image data used for testing the model performance was not recorded in the same region nor at the same time as the training/validation data set. For this purpose, nine different testing regions were defined (see
Figure 1). During model training, successively data from all combinations of each of these areas and each flight date were completely excluded from the training process and used exclusively for testing.
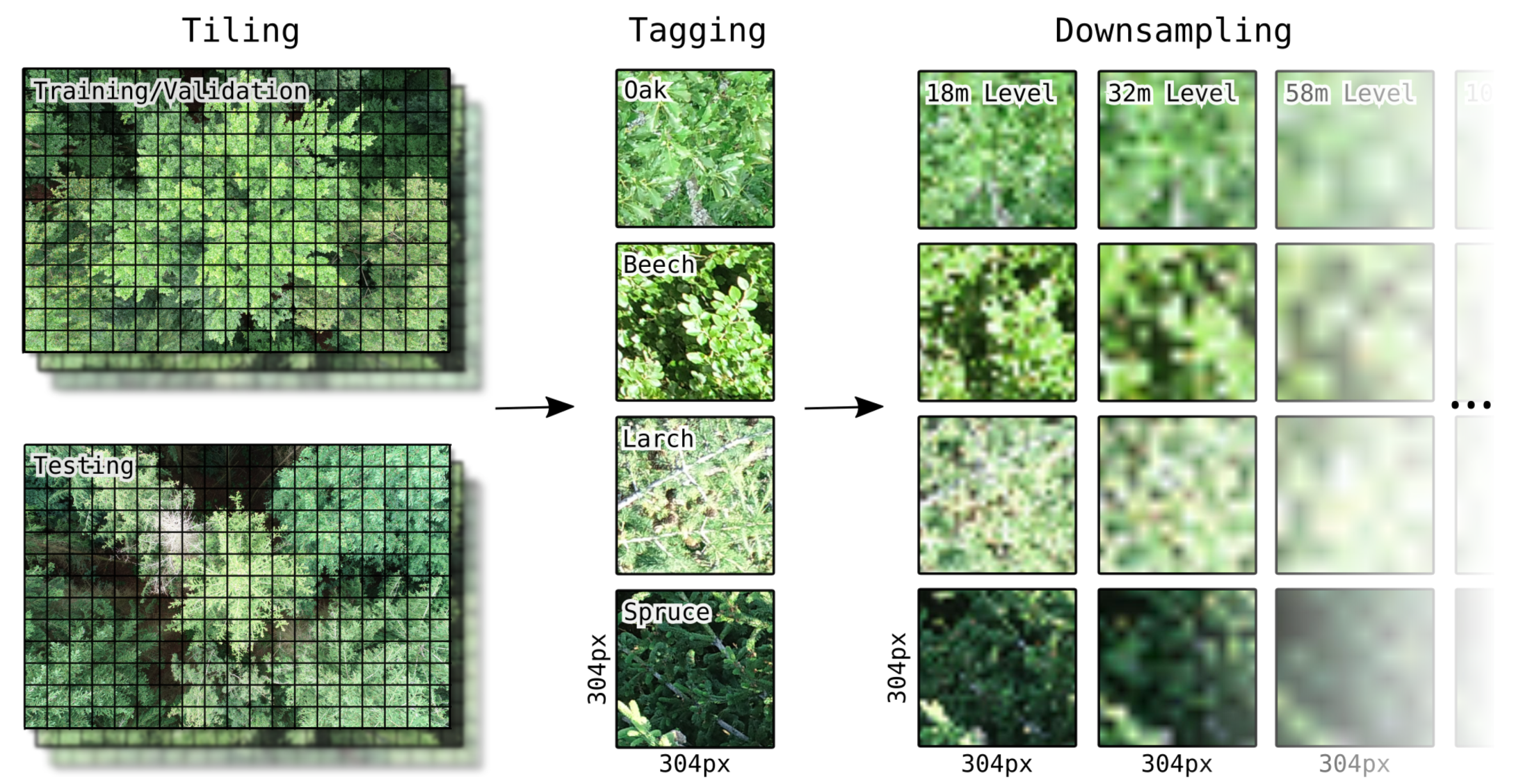
A complete pixel-accurate segmentation of tree species within the UAV photos is not necessary from a remote sensing point of view, since a resolution in the range is not useful for the creation of a spatially explicit product on tree species occurrence. Therefore, and in order to simplify the manual tagging process, to keep the computational effort within limits and to be able to generate a larger training data set, each photo was divided into 216 tiles with an edge length of 304 pixels prior to further processing. An edge length of 304 pixels was chosen as a reasonable compromise for the trade-off between recognizability of leaf and branch structure (smaller tile sizes would have resulted in too few species-specific leaf and branch structures within a tile, especially in less leafy trees) and the assignability of a tile to one specific tree (larger tile sizes would have resulted in many tiles containing several different tree species).
It should also be mentioned here that, in contrast to existing classification approaches, we did not first georeference the original data and then perform the classification task on the resulting orthophoto. Instead, we classified the image data in its original resolution in order to exploit the maximum information content of the original data and to avoid distortion effects during orthophoto generation.
The previously generated tiles were then manually grouped into the four tree species categories oak, beech, larch and spruce. A complex and error-prone co-localization of training and remote sensing data was virtually unnecessary during this process, since the tiles could be tagged almost exclusively on the basis of the leaf and branch structures recognizable in the tiles themselves. Since individual tree locations were also visited during the flight missions on site, the type of the centrally located tree of each photo could explicitly be linked to the respective photo. Where a clear tree species assignment on the basis of a single tile was not possible, the information from the photo belonging to the respective tile could thus be used to tag the tile. Only at the tree crown edges a clear assignment of a tile to one specific tree species was not always feasible. These tiles were excluded from the study. In total, 59,987 tiles were tagged with this approach. Detailed numbers per category and flight date are listed in
Table 1.
To assess the influence of the spatial resolution of the image data on the model performance, all tiles were downsampled with bilinear interpolation to ten different resolution levels corresponding to flight altitudes from 10 to 2000 above canopy level in logarithmic increments. These flight altitudes correspond to ground sampling densities between /px and /px. With each increment, the pixel count per sample decreases at a rate of 69%, starting from 92,416 px to 1 px. At higher flight altitudes than 2000 , the original tile contents at 10 flight altitude would be smaller than a single pixel in the image data which makes further downsampling pointless.
Differently sized dimensions in the input data require different CNN model architectures. To ensure that differences in the model performance are not due to changes in the model architecture but exclusively due to changes in the input data, the artificially downsampled tiles were thus scaled to the original tile resolution of 304 × 304 on all levels. An overview of all data preparation steps described here is shown in
Figure 3.
2.4. Classification
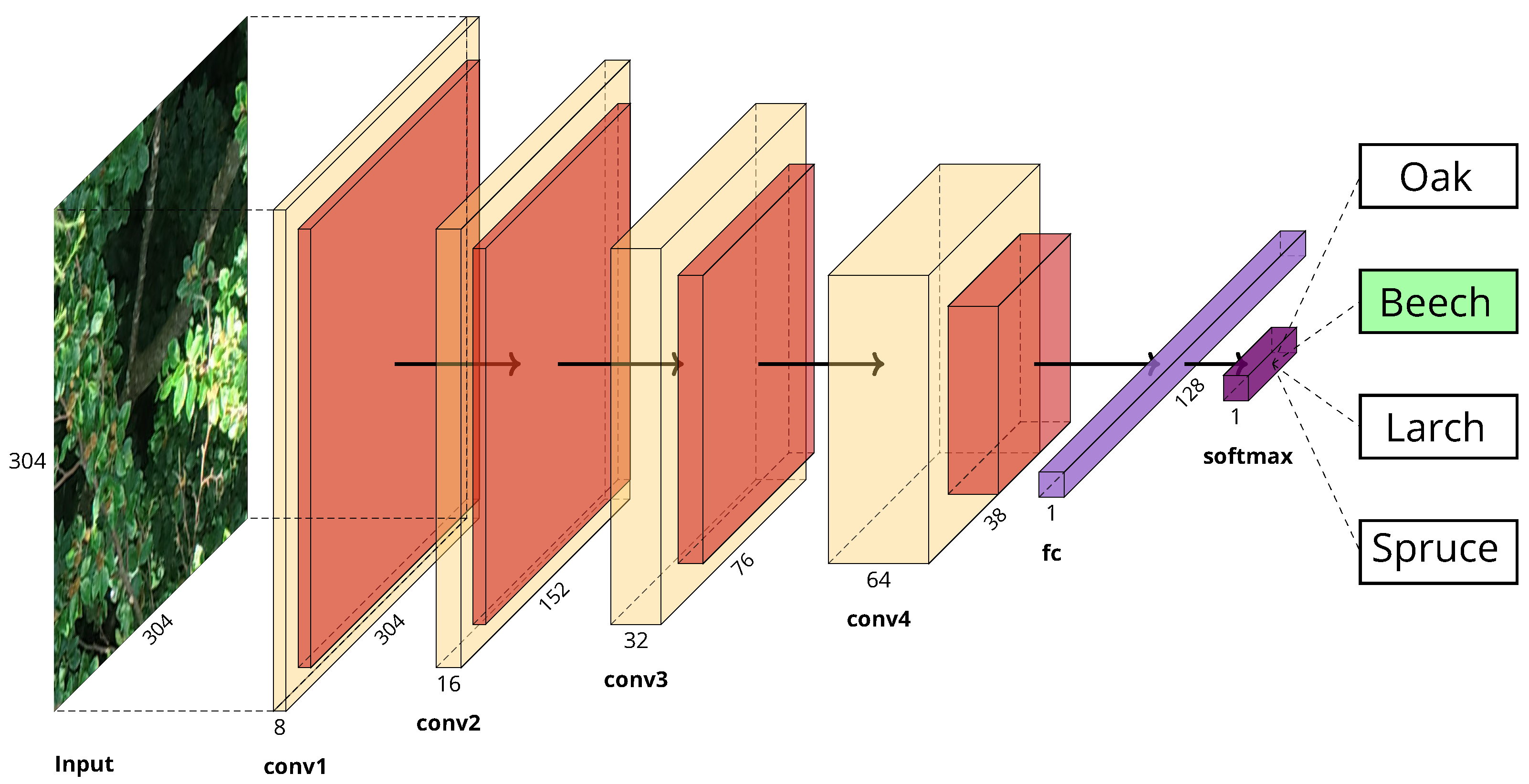
Current state-of-the-art CNN-models such as VGG16/19, Inception or ResNet that are widely used for general image classification tasks (>1000 classes) are overly complex and thus unnecessarily computationally expensive when it comes to classification problems with very few classes (4 in our case). Since it has also been shown that these large CNN models do not perform better in classification tasks used in forestry applications than smaller models [
37], a more compact model architecture was designed for the classification of the UAV image tiles. After testing several different architectures, a sequential model design with four consecutive convolution/pooling layers has proven to be the one with the best performance based on the available data set. All convolutional layers as well as the fully connected layer use ReLu activation functions whereas the last classification layer uses a softmax activation function to classify the tiles into the four tree species. With 2,982,484 trainable parameters, the model is much smaller and computationally less heavy than its large counterparts, the smallest of which comprises more than 25 million trainable parameters. The model implementation and its training were realized with the large-scale machine learning library TensorFlow [
38] and the deep learning API Keras [
39]. The complete model architecture is shown in
Figure 4.
For all model training/validation runs in this study, Adam optimization with a learning rate of was used as gradient descent method. Kernel regularizers were used in each convolutional layer to prevent the model from overfitting to the training data. Image tiles were augmented with horizontal and vertical flips. Models were trained in batches of 128 tiles for 50 epochs. Further training did not result in any enhancement of the validation accuracy.
The models were trained on a platform with twelve Intel(R) Xeon(R) CPU E5-1650 v4 units each with and four GeForce GTX TITAN X graphics cards. With this setup, about 120 of computing time were needed for one epoch which resulted in training times of about 100 per model.
To be able to assess the generalizability of the model performance and to test hypothesis H1, the model training and testing procedure was iteratively repeated for each combination of the nine testing areas and all flight dates as described in
Section 2.3. As photos were not taken for each of all possible 54 area/date combinations, in total only 34 different training/testing runs were conducted. In each run, a new model was trained using only data that was not collected within the respective testing area nor at the respective testing date. Sample counts for the train/validation data set were equally distributed and limited to 10,000 per category but could be lower for certain area/date-combinations with a minimum value of 5875. Sample counts of the testing data set varied between 191 and 3039 tiles. Model weights were saved after every training epoch. Each model was then tested using the data from within the respective testing area that was collected on the respective testing date. For testing, the weights with the best performance in terms of validation accuracy were loaded.
To assess the influence of the spatial resolution of the image data on the accuracy of the classification results and to test hypothesis H2, in a second step the model was iteratively trained, validated and tested with artificially downsampled image data as described in
Section 2.3. To this end, the image data set was randomly split into training, validation and testing subsets of size 8000, 2000 and 1000 tiles for each category.
To get a benchmark for the model performance, a subset of the testing data set was also manually classified by people who were not directly involved in the project. The subset consisted of 10 tiles per resolution level per person. With a total of 20 participants this resulted in 200 tiles per resolution level and 2000 tiles across all levels. During the manual tagging process, the participants did not have any information about the geo-location of the tiles, nor did they get any information about the original photos the tiles were clipped from. The only help was provided by showing 10 sample images per tree species from the training data set for each resolution level to the participants prior to the tagging process. To be able to compare the results of the human assessment directly with the model performance, the model was additionally applied to the tiles of the subset only.
2.5. Grid Application
To demonstrate the usability of the presented approach for the derivation of spatially explicit tree species information, a gridded product was generated for an exemplary region within the study area (see
Figure 1). The region was selected in such a way that all tree species considered in this study occurred in it in sufficient abundance. Image data were recorded during an automated flight in a 5 m × 5 m grid 10 m above canopy level on 27 August 2020. An orthophoto of the region that was recorded during a preliminary UAV survey on 2 July 2020 is shown in
Figure 5 together with the 5 m × 5 m grid flight pattern.
To minimize the offset between real-world coordinates and image pixels, the images were clipped to the central 1216 pixel × 1216 pixel area which roughly corresponds to a 5 m × 5 m area at a flight altitude of 10 m above canopy level. This central image area was then cut into 16 square tiles with the previously chosen 304 pixel edge length. A CNN was trained using only image data that were not taken on 27 August 2020 and outside the chosen region. This model was then applied to classify all 16 central tiles of each image taken within the region. In this way, a final product with 120 pixel × 80 pixel at 1.25 m spatial resolution was generated. In order to be able to evaluate the quality of the product, all tiles were also manually tagged and compared with the model predictions. Tiles that could not be assigned to any of the four tree species under consideration were not included in this analysis.
3. Results and Discussion
3.1. Generalizability
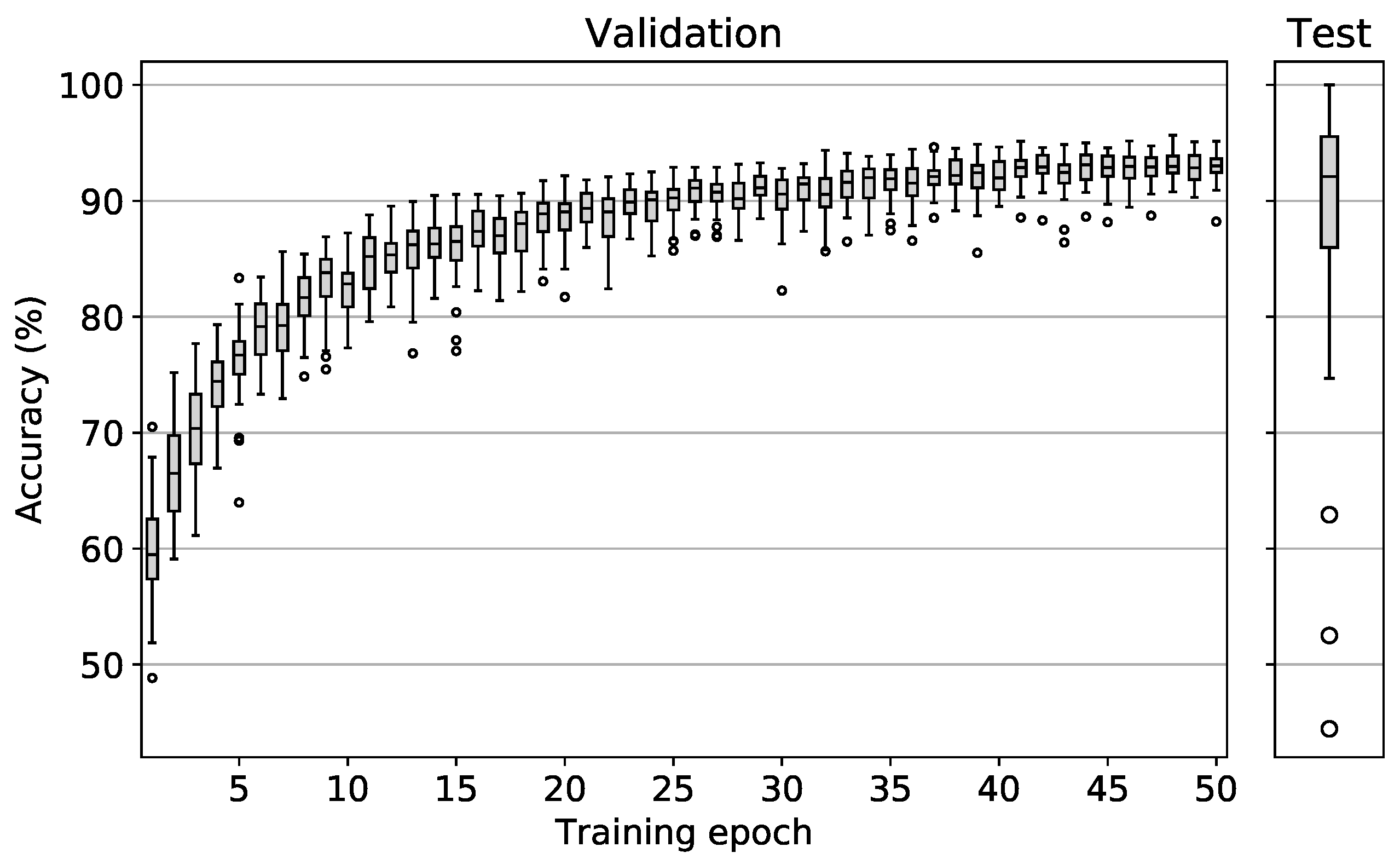
The results of the model training and testing procedure for all testing area and flight date combinations are shown in
Figure 6. Validation accuracies of all 34 runs are depicted in the left plot. After approximately 20 epochs, only slight accuracy enhancements could be observed. After 50 epochs, average validation accuracy values peaked at 93% with the best model reaching 96% and the worst model reaching 92%. It should be noted that the best model performance did not necessarily occur after epoch 50, but often earlier with the earliest peak performance occurring after epoch 38. These results show that the chosen model architecture is essentially very well suited to differentiate the tree species treated in this study on the basis of simple RGB images. It can also be deduced from the plot that the variation of the validation accuracy becomes increasingly smaller as training progresses. This means that regardless of the date/area combination, the model was able to derive the essential information for the differentiation of tree species from the data.
Similar studies in the literature report highly varying accuracy results with values between 59% and 95% [
28]. With an average validation accuracy of 93% after 50 epochs our model thus lies in the upper range. However, most of these study results cannot be directly compared with the results of this study, as too little focus was put on spatial and temporal independence in the validation procedure. The only work we are aware of, which is also based on RGB data collected with a UAV and which was validated spatially and temporally independently, is the work of Natesan et al. [
33]. Here an average accuracy of 80% is reported.
To be able to assess the true spatiotemporal generalizability of the model, the testing accuracies must be analyzed. The right plot shows the testing accuracy results which were generated by loading the best weights for each model and then applying it to its respective testing data set. In comparison to the validation accuracies, here a similar median value of 92% was observed. However, the value range of the testing accuracies is much larger than the value range of the validation accuracies of the best models with a minimum of 44% and a maximum of 100%.
The median value of 92% in the test accuracies is very promising and shows that trained models can indeed be successfully applied in temporally and spatially independent areas where they are able to achieve prediction accuracies of 92% on average. However, the large value range of 56 pp indicates that in a few exceptional cases significantly higher/lower accuracy values are reached. The maximum accuracy of 100% was achieved for testing area 8 on 27 August 2020. However, only 191 test tiles were available here, all of which fell into the oak category, thus making this test sample unrepresentative. The same applies to the three lower outliers with accuracy values between 44% and 63%. With average tile counts of only 526 and a maximum of three tree species categories being represented, these test samples are not representative either.
In summary, it can be stated that the presented method made it possible to differentiate between the four tree species oak, beech, larch and spruce. Apart from a few non-representative test cases, the CNN models were able to achieve promising classification accuracies temporally and spatially independently. Hypothesis H1 is thus accepted.
3.2. Resolution Dependence
The model performance data collected during training of the model at different resolution levels of the image data are shown in
Figure 7 and
Figure 8.
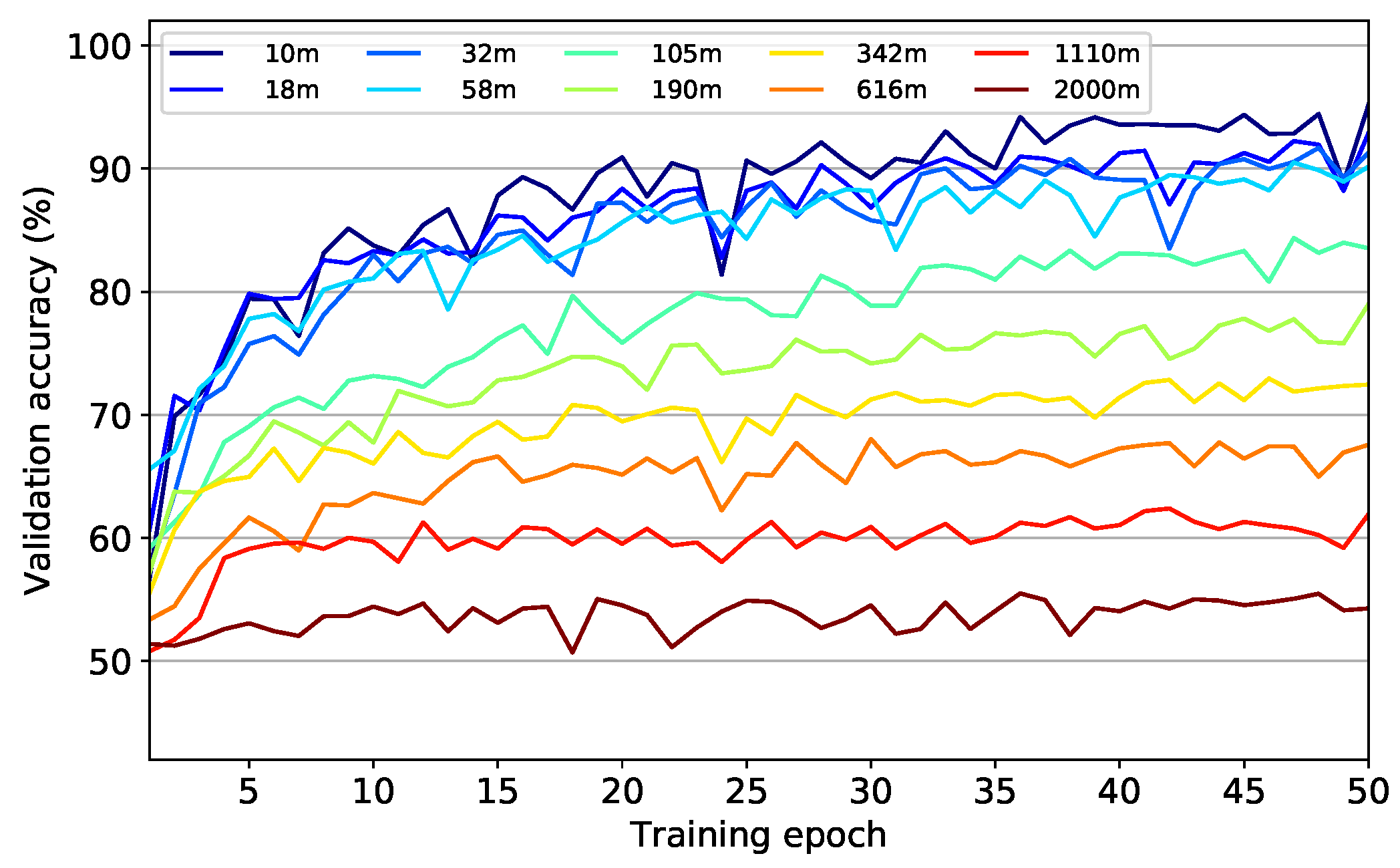
Figure 7 shows the model accuracies after each training epoch. Starting from accuracy values between 50% and 60% after the first training epoch, all models were able to improve till the end of the training procedure. However, with increasing flight altitude and thus with lower information content in the image data, it became increasingly difficult for the models to classify the image data into the correct categories. This manifests itself in a successive flattening of the learning curves with maxima being achieved after less training epochs but at considerably lower levels. While for the original resolution at 10 m flight altitude a maximum validation accuracy of 95% was achieved after 50 epochs, at a theoretical flight altitude of 2000 m a maximum validation accuracy value of only 56% was reached already after 36 epochs.
The reason for the early reaching of maximum performance on a low level for models with low-resolution data is presumably the reduced information content in the data. This allows the models to learn the complete information content that is still contained in the data more quickly. However, this is also inevitably accompanied by a decrease in maximum accuracy values. On the other hand, models based on high-resolution data need longer to reach maximum performance due to the larger information content within the data. Nevertheless, the higher information content also allows the models to achieve higher maximum accuracies.
It should be pointed out that even at the maximum theoretical flight altitude of 2000 m accuracy values of 56% are still achieved. Since only one color value per tile remains, i.e., 3 features per sample in contrast to 277,248 features in the original samples, it can be concluded that the mean spectral values of the tiles already provide an explanatory content of 31%, assuming a base rate accuracy of 25% of a random model with 4 equally distributed classes. Beyond this level, another 39% of additional explanatory content can be obtained from the spatial structure in the image data, resulting in maximum accuracies of 95% at the highest resolution level.
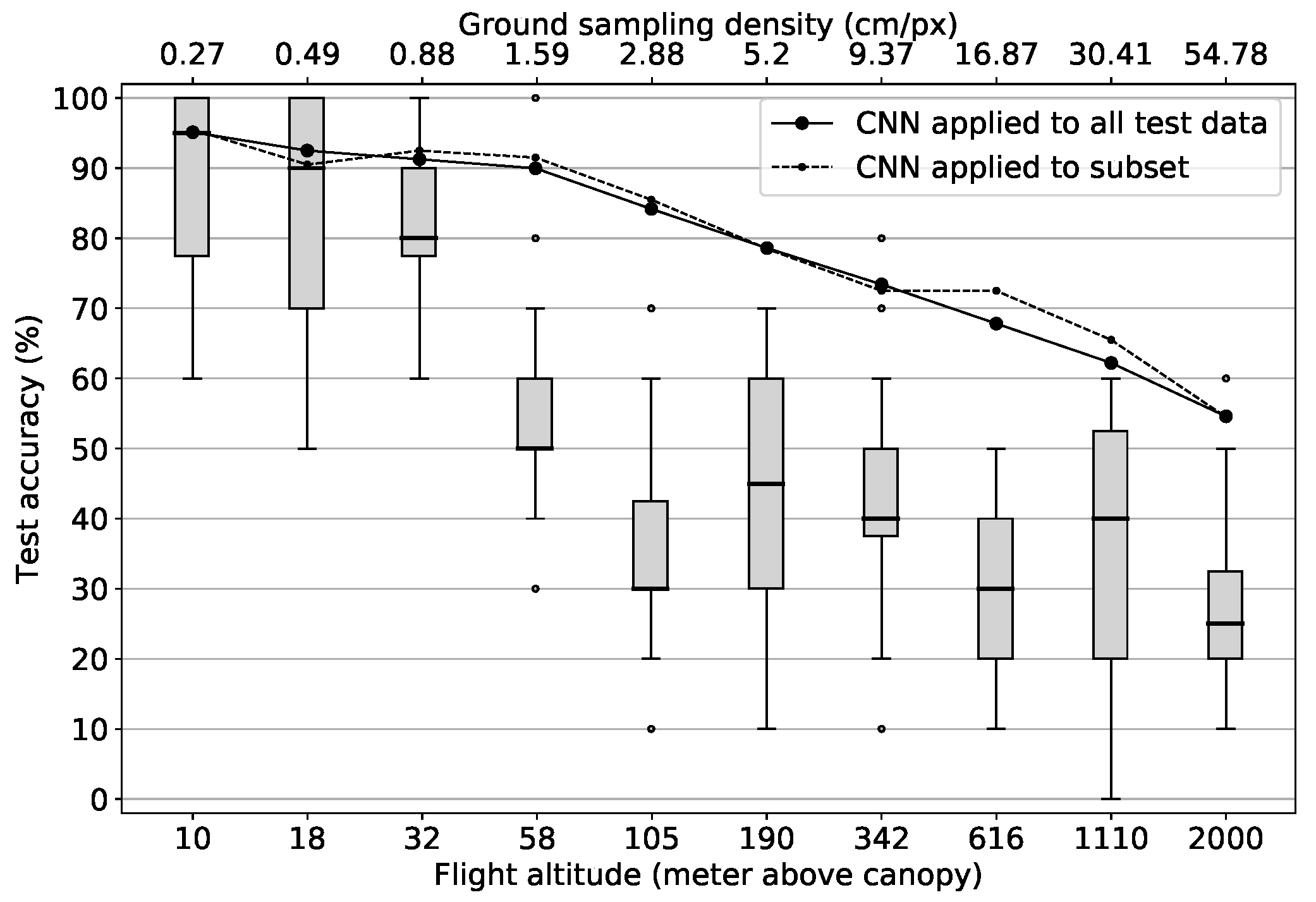
In
Figure 8 accuracy values are shown that were obtained by applying each run’s best model to the test data together with the human performance results. The solid line depicts the performance results of the models when they were tested against the complete testing data set of 4000 tiles per resolution level. The dashed line marks model results when they were tested against the subset that was used in the “human” tagging process (200 tiles per resolution level). With the only exceptions of flight altitude levels 616 m and 1110 m which correspond to ground sampling densities of 16.87 cm/px and 30.41 cm/px, the deviation between the two curves is minimal. These slight deviations can be explained by the size of the subset with only 200 samples per altitude level compared to 4000 in the complete testing data set. In general, both curves follow the same scheme:
The first four models trained with theoretical data acquisition altitudes of up to 58 m all reach accuracy values above 90%, only showing a very slow accuracy decrease of about 1.6 pp per flight altitude level increase. This can probably be explained by the fact that many images in the data set at these high resolution levels contain redundant information regarding the determinability of the four tree species investigated. For instance, the leaf shape is still “over-resolved” at these levels. Thus, despite the absolute loss of information of 97.1% from the 10 m level to the 58 m level (277,248 vs. 8112 features) the models are still able to distinguish between the four categories in most cases.
Starting from 105 m theoretical flight altitude and above, which is equivalent to a ground sampling density of 2.88 cm/px, accuracy values decrease more rapidly at a rate of about 5 pp per logarithmically increased altitude level. A possible explanation for this is that from this resolution on, important information in the image data required for the determination of tree species, such as the leaf and branch structure of the trees, can no longer be recognized by the models. Thus, the models can only rely on coarser structures and average spectral values, which do not allow for a clear identification anymore. Nevertheless, the decrease of the model performance is much slower, than a decrease proportional to the number of features per sample on all resolution levels. This again emphasizes the suitability of the CNN application to the tree species classification problem investigated in this work which becomes particularly obvious when comparing the model results with the results of the human classification:
Results of the manual tagging process are depicted as boxplots in
Figure 8. Despite large variations at each level, the same basic pattern as for the models can be observed: The human classification results are at a similarly high level as the model results at the beginning and decrease with increasing theoretical flight altitudes. However, there are some differences to the model performance: The human classification accuracy significantly decreases already at the flight altitude level of 32 m (0.88 cm/px ground sampling density) where it drops relatively abruptly from median accuracy values of 80% over the 58 m level with 50% to 30% at level 105 m (2.88 cm/px ground sampling density). From this flight altitude level up to the maximum flight altitude level of 2000 m the classification accuracy then varies between median values of 45% and 25% with a slight downward trend towards coarser resolution levels.
This shows that humans can keep up with the models in the beginning where they reach a median accuracy value of 95% at the 10 m level. However, as soon as the visual characteristics essential for humans to differentiate between tree species can no longer be recognized, the performance drops abruptly and settles at values just above a random hit probability of 25%. Thus, humans are not able to draw meaningful information from the mean spectral characteristics and the coarse spatial structures, which are still recognizable at the coarser resolution levels, in order to make a meaningful differentiation. Besides this, the trend of the human performance supports the assumption that at a flight altitude level between 32 m and 105 m the recognition of important spatial-structural properties for the differentiation of tree species is no longer possible, given the UAV setup used in this study.
In summary, it can be stated that the predictive power of humans and the CNN models continuously decreases with decreasing resolution in the image data. The fact that the model performance decrease with logarithmically increasing flight altitudes is steeper for flight altitudes above 58 m (1.59 cm/px ground sampling density) and that the human performance drops significantly around this level too, indicates that essential spatial-structural information needed for the classification of tree species can no longer be recognized above this level. Hypothesis H2 can thus also be accepted. It can further be concluded that with the given UAV and camera sensor technology, acquisition altitudes of about 58 m above canopy level are sufficient to still achieve very good accuracy results with values of more than 90%. This is particularly noteworthy as at flight altitudes of around 58 m above canopy a much larger area can be covered per flight than at 10 m flight altitudes.
While Fassnacht et al. [
5] hypothesize that classification accuracies of non-pixel-based approaches are likely to benefit from higher spatial resolutions, they emphasize that more systematic research is needed in this area. Most importantly, until now it remained unclear, where a possible upper limit of the resolution enhancement with respect to the classification accuracy improvement may be found. With the help of the results of this study, this upper limit can now be assumed to be somewhere around a ground sampling density of 1.6 cm/px which corresponds to flight altitudes of about 60 m with the UAV setup used in this study.
3.3. Grid Application
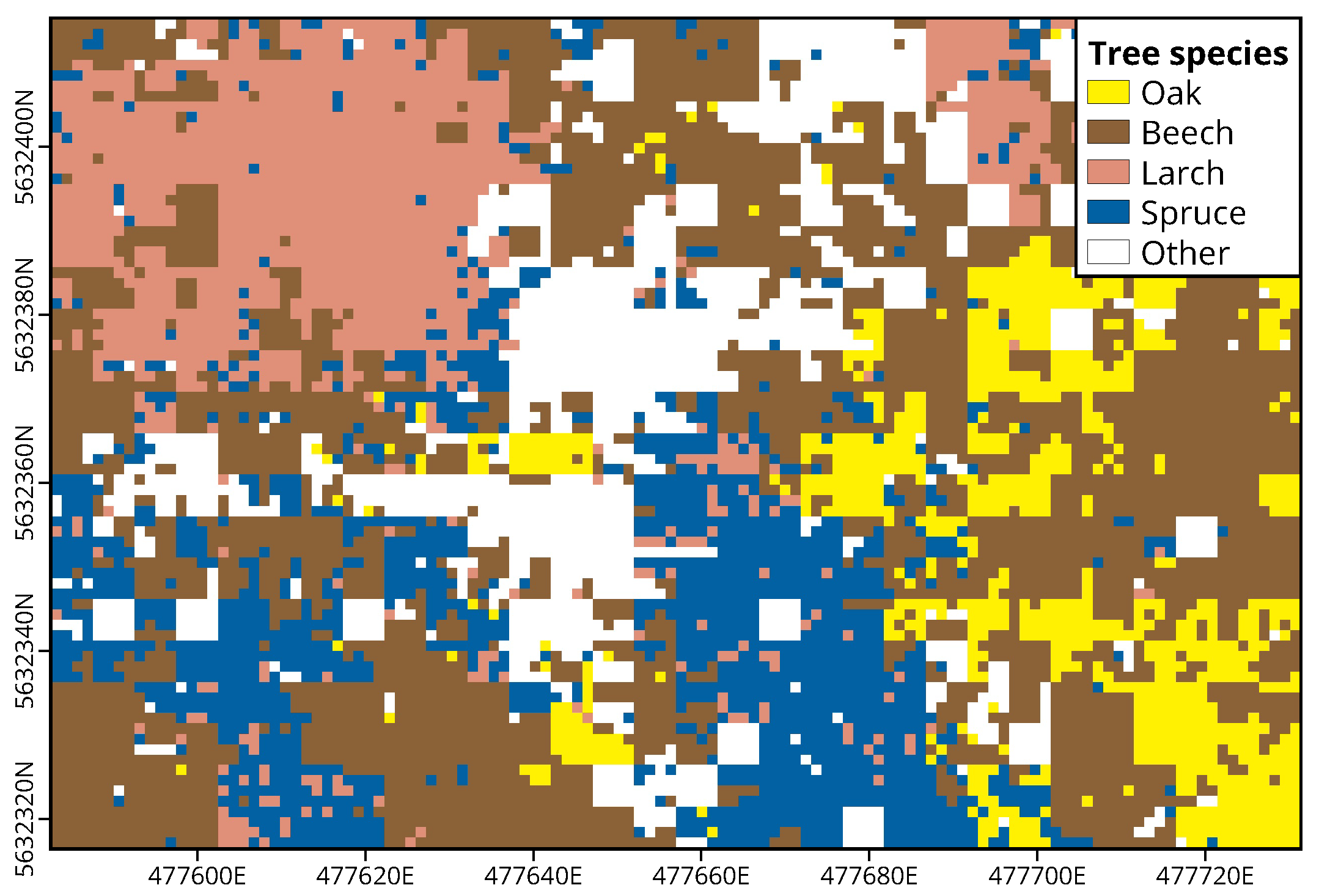
The result of the grid-based application of the CNN approach to an exemplary region is shown in
Figure 9. Areas covered by oaks, beeches, larches and spruces are colored yellow, brown, coral and blue, which are the colors used for these tree species in map illustrations of the Hessian forestry management. White areas are regions that could not be assigned to one of the four categories either due to image data deficiencies such as motion blur or because the respective image tile did not contain any of the four tree species. This was the case when either no tree was visible in the tile at all (e.g., forest floor or shrubs) or because the tile showed another tree species than the four considered here. Apart from the white areas in the central part of the domain resulting from the presence of ash trees (
Fraxinus excelsior), the optical comparison with the orthophoto shows that the approach has produced reasonable results. Beeches are relatively evenly distributed over the complete domain. Larch trees, on the other hand, are concentrated in the northwestern part, while oak trees are most common in the southeast. Spruces form a strip from the southern to the central part of the domain and show a few scattered occurrences in the southwest.
The validation against manually tagged tiles which were visually assignable to one of the four tree species resulted in an accuracy of 88%. This is within the range of the third quartile of the testing accuracies from the generalizability analysis in
Section 3.1 and shows that the grid product achieves a good overall spatially explicit classification accuracy. It must be mentioned, however, that the geographical location of the individual pixels of the product is not very accurate. Although only the central section of each UAV photo was used for the classification procedure, an exact assignment of an image tile to a product pixel was not always unambiguously possible. This was due to optical distortions towards the edge of the image, irregularities in the height of the canopy and because of deviations between the measured and the actual position and orientation of the UAV.
Furthermore, due to the low flight altitude of the UAV, this approach cannot yet be applied on a large spatial scale. Nevertheless, the procedure may be used to generate spatially explicit data on tree species as seen from the aerial perspective for selected example areas or it can help to get an overview of tree species distribution during local forest inventories. Independently validated spatially explicit reference data on tree species occurrence are hardly available due to the difficulties in locating exact tree positions and crown extensions [
5]. Locally gathered and gridded products using the method proposed in this study, could help to close this gap. These data sets could then also be used as “ground truth” to support spatial upscaling procedures based on coarser satellite data in order to produce large scale spatially explicit data on tree species occurrence.
4. Conclusions
In this study, we presented a novel CNN-employing tree species classification approach based on low-cost UAV-RGB image data. It was shown that validation results of 92% could be achieved with this technique on spatially and temporally independent data. This demonstrates that, in contrast to existing pixel-based classification methods, this approach enables the determination of tree species largely independent of the prevailing illumination conditions, i.e., independent of daytime and season, phenological stage and prevailing weather conditions. However, since only four tree species were classified in this work, a reasonable next step is to extend the data set by further tree species.
An analysis of the effect of the spatial resolution on the classification accuracy has shown that human and model performance continuously decrease with decreasing spatial resolution. However, it could be shown that up to a theoretical flight altitude of about 60 m, equivalent to a ground sampling density of 1.6 cm/px, the model’s classification accuracy remained above 90% and that only at higher flight altitudes or lower spatial resolutions the classification accuracy decreased significantly. Although human performance shows large variances in the individual resolution levels, a clear trend is discernible which shows that human classification accuracy drops faster and earlier than the model’s accuracy at decreasing resolution in the input image data. It should be added that the resolution analysis was only performed on the basis of artificially downsampled image data. Thus, to confirm our results, they should be compared to classification results generated on the basis of original data gathered in exemplary flights performed at the respective flight altitude levels.
Finally, it was shown that the presented approach can be used for the generation of gridded tree species products, which in turn could be made available as ground truth reference data for various further applications. A remaining deficiency of the grid generation is the inaccurate assignment of image tiles to grid pixels. This assignment could be improved by using RTK-UAVs, which can measure their location with high accuracy, or by including a simultaneously generated orthophoto in the post-processing of the collected image data for automatic georeferencing.
As the RGB image data required for the classification can be obtained at reasonable costs and since the UAV employed was an easy-to-use consumer model, the input data could potentially be directly collected by people who do not need to have remote sensing expertise, e.g., forest managers or rangers. This again increases the probability of a broad applicability of the approach in many different contexts.