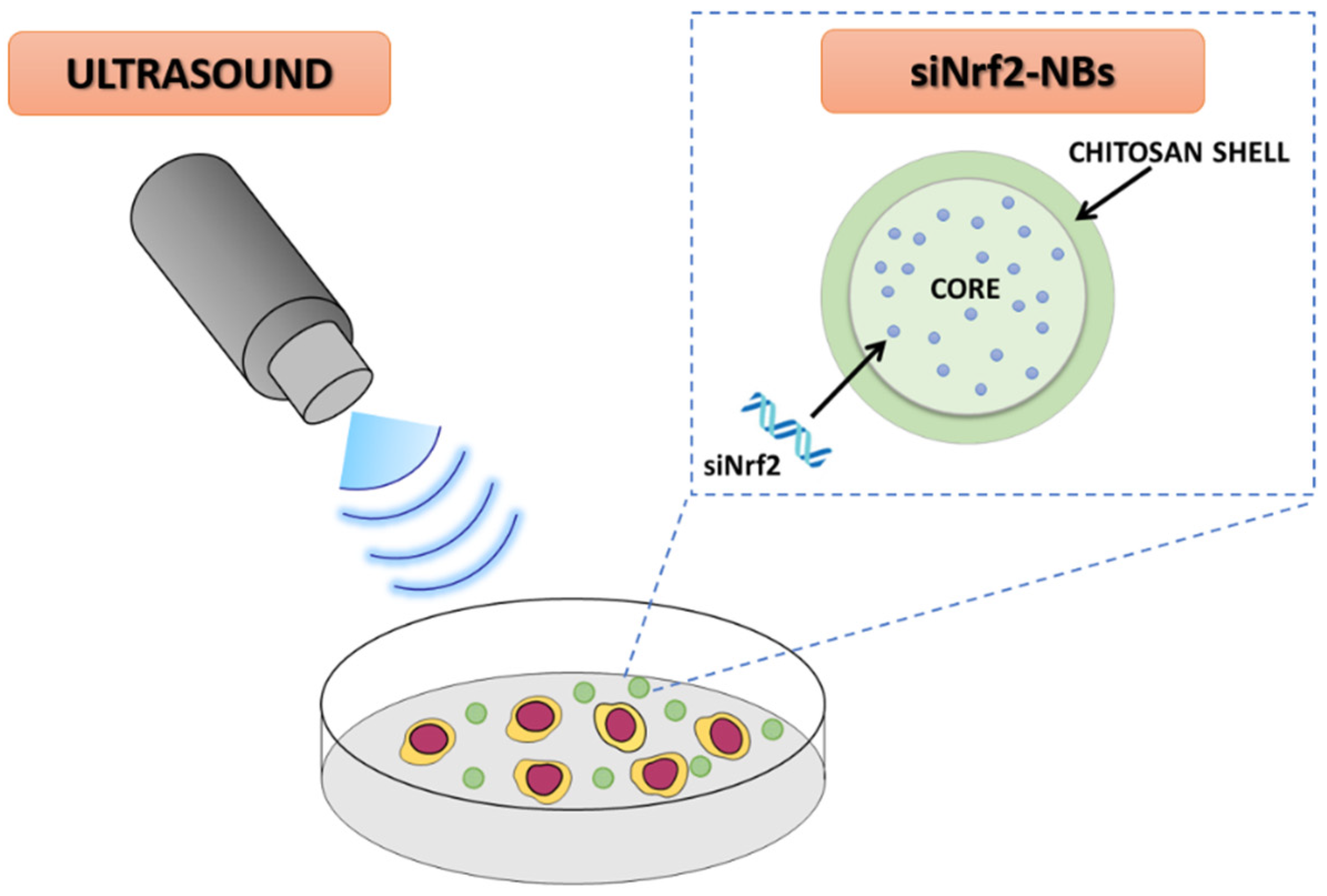
Ultrasound-Responsive Nrf2-Targeting siRNA-Loaded Nanobubbles for Enhancing the Treatment of Melanoma
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Preparation of siNrf2-Nanobubble Formulations
2.3. Characterization of siNrf2-Nanobubble Formulations
2.4. In Vitro Release Studies
2.5. In Vitro Stability Studies
2.6. Evaluation of Nanobubble Haemolytic Activity
2.7. Cell Line
2.8. Evaluation of Nanobubble Cellular Internalization in M14 Cells under Fluorescence Miscroscopy
2.9. siNrf2 Transfection with a Traditional Protocol
2.10. Western Blot
2.11. Viability Analysis in M14 Cell Line after siNrf2-Nanobubble Treatment
2.12. Viability Analysis in M14 Cell Line after Ultrasound Irradiation
2.13. Cytofluorimetric Evaluation of NB Cellular Internalizazion in M14 Cells after Ultrasound Irradiation
2.14. Transfection Efficiency of siNrf2-NB M14 Cells after Ultrasound Irradiation
2.15. Echogenic Properties of NB Formulations
2.16. Statistical Analysis
3. Results and Discussion
3.1. Characterization of siNrf2-Nanobubble Formulations
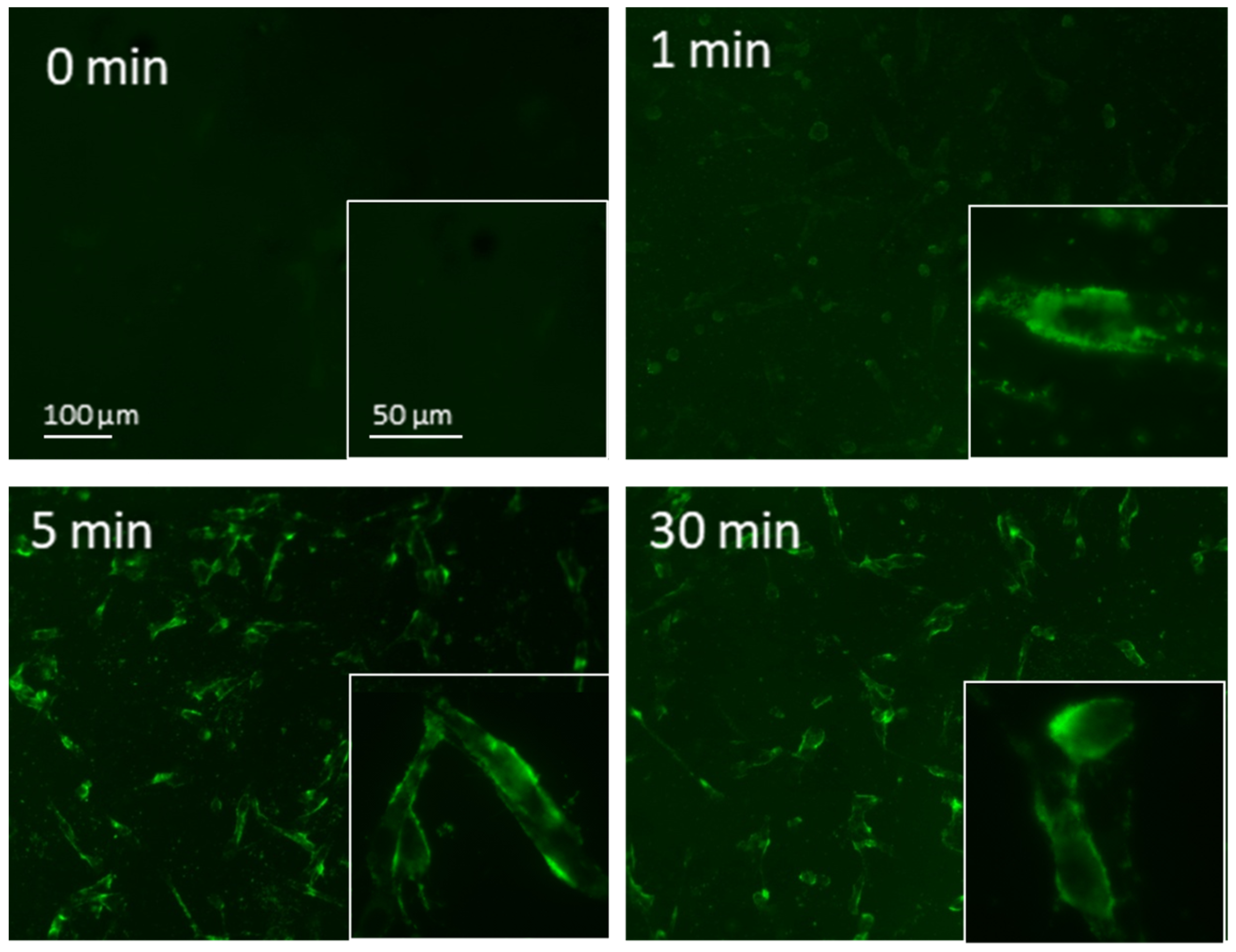
3.2. Evaluation of NB Cellular Internalization
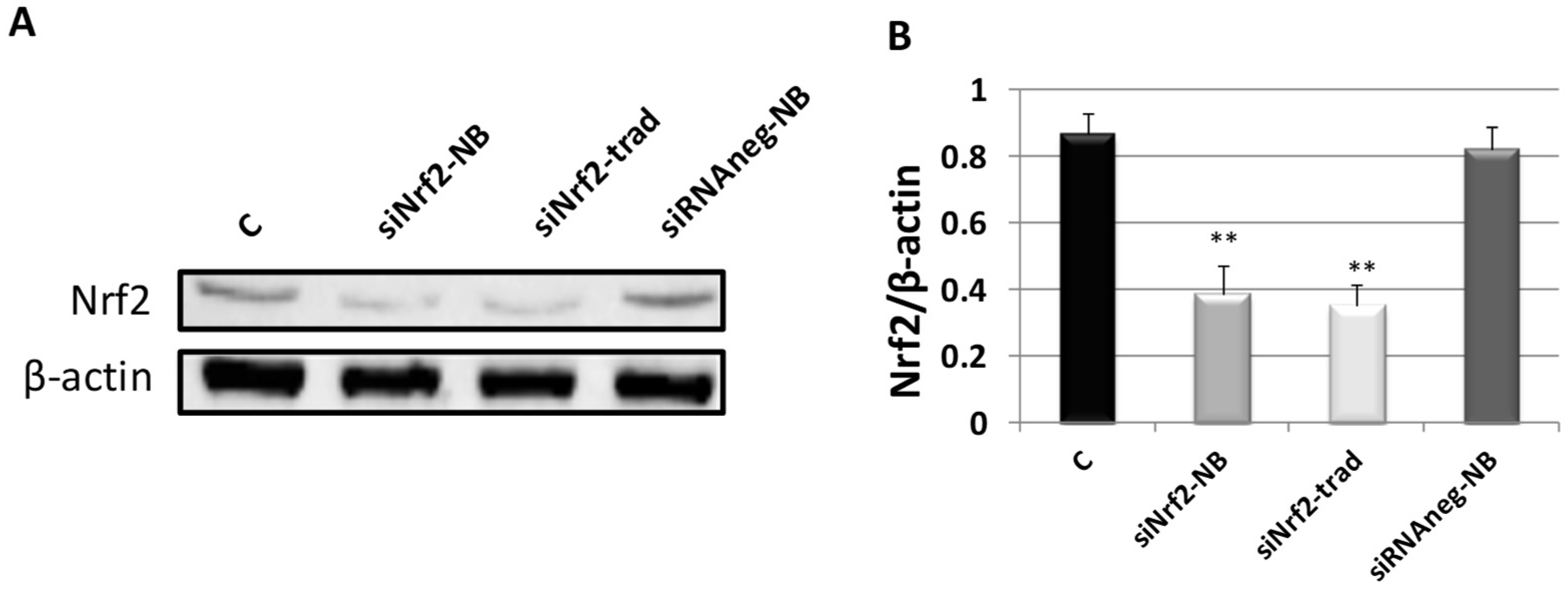
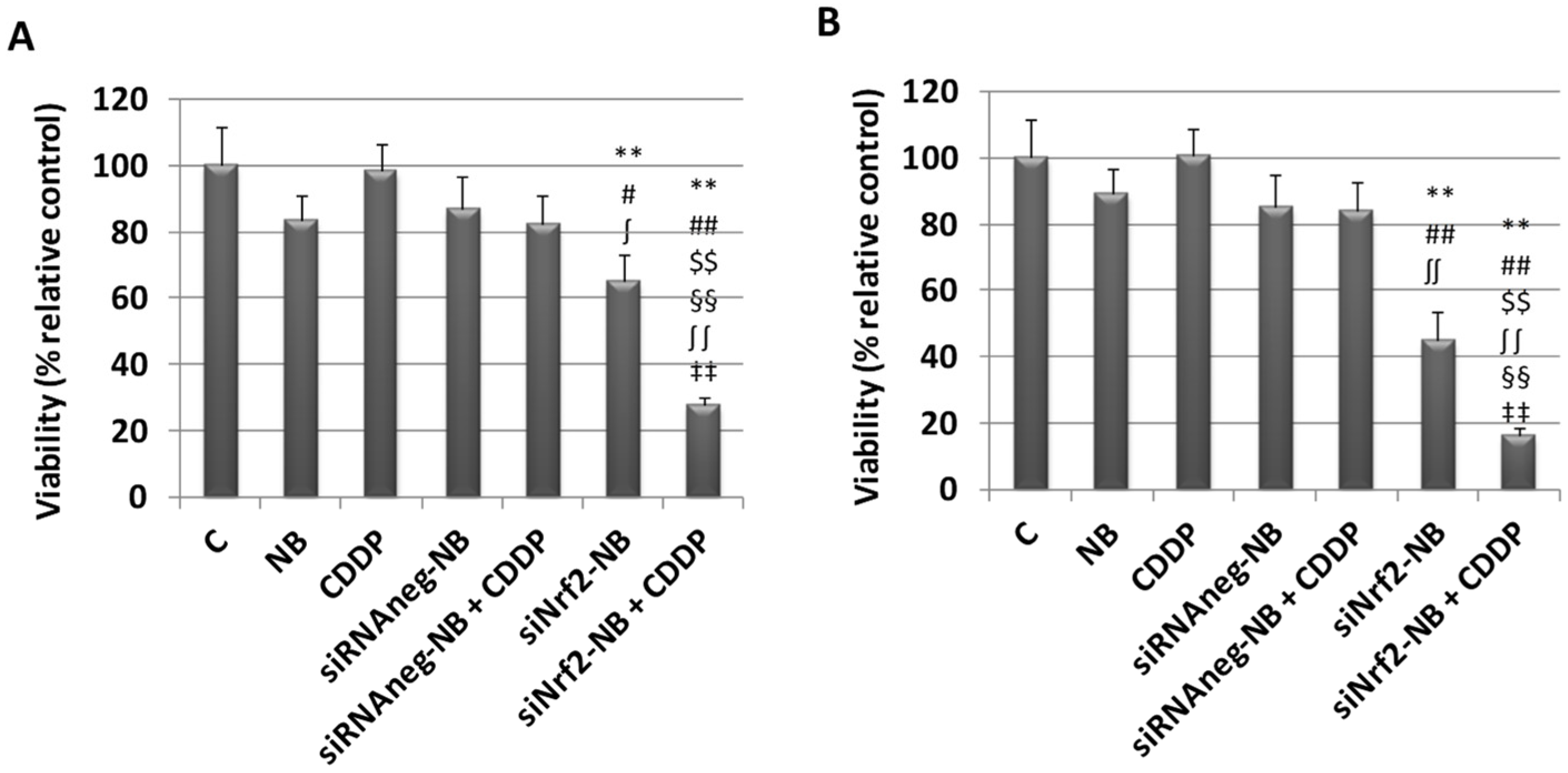
3.3. Biological Evaluation of siNrf2-NB on M14 Human Melanoma Cell Line
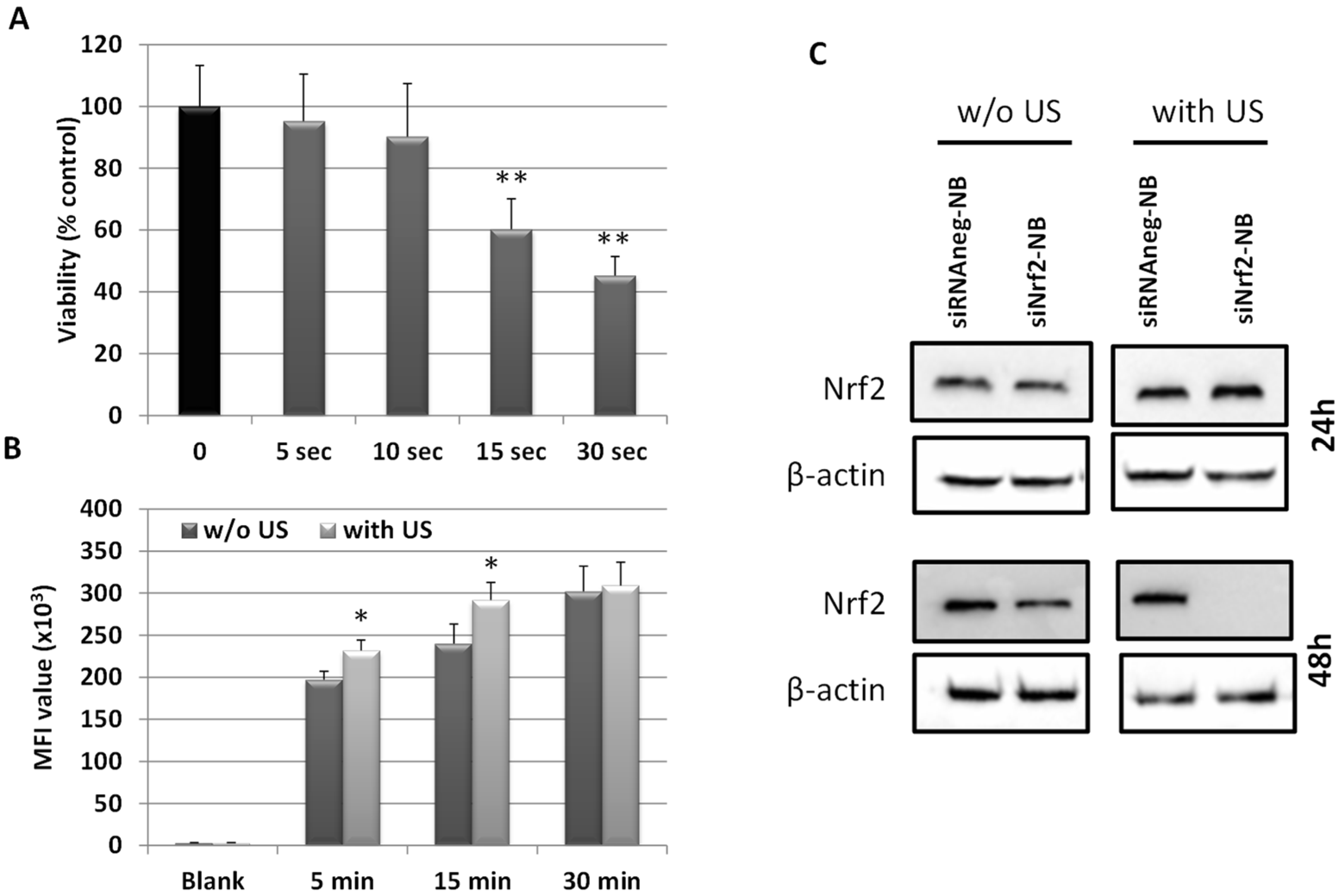
3.4. Ultrasound-Mediated siNrf2-NB Delivery in M14 Cell Line
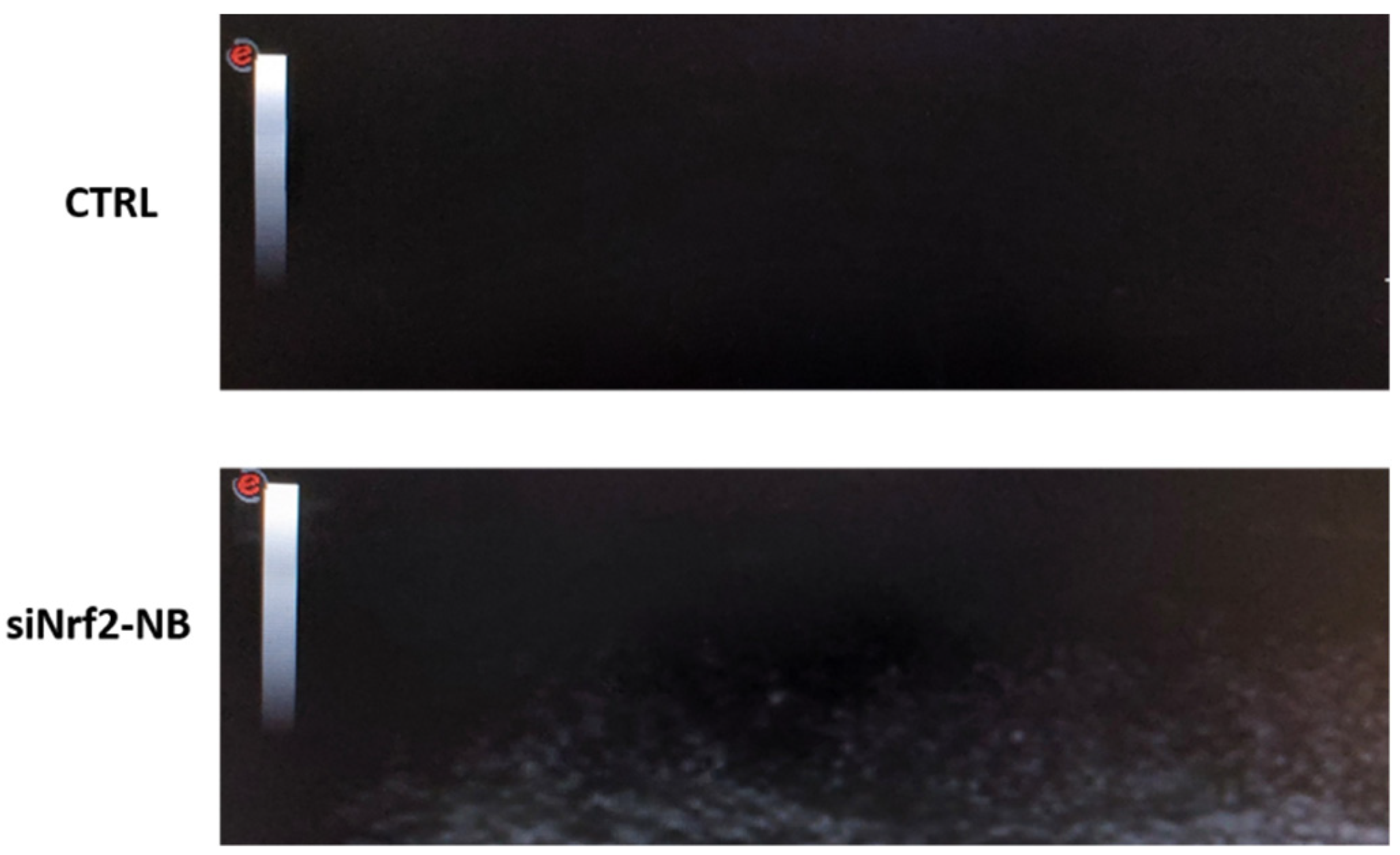
3.5. Echogenic Properties of NB Formulations
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Matthews, N.H.; Li, W.Q.; Qureshi, A.A.; Weinstock, M.A.; Cho, E. Epidemiology of melanoma. In Cutaneous Melanoma: Etiology and Therapy; Ward, W.H., Farma, J.M., Eds.; Codon Publications: Brisbane, Australia, 2017; Chapter 1. [Google Scholar]
- Davis, L.E.; Shalin, S.C.; Tackett, A.J. Current state of melanoma diagnosis and treatment. Cancer Biol. Ther. 2019, 20, 1366–1379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Menzies, A.M.; Long, G.V. Recent advances in melanoma systemic therapy. BRAF inhibitors, CTLA4 antibodies and beyond. Eur. J. Cancer 2013, 49, 3229–3241. [Google Scholar] [CrossRef]
- Battaglia, L.; Scomparin, A.; Dianzani, C.; Milla, P.; Muntoni, E.; Arpicco, S.; Cavalli, R. Nanotechnology Addressing Cutaneous Melanoma: The Italian Landscape. Pharmaceutics 2021, 13, 1617. [Google Scholar] [CrossRef]
- Hodis, E.; Watson, I.R.; Kryukov, G.V.; Arold, S.T.; Imielinski, M.; Theurillat, J.P.; Nickerson, E.; Auclair, D.; Li, L.; Place, C.; et al. A landscape of driver mutations in melanoma. Cell 2012, 150, 251–263. [Google Scholar] [CrossRef] [Green Version]
- Robert, C.; Karaszewska, B.; Schachter, J.; Rutkowski, P.; Mackiewicz, A.; Stroiakovski, D.; Lichinitser, M.; Dummer, R.; Grange, F.; Mortier, L.; et al. Improved overall survival in melanoma with combined dabrafenib and trametinib. N. Engl. J. Med. 2015, 372, 30–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albittar, A.A.; Alhalabi, O.; Glitza Oliva, I.C. Immunotherapy for Melanoma. Adv. Exp. Med. Biol. 2020, 1244, 51–68. [Google Scholar] [CrossRef]
- Rizos, H.; Menzies, A.M.; Pupo, G.M.; Carlino, M.S.; Fung, C.; Hyman, J.; Haydu, L.E.; Mijatov, B.; Becker, T.M.; Boyd, S.C.; et al. BRAF inhibitor resistance mechanisms in metastatic melanoma: Spectrum and clinical impact. Clin. Cancer Res. 2014, 20, 1965–1977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vukadin, S.; Khaznadar, F.; Kizivat, T.; Vcev, A.; Smolic, M. Molecular Mechanisms of Resistance to Immune Checkpoint Inhibitors in Melanoma Treatment: An Update. Biomedicines 2021, 9, 835. [Google Scholar] [CrossRef]
- Moloney, J.N.; Cotter, T.G. ROS signalling in the biology of cancer. Semin. Cell Dev. Biol. 2018, 80, 50–64. [Google Scholar] [CrossRef]
- Pizzimenti, S.; Ribero, S.; Cucci, M.A.; Grattarola, M.; Monge, C.; Dianzani, C.; Barrera, G.; Muzio, G. Oxidative Stress-Related Mechanisms in Melanoma and in the Acquired Resistance to Targeted Therapies. Antioxidants 2021, 10, 1942. [Google Scholar] [CrossRef]
- Harris, I.S.; DeNicola, G.M. The Complex Interplay between Antioxidants and ROS in Cancer. Trends Cell Biol. 2020, 30, 440–451. [Google Scholar] [CrossRef] [PubMed]
- Rojo de la Vega, M.; Chapman, E.; Zhang, D.D. NRF2 and the Hallmarks of Cancer. Cancer Cell 2018, 34, 21–43. [Google Scholar] [CrossRef] [PubMed]
- Friedmann Angeli, J.P.; Meierjohann, S. NRF2-dependent stress defense in tumor antioxidant control and immune evasion. Pigment Cell Melanoma Res. 2021, 34, 268–279. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Ye, W.; Zhang, M.; Liang, J. The effects of nrf2 on tumor angiogenesis: A review of the possible mechanisms of action. Crit. Rev. Eukaryot. Gene Expr. 2012, 22, 149–160. [Google Scholar] [CrossRef]
- Lau, A.; Villeneuve, N.F.; Sun, Z.; Wong, P.K.; Zhang, D.D. Dual roles of Nrf2 in cancer. Pharmacol Res. 2008, 58, 262–270. [Google Scholar] [CrossRef] [PubMed]
- Furfaro, A.L.; Traverso, N.; Domenicotti, C.; Piras, S.; Moretta, L.; Marinari, U.M.; Pronzato, M.A.; Nitti, M. The Nrf2/HO-1 Axis in Cancer Cell Growth and Chemoresistance. Oxid. Med. Cell. Longev. 2016, 2016, 1958174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, Y.; Zhao, Z.; Meng, X.; Chen, H.; Fu, G. Migration and invasion in B16-F10 mouse melanoma cells are regulated by Nrf2 inhibition during treatment with ionizing radiation. Oncol. Lett. 2018, 16, 1959–1966. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hintsala, H.R.; Jokinen, E.; Haapasaari, K.M.; Moza, M.; Ristimäki, A.; Soini, Y.; Koivunen, J.; Karihtala, P. Nrf2/Keap1 Pathway and Expression of Oxidative Stress Lesions 8-hydroxy-2′-deoxyguanosine and Nitrotyrosine in Melanoma. Anticancer Res. 2016, 36, 1497–1506. [Google Scholar]
- Miura, S.; Shibazaki, M.; Kasai, S.; Yasuhira, S.; Watanabe, A.; Inoue, T.; Kageshita, Y.; Tsunoda, K.; Takahashi, K.; Akasaka, T.; et al. A somatic mutation of the KEAP1 gene in malignant melanoma is involved in aberrant NRF2 activation and an increase in intrinsic drug resistance. J. Investig. Dermatol. 2014, 134, 553–556. [Google Scholar] [CrossRef] [Green Version]
- Rocha, C.R.; Kajitani, G.S.; Quinet, A.; Fortunato, R.S.; Menck, C.F. NRF2 and glutathione are key resistance mediators to temozolomide in glioma and melanoma cells. Oncotarget 2016, 7, 48081–48092. [Google Scholar] [CrossRef] [Green Version]
- Khamari, R.; Trinh, A.; Gabert, P.E.; Corazao-Rozas, P.; Riveros-Cruz, S.; Balayssac, S.; Malet-Martino, M.; Dekiouk, S.; Joncquel Chevalier Curt, M.; Maboudou, P.; et al. Glucose metabolism and NRF2 coordinate the antioxidant response in melanoma resistant to MAPK inhibitors. Cell Death Dis. 2018, 9, 325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, M.; Shi, G.; Bian, C.; Nisar, M.F.; Guo, Y.; Wu, Y.; Li, W.; Huang, X.; Jiang, X.; Bartsch, J.W.; et al. UVA Irradiation Enhances Brusatol-Mediated Inhibition of Melanoma Growth by Downregulation of the Nrf2-Mediated Antioxidant Response. Oxid. Med. Cell. Longev. 2018, 2018, 9742154. [Google Scholar] [CrossRef] [PubMed]
- Cai, S.J.; Liu, Y.; Han, S.; Yang, C. Brusatol, an NRF2 inhibitor for future cancer therapeutic. Cell Biosci. 2019, 9, 45. [Google Scholar] [CrossRef] [Green Version]
- Daga, M.; Pizzimenti, S.; Dianzani, C.; Cucci, M.A.; Cavalli, R.; Grattarola, M.; Ferrara, B.; Scariot, V.; Trotta, F.; Barrera, G. Ailanthone inhibits cell growth and migration of cisplatin resistant bladder cancer cells through down-regulation of Nrf2, YAP, and c-Myc expression. Phytomedicine 2019, 56, 156–164. [Google Scholar] [CrossRef]
- Cucci, M.A.; Grattarola, M.; Dianzani, C.; Damia, G.; Ricci, F.; Roetto, A.; Trotta, F.; Barrera, G.; Pizzimenti, S. Ailanthone increases oxidative stress in CDDP-resistant ovarian and bladder cancer cells by inhibiting of Nrf2 and YAP expression through a post-translational mechanism. Free Radic. Biol. Med. 2020, 150, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Mirzaei, S.; Mahabady, M.K.; Zabolian, A.; Abbaspour, A.; Fallahzadeh, P.; Noori, M.; Hashemi, F.; Hushmandi, K.; Daneshi, S.; Kumar, A.P.; et al. Small interfering RNA (siRNA) to target genes and molecular pathways in glioblastoma therapy: Current status with an emphasis on delivery systems. Life Sci. 2021, 275, 119368. [Google Scholar] [CrossRef]
- Nikam, R.R.; Gore, K.R. Journey of siRNA: Clinical Developments and Targeted Delivery. Nucleic Acid Ther. 2018, 28, 209–224. [Google Scholar] [CrossRef]
- Ku, S.H.; Jo, S.D.; Lee, Y.K.; Kim, K.; Kim, S.H. Chemical and structural modifications of RNAi therapeutics. Adv. Drug Deliv. Rev. 2016, 104, 16–28. [Google Scholar] [CrossRef]
- Mainini, F.; Eccles, M.R. Lipid and Polymer-Based Nanoparticle siRNA Delivery Systems for Cancer Therapy. Molecules 2020, 25, 2692. [Google Scholar] [CrossRef]
- Hu, B.; Zhong, L.; Weng, Y.; Peng, L.; Huang, Y.; Zhao, Y.; Liang, X.J. Therapeutic siRNA: State of the art. Signal Transduct. Target. Ther. 2020, 5, 101. [Google Scholar] [CrossRef]
- Dong, Y.; Siegwart, D.J.; Anderson, D.G. Strategies, design, and chemistry in siRNA delivery systems. Adv. Drug Deliv. Rev. 2019, 144, 133–147. [Google Scholar] [CrossRef] [PubMed]
- Subhan, M.A.; Torchilin, V.P. Efficient nanocarriers of siRNA therapeutics for cancer treatment. Transl. Res. 2019, 214, 62–91. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; An, K.; Duan, X.; Xu, H.; Li, F.; Xu, F. Recent advances in siRNA delivery for cancer therapy using smart nanocarriers. Drug Discov. Today 2018, 23, 900–911. [Google Scholar] [CrossRef] [PubMed]
- Ambrosio, L.; Argenziano, M.; Cucci, M.A.; Grattarola, M.; de Graaf, I.A.M.; Dianzani, C.; Barrera, G.; Sánchez Nieves, J.; Gomez, R.; Cavalli, R.; et al. Carbosilane Dendrimers Loaded with siRNA Targeting Nrf2 as a Tool to Overcome Cisplatin Chemoresistance in Bladder Cancer Cells. Antioxidants 2020, 9, 993. [Google Scholar] [CrossRef]
- Yonezawa, S.; Koide, H.; Asai, T. Recent advances in siRNA delivery mediated by lipid-based nanoparticles. Adv. Drug Deliv. Rev. 2020, 154-155, 64–78. [Google Scholar] [CrossRef]
- Cavalli, R.; Soster, M.; Argenziano, M. Nanobubbles: A promising efficient tool for therapeutic delivery. Ther. Deliv. 2016, 7, 117–138. [Google Scholar] [CrossRef]
- Bisazza, A.; Civra, A.; Donalisio, M.; Lembo, D.; Cavalli, R. The in vitro characterization of dextran-based nanobubbles as possible DNA transfection agents. Soft Matter 2011, 7, 10590–10593. [Google Scholar] [CrossRef]
- Cavalli, R.; Bisazza, A.; Trotta, M.; Argenziano, M.; Civra, A.; Donalisio, M.; Lembo, D. New chitosan nanobubbles for ultrasound-mediated gene delivery: Preparation and in vitro characterization. Int. J. Nanomed. 2012, 7, 3309–3318. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Shen, X.; Yan, J.; Xie, X.; Chen, Z.; Li, T.; Li, S.; Qin, X.; Wu, C.; Liu, Y. Charge-reversal-functionalized PLGA nanobubbles as theranostic agents for ultrasonic-imaging-guided combination therapy. Biomater. Sci. 2018, 6, 2426–2439. [Google Scholar] [CrossRef]
- Yuan, Y.; Li, E.; Zhao, J.; Wu, B.; Na, Z.; Cheng, W.; Jing, H. Highly penetrating nanobubble polymer enhances LINC00511-siRNA delivery for improving the chemosensitivity of triple-negative breast cancer. Anticancer Drugs 2021, 32, 178–188. [Google Scholar] [CrossRef]
- Su, C.; Ren, X.; Nie, F.; Li, T.; Lw, W.; Li, H.; Zhang, Y. Current advances in ultrasound-combined nanobubbles for cancer-targeted therapy: A review of the current status and future perspectives. RSC Adv. 2021, 11, 12915–12928. [Google Scholar] [CrossRef]
- Su, C.; Ren, X.; Yang, F.; Li, B.; Wu, H.; Li, H.; Nie, F. Ultrasound-sensitive siRNA-loaded nanobubbles fabrication and antagonism in drug resistance for NSCLC. Drug Deliv. 2022, 29, 99–110. [Google Scholar] [CrossRef]
- Endo-Takahashi, Y.; Negishi, Y. Microbubbles and Nanobubbles with Ultrasound for Systemic Gene Delivery. Pharmaceutics 2020, 12, 964. [Google Scholar] [CrossRef] [PubMed]
- Cai, W.; Lv, W.; Feng, Y.; Yang, H.; Zhang, Y.; Yang, G.; Duan, Y.; Wang, J. The therapeutic effect in gliomas of nanobubbles carrying siRNA combined with ultrasound-targeted destruction. Int. J. Nanomed. 2018, 13, 6791–6807. [Google Scholar] [CrossRef] [PubMed]
- Ciamporcero, E.; Daga, M.; Pizzimenti, S.; Roetto, A.; Dianzani, C.; Compagnone, A.; Palmieri, A.; Ullio, C.; Cangemi, L.; Pili, R.; et al. Crosstalk between Nrf2 and YAP contributes to maintaining the antioxidant potential and chemoresistance in bladder cancer. Free Radic. Biol. Med. 2018, 115, 447–457. [Google Scholar] [CrossRef] [PubMed]
- Rosenblum, D.; Joshi, N.; Tao, W.; Karp, J.M.; Peer, D. Progress and challenges towards targeted delivery of cancer therapeutics. Nat. Commun. 2018, 9, 1410. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Xi, W.S.; Su, Q.; Li, Y.; Yan, G.H.; Liu, Y.; Wang, H.; Cao, A. Unexpected Size Effect: The Interplay between Different-Sized Nanoparticles in Their Cellular Uptake. Small 2019, 15, e1901687. [Google Scholar] [CrossRef]
- Argenziano, M.; Bressan, B.; Luganini, A.; Finesso, N.; Genova, T.; Troia, A.; Giribaldi, G.; Banche, G.; Mandras, N.; Cuffini, A.M.; et al. Comparative Evaluation of Different Chitosan Species and Derivatives as Candidate Biomaterials for Oxygen-Loaded Nanodroplet Formulations to Treat Chronic Wounds. Mar. Drugs 2021, 19, 112. [Google Scholar] [CrossRef]
- Pavinatto, F.J.; Caseli, L.; Oliveira, O.N. Chitosan in nanostructured thin films. Biomacromolecules 2010, 11, 1897–1908. [Google Scholar] [CrossRef]
- Pavinatto, A.; Pavinatto, F.J.; Delezuk, J.A.; Nobre, T.M.; Souza, A.L.; Campana-Filho, S.P.; Oliveira, O.N., Jr. Low molecular-weight chitosans are stronger biomembrane model perturbants. Colloids Surf. B Biointerfaces 2013, 104, 48–53. [Google Scholar] [CrossRef]
- Ashrafizadeh, M.; Delfi, M.; Hashemi, F.; Zabolian, A.; Saleki, H.; Bagherian, M.; Azami, N.; Farahani, M.V.; Sharifzadeh, S.O.; Hamzehlou, S.; et al. Biomedical application of chitosan-based nanoscale delivery systems: Potential usefulness in siRNA delivery for cancer therapy. Carbohydr. Polym. 2021, 260, 117809. [Google Scholar] [CrossRef] [PubMed]
- Cavalli, R.; Bisazza, A.; Lembo, D. Micro- and nanobubbles: A versatile non-viral platform for gene delivery. Int. J. Pharm. 2013, 456, 437–445. [Google Scholar] [CrossRef] [PubMed]
- Serrano-Sevilla, I.; Artiga, Á.; Mitchell, S.G.; De Matteis, L.; de la Fuente, J.M. Natural Polysaccharides for siRNA Delivery: Nanocarriers Based on Chitosan, Hyaluronic Acid, and Their Derivatives. Molecules 2019, 24, 2570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ragelle, H.; Vandermeulen, G.; Préat, V. Chitosan-based siRNA delivery systems. J. Control. Release 2013, 172, 207–218. [Google Scholar] [CrossRef]
- Cavallaro, G.; Sardo, C.; Craparo, E.F.; Porsio, B.; Giammona, G. Polymeric nanoparticles for siRNA delivery: Production and applications. Int. J. Pharm. 2017, 525, 313–333. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Hart, S.L.; Du, Z. Assembly strategy of liposome and polymer systems for siRNA delivery. Int. J. Pharm. 2021, 592, 120033. [Google Scholar] [CrossRef]
- Cavalli, R.; Primo, L.; Sessa, R.; Chiaverina, G.; di Blasio, L.; Alongi, J.; Manfredi, A.; Ranucci, E.; Ferruti, P. The AGMA1 polyamidoamine mediates the efficient delivery of siRNA. J. Drug Target. 2017, 25, 891–898. [Google Scholar] [CrossRef]
- Cao, Y.; Tan, Y.F.; Wong, Y.S.; Liew, M.W.J.; Venkatraman, S. Recent Advances in Chitosan-Based Carriers for Gene Delivery. Mar. Drugs 2019, 17, 381. [Google Scholar] [CrossRef] [Green Version]
- Singh, A.; Trivedi, P.; Jain, N.K. Advances in siRNA delivery in cancer therapy. Artif. Cells Nanomed. Biotechnol. 2018, 46, 274–283. [Google Scholar] [CrossRef]
- Donalisio, M.; Argenziano, M.; Rittà, M.; Bastiancich, C.; Civra, A.; Lembo, D.; Cavalli, R. Acyclovir-loaded sulfobutyl ether-β-cyclodextrin decorated chitosan nanodroplets for the local treatment of HSV-2 infections. Int. J. Pharm. 2020, 587, 119676. [Google Scholar] [CrossRef]
- Marano, F.; Argenziano, M.; Frairia, R.; Adamini, A.; Bosco, O.; Rinella, L.; Fortunati, N.; Cavalli, R.; Catalano, M.G. Doxorubicin-Loaded Nanobubbles Combined with Extracorporeal Shock Waves: Basis for a New Drug Delivery Tool in Anaplastic Thyroid Cancer. Thyroid 2016, 26, 705–716. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Guo, D.; Mao, X.; Shan, X.; He, X.; Yu, C. Perfluoropentane-filled chitosan poly-acrylic acid nanobubbles with high stability for long-term ultrasound imaging in vivo. Nanoscale 2021, 13, 5333–5343. [Google Scholar] [CrossRef] [PubMed]
- Mirzaei, S.; Gholami, M.H.; Hashemi, F.; Zabolian, A.; Hushmandi, K.; Rahmanian, V.; Entezari, M.; Girish, Y.R.; Sharath Kumar, K.S.; Aref, A.R.; et al. Employing siRNA tool and its delivery platforms in suppressing cisplatin resistance: Approaching to a new era of cancer chemotherapy. Life Sci. 2021, 277, 119430. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Yuan, Y.; Han, X.; Wang, Q.; Shang, H.; Liang, X.; Jing, H.; Cheng, W. Structure of LINC00511-siRNA-conjugated nanobubbles and improvement of cisplatin sensitivity on triple negative breast cancer. FASEB J. 2020, 34, 9713–9726. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Jiang, Y.; Lin, M.; Zhang, J.; Guo, H.; Yang, F.; Leung, W.; Xu, C. Ultrasound-Responsive Materials for Drug/Gene Delivery. Front. Pharmacol. 2020, 10, 1650. [Google Scholar] [CrossRef] [PubMed]
- Kripfgans, O.D.; Fabiilli, M.L.; Carson, P.L.; Fowlkes, J.B. On the acoustic vaporization of micrometer-sized droplets. J. Acoust. Soc. Am. 2004, 116, 272–281. [Google Scholar] [CrossRef] [PubMed]
- Zullino, S.; Argenziano, M.; Ansari, S.; Ciprian, R.; Nasi, L.; Albertini, F.; Cavalli, R.; Guiot, C. Superparamagnetic Oxygen-Loaded Nanobubbles to Enhance Tumor Oxygenation During Hyperthermia. Front. Pharmacol. 2019, 10, 1001. [Google Scholar] [CrossRef]
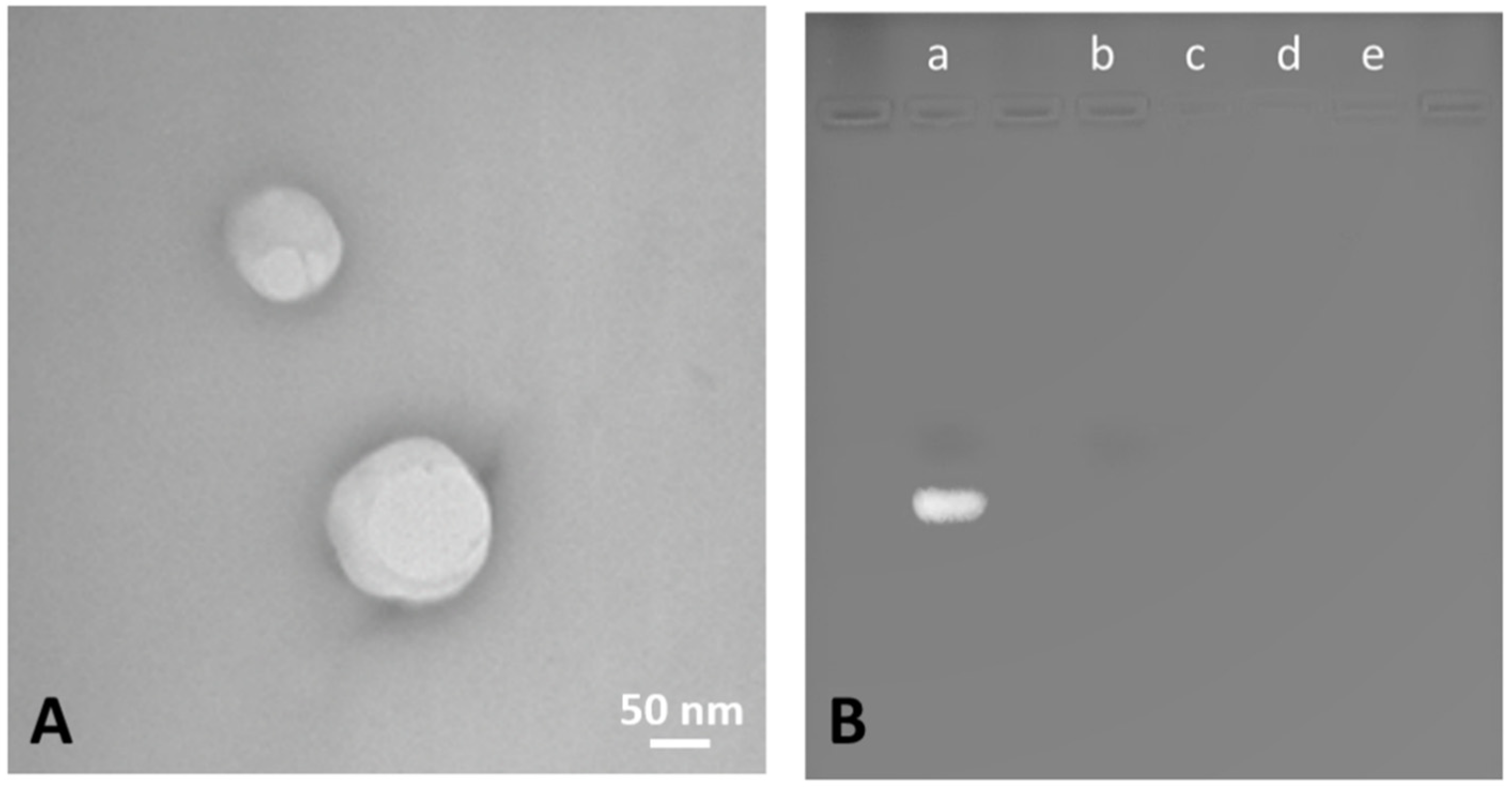
| Formulation | Average Diameter ± SD (nm) | Polydispersity Index (PDI) ± SD | Zeta Potential ± SD (mV) | Osmolarity (mOsm) |
|---|---|---|---|---|
| NBs | 102.3 ± 2.3 | 0.221 ± 0.02 | 27.5 ± 2.5 | 280 |
| Fluorescent NBs | 98.4 ± 5.0 | 0.220 ± 0.03 | 26.8 ± 2.2 | 282 |
| siNrf2-NBs | 97.5 ± 4.6 | 0.218 ± 0.02 | 26.2 ± 1.8 | 280 |
| siRNAneg-NBs | 100.2 ± 3.1 | 0.222 ± 0.01 | 26.4 ± 2.1 | 278 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Argenziano, M.; Bessone, F.; Dianzani, C.; Cucci, M.A.; Grattarola, M.; Pizzimenti, S.; Cavalli, R. Ultrasound-Responsive Nrf2-Targeting siRNA-Loaded Nanobubbles for Enhancing the Treatment of Melanoma. Pharmaceutics 2022, 14, 341. https://doi.org/10.3390/pharmaceutics14020341
Argenziano M, Bessone F, Dianzani C, Cucci MA, Grattarola M, Pizzimenti S, Cavalli R. Ultrasound-Responsive Nrf2-Targeting siRNA-Loaded Nanobubbles for Enhancing the Treatment of Melanoma. Pharmaceutics. 2022; 14(2):341. https://doi.org/10.3390/pharmaceutics14020341
Chicago/Turabian StyleArgenziano, Monica, Federica Bessone, Chiara Dianzani, Marie Angèle Cucci, Margherita Grattarola, Stefania Pizzimenti, and Roberta Cavalli. 2022. "Ultrasound-Responsive Nrf2-Targeting siRNA-Loaded Nanobubbles for Enhancing the Treatment of Melanoma" Pharmaceutics 14, no. 2: 341. https://doi.org/10.3390/pharmaceutics14020341
APA StyleArgenziano, M., Bessone, F., Dianzani, C., Cucci, M. A., Grattarola, M., Pizzimenti, S., & Cavalli, R. (2022). Ultrasound-Responsive Nrf2-Targeting siRNA-Loaded Nanobubbles for Enhancing the Treatment of Melanoma. Pharmaceutics, 14(2), 341. https://doi.org/10.3390/pharmaceutics14020341








