Endogenous Bornavirus-like Elements in Bats: Evolutionary Insights from the Conserved Riboviral L-Gene in Microbats and Its Antisense Transcription in Myotis daubentonii
Abstract
1. Introduction
2. Materials and Methods
2.1. Bat Genome References and Gene Annotation Construction
2.2. Processing of RNA-Seq Data, Refinement of CV.10-MyoDau Annotation, and Differential Gene Expression
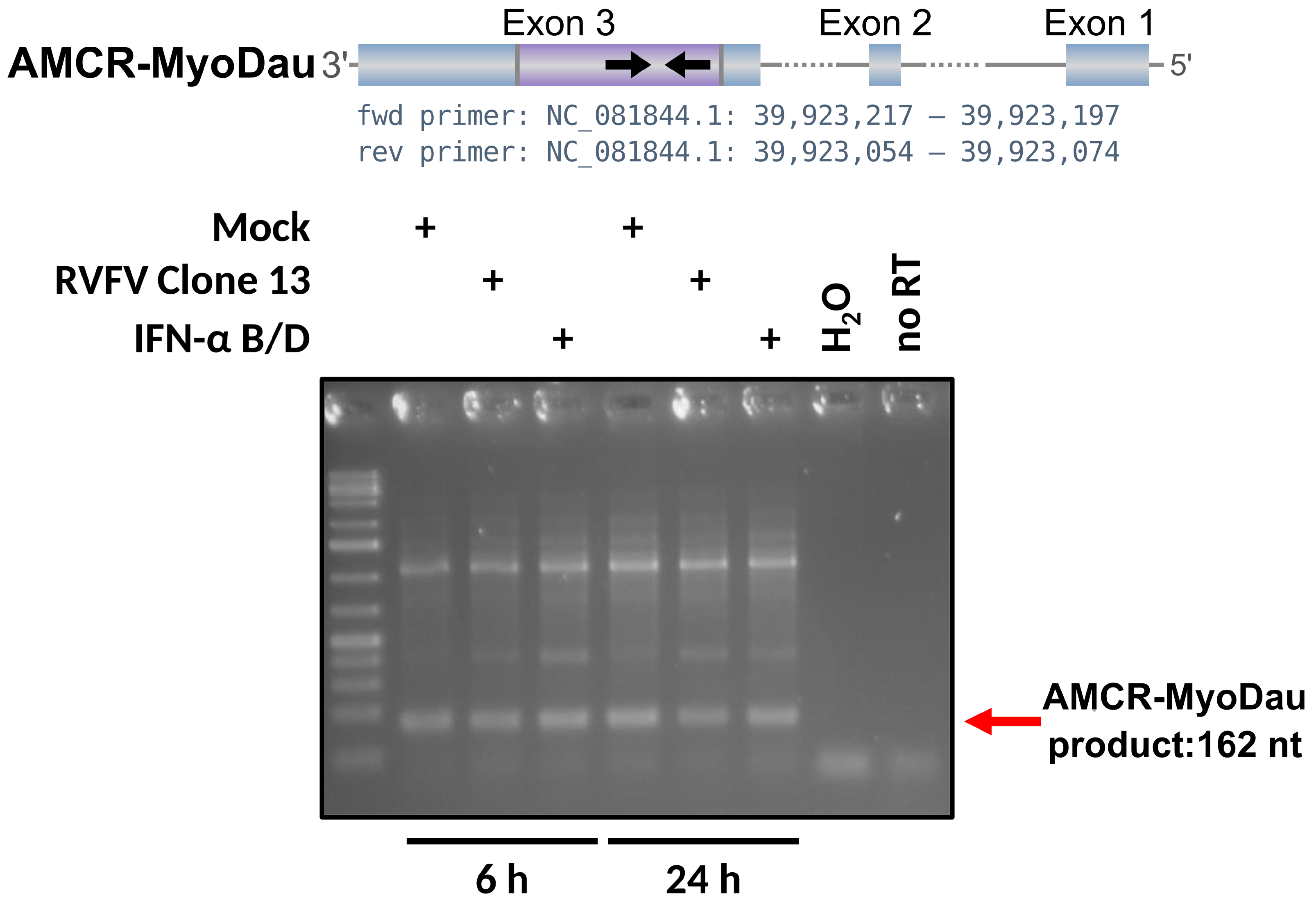
2.3. In Vitro Validation of EVE Candidate
2.4. Synteny Analysis to Confirm the Conservation of AMCR-MyoDau and CV.10-MyoDau
2.5. Identification and Further Investigation of the Previously Described EBLL-IG
2.6. Multiple Sequence Alignments of the Three Exons of AMCR in Various Bat Species
3. Results
3.1. A Novel EVE and Its Antisense Transcription in Myotis daubentonii: EBLL-Cultervirus.10-MyoDau and AMCR-MyoDau
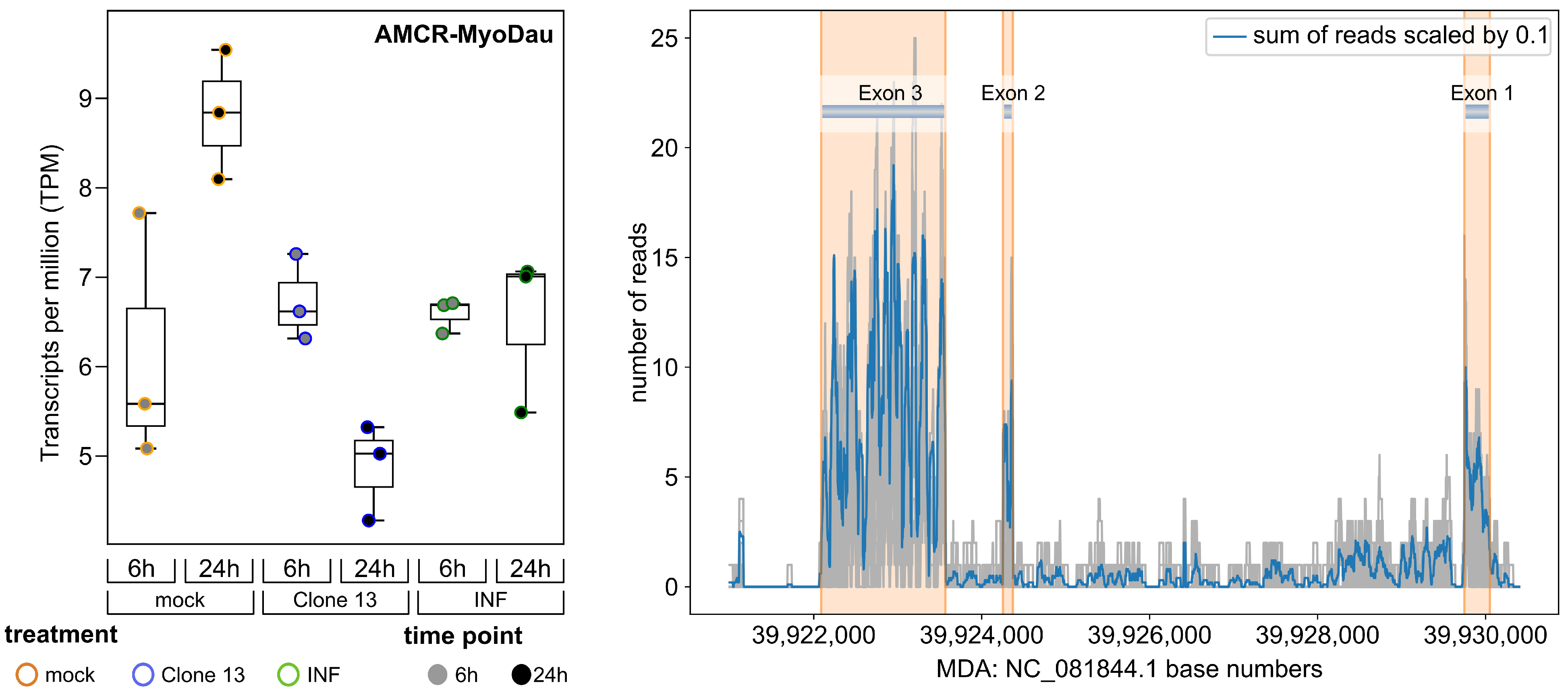
3.2. AMCR-MyoDau Shows Weak but Constant Expression in a Myotis daubentonii Cell Line under Mock, Interferon-Induced, and Virus-Infected Conditions
3.3. Synteny Patterns of AMCR and CV.10-MyoDau across Various Bat Species
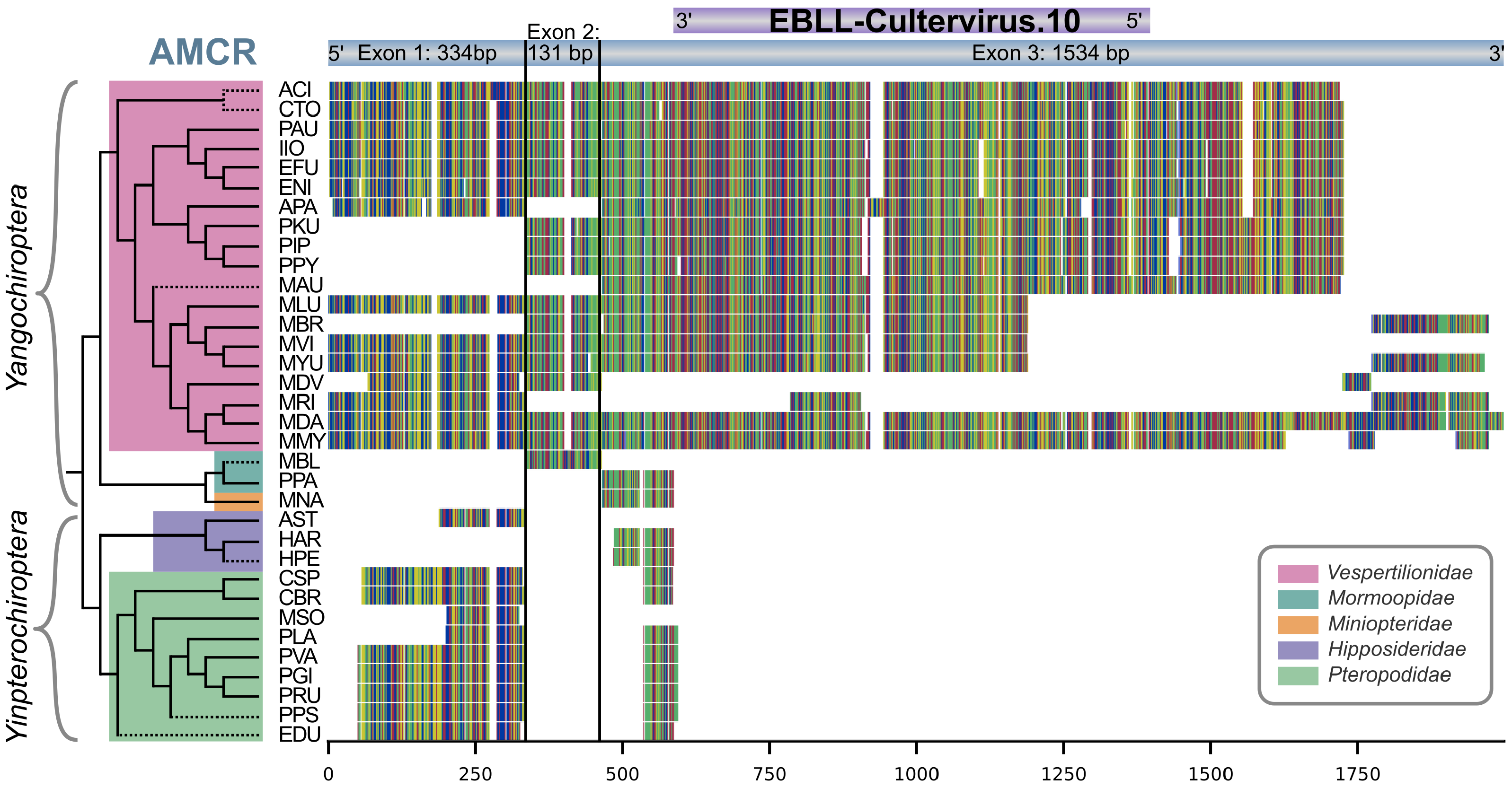
3.4. Comparative Genome Analysis Shows AMCR Sequence Similarities in 34 Bat Species
3.5. Partial Occurrence of AMCR across Multiple Bat Families and Evolutionary Conservation of CV.10 in Vespertilionidae
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Calisher, C.H.; Childs, J.E.; Field, H.E.; Holmes, K.V.; Schountz, T. Bats: Important reservoir hosts of emerging viruses. Clin. Microbiol. Rev. 2006, 19, 531–545. [Google Scholar] [CrossRef]
- Teeling, E.C.; Vernes, S.C.; Dávalos, L.M.; Ray, D.A.; Gilbert, M.T.P.; Myers, E.; Consortium, B. Bat biology, genomes, and the Bat1K Project: To generate Chromosome-Level genomes for all living bat species. Annu. Rev. Anim. Biosci. 2018, 6, 23–46. [Google Scholar] [CrossRef]
- Simmons, N.B. An Eocene big bang for bats. Science 2005, 307, 527–528. [Google Scholar] [CrossRef]
- Prat, Y.; Azoulay, L.; Dor, R.; Yovel, Y. Crowd vocal learning induces vocal dialects in bats: Playback of conspecifics shapes fundamental frequency usage by pups. PLoS Biol. 2017, 15, e2002556. [Google Scholar] [CrossRef]
- Huang, Z.; Jebb, D.; Teeling, E.C. Blood miRNomes and transcriptomes reveal novel longevity mechanisms in the long-lived bat, Myotis myotis. BMC Genom. 2016, 17, 906. [Google Scholar] [CrossRef]
- Foley, N.M.; Hughes, G.M.; Huang, Z.; Clarke, M.; Jebb, D.; Whelan, C.V.; Petit, E.J.; Touzalin, F.; Farcy, O.; Jones, G.; et al. Growing old, yet staying young: The role of telomeres in bats’ exceptional longevity. Sci. Adv. 2018, 4, eaao0926. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Whelan, C.V.; Foley, N.M.; Jebb, D.; Touzalin, F.; Petit, E.J.; Puechmaille, S.J.; Teeling, E.C. Longitudinal comparative transcriptomics reveals unique mechanisms underlying extended healthspan in bats. Nat. Ecol. Evol. 2019, 3, 1110–1120. [Google Scholar] [CrossRef] [PubMed]
- Smith, I.; Wang, L.F. Bats and their virome: An important source of emerging viruses capable of infecting humans. Curr. Opin. Virol. 2013, 3, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Baker, M.L.; Kulcsar, K.; Misra, V.; Plowright, R.; Mossman, K. Novel insights into immune systems of bats. Front. Immunol. 2020, 11, 26. [Google Scholar] [CrossRef]
- Olival, K.J.; Hosseini, P.R.; Zambrana-Torrelio, C.; Ross, N.; Bogich, T.L.; Daszak, P. Host and viral traits predict zoonotic spillover from mammals. Nature 2017, 546, 646–650. [Google Scholar] [CrossRef]
- Wang, L.F.; Walker, P.J.; Poon, L.L. Mass extinctions, biodiversity and mitochondrial function: Are bats “special” as reservoirs for emerging viruses? Curr. Opin. Virol. 2011, 1, 649–657. [Google Scholar] [CrossRef]
- Brook, C.E.; Dobson, A.P. Bats as “special” reservoirs for emerging zoonotic pathogens. Trends Microbiol. 2015, 23, 172–180. [Google Scholar] [CrossRef] [PubMed]
- Hölzer, M.; Krähling, V.; Amman, F.; Barth, E.; Bernhart, S.H.; Carmelo, V.A.O.; Collatz, M.; Doose, G.; Eggenhofer, F.; Ewald, J.; et al. Differential transcriptional responses to Ebola and Marburg virus infection in bat and human cells. Sci. Rep. 2016, 6, 34589. [Google Scholar] [CrossRef] [PubMed]
- Anthony, S.J.; Johnson, C.K.; Greig, D.J.; Kramer, S.; Che, X.; Wells, H.; Hicks, A.L.; Joly, D.O.; Wolfe, N.D.; Daszak, P.; et al. Global patterns in coronavirus diversity. Virus Evol. 2017, 3. [Google Scholar] [CrossRef] [PubMed]
- Andersen, K.G.; Rambaut, A.; Lipkin, W.I.; Holmes, E.C.; Garry, R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020, 26, 450–452. [Google Scholar] [CrossRef]
- Wu, Z.; Han, Y.; Wang, Y.; Liu, B.; Zhao, L.; Zhang, J.; Su, H.; Zhao, W.; Liu, L.; Bai, S.; et al. A comprehensive survey of bat sarbecoviruses across China in relation to the origins of SARS-CoV and SARS-CoV-2. Natl. Sci. Rev. 2023, 10, nwac213. [Google Scholar] [CrossRef] [PubMed]
- Papenfuss, A.T.; Baker, M.L.; Feng, Z.P.; Tachedjian, M.; Crameri, G.; Cowled, C.; Ng, J.; Janardhana, V.; Field, H.E.; Wang, L.F. The immune gene repertoire of an important viral reservoir, the Australian black flying fox. BMC Genom. 2012, 13, 261. [Google Scholar] [CrossRef] [PubMed]
- Power, M.L.; Foley, N.M.; Jones, G.; Teeling, E.C. Taking flight: An ecological, evolutionary and genomic perspective on bat telomeres. Mol. Ecol. 2022, 31, 6053–6068. [Google Scholar] [CrossRef] [PubMed]
- Luis, A.D.; Hayman, D.T.; O’Shea, T.J.; Cryan, P.M.; Gilbert, A.T.; Pulliam, J.R.; Mills, J.N.; Timonin, M.E.; Willis, C.K.; Cunningham, A.A.; et al. A comparison of bats and rodents as reservoirs of zoonotic viruses: Are bats special? Proc. R. Soc. B Biol. Sci. 2013, 280, 20122753. [Google Scholar] [CrossRef]
- Wang, L.F.; Anderson, D.E. Viruses in bats and potential spillover to animals and humans. Curr. Opin. Virol. 2019, 34, 79–89. [Google Scholar] [CrossRef]
- Skirmuntt, E.C.; Escalera-Zamudio, M.; Teeling, E.C.; Smith, A.; Katzourakis, A. The potential role of endogenous viral elements in the evolution of bats as reservoirs for zoonotic viruses. Annu. Rev. Virol. 2020, 7, 103–119. [Google Scholar] [CrossRef] [PubMed]
- O’shea, T.J.; Cryan, P.M.; Cunningham, A.A.; Fooks, A.R.; Hayman, D.T.; Luis, A.D.; Peel, A.J.; Plowright, R.K.; Wood, J.L. Bat flight and zoonotic viruses. Emerg. Infect. Dis. 2014, 20, 741. [Google Scholar] [CrossRef] [PubMed]
- Hölzer, M.; Schoen, A.; Wulle, J.; Müller, M.A.; Drosten, C.; Marz, M.; Weber, F. Virus-and interferon alpha-induced transcriptomes of cells from the microbat Myotis daubentonii. Iscience 2019, 19, 647–661. [Google Scholar] [CrossRef]
- Pavlovich, S.S.; Lovett, S.P.; Koroleva, G.; Guito, J.C.; Arnold, C.E.; Nagle, E.R.; Kulcsar, K.; Lee, A.; Thibaud-Nissen, F.; Hume, A.J.; et al. The Egyptian rousette genome reveals unexpected features of bat antiviral immunity. Cell 2018, 173, 1098–1110. [Google Scholar] [CrossRef] [PubMed]
- Hölzer, M.; Marz, M. De novo transcriptome assembly: A comprehensive cross-species comparison of short-read RNA-Seq assemblers. GigaScience 2019, 8, giz039. [Google Scholar] [CrossRef] [PubMed]
- Jayaprakash, A.D.; Ronk, A.J.; Prasad, A.N.; Covington, M.F.; Stein, K.R.; Schwarz, T.M.; Hekmaty, S.; Fenton, K.A.; Geisbert, T.W.; Basler, C.F.; et al. Filovirus infection induces an anti-inflammatory state in Rousettus bats. bioRxiv 2020. [Google Scholar] [CrossRef]
- Subudhi, S.; Rapin, N.; Misra, V. Immune system modulation and viral persistence in bats: Understanding viral spillover. Viruses 2019, 11, 192. [Google Scholar] [CrossRef]
- Cowled, C.; Stewart, C.R.; Likic, V.A.; Friedländer, M.R.; Tachedjian, M.; Jenkins, K.A.; Tizard, M.L.; Cottee, P.; Marsh, G.A.; Zhou, P.; et al. Characterisation of novel microRNAs in the Black flying fox (Pteropus alecto) by deep sequencing. BMC Genom. 2014, 15, 682. [Google Scholar] [CrossRef]
- Mostajo Berrospi, N.; Lataretu, M.; Krautwurst, S.; Mock, F.; Desirò, D.; Lamkiewicz, K.; Collatz, M.; Schoen, A.; Weber, F.; Marz, M.; et al. A comprehensive annotation and differential expression analysis of short and long non-coding RNAs in 16 bat genomes. NAR Genom. Bioinform. 2019, 2, lqz006. [Google Scholar]
- Edwards, M.R.; Liu, H.; Shabman, R.S.; Ginell, G.M.; Luthra, P.; Ramanan, P.; Keefe, L.J.; Köllner, B.; Amarasinghe, G.K.; Taylor, D.J.; et al. Conservation of structure and immune antagonist functions of filoviral VP35 homologs present in microbat genomes. Cell Rep. 2018, 24, 861–872.e6. [Google Scholar] [CrossRef]
- Ophinni, Y.; Palatini, U.; Hayashi, Y.; Parrish, N.F. piRNA-guided CRISPR-like immunity in eukaryotes. Trends Immunol. 2019, 40, 998–1010. [Google Scholar] [CrossRef]
- Feschotte, C.; Gilbert, C. Endogenous viruses: Insights into viral evolution and impact on host biology. Nat. Rev. Genet. 2012, 13, 283–296. [Google Scholar] [CrossRef] [PubMed]
- Belyi, V.A.; Levine, A.J.; Skalka, A.M. Unexpected inheritance: Multiple integrations of ancient bornavirus and ebolavirus/marburgvirus sequences in vertebrate genomes. PLoS Pathog. 2010, 6, e1001030. [Google Scholar] [CrossRef]
- Weiss, R. Spontaneous virus production from “non-virus producing” Rous sarcoma cells. Virology 1967, 32, 719–723. [Google Scholar] [CrossRef]
- Johnson, W.E. Origins and evolutionary consequences of ancient endogenous retroviruses. Nat. Rev. Microbiol. 2019, 17, 355–370. [Google Scholar] [CrossRef]
- Bellas, C.; Hackl, T.; Plakolb, M.S.; Koslová, A.; Fischer, M.G.; Sommaruga, R. Large-scale invasion of unicellular eukaryotic genomes by integrating DNA viruses. Proc. Natl. Acad. Sci. USA 2023, 120, e2300465120. [Google Scholar] [CrossRef]
- Klenerman, P.; Hengartner, H.; Zinkernagel, R.M. A non-retroviral RNA virus persists in DNA form. Nature 1997, 390, 298–301. [Google Scholar] [CrossRef]
- Taylor, D.J.; Bruenn, J. The evolution of novel fungal genes from non-retroviral RNA viruses. BMC Biol. 2009, 7, 88. [Google Scholar] [CrossRef]
- Horie, M.; Honda, T.; Suzuki, Y.; Kobayashi, Y.; Daito, T.; Oshida, T.; Ikuta, K.; Jern, P.; Gojobori, T.; Coffin, J.M.; et al. Endogenous non-retroviral RNA virus elements in mammalian genomes. Nature 2010, 463, 84–87. [Google Scholar] [CrossRef] [PubMed]
- Horie, M.; Tomonaga, K. Non-retroviral fossils in vertebrate genomes. Viruses 2011, 3, 1836–1848. [Google Scholar] [CrossRef]
- Fort, P.; Albertini, A.; Van-Hua, A.; Berthomieu, A.; Roche, S.; Delsuc, F.; Pasteur, N.; Capy, P.; Gaudin, Y.; Weill, M. Fossil rhabdoviral sequences integrated into arthropod genomes: Ontogeny, evolution, and potential functionality. Mol. Biol. Evol. 2012, 29, 381–390. [Google Scholar] [CrossRef]
- Takahashi, H.; Fukuhara, T.; Kitazawa, H.; Kormelink, R. Virus latency and the impact on plants. Front. Microbiol. 2019, 10, 2764. [Google Scholar] [CrossRef]
- Kapusta, A.; Suh, A. Evolution of bird genomes—A transposon’s-eye view. Ann. N. Y. Acad. Sci. 2017, 1389, 164–185. [Google Scholar] [CrossRef]
- Wallau, G.L. RNA virus EVEs in insect genomes. Curr. Opin. Insect Sci. 2022, 49, 42–47. [Google Scholar] [CrossRef]
- Horie, M.; Kobayashi, Y.; Suzuki, Y.; Tomonaga, K. Comprehensive analysis of endogenous bornavirus-like elements in eukaryote genomes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013, 368, 20120499. [Google Scholar] [CrossRef]
- Afonso, C.L.; Amarasinghe, G.K.; Bányai, K.; Bào, Y.; Basler, C.F.; Bavari, S.; Bejerman, N.; Blasdell, K.R.; Briand, F.X.; Briese, T.; et al. Taxonomy of the order Mononegavirales: Update 2016. Arch. Virol. 2016, 161, 2351–2360. [Google Scholar] [CrossRef]
- Horie, M.; Kobayashi, Y.; Honda, T.; Fujino, K.; Akasaka, T.; Kohl, C.; Wibbelt, G.; Mühldorfer, K.; Kurth, A.; Müller, M.A.; et al. An RNA-dependent RNA polymerase gene in bat genomes derived from an ancient negative-strand RNA virus. Sci. Rep. 2016, 6, 25873. [Google Scholar] [CrossRef]
- Kawasaki, J.; Kojima, S.; Mukai, Y.; Tomonaga, K.; Horie, M. 100-My history of bornavirus infections hidden in vertebrate genomes. Proc. Natl. Acad. Sci. USA 2021, 118, e2026235118. [Google Scholar] [CrossRef]
- Garcia, B.C.B.; Mukai, Y.; Tomonaga, K.; Horie, M. The hidden diversity of ancient bornaviral sequences from X and P genes in vertebrate genomes. Virus Evol. 2023, 9, vead038. [Google Scholar] [CrossRef]
- Mukai, Y.; Horie, M.; Kojima, S.; Kawasaki, J.; Maeda, K.; Tomonaga, K. An endogenous bornavirus-like nucleoprotein in miniopterid bats retains the RNA-binding properties of the original viral protein. FEBS Lett. 2022, 596, 323–337. [Google Scholar] [CrossRef]
- Frank, J.A.; Feschotte, C. Co-option of endogenous viral sequences for host cell function. Curr. Opin. Virol. 2017, 25, 81–89. [Google Scholar] [CrossRef]
- Roberts, R.M.; Ezashi, T.; Schulz, L.C.; Sugimoto, J.; Schust, D.J.; Khan, T.; Zhou, J. Syncytins expressed in human placental trophoblast. Placenta 2021, 113, 8–14. [Google Scholar] [CrossRef]
- Lokossou, A.G.; Toudic, C.; Barbeau, B. Implication of human endogenous retrovirus envelope proteins in placental functions. Viruses 2014, 6, 4609–4627. [Google Scholar] [CrossRef]
- Boyd, M.T.; Bax, C.M.; Bax, B.E.; Bloxam, D.L.; Weiss, R.A. The human endogenous retrovirus ERV-3 is upregulated in differentiating placental trophoblast cells. Virology 1993, 196, 905–909. [Google Scholar] [CrossRef]
- Venables, P.J.; Brookes, S.M.; Griffiths, D.; Weiss, R.A.; Boyd, M.T. Abundance of an endogenous retroviral envelope protein in placental trophoblasts suggests a biological function. Virology 1995, 211, 589–592. [Google Scholar] [CrossRef]
- Robertson, W.; Warner, B. The ultrastructure of the human placental bed. J. Pathol. 1974, 112, 203–211. [Google Scholar] [CrossRef]
- Hertig, A.T. The primary human oocyte: Some observations on the fine structure of Balbiani’s vitelline body and the origin of the annulate lamellae. Am. J. Anat. 1968, 122, 107–137. [Google Scholar] [CrossRef]
- NCBI, R.C. Database Resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2017, 45, D12. [Google Scholar]
- Shumate, A.; Salzberg, S.L. Liftoff: Accurate mapping of gene annotations. Bioinformatics 2021, 37, 1639–1643. [Google Scholar] [CrossRef]
- Agnarsson, I.; Zambrana-Torrelio, C.M.; Flores-Saldana, N.P.; May-Collado, L.J. A time-calibrated species-level phylogeny of bats (Chiroptera, Mammalia). PLoS Curr. 2011, 3. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Thorvaldsdóttir, H.; Robinson, J.T.; Mesirov, J.P. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 2013, 14, 178–192. [Google Scholar] [CrossRef]
- Lataretu, M.; Hölzer, M. RNAflow: An effective and simple RNA-seq differential gene expression pipeline using nextflow. Genes 2020, 11, 1487. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef]
- Stephen, F.A. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar]
- Mukai, Y.; Horie, M.; Tomonaga, K. Systematic estimation of insertion dates of endogenous bornavirus-like elements in vesper bats. J. Vet. Med. Sci. 2018, 80, 1356–1363. [Google Scholar] [CrossRef]
- Yamada, K.D.; Tomii, K.; Katoh, K. Application of the MAFFT sequence alignment program to large data—Reexamination of the usefulness of chained guide trees. Bioinformatics 2016, 32, 3246–3251. [Google Scholar] [CrossRef]
- Tumescheit, C.; Firth, A.E.; Brown, K. CIAlign: A highly customisable command line tool to clean, interpret and visualise multiple sequence alignments. PeerJ 2022, 10, e12983. [Google Scholar] [CrossRef]
- Cui, J.; Wang, L.F. Genomic mining reveals deep evolutionary relationships between Bornaviruses and bats. Viruses 2015, 7, 5792–5800. [Google Scholar] [CrossRef] [PubMed]
- Blanco-Melo, D.; Campbell, M.A.; Zhu, H.; Dennis, T.P.; Modha, S.; Lytras, S.; Hughes, J.; Gatseva, A.; Gifford, R.J. A novel approach to exploring the dark genome and its application to mapping of the vertebrate virus fossil record. Genome Biol. 2024, 25, 120. [Google Scholar] [CrossRef] [PubMed]
- Olschewski, S.; Cusack, S.; Rosenthal, M. The cap-snatching mechanism of bunyaviruses. Trends Microbiol. 2020, 28, 293–303. [Google Scholar] [CrossRef]
- Gifford, R.J.; Blomberg, J.; Coffin, J.M.; Fan, H.; Heidmann, T.; Mayer, J.; Stoye, J.; Tristem, M.; Johnson, W.E. Nomenclature for endogenous retrovirus (ERV) loci. Retrovirology 2018, 15, 59. [Google Scholar] [CrossRef]
- Teeling, E.C.; Springer, M.S.; Madsen, O.; Bates, P.; O’brien, S.J.; Murphy, W.J. A molecular phylogeny for bats illuminates biogeography and the fossil record. Science 2005, 307, 580–584. [Google Scholar] [CrossRef] [PubMed]


| Species | Abb. | #Con|#Chr | N50 | Size | NCBI Acc. | Year | Family |
|---|---|---|---|---|---|---|---|
| [Mb] | [Gb] | ||||||
| Yangochiroptera | |||||||
| Aeorestes cinereus | ACI | 2536|0 | 35.1 | 2.1 | GCA_011751065.1 | 2020 | Vespertilionidae |
| Corynorhinus townsendii | CTO | 182|0 | 177.8 | 2.0 | GCA_026230045.1 | 2022 | Vespertilionidae |
| Plecotus auritus | PAU | 5570|0 | 186.5 | 2.2 | GCA_963455325.1 | 2023 | Vespertilionidae |
| Nycticeius humeralis | NHU | 1,676,240|0 | 0.015 | 2.8 | GCA_007922795.1 | 2019 | Vespertilionidae |
| Ia io | IIO | 2008|0 | 105.8 | 2.1 | GCA_025583905.1 | 2022 | Vespertilionidae |
| Eptesicus fuscus | EFU | 48|25 | 102.8 | 2.0 | GCF_027574615.1 * | 2023 | Vespertilionidae |
| Eptesicus nilssonii | ENI | 726|0 | 102.4 | 2.0 | GCA_030846915.1 | 2023 | Vespertilionidae |
| Lasiurus borealis | LBO | 518,900|0 | 0.039 | 2.9 | GCA_004026805.1 | 2019 | Vespertilionidae |
| Antrozous pallidus | APA | 93|23 | 114.6 | 2.1 | GCA_027563665.1 | 2023 | Vespertilionidae |
| Pipistrellus kuhlii | PKU | 202|0 | 80.2 | 1.8 | GCF_014108245.1 * | 2020 | Vespertilionidae |
| Pipistrellus pipistrellus | PIP | 323|0 | 94.9 | 1.8 | GCA_903992545.1 | 2020 | Vespertilionidae |
| Pipistrellus pygmaeus | PPY | 243|0 | 89.5 | 1.9 | GCA_949987585.1 | 2023 | Vespertilionidae |
| Murina aurata feae | MAU | 880,177|0 | 0.026 | 2.3 | GCA_004026665.1 | 2019 | Vespertilionidae |
| Myotis lucifugu | MLU | 11,654|0 | 4.3 | 2.0 | GCF_000147115.1 * | 2010 | Vespertilionidae |
| Myotis brandtii | MBR | 169,750|0 | 3.2 | 2.1 | GCF_000412655.1 * | 2013 | Vespertilionidae |
| Myotis vivesi | MVI | 64,503|0 | 91.8 | 2.1 | GCA_035771395.1 | 2024 | Vespertilionidae |
| Myotis yumanensis | MYU | 476|0 | 99.1 | 2.0 | GCA_028538775.1 | 2023 | Vespertilionidae |
| Myotis davidii | MDV | 101,769|0 | 3.5 | 2.1 | GCF_000327345.1 * | 2012 | Vespertilionidae |
| Myotis ricketti | MRI | 105|0 | 80 | 2.0 | GCA_036010255.1 | 2024 | Vespertilionidae |
| Myotis daubentonii | MDA | 121|23 | 102.2 | 2.1 | GCF_963259705.1 * | 2023 | Vespertilionidae |
| Myotis myotis | MMY | 93|0 | 94.4 | 2.0 | GCF_014108235.1 * | 2020 | Vespertilionidae |
| Molossus nigricans | MNI | 146|0 | 81.9 | 2.4 | GCA_026936385.1 | 2022 | Molossidae |
| Molossus alvarezi | MAL | 187|0 | 113.9 | 2.4 | GCA_031001765.1 | 2023 | Molossidae |
| Molossus molossus | MMO | 60|0 | 110.7 | 2.3 | GCF_014108415.1 * | 2020 | Molossidae |
| Tadarida brasiliensis | TBR | 148|25 | 111.1 | 2.3 | GCA_030848825.1 | 2023 | Molossidae |
| Rhynchonycteris naso | RNA | 50|0 | 286.1 | 2.4 | GCA_031021685.1 | 2023 | Eallonurida |
| Sturnira hondurensis | SHO | 25,881|0 | 10.2 | 2.1 | GCF_014824575.3 * | 2022 | Phyllostomidae |
| Tonatia saurophila | TSA | 249,810 | 0.166 | 2.1 | GCA_004024845.1 | 2019 | Phyllostomidae |
| Trachops cirrhosus | TCI | 396,519 | 124.5 | 2.2 | GCA_028533065.1 | 2023 | Phyllostomidae |
| Micronycteris hirsuta | MHI | 550,090|0 | 0.069 | 2.3 | GCA_004026765.1 | 2019 | Phyllostomidae |
| Carollia perspicillata | CPE | 1,925,339|0 | 0.010 | 2.7 | GCA_004027735.1 | 2019 | Phyllostomidae |
| Anoura caudifer | ACU | 337,255|0 | 0.143 | 2.2 | GCA_004027475.1 | 2019 | Phyllostomidae |
| Desmodus rotundus | DRO | 573|14 | 160.1 | 2.1 | GCF_022682495.1 * | 2022 | Phyllostomidae |
| Phyllostomus discolor | PDI | 78|17 | 171.7 | 2.1 | GCF_004126475.2 * | 2020 | Phyllostomidae |
| Phyllostomus hastatus | PHA | 534|0 | 39.2 | 2.1 | GCF_019186645.2 * | 2021 | Phyllostomidae |
| Macrotus californicus | MCA | 1,128,787|0 | 0.017 | 2.2 | GCA_007922815.1 | 2019 | Phyllostomidae |
| Artibeus jamaicensis | AJA | 868|0 | 22.1 | 2.1 | GCF_021234435.1 * | 2021 | Phyllostomidae |
| Pteronotus parnellii | PPA | 333|0 | 31.5 | 2.1 | GCF_021234165.1 * | 2021 | Mormoopidae |
| Mormoops blainvillei | MBL | 205,259|0 | 0.156 | 2.1 | GCA_004026545.1 | 2019 | Mormoopidae |
| Noctilio leporinus | NLE | 298,222|0 | 0.136 | 2.1 | GCA_004026585.1 | 2019 | Noctilionidae |
| Miniopterus natalensis | MNA | 1269|0 | 4.3 | 1.8 | GCF_001595765.1 * | 2016 | Miniopteridae |
| Miniopterus schreibersii | MSC | 177,620|0 | 0.109 | 1.8 | GCA_004026525.1 | 2019 | Miniopteridae |
| Yinpterochiroptera | |||||||
| Megaderma lyra | MLY | 1,902,801|0 | 0.072 | 2.6 | GCA_004026885.1 | 2019 | Megadermatidae |
| Craseonycteris thonglongyai | CTH | 1,224,256|0 | 0.026 | 2.3 | GCA_004027555.1 | 2019 | Craseonycteridae |
| Aselliscus stoliczkanus | AST | 191|16 | 162 | 2.2 | GCA_033961575.1 | 2023 | Hipposideridae |
| Hipposideros pendleburyi | HPE | 28,685|0 | 15.4 | 2.2 | GCA_021464545.1 | 2022 | Hipposideridae |
| Hipposideros armiger | HAR | 7571|0 | 2.3 | 2.2 | GCF_001890085.2 * | 2016 | Hipposideridae |
| Hipposideros larvatus | HLA | 69|18 | 185.5 | 2.3 | GCA_031876335.1 | 2023 | Hipposideridae |
| Rhinolophus ferrumequinum | RFE | 50|0 | 92 | 2.1 | GCA_014108255.1 | 2020 | Rhinolophidae |
| Cynopterus sphinx | CSP | 181|17 | 145.2 | 1.9 | GCA_030015415.1 | 2023 | Pteropodidae |
| Cynopterus brachyotis | CBR | 48,006|0 | 0.251 | 1.8 | GCA_009793145.1 | 2019 | Pteropodidae |
| Macroglossus sobrinus | MSO | 171,985|0 | 0.453 | 1.9 | GCA_004027375.1 | 2019 | Pteropodidae |
| Pteropus alecto | PLA | 65,598|0 | 6 | 2.0 | GCF_000325575.1 * | 2013 | Pteropodidae |
| Pteropus vampyrus | PVA | 36,094|0 | 6 | 2.2 | GCF_000151845.1 * | 2014 | Pteropodidae |
| Pteropus giganteus | PGI | 16,113|0 | 18.9 | 2.0 | GCF_902729225.1 * | 2020 | Pteropodidae |
| Pteropus rufus | PRU | 469,091|19 | 110.5 | 2.1 | GCA_028533765.1 | 2023 | Pteropodidae |
| Pteropus pselaphon | PPS | 7513|0 | 0.770 | 1.9 | GCA_014363405.1 | 2020 | Pteropodidae |
| Eidolon dupreanum | EDU | 1,191,098|17 | 101.6 | 2.3 | GCA_028627145.1 | 2023 | Pteropodidae |
| Eidolon helvum | EHE | 133,538|0 | 0.028 | 1.8 | GCA_000465285.1 | 2013 | Pteropodidae |
| Eonycteris spelaea | ESP | 4469|0 | 8 | 2.0 | GCA_003508835.1 | 2018 | Pteropodidae |
| Rousettus madagascariensis | RMA | 1,467,186|18 | 85.8 | 2.3 | GCA_028533395.1 | 2023 | Pteropodidae |
| Rousettus leschenaultii | RLE | 8141|0 | 32.7 | 1.9 | GCA_015472975.1 | 2020 | Pteropodidae |
| Rousettus aegyptiacus | RAE | 29|0 | 113.8 | 1.9 | GCF_014176215.1 * | 2020 | Pteropodidae |
| Name EVE | Species | NCBI acc. | Start-Stop | Literature |
|---|---|---|---|---|
| CV.10-MyoDau | Myotis daubentonii | NC_081844.1 | 39,922,667–39,923,434 | |
| CV.10-AeqCin | Aeorestes cinereus | JAAGEH010000014.1 | 12,648,154–12,648,930 | - |
| CV.10-CorTow | Corynorhinus townsendii | JAPDVU010000006.1 | 98,426,944–98,427,716 | - |
| CV.10-PleAur | Plecotus auritus | CAUOHH010000436.1 | 107,056–107,829 | - |
| CV.10-IaIo | Ia io | JAJQQW010000006.1 | 73,804,507–73,805,270 | - |
| CV.10-EptFus | Eptesicus fuscus | NC_072478.1 | 36,387,431–36,388,196 | [48,72] |
| CV.10-EptNil | Eptesicus nilssonii | JAULJE010000005.1 | 36,472,464–36,473,231 | - |
| CV.10-AntPal | Antrozous pallidus | CM050516.1 | 75,692,535–75,693,314 | [72] |
| CV.10-PipKuh | Pipistrellus kuhlii | NW_023425584.1 | 64,293,707–64,294,472 | [72] |
| CV.10-PipPip | Pipistrellus pipistrellus | LR862361.1 | 31,459,988–31,460,753 | [72] |
| CV.10-PipPym | Pipistrellus pygmaeus | OX465307.1 | 64,495,766–64,496,530 | - |
| CV.10-MurAur | Murina aurata feae | PVJC01025922.1 | 14,138–14,917 | [72] |
| CV.10-MyoLuc | Myotis lucifugus | NW_005871049.1 | 34,243,866–34,244,427 | [48,72] |
| CV.10-MyoBra | Myotis brandtii | NW_005370908.1 | 1,501,243–1,501,804 | [48,72] |
| CV.10-MyoViv | Myotis vivesi | JAWPEG010000011.1 | 69,888,777–69,889,333 | - |
| CV.10-MyoYum | Myotis yumanensis | JAPQVT010000005.1 | 70,588,059–70,588,617 | - |
| CV.10-MyoRic | Myotis ricketti | JASKON010000005.1 | 71,087,272–71,087,392 | - |
| CV.10-MyoMyo | Myotis myotis | NW_023416368.1 | 71,971,796–71,972,569 | [72] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ritsch, M.; Eulenfeld, T.; Lamkiewicz, K.; Schoen, A.; Weber, F.; Hölzer, M.; Marz, M. Endogenous Bornavirus-like Elements in Bats: Evolutionary Insights from the Conserved Riboviral L-Gene in Microbats and Its Antisense Transcription in Myotis daubentonii. Viruses 2024, 16, 1210. https://doi.org/10.3390/v16081210
Ritsch M, Eulenfeld T, Lamkiewicz K, Schoen A, Weber F, Hölzer M, Marz M. Endogenous Bornavirus-like Elements in Bats: Evolutionary Insights from the Conserved Riboviral L-Gene in Microbats and Its Antisense Transcription in Myotis daubentonii. Viruses. 2024; 16(8):1210. https://doi.org/10.3390/v16081210
Chicago/Turabian StyleRitsch, Muriel, Tom Eulenfeld, Kevin Lamkiewicz, Andreas Schoen, Friedemann Weber, Martin Hölzer, and Manja Marz. 2024. "Endogenous Bornavirus-like Elements in Bats: Evolutionary Insights from the Conserved Riboviral L-Gene in Microbats and Its Antisense Transcription in Myotis daubentonii" Viruses 16, no. 8: 1210. https://doi.org/10.3390/v16081210
APA StyleRitsch, M., Eulenfeld, T., Lamkiewicz, K., Schoen, A., Weber, F., Hölzer, M., & Marz, M. (2024). Endogenous Bornavirus-like Elements in Bats: Evolutionary Insights from the Conserved Riboviral L-Gene in Microbats and Its Antisense Transcription in Myotis daubentonii. Viruses, 16(8), 1210. https://doi.org/10.3390/v16081210






