Abstract
The COVID-19 pandemic made the medical community realize how large a problem it would face. The epidemiological situation forced the opening of additional wards, the so-called “COVID wards”, where an increase in the rate of coexisting bacterial infections was observed. We report a hospital outbreak due to New Delhi carbapenemases producing K. pneumoniae clones. Twenty-eight K. pneumoniae strains were analyzed from patients with primary COVID-19 infection. The drug susceptibility of the strains was determined by the diffusion–circulation method and E-test. Phenotypic and PCR methods confirmed the production of carbapenemases. The phylogenetic similarity of the obtained strains was examined using pulsed-field electrophoresis. Most strains were isolated from bronchoalveolar lavage. All isolates obtained were resistant to β-lactams and fluoroquinolones. All strains produced New Delhi carbapenemases and were classified into two genetic clusters, A and B. Eight risk factors for secondary bacterial infection were analyzed. Following an intervention involving hand hygiene, strict contact prevention, and cleaning of the hospital environment and medical devices, this outbreak was successfully brought under control.
1. Introduction
For many years, viruses of the Coronaviridae family were not a significant topic of research. It was believed that they are not very dangerous to the human body and cause mild respiratory infections. The breakthrough of increased interest in these pathogens came with the emergence of new, hazardous types that cause life-threatening infections in children, the elderly, and patients with immune deficiencies [1]. Coronaviridae shows tropism to cells of the respiratory and gastrointestinal tracts and the central nervous system in humans, farm animals, birds, and other wildlife. We have already had outbreaks of two pandemics, SARS in 2002/2003 and MERS in 2012. In 2019, there was an outbreak of a pandemic caused by the SARS-CoV-2 virus in the city of Wuhan, Hubei Province, which the world is still dealing with [2]. The National Health Commission of China reported on 20 January 2020 that SARS-CoV-2 can be transmitted from human to human [3]. Infection most often occurs via the droplet route or through direct contact with a sick animal or human [4]. Feces, in which viral RNA has been detected, are also a potentially infectious material. Many patients with COVID-19 develop diarrhea and vomiting approximately two days before the onset of symptoms. Transmission of the virus via the fecal–oral route is particularly crucial in areas with poor sanitation [5]. Individuals most susceptible to SARS-CoV-2 infection are elderly individuals or those with impaired immune systems, such as those with AIDS or those under immunosuppression. People with respiratory diseases, such as asthma or chronic obstructive pulmonary disease, cardiovascular disease, cancer, and diabetes, are also much more susceptible [6]. The most dangerous stage of the disease is the last stage, which is accompanied by acute respiratory distress syndrome (ARDS), with the characteristic presence of septic shock and multiple organ failure (MODS), especially acute renal and hepatic failure. In addition to ARDS and MODS, concurrent bacterial and fungal infections contribute to the increased deaths in COVID-19 patients. The most commonly isolated strains are Streptococcus pneumoniae, Klebsiella pneumoniae, Haemophilus influenzae, and Aspergillus fumigatus [7]. At this stage, patients require high-flow oxygen therapy and sometimes mechanical ventilation and transfer to the ICU. Empiric broad-spectrum antibiotics are usually used. The mortality rate of patients in stage 4 due to COVID-19 in the ICU is approximately 70% [8,9]. According to many sources, K. pneumoniae is, after Escherichia coli, the most common bacterium of the Enterobacteriaceae family, causing infections in the ICU [10,11]. During the COVID-19 pandemic, it was realized how much of a problem multidrug-resistant microorganisms pose for patients dealing with other medical conditions. The incidence of colonization with carbapenem-resistant Enterobacterales rose from 6.7% in 2019 to as high as 50% in the second quarter of 2020. Among patients hospitalized for COVID-19, Acinetobacter baumannii was the leading cause of bacterial infections, followed closely by K. pneumoniae. Of concern was the fact that resistance to carbapenems was demonstrated in nearly 75.5% of K. pneumoniae strains [12]. The frequency of infection in patients with coronavirus varies. Estimates range up to 50% in nonsurvivors. The risk of healthcare-associated infection increases with the severity of SARS-CoV-2 infection and the length of hospitalization [12,13].
The invention and introduction of antimicrobial drugs revolutionized the medical world. The widespread use and access to pharmaceuticals, especially antibiotics, have also brought negative consequences, such as the spread of drug resistance [14]. Currently, Gram-negative bacteria that produce carbapenemases cause the most problems. Carbapenemase-producing K. pneumoniae is a multidrug-resistant bacterium and poses an enormous threat to human life. An additional threat is that it can easily pass on resistance genes to other bacteria in the Enterobacteriaceae family [15]. Currently, the most commonly detected in K. pneumoniae is the production of NDM-type carbapenemases (New Delhi metallo-β-lactamases) [16]. The genes encoding carbapenemases are located on a plasmid, which often carries other genes encoding resistance to other groups of antibiotics. The rapid spread and lack of effective therapy have led to the designation of NDM-positive K. pneumoniae as “superbugs” [17].
The phenomenon of isolating NDM-positive strains from clinical specimens particularly intensified during the COVID-19 pandemic, as SARS-CoV-2 cases required additional hospital wards. Virus-infected patients often require prolonged hospitalization and, due to complications, broad-spectrum antibiotic therapy. These factors directly increased the risk of secondary infection and colonization with hospital strains. Therefore, monitoring the transmission of such strains in the hospital environment, both as etiological agents of infection and asymptomatically colonizing patients admitted to the wards, is a direct epidemiological tool. Our study aimed to analyze the phylogenetic similarity of K. pneumoniae NDM isolated from patients diagnosed with COVID-19.
2. Materials and Methods
2.1. Hospital Settings
The outbreak occurred during the COVID-19 pandemic between 2 December 2020 and 20 February 2021 in a temporary ward dedicated to patients with confirmed COVID-19 infection at Szczecin University Hospital. According to the guidelines of the Hospital Infection Control Committee (HICC), surveillance for colonization with alert strains was routinely conducted once a week in high-risk patients hospitalized in the temporary ward, all with symptoms of infection and all transferred from another hospital. Patients identified as carrying alert pathogens or infected (or both) were isolated/coordinated and managed by individual nursing staff. Infection control measures, including hand hygiene, cleaning, and disinfection protocols, as well as clinical and microbiological data, were routinely monitored every quarter of the year according to procedures implemented by the Hospital Infections Control Team. Antibiotic protocols and microbiological methods used were performed in accordance with these principles.
2.2. Characteristics of the Strains
Positive K. pneumoniae cultures were obtained from 28 patients. The bacteria were isolated from various clinical materials. The healthcare personnel had no hand or other specimens taken. The site of the study was the Department of Microbiology, Immunology, and Laboratory Medicine, part of the Pomeranian Medical University in Szczecin.
2.3. Microbiological Identification
Isolated K. pneumoniae were seeded on McConkey agar (bioMérieux, Marcy-l’Étoile, France). Species analysis on the basis of the biochemical characteristics of the obtained isolates was carried out using GN tests of VITEK 2 (bioMérieux, France) and Maldi-Tof MS (Brucker, Karlsruhe, Germany). Both methods yielded identical results. All strains were identified as K. pneumoniae.
2.4. Drug Susceptibility
Antimicrobial susceptibility testing (AST) for amoxicillin–clavulanate (20/10 µg), piperacillin–tazobactam (100/10 µg), cefotaxime (30 µg), ceftazidime (30 µg), cefepime (30 µg), gentamicin (10 µg), amikacin (30 µg), imipenem (10 µg), meropenem (10 µg), trimethoprim–sulfamethoxazole (1.25/23.75 µg), ciprofloxacin (5 µg), colistin, and tigecycline was performed using the Kirby–Bauer agar disk diffusion method and E-test method according to the EUCAST (European Committee on Antimicrobial Susceptibility Testing) guidelines [18].
2.5. Carbapenemase Detection
Carbapenemases were detected by culture on CarbaId agar (bioMerieux, France). Growth in the form of green colonies indicated the presence of a strain producing carbapenemases. Confirmation of the presence of carbapenemases in K. pneumoniae was also carried out by real-time PCR (GeneXpert, Cepheid, Solna, Sweden)
2.6. Confirmation of Phylogenetic Similarity of K. Pneumoniae Strains Using the PFGE Method
DNA was isolated using the CHEF Bacterial Genomic DNA Plug Kit (Bio-Rad, Hercules, CA, USA). Restriction digestion was performed using Tango buffer and XbaI enzyme (Thermo Scientific, Waltham, MA, USA). A GelDoc-It2 Imager system (Upland, CA, USA) was used to read the results. The classification of individual restriction patterns for particular genetic profiles was carried out using the Unweighted Pair Group Method with Arithmetic Mean (UPGMA) method (SAB value = 82.5%) and the Dice coefficient (2.0%).
2.7. Statistics
Data were organized and analyzed using GraphPad Prism 5.02 (GraphPad Software Inc., San Diego, CA, USA) software. Then, they were analyzed with the nonparametric tests (Chi-square continuity test with Yates correction). Statistical significance was set at p < 0.05.
3. Results
3.1. Outbreak Description
The NDM K. pneumoniae outbreak persisted in the temporary ward for COVID-19 patients for more than two months. During this period, 47 patients were hospitalized. In 28 patients (59.6%), K. pneumoniae NDM was cultured from clinical specimens. All patients had been on the ward for more than 14 days. Among them, 23 patients showed signs of ongoing secondary bacterial infection. Rectal colonization without other evidence of infection was confirmed in five patients. There were five deaths during the outbreak period. The remaining patients were successfully treated and discharged home. The first NDM-positive strain of K. pneumoniae was isolated from bronchoalveolar lavage from a patient with secondary pneumonia within 6 days after hospitalization. The patient was not rectally colonized with K. pneumoniae NDM. An analysis of the epidemic has been initiated to determine risk factors and the spread of cases. A retrospective analysis was conducted to determine risk factors. Patient data, including age, gender, underlying diseases, invasive procedures, and other risk factors were collected from electronic patient records and medical staff. Infection control measures were also immediately applied. The characteristics of the patients are shown in Table 1.

Table 1.
Characteristics of patients with laboratory-confirmed carbapenemases-producing Klebsiella pneumoniae during the outbreak (from 2 December 2020 to 20 February 2021).
3.2. Risk Factors Analysis
Risk factors for K. pneumoniae NDM infection/colonization among patients were analyzed. Patients were divided into two groups: those in whom K. pneumoniae NDM was isolated within 14 days after the start of hospitalization, and patients whose first K. pneumoniae NDM isolation occurred more than 14 days after the start of hospitalization. Data included gender, underlying diseases, intra-abdominal surgical procedures, invasive catheterization, mechanical ventilation, carbapenem use, and rectal colonization. No risk factor was statistically significant (Table 2).

Table 2.
Analysis of risk factors for NDM–positive K. pneumoniae.
3.3. Environmental Contamination Control
All environmental samples collected during the outbreak were negative for K. pneumoniae NDM. From the environmental cultures, the presence of coagulase-negative staphylococci (CNS) and aerobic spore-forming bacilli was isolated from three samples collected from the sink. The point source remained unidentified when environmental samples were examined. Microbiological cultures of swabs collected from medical toilets were sterile or contaminated with microorganisms, with no significance for K. pneumoniae.
3.4. Isolation Sites
A total of 28 K. pneumoniae strains were analyzed. Isolates were cultured from a variety of clinical materials, both from infections and screening, from 28 patients. Thirteen strains were cultured from bronchoalveolar lavage fluid, five from rectal swabs, two from swabs of surgical sites, four from blood, and four from urine.
3.5. Drug Susceptibility
All K. pneumoniae strains were resistant to piperacillin, amoxicillin with clavulanic acid, all antibiotics of the cephalosporin group, carbapenems, and ciprofloxacin. Two strains showed sensitivity to trimethoprim–sulfamethoxazole. Isolates showed minor susceptibility to aminoglycosides and were fully susceptible to colistin and tigecycline (Figure 1).
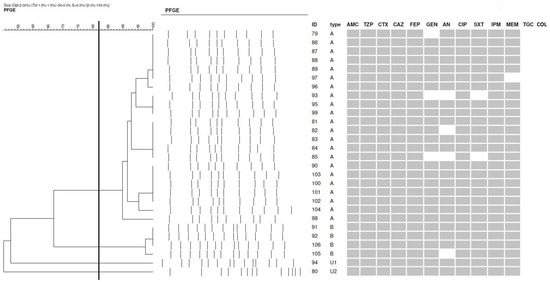
Figure 1.
Dendrogram representing the relatedness of 28 K. pneumoniae (cut-off point = 82.5%; Dice) and drug susceptibility (dark field—resistant, bright field—susceptible). The clusters were marked with the letters A and B, whereas the unique genotypes were marked with the letter U. PFGE—pulsed-field gel electrophoresis; AMC—amoxicillin with clavulanic acid; PIP—piperacillin/tazobactam; CTX—cefotaxime; CAZ—ceftazidime; FEP—cefepime; AN—amikacin; GE—gentamicin; CIP—ciprofloxacin; SXT—trimethoprim–sulfamethoxazole; MEM—meropenem; IMP—imipenem; COL—colistin; TGC—tigecycline.
3.6. Detection of Carbapenemases
On CarbaId agar (Biomerieux, Craponne, France), all strains grew green colonies, which meant that all isolates were carbapenemase producers. RT–PCR-based tests (GeneExpert, Cepheid, Solna, Sweden) confirmed the presence of genes encoding a carbapenemase belonging to the NDM family in all K. pneumoniae strains tested, while no genes specific to other families of enzymes (KPC, VIM, IMP, OXA-48) were detected
3.7. Phylogenetic Similarity Analysis
The phylogenetic similarity of K. pneumoniae strains was assessed by chromosomal DNA restriction analysis combined with pulsed-field gel electrophoresis. Isolates were classified into two clusters, designated A and B.
Cluster A included the most significant number of isolates (22/28), while Cluster B contained only four strains. Two strains were marked as unique (Figure 1).
4. Discussion
During the COVID-19 pandemic, patients were admitted to intensive care units whose condition involved acute respiratory failure and the need for oxygen therapy or mechanical ventilation. Patients often developed bacterial infections, which contributed significantly to debilitation and death. Mortality rates in ICUs reached up to 70%. One of the most common coinfections immediately after pneumococcal infection was that caused by K. pneumoniae. The reason is usually the use of empirical broad-spectrum antibiotic therapy, which at the same time contributes to the spread of drug resistance [19]. K. pneumoniae is a dangerous bacterium that, in addition to its virulence characteristics, can have a wide range of drug resistance. It is often the cause of hospital-acquired endemic outbreaks, mainly because it can asymptomatically colonize staff and patients. Therapy of infections with carbapenemase-producing K. pneumoniae is the most difficult, as carbapenems often represent the last chance for effective therapy. K. pneumoniae most often causes infections of the urinary tract, blood, cardiovascular system, and respiratory system. It is also a common agent of intraoperative and postoperative infections [20]. In the team of Li et al., who conducted a study in central China, most of the strains were from the respiratory tract (62.9%) [21]. In the study by Pruss et al., most isolates came from urine (46.4%) [22]. Campos et al. grew the most strains from rectal swabs and urine, while 21% of isolates in their study came from bronchoalveolar lavage [23]. Similar results were obtained by Aires-de-Sousa, where the leading materials from which K. pneumoniae strains were obtained were rectal swabs (43.8%) and urine (32.6%) [24]. In our study, the highest number of strains was cultured from bronchoalveolar lavage (47%). Swabs were collected from patients diagnosed with COVID-19 between December 2020 and February 2021 in specially created “COVID wards”. Thus, we can conclude that SARS-CoV-2 virus infection largely coexisted with respiratory bacterial infection with K. pneumoniae. The lowest percentage of strains was obtained from swabs from perioperative sites (7%), which may indicate proper asepsis during surgical procedures. Treatment of critically ill patients often requires various invasive procedures, including mechanical respiratory support, catheter insertion, etc., and the potential risks of these procedures are secondary to other microbial infections, which are usually respiratory and bloodstream infections [25]. In our study, invasive mechanical ventilation, urinary catheterization, deep venous catheterization, intra-abdominal surgery, or prior carbapenem therapy, among others, were analyzed as risk factors for secondary bacterial infections. Microbial infections can lead to further deterioration of the patient’s condition, which is also a major reason for treatment failure.
Multidrug-resistant bacteria, which include K. pneumoniae, pose an enormous threat, especially to immunocompromised patients who are highly susceptible to infection. The outbreak of the COVID-19 pandemic has contributed to an increase in infections caused by the bacteria mentioned above. The situation necessitated the opening of temporary hospitals and additional ICUs, often staffed by people without proper training. The widespread use of broad-spectrum antibiotics also had a negative impact. The widespread use of broad-spectrum antibiotics has also had a negative impact. K. pneumoniae, which produces carbapenemases, is resistant to combinations of β-lactamase inhibitors with penicillins and cephalosporins, often with concomitant resistance to fluoroquinolones and aminoglycosides [26]. Depending on the region of the world, different families of carbapenemases predominate. Strains spread easily, resulting in rapid changes in the predominance of a particular enzyme. A 2019 article published by Ojdana et al. also outlined the epidemiological situation in Poland. The authors showed that Poland is dominated by K. pneumoniae NDM, which accounts for 71% of the strains [27]. Kazi et al. studied the epidemiological status in India. The most commonly produced enzymes were found to be NDM (76.57%), followed by OXA-48 (4.5%) [28]. Within China, K. pneumoniae, producing KPC-type carbapenemases, is leading (52%), followed by the NDM variant (11%) and OXA-48 (37%) [29]. In our study, all analyzed strains produced NDM-type enzymes, aligning with literature data on the predominant type of carbapenemases in Poland. Increasing resistance to antibiotics is becoming an increasingly serious global problem in the medical community. Infections caused by carbapenem-resistant Enterobacteriaceae (CRE) are much more serious than those caused by susceptible microorganisms. The result is increased treatment time for patients, resulting in higher healthcare costs. The biggest and most dramatic consequence is the increased mortality of infected patients [30]. The isolates we analyzed were resistant to β-lactam antibiotics, including carbapenems. In addition, they showed resistance to ciprofloxacin. We recorded a low percentage of susceptibility only to aminoglycosides and trimethoprim–sulfamethoxazole. In an analysis by Durdu et al., isolates were 55.1% resistant to amikacin and 55.3% resistant to gentamicin [31]. K. pneumoniae is a microorganism that, especially now in an era of increasing viral infections, poses a major threat. Through the overuse of antibiotics, drug resistance is spreading. It is the most dangerous phenomenon in hospital environments, where there are patients with reduced resistance, who are most susceptible to infection. Staff and patients who are asymptomatic carriers unwittingly contribute to the spread of the bacteria. The ease of transmission of bacteria from patient to patient and from ward to ward results in hospital outbreaks. The main goal of assessing the clonal similarity of strains is to determine the etiological agent and the pathogen’s route of transmission, helping prevent further spread [32]. A thorough understanding of how microorganisms are spread and their potential vectors is the first step toward reducing nosocomial infections. Microbiological research plays a major role here. Determining which microorganism is most prevalent in a ward allows rapid intervention and the use of targeted therapy. The basic method of identifying bacteria is the phenotypic method. A much more accurate analysis can be carried out using molecular biology methods. They allow precise identification and differentiation of bacteria. These are the methods used in epidemiological investigations to analyze the phylogenetic similarity of isolates [33]. In 2016/2017, there was an outbreak of K. pneumoniae in the burn units of a hospital in China. The outbreak was analyzed using the PFGE method. Researchers determined that the outbreak was caused by a single clone [34]. Another outbreak in China was examined by Kong et al. The outbreak occurred in the neonatal ward in 2015/2016, and the isolated K. pneumoniae appeared to be clonally related. It can be concluded that the reason for the outbreak of K. pneumoniae producing NDM-type carbapenemases was the rapid spread of clones, the source of which was likely to be the first newborn admitted to the ward [35]. Pruss et al. isolated 32 K. pneumoniae strains belonging to a single clonal type in a neonatal ward, further demonstrating the high potential for transmission of this bacterium in the hospital environment [32]. Mansour et al. isolated K. pneumoniae strains from patients in different wards in a hospital in Tunisia. All isolates showed resistance to colistin and appeared to be genetically related. The strains had similar PFGE profiles [36]. In their study, Duman et al. analyzed K. pneumoniae that coproduced two enzymes, OXA-48 and NDM, and caused an outbreak in an ICU in Turkey. Their affinity was determined by electrophoresis in an alternating electric field [37]. Lixandru et al., who conducted a study throughout Romania, isolated strains from a hospital and then determined their genetic similarity. It turned out that all isolates belonged to the same clone and were the cause of hospital outbreaks [38]. In our study, conducted by means of electrophoresis in an alternating field, it was shown that among the isolates, as many as 22 out of 28 were bacteria that were classified into a single genetic type (cluster A). It included strains that were isolated primarily from infections. Cluster B included four isolates, two of which were isolated from urine and two from bronchoalveolar lavage. There were two strains from rectal swabs which were identified as unique. Although we were unable to determine the source of K. pneumoniae NDM from Cluster A and Cluster B, the results seem to support the fact that most of the related strains were from patients with confirmed secondary bacterial infections, in whom antibiotic therapy was undertaken due to complicated SARS-CoV-2 virus infections.
As of February 2021, infection control measures have been increased at our hospital, considering that it could be an epidemic. Staff training has been implemented on hand hygiene and environmental and equipment cleaning, and new environmental cleaning procedures have been prepared. Isolation of these patients has been ensured. From March to September, no new carbapenem-resistant K. pneumoniae infections were reported.
Epidemiological investigation plays a critical role in curbing nosocomial infections. Hospitalized patients are usually ailing or after surgery. An organism burdened with inflammation of various origins is more susceptible to secondary infections. Monitoring the spread of bacterial infections prevents epidemic outbreaks. Efficient identification of the source of the pathogen allows for its rapid elimination and inhibition of further spread.
5. Conclusions
During the COVID-19 pandemic, patients with primary viral infection often required hospitalization as a result of complications. The most common complication was secondary bacterial infection. The outbreak described retrospectively included 28 patients—23 infected and five colonized with K. pneumoniae NDM strains belonging to two genetic clones. Immediate tightening of hospital hygiene rules, screening of all hospitalized patients, and isolation/cohorting of positive patients proved effective in controlling and ending the two-month outbreak. The absence of K. pneumoniae NDM in the environment suggests that the outbreak was transmitted by colonized hospital staff. However, the colonization rate of hospital staff has not been determined. Screening for colonization of hospital staff can probably provide further details on the transmission pathway and effective control of the outbreak.
Author Contributions
Conceptualization, A.P.; methodology, A.P. and K.M.; validation, A.P. and H.M.; formal analysis, A.P., P.K. and H.M.; investigation, A.P. and K.M.; data curation, A.P.; writing—original draft preparation, A.P., K.M., P.K., J.J.-K., B.W. and S.G.-K.; visualization, A.P., H.M. and P.K.; supervision, A.P.; funding acquisition, B.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
In the current study, informed consent for swab and strain collection was not required as the isolates examined in the current study were collected during routine microbiological diagnostics by qualified medical staff.
Data Availability Statement
Data and materials are available on request.
Conflicts of Interest
The authors confirm that there are no known conflicts of interest associated with this publication.
References
- Santacroce, L.; Charitos, I.A.; Carretta, D.M.; De Nitto, E.; Lovero, R. The human coronaviruses (HCoVs) and the molecular mechanisms of SARS-CoV-2 infection. J. Mol. Med. 2021, 99, 93–106. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Liu, Q.; Guo, D. Emerging coronaviruses: Genome structure, replication, and pathogenesis. J. Med. Virol. 2020, 92, 418–423. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.J.; Shan, J. 2019 Novel coronavirus: Where we are and what we know. Infection 2020, 48, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zhang, N. Focus on the 2019 novel coronavirus (SARS-CoV-2). Future Microbiol. 2020, 15, 905–918. [Google Scholar] [CrossRef] [PubMed]
- Alexander, S.P.H.; Armstrong, J.F.; Davenport, A.P.; Davies, J.A.; Faccenda, E.; Harding, S.D.; Levi-Schaffer, F.; Maguire, J.J.; Pawson, A.J.; Southan, C.; et al. A rational roadmap for SARS-CoV-2/COVID-19 pharmacotherapeutic research and development: IUPHAR Review 29. Br. J. Pharmacol. 2020, 177, 4942–4966. [Google Scholar] [CrossRef]
- WHO. COVID-19 Situation Update for the WHO European Region, Data for the week of 9–15 March 2020 (Epi Week 11). Available online: https://www.who.int/europe/emergencies/situations/covid-19/who-european-region-operational-updates (accessed on 6 July 2020).
- Marcinkiewicz, J. Increase in the incidence of invasive bacterial infections following the COVID-19 pandemic: Potential links with decreased herd trained immunity—A novel concept in medicine. Pol. Arch. Intern. Med. 2024, 134, 16794. [Google Scholar] [CrossRef] [PubMed]
- Flisiak, R.; Horban, A.; Jaroszewicz, J.; Kozielewicz, D.; Mastalerz-Migas, A.; Owczuk, R.; Parczewski, M.; Pawłowska, M.; Piekarska, A.; Simon, K.; et al. Management of SARS-CoV-2 infection: Recommendations of the Polish Association of Epidemiologists and Infectiologists. Pol. Arch. Intern. Med. 2022, 132, 16230. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.Z.; Yang, M.; Lai, C.L. Post-COVID-19 Syndrome Comprehensive Assessment: From Clinical Diagnosis to Imaging and Biochemical-Guided Diagnosis and Management. Viruses 2023, 15, 533. [Google Scholar] [CrossRef] [PubMed]
- Lockhart, S.R.; Abramson, M.A.; Beekmann, S.E.; Gallagher, G.; Riedel, S.; Diekema, D.J.; Quinn, J.P.; Doern, G.V. Antimicrobial resistance among Gram-negative bacilli causing infections in intensive care unit patients in the United States between 1993 and 2004. J. Clin. Microbiol. 2007, 45, 3352–3359. [Google Scholar] [CrossRef] [PubMed]
- Preethika, R.; Ravindranath, C.; Deepa, S. Antibiotic susceptibility pattern of gram-negative bacterial isolates with special mention on colistin resistance from Intensive Care Unit of a tertiary care hospital: A prospective study assessing the impact of microbial resistance on clinical outcomes. Int. J. Res. Med. Sci. 2023, 11, 2206–2213. [Google Scholar] [CrossRef]
- Lai, C.C.; Chen, S.Y.; Ko, W.C.; Hsueh, P.R. Increased antimicrobial resistance during the COVID-19 pandemic. Int. J. Antimicrob. Agents 2021, 57, 106324. [Google Scholar] [CrossRef] [PubMed]
- O’Toole, R.F. The interface between COVID-19 and bacterial healthcare-associated infections. Clin. Microbiol. Infect. 2021, 27, 1772–1776. [Google Scholar] [CrossRef] [PubMed]
- Munita, J.M.; Arias, C.A. Mechanisms of Antibiotic Resistance. Microbiol. Spectr. 2016, 4. [Google Scholar] [CrossRef] [PubMed]
- Meini, S.; Tascini, C.; Cei, M.; Sozio, E.; Rossolini, G.M. AmpC β-lactamase-producing Enterobacterales: What a clinician should know. Infection 2019, 47, 363–375. [Google Scholar] [CrossRef]
- Lazar, D.S.; Nica, M.; Dascalu, A.; Oprisan, C.; Albu, O.; Codreanu, D.R.; Kosa, A.G.; Popescu, C.P.; Florescu, S.A. Carbapenem-Resistant NDM and OXA-48-like Producing K. pneumoniae: From Menacing Superbug to a Mundane Bacteria; A Retrospective Study in a Romanian Tertiary Hospital. Antibiotics 2024, 13, 435. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Xiang, T.; Chen, C.; Wen, J.; Liu, Y.; Zhang, Q.; Cheng, N.; Wu, X.; Zhang, W. Resistance of Klebsiella pneumoniae Strains Carrying blaNDM-1 Gene and the Genetic Environment of blaNDM-1. Front. Microbiol. 2020, 11, 700. [Google Scholar] [CrossRef] [PubMed]
- European Committee on Antimicrobial Susceptibility Testing: Breakpoints Tables for Interpretation of MICs and Zone Diameters. Version 11.0. Available online: https://www.eucast.org (accessed on 1 January 2021).
- Yahya, R.O. Problems Associated with Co-Infection by Multidrug-Resistant Klebsiella pneumoniae in COVID-19 Patients: A Review. Healthcare 2022, 10, 2412. [Google Scholar] [CrossRef] [PubMed]
- Hosoda, T.; Harada, S.; Okamoto, K.; Ishino, S.; Kaneko, M.; Suzuki, M.; Ito, R.; Mizoguchi, M. COVID-19 and Fatal Sepsis Caused by Hypervirulent Klebsiella pneumoniae, Japan. Emerg. Infect. Dis. 2021, 27, 556–559. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Shen, H.; Zhu, C.; Yu, Y. Carbapenem-Resistant Klebsiella pneumoniae Infections among ICU Admission Patients in Central China: Prevalence and Prediction Model. Biomed. Res. Int. 2019, 2019, 9767313. [Google Scholar] [CrossRef]
- Pruss, A.; Kwiatkowski, P.; Sienkiewicz, M.; Masiuk, H.; Łapińska, A.; Kot, B.; Kilczewska, Z.; Giedrys-Kalemba, S.; Dołęgowska, B. Similarity Analysis of Klebsiella pneumoniae Producing Carbapenemases Isolated from UTI and Other Infections. Antibiotics 2023, 12, 1224. [Google Scholar] [CrossRef]
- Campos, A.C.; Albiero, J.; Ecker, A.B.; Kuroda, C.M.; Meirelles, L.E.; Polato, A.; Tognim, M.C.; Wingeter, M.A.; Teixeira, J.J. Outbreak of Klebsiella pneumoniae carbapenemase-producing K pneumoniae: A systematic review. Am. J. Infect. Control 2016, 44, 1374–1380. [Google Scholar] [CrossRef] [PubMed]
- Aires-de-Sousa, M.; Ortiz de la Rosa, J.M.; Goncalves, M.L.; Pereira, A.L.; Nordmann, P.; Poirel, L. Epidemiology of Carbapenemase-Producing Klebsiella pneumoniae in a Hospital, Portugal. Emerg. Infect. 2019, 25, 1632–1638. [Google Scholar] [CrossRef]
- Torres, A.; Niederman, M.S.; Chastre, J.; Ewig, S.; Fernandez-Vandellos, P.; Hanberger, H.; Kollef, M.; Li Bassi, G.; Luna, C.M.; Martin-Loeches, I.; et al. International ERS/ESICM/ESCMID/ALAT guidelines for the management of hospital-acquired pneumonia and ventilator-associated pneumonia: Guidelines for the management of hospital-acquired pneumonia (HAP)/ventilator-associated pneumonia (VAP) of the European Respiratory Society (ERS), European Society of Intensive Care Medicine (ESICM), European Society of Clinical Microbiology and Infectious Diseases (ESCMID) and Asociación Latinoamericana del Tórax (ALAT). Eur. Respir. J. 2017, 50, 1700582. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Liu, Y.; Wang, R.; Wang, Q.; Jin, L.; Wang, H. The transferability and evolution of NDM-1 and KPC-2 co-producing Klebsiella pneumoniae from clinical settings. EBioMedicine 2020, 51, 102599. [Google Scholar] [CrossRef] [PubMed]
- Ojdana, D.; Sacha, P.; Gutowska, A.; Majewski, P.; Wieczorek, P.; Tryniszewska, E. Antibiotic resistance profiles of KPC and NDM carbapenemases-producing Klebsiella pneumoniae. Zakażenia XXI wieku 2019, 2, 131–137. [Google Scholar] [CrossRef]
- Kazi, M.; Drego, L.; Nikam, C.; Ajbani, K.; Soman, R.; Shetty, A.; Rodrigues, C. Molecular characterization of carbapenem-resistant Enterobacteriaceae at a tertiary care laboratory in Mumbai. Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 467–472. [Google Scholar] [CrossRef] [PubMed]
- Han, R.; Shi, Q.; Wu, S.; Yin, D.; Peng, M.; Dong, D.; Zheng, Y.; Guo, Y.; Zhang, R.; Hu, F. China Antimicrobial Surveillance Network (CHINET) Study Group. Dissemination of Carbapenemases (KPC, NDM, OXA-48, IMP, and VIM) Among Carbapenem-Resistant Enterobacteriaceae Isolated From Adult and Children Patients in China. Front. Cell Infect. Microbiol. 2020, 10, 314. [Google Scholar] [CrossRef]
- Lutgring, J.D. Carbapenem-resistant Enterobacteriaceae: An emerging bacterial threat. Semin. Diagn. Pathol. 2019, 36, 182–186. [Google Scholar] [CrossRef]
- Durdu, B.; Meric Koc, M.; Hakyemez, I.N.; Akkoyunlu, Y.; Daskaya, H.; Sumbul Gultepe, B.; Aslan, T. Risk Factors Affecting Patterns of Antibiotic Resistance and Treatment Efficacy in Extreme Drug Resistance in Intensive Care Unit-Acquired Klebsiella Pneumoniae Infections: A 5-Year Analysis. Med. Sci. Monit. 2019, 25, 174–183. [Google Scholar] [CrossRef] [PubMed]
- Pruss, A.; Kwiatkowski, P.; Masiuk, H.; Bilska, I.; Giedrys-Kalemba, S.; Dołęgowska, B. Epidemiological Analysis of Extended-Spectrum β-Lactamase-Producing Klebsiella pneumoniae Outbreak in a Neonatal Clinic in Poland. Antibiotics 2022, 12, 50. [Google Scholar] [CrossRef] [PubMed]
- Adzitey, F.; Huda, N.; Ali, G.R. Molecular techniques for detecting and typing of bacteria, advantages and application to foodborne pathogens isolated from ducks. 3 Biotech 2013, 3, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Li, M.; Shen, X.; Wang, L.; Liu, L.; Hao, Z.; Duan, J.; Yu, F. Outbreak of blaNDM-5-Harboring Klebsiella pneumoniae ST290 in a Tertiary Hospital in China. Microb. Drug Resist. 2019, 25, 1443–1448. [Google Scholar] [CrossRef]
- Kong, Z.; Cai, R.; Cheng, C.; Zhang, C.; Kang, H.; Ma, P.; Gu, B. First Reported Nosocomial Outbreak Of NDM-5-Producing Klebsiella pneumoniae In A Neonatal Unit In China. Infect. Drug Resist. 2019, 12, 3557–3566. [Google Scholar] [CrossRef] [PubMed]
- Mansour, W.; Haenni, M.; Saras, E.; Grami, R.; Mani, Y.; Ben Haj Khalifa, A.; El Atrouss, S.; Kheder, M.; Fekih Hassen, M.; Boujâafar, N.; et al. Outbreak of colistin-resistant carbapenemase-producing Klebsiella pneumoniae in Tunisia. J. Glob. Antimicrob. Resist. 2017, 10, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Duman, Y.; Ersoy, Y.; Gursoy, N.C.; Altunisik Toplu, S.; Otlu, B. A silent outbreak due to Klebsiella pneumoniae that co-produced NDM-1 and OXA-48 carbapenemases, and infection control measures. Iran. J. Basic. Med. Sci. 2020, 23, 46–50. [Google Scholar] [CrossRef] [PubMed]
- Lixandru, B.E.; Cotar, A.I.; Straut, M.; Usein, C.R.; Cristea, D.; Ciontea, S.; Tatu-Chitoiu, D.; Codita, I.; Rafila, A.; Nica, M.; et al. Carbapenemase-Producing Klebsiella pneumoniae in Romania: A Six-Month Survey. PLoS ONE 2015, 10, e0143214. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).