Abstract
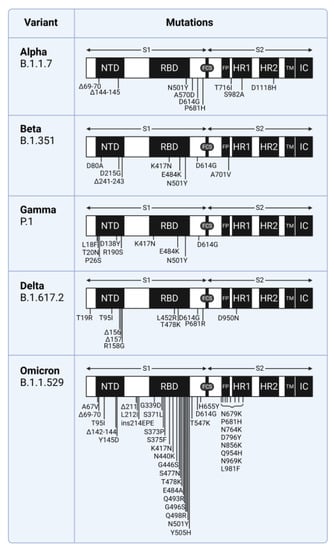
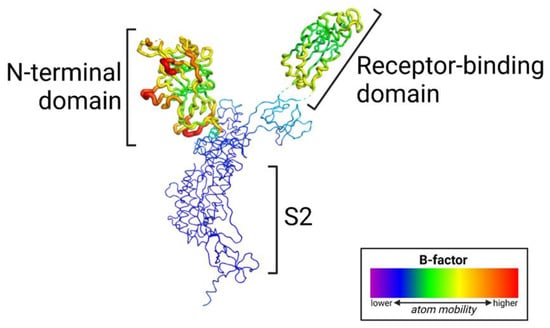
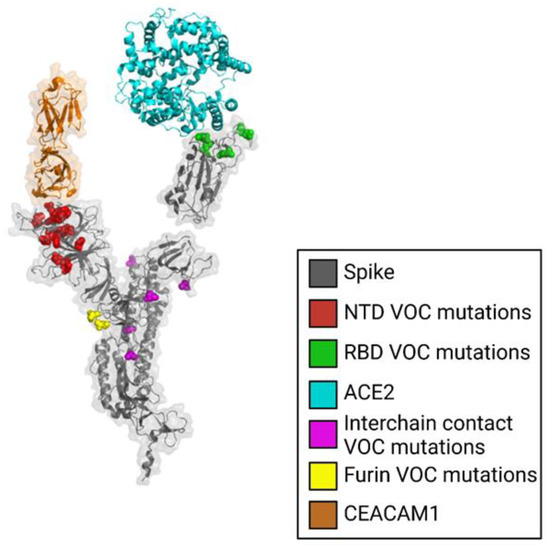
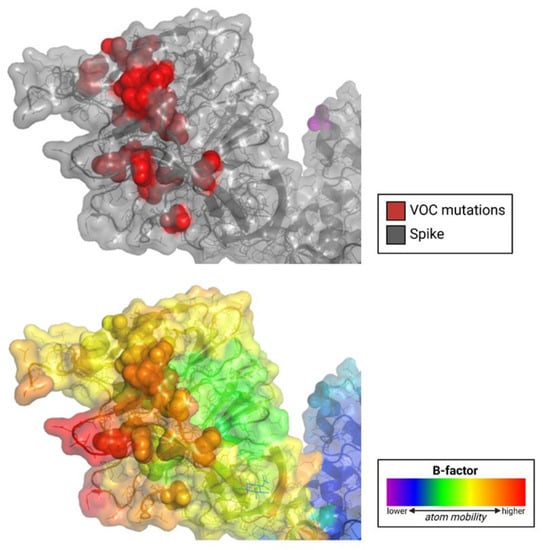
Evidence suggests that the N-terminal domain (NTD) of the SARS-CoV-2 spike protein interacts with host coreceptors that participate in viral entry. Resolving the identity of coreceptors has important clinical implications as it may provide the basis for the development of antiviral drugs and vaccine candidates. The majority of characteristic mutations in variants of concern (VOCs) have occurred in the NTD and receptor binding domain (RBD). Unlike the RBD, mutations in the NTD have clustered in the most flexible parts of the spike protein. Many possible coreceptors have been proposed, including various sugars such as gangliosides, sialosides, and heparan sulfate. Protein coreceptors, including neuropilin-1 and leucine-rich repeat containing 15 (LRRC15), are also proposed coreceptors that engage the NTD.
2. Sugars as SARS-CoV-2 Spike Protein Ligands
2.1. Sialosides
As mentioned before, a dual-receptor mechanism binding to DPP4 and host sialoside sugars was proposed to be used by MERS-CoV for the infection of host cells. Awasthi et al. noted that the SARS-CoV-2 S1-NTD has three divergent loop regions that structurally resemble the MERS-CoV sialoside-binding pocket, as confirmed by a cryo-EM study [4,10]. Specifically, the β14–β15 loop of the NTD is implicated in sialoside binding [4].
Computational binding studies found that diverse sialosides interacted with and localized to the proposed sialoside-binding pocket in the NTD of both MERS-CoV and SARS-CoV-2. SARS-CoV was not found to interact similarly with sialosides, demonstrating binding in various locations of the NTD, which was the expected result as SARS-CoV is not known to bind to sialosides [4]. Further, Milanetti et al. also predicted a sialoside-binding pocket in the SARS-CoV-2 NTD by surface iso-electron density mapping [11].
Awasthi et al. also propose that the predicted ability of SARS-CoV-2 to engage diverse sialosides could explain its high infectivity with broad tissue tropism [4]. The distribution of sialosides in the respiratory tract could explain why SARS-CoV-2 exhibits high infectivity for these cells despite their limited ACE2 expression.
Another proposed function of sialoside binding is the facilitation of viral surfing over the host cell surface [11,12,13,14]. Viral surfing describes the movement of SARS-CoV-2 virions across a cell surface’s sialic acid layer to find and attach to ACE2. Similarly, Milanetti et al. proposed a dual or even triple binding of SARS-CoV-2 to ACE2 and gangliosides present in lipid rafts [11]. A glycan-binding domain could bind to certain glycosphingolipids present in lipid rafts. Lipid raft coalescence is proposed to lead to the recruitment of ACE2, and ganglioside expression is higher in epithelial intestinal and brain cells, supporting the potential role of gangliosides in SARS-CoV-2 infectivity.
Sialoglycan microarrays were used to test the spike protein’s sialic acid binding capacity. Hao et al. did not determine significant fluorescent signals when recombinant SARS-CoV-2 S protein was incubated with sialic acid-containing oligosaccharides on an array chip [15]. Though, these immobilized sialic acids may not model the native presentation of sialic acids on the cell surface in vivo, where sialic acids cluster in the flexible plasma membrane. Baker et al. used polymer-stabilized gold nanoparticle bearing sialic acids to confirm the binding of single sialic acid to the S protein, but not sialyllactoses [16].
Yang et al. investigated the role of glycans containing sialic acids on the ACE2 receptor for SARS-CoV-2 infection [17]. They found that glycans did not greatly contribute to the binding of the S protein to ACE2 and that ACE2′s sialic acids actually shielded cells from pseudovirus binding. Chu et al. then treated epithelial cells with neuraminidase to remove cell surface sialic acids [18]. They found that this treatment reduced MERS-CoV entry by 86%, which was expected since MERS-CoV uses DPP4 and sialic acids as coreceptors to facilitate binding. However, this sialic acid-removing treatment increased SARS-CoV and SARS-CoV-2 infection by 492% and 80.3%, respectively [19]. These results indicate that the presence of cell-surface sialic acids prevents ACE2-S protein binding, thus inhibiting virus entry.
2.2. Gangliosides
Though Yang et al. and Chu et al. reveal a decreasing likelihood that sialic acids potentiate SARS-CoV-2-cell binding, a preprint study by Nguyen et al. suggested that the RBD had an affinity for monosialylated gangliosides [17,18,20]. Using an artificial membrane embedded within gangliosides, they found that three monosialylated gangliosides (GM1, GM2, and GM3) were recognized by the RBD [20]. The affinity was similar to that of the glycan heparan sulfate, another proposed ligand of the RBD. Next, by depleting cell surface sialic acids using sialyltransferase inhibition, genetic knockout of sialic acid biosynthesis, and neuraminidase treatment of ACE2-expressing cells, they demonstrated decreased binding and infection.
Functions of gangliosides supporting their theoretical implication in SARS-CoV-2 infection include their negatively charged flat surface that attracts the electropositive tip of virus envelope proteins, ability to facilitate the recruitment of virus protein receptors from lipid rafts, association with cholesterol to form lipid rafts that could enhance fusion and activation of viral proteins through membrane chaperone properties [19].
Fantini et al. proposed a ganglioside binding domain at the top of the NTD, positing that it allows the S protein to interact with lipid rafts independently of the RBD [21]. The study found that neutralizing antibodies directed against the NTD’s tip did, in fact, prevent access of the S protein to lipid rafts independently of RBD-ACE2 interactions [21]. Though, Fantini et al.’s data led to the proposal that hydroxychloroquine and other antimalarials could block the interaction between the S protein and cell surface gangliosides due to their affinity for gangliosides. The long-term lack of clinical validation for this strategy is not supportive of this theory [22].
2.3. Heparan Sulfate
Heparan sulfate is a highly conserved, negatively charged linear polysaccharide and cellular receptor found in almost all mammalian cells. In its proteoglycan form, HS binds to a variety of extracellular proteins, with wide-ranging functions related to development, inflammation, coagulation, angiogenesis, and viral entry. Heparan sulfate proteoglycans (HSPGs) are known to serve as coreceptors for many viruses [23].
Clausen et al. determined via molecular docking that the RBD likely contains a positively charged site adjacent to the ACE2-binding site that binds negatively charged heparan sulfate [24]. Subsequent competition studies, enzymatic removal of HS, and genetic studies showed that recombinant and pseudoviral S proteins, as well as authentic SARS-CoV-2 virions, bind to cell surface HS cooperatively with ACE2 [24].
A ternary complex of ACE2, heparin and the S protein using heparin as a scaffold was observed by Clausen et al. in vitro. Kearns et al. demonstrated a polyanionic HS-binding site that starts at the RBD and runs between the RBD and NTD down to the furin cleavage site [25]. Electron micrographs suggested that binding to heparin enhances the open conformation of the RBD, which supports ACE2 binding. Further, in vitro treatment of cells with heparin lyases that degrade HS significantly reduced infection. Clausen et al. concluded that HS not only supports infection but is an essential factor in infection that enables the RBD to bind ACE2. This proposed role has since been supported by a few studies (including Liu et al. [26]).
HSPGs have previously been proposed as coreceptors for HCoV-NL63 [27] and SARS-CoV [28]. In both cases, HSPGs were also proposed as necessary adhesion molecules for HCoV-NL63 and SARS-CoV infection. In fact, the basis of treatment of SARS with lactoferrin, an innate immunity protein, is lactoferrin’s colocalization with HSPGs, blocking S protein-HSPG binding, and thus infection [28,29]. Also, HS’s role in coagulation could be responsible for thrombotic complications seen in critically ill COVID-19 patients since the S protein could outcompete antithrombin and heparin cofactor II for HS binding [30].
2.4. Summary of Sugars as SARS-CoV-2 Spike Protein Receptors
Experimental data has yet to reveal anything conclusive regarding the role of sialosides or gangliosides in SARS-CoV-2-cell binding. Studies offer conflicting data regarding the enabling or prevention of virus entry due to these cell surface sugars. Due to the structural basis for the spike protein’s proposed affinity for sialosides, including divergent loop regions in the NTD resembling sialoside-binding pockets and locations with an affinity for gangliosides in the NTD and RBD, more investigation into spike-sialoside interactions is warranted. If sialosides or gangliosides do, in fact, play a role in coronavirus host-cell binding, it is likely to use a novel mechanism yet unknown.
More likely is the possibility of heparan sulfate and its proteoglycan form as an important coreceptor for SARS-CoV-2. Strong but limited experimental data supports its role as a necessary or supporting factor for cell-virus adhesion, as its binding to the S protein prompts the open conformation of the RBD to accommodate subsequent ACE2 binding and infection [17].
3. Other Molecules
3.1. LRRC15 as a SARS-CoV-2 Spike Protein Ligand
Leucine-rich repeat (LRR) proteins are a family of functionally unrelated α/β horseshoe-shaped proteins that contain tandem repeats of 20–30 amino acids with an unusually high composition of leucine [31]. Leucine-rich repeat-containing protein 15 (LRRC15) is a member of the LRR family with many known functions, including innate immunity and nervous system development [32]. LRRC15 is known to be involved in the negative regulation of protein localization to the plasma membrane [33].
Multiple late-2021 preprints independently identified LRRC15 as a ligand of the SARS-CoV-2 spike protein (Figure 5). Shilts et al. employed two strategies to determine possible ligands of the spike protein [34]. First, HEK293 cells were individually transfected with one of 2363 genes encoding cell surface membrane proteins, measuring protein-spike binding with flow cytometry. Second, a genome-wide CRISPR activation library was used in RPE1 cells to identify which genes, when upregulated, induced the binding of the spike protein. Both systematic cell-based screens identified LRRC15 as SARS-CoV-2 spike protein ligands, though structural details of this interaction were unclear [34].
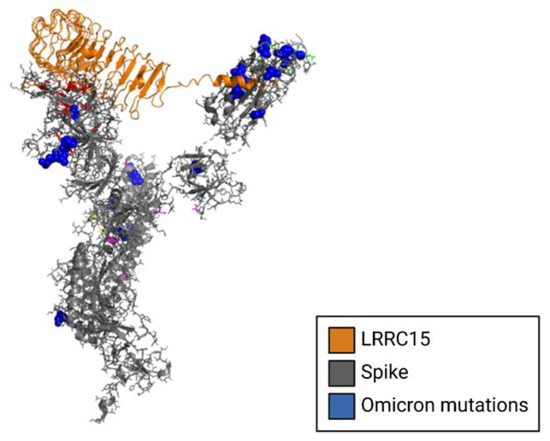
Figure 5.
Structural model of LRRC15 complexed to the NTD of the SARS-CoV-2 spike glycoprotein. The cryo-EM structure of the spike protein is shown in gray complexed to LRRC15, shown in gold. Spike mutations from the omicron variant are shown in blue.
Loo et al. also identified LRRC15 as a spike protein ligand using a CRISPR activation strategy similar to Shilts et al. [34,35]. Further, LRRC15 sequestered virions and functioned as a negative receptor that suppressed live SARS-CoV-2 infection in trans [35]. Since LRRC15 and ACE2 expression are mutually exclusive (not expressed in the same cell types) in lung cells, it follows that LRRC15′s virion sequestration may prevent virions from reaching ACE2-expressing cells.
Additionally, LRRC15 expression could regulate the reaction of fibroblasts to infection. It is found in collagen-producing myofibroblasts, where it regulates collagen production. Since it is known to be upregulated by proinflammatory cytokines, including TNFα, IL-1β, and IFNγ [36], LRRC15 could suppress lung fibrosis during virus-induced inflammation, independent of virion sequestration and immobilization. LRRC15 could potentially help fibroblasts transport virions to antigen-presenting cells.
A subsequent decrease in LRRC15 levels following inflammation could promote collagen production to support lung repair. It is posited by Loo et al. that dysregulation of this system caused by chronic lung infection could cause inappropriate collagen production, contributing to lung fibrosis seen in “long-haul” COVID patients [35].
Interestingly, when compared to the earlier D614G variant of SARS-CoV-2, Loo et al. found that the mutations in the Delta variant’s spike protein reduced LRRC15′s antiviral activity. This indicates that the numerous mutations of the spike protein in highly contagious variants could be attributed to an adaptation against the anti-SARS-CoV-2 activity of LRRC15.
A third preprint, by Song et al., affirmed the trans-inhibition of viral entry of SARS-CoV-2 by LRRC15 [37]. The interaction of LRRC15 with the RBD was not found to compete with the interaction of ACE2 with the RBD. LRRC15 did, in fact, inhibit spike-mediated viral entry in LRRC15-expressing ACE2-negative cells as well as neighboring ACE2-positive cells in trans. LRRC15 had a specific inhibitory effect on multiple variants of SARS-CoV-2 and SARS-CoV-1 but not MERS-CoV, which Song et al. noted as a suggestion of an evolutionary arms race between humans and coronaviruses. Song et al. also confirmed by analysis of human lung single cell RNA sequencing that LRRC15 expression is primarily detected in fibroblasts, including those pathologically enriched in COVID-19 patients.
3.2. Summary of LRRC15 Relating to SARS-CoV-2
These preprints display convincing experimental evidence that LRRC15 binds to the spike protein, offering mechanistic theories with plausibility established by experimental observations. The role of LRRC15 is currently best explained as SARS-CoV-2-virion-sequestering, protecting nearby ACE2-positive cells from infection. Further speculation reveals a possible mechanism by which LRRC15′s role in collagen production regulation on fibroblasts could contribute to “long-haul” COVID. These roles have earned the protein its characterization as a “master regulator” of SARS-CoV-2 infection and collagen production associated with “long-haul” COVID-related lung fibrosis.
More experimental investigation into the spike-LRRC15 interaction is warranted to further establish the theories about its potential role in preventing infection. Resolving the structure of the spike-LRRC15 complex could offer insight into the exact binding location of LRRC15 on the spike protein.
3.3. Neuropilin-1 as a SARS-CoV-2 Spike Protein Ligand
Neuropilin-1 is a glycoprotein receptor found in neurons that have functions related to angiogenesis, neuronal development, and immune response regulation [38].
Some studies have identified NRP-1 as a coreceptor for SARS-CoV-2 entry [38,39,40]. The presence of NRP-1 on the host cell membrane has been shown to increase infection and spread of SARS-CoV-2, as shown by Cantuti-Castelvetri et al. [39]. Particularly, NRP-1 is implicated in neurological manifestations of COVID-19, as it is thought to enable SARS-CoV-2 to enter the central nervous system through the respiratory and olfactory epithelia—where NRP-1 is highly expressed—including those of the nasal cavity.
Daly et al. explains that NRP-1 binds furin-cleaved substrates such as the one in the spike protein (Figure 6) [40]. Daly et al. successfully demonstrated that the furin-cleaved spike protein binds directly to cell surface NRP-1. Further, blocking the S1-NRP-1 interaction with small-molecule inhibitors and monoclonal antibodies reduced viral infection in vitro. This finding has significant implications for future antiviral therapeutics.
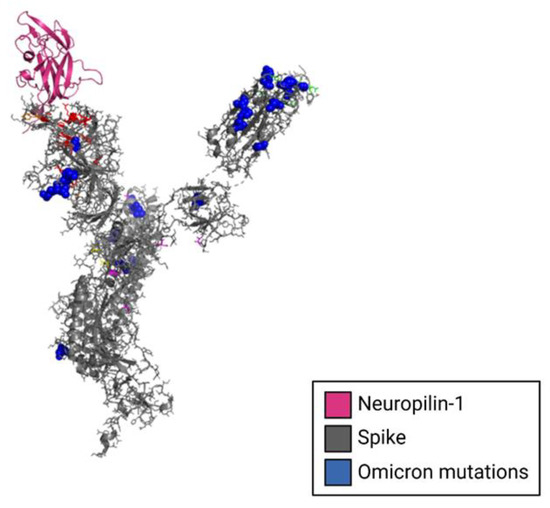
Figure 6.
Structural model of Neuropilin-1 complexed to the NTD of the SARS-CoV-2 spike glycoprotein monomer. The cryo-EM structure of the spike protein monomer is shown in gray complex to neuropilin-1, shown in magenta. Spike mutations from the omicron variant are shown in blue.
4. Non-Receptor Spike-Host Cell Protein Interactions
4.1. SARS-CoV-2 Spike Protein Cleavage and Restriction Factors
The SARS-CoV-2 spike protein relies on cleavage by proteases on the cell surface to transform into its infectious conformation that enables cell entry through plasma membrane fusion [41]. Spike protein cleavage/fusion at (or in close proximity to) the cell surface is crucial to successful infection. Spike protein fusion with host cell membranes near the cell surface permits avoidance of restriction factors located in early and late endosomes within the cell. The furin cleavage site where the spike is cleaved is thought to be an advantage of SARS-CoV-2, as viruses that lack such a cleavage site generally must enter cells through the restriction-factor-containing endosome [42].
Restriction factors are proteins that host cells use as the first line of defense against viruses [42]. Some interfere with viral replication and propagation, while others are sensors that trigger innate immune responses. They are generally induced by interferons.
Experimental evidence showed that SARS-CoV-2 is predominantly sensitive to IFITM2, a restriction factor that inhibits virus-host cell fusion [43]. The same study observed decreased infectivity when removing the furin cleavage site from pre-Omicron variants, supporting the theory that this cleavage is an advantage of the virus. Additionally, SARS-CoV-2 was found to be highly sensitive to IFN-β and IFN-γ. IFN-α and IFN-λ were less effective but still demonstrated restrictive properties in various cell types. It is not yet elucidated how changes in Omicron’s furin cleavage site (and consequently its protease-based infection cascade) may affect its interactions with restriction factors.
4.2. Membrane-Associated Serine Proteases
The spike protein must undergo a conformational change at the RBD to interact with ACE2. TMPRSS2 has been known as the first host cell-surface protease used by the spike protein to cleave its furin cleavage site prompting the conformational changes necessary to enter the cell [41]. Following cleavage by TMPRSS2, the spike is further cleaved and processed by furin and cathepsin, then fusing with the cell and provoking viral replication.
Currently, Omicron variants inefficiently use TMPRSS2, causing the spike to rely on cell entry through endocytosis [44]. This caused a change in the cellular tropism of SARS-CoV-2, moving away from TMPRSS2-expressing cells. However, BA.5 has recently shown efficient use of TMPRSS2, indicating a possible shift back to pre-Omicron tropism and infectious mechanism [45].
5. Summary
The identity of a putative SARS-CoV-2 coreceptor has yet to be determined. Though, few proposed candidates increasingly demonstrate activity as potential ligands of the spike protein (Table 1). Further experimentation is warranted to elucidate the roles of these candidates in the SARS-CoV-2 infection mechanism. Significant improvements in the prevention and treatment of COVID-19 may be achieved by understanding how to modulate the interactions between the S1-NTD/RBD and coreceptors, antibodies, and restriction factors.

Table 1.
Summary of potential coreceptor ligands of the SARS-CoV-2 spike protein.
6. Methods
6.1. SARS-CoV-2 Mutations in Variants of Concern
Mutations in SARS-CoV-2 variants of concern are shown with the Wuhan-Hu-1 sequence as a reference (GENBANK accession number MN908947). The Centers for Disease Control and Prevention classification and definitions of mutations that distinguish variants of concern were used: https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-classifications.html (accessed on 26 April 2022) [48].
6.2. Mapping Mutations and Structural Variability on Structure of the Spike Protein Monomer
The prefusion SARS-CoV-2 spike glycoprotein with a single receptor-binding domain up (PDB 6VSB) [49] was used for mapping mutations and structural variability (B-factor) in variants of concern.
6.3. Modeling Interactions between LRRC15, Neuropilin-1 and the SARS-CoV-2 Spike Protein NTD
The crystal structure of Neuropilin-1 (PDB 7JJC) [40] was docked to the SARS-CoV-2 spike protein NTD using the program HDOCK [50]. A structural model of human LRRC15 was generated by SWISS-MODEL [51] and docked to the SARS-CoV-2 spike protein NTD using the program HDOCK [50].
Author Contributions
Writing—Original draft preparation, R.L.B.; Writing—Review and editing, D.A.O.; supervision, D.A.O. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Institute of Allergy and Infectious Diseases, grant number R01AI170187. The APC was funded by David A. Ostrov.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
Figures created using BioRender.com.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Li, F. Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu. Rev. Virol. 2016, 3, 237–261. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Yang, C.; Xu, X.; Xu, W.; Liu, S. Structural and functional properties of SARS-CoV-2 spike protein: Potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020, 41, 1141–1149. [Google Scholar] [CrossRef] [PubMed]
- Eslami, N.; Aghbash, P.S.; Shamekh, A.; Entezari-Maleki, T.; Nahand, J.S.; Sales, A.J.; Baghi, H.B. SARS-CoV-2: Receptor and Co-receptor Tropism Probability. Curr. Microbiol. 2022, 79, 133. [Google Scholar] [CrossRef] [PubMed]
- Awasthi, M.; Gulati, S.; Sarkar, D.P.; Tiwari, S.; Kateriya, S.; Ranjan, P.; Verma, S.K. The Sialoside-Binding Pocket of SARS-CoV-2 Spike Glycoprotein Structurally Resembles MERS-CoV. Viruses 2020, 12, 909. [Google Scholar] [CrossRef]
- Ostrov, D.A.; Knox, G.W. Emerging mutation patterns in SARS-CoV-2 variants. Biochem. Biophys. Res. Commun. 2022, 586, 87–92. [Google Scholar] [CrossRef]
- Wang, K.; Chen, W.; Zhang, Z.; Deng, Y.; Lian, J.-Q.; Du, P.; Wei, D.; Zhang, Y.; Sun, X.-X.; Gong, L.; et al. CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal Transduct. Target. Ther. 2020, 5, 283. [Google Scholar] [CrossRef]
- Jasinska, A.J.; Pandrea, I.; Apetrei, C. CCR5 as a Coreceptor for Human Immunodeficiency Virus and Simian Immunodeficiency Viruses: A Prototypic Love-Hate Affair. Front. Immunol. 2022, 13, 835994. [Google Scholar] [CrossRef]
- Liu, Y.; Soh, W.T.; Kishikawa, J.; Hirose, M.; Nakayama, E.E.; Li, S.; Sasai, M.; Suzuki, T.; Tada, A.; Arakawa, A.; et al. An infectivity-enhancing site on the SARS-CoV-2 spike protein targeted by antibodies. Cell 2021, 184, 3452–3466.e18. [Google Scholar] [CrossRef]
- Wu, L.; Zhou, L.; Mo, M.; Liu, T.; Wu, C.; Gong, C.; Lu, K.; Gong, L.; Zhu, W.; Xu, Z. SARS-CoV-2 Omicron RBD shows weaker binding affinity than the currently dominant Delta variant to human ACE2. Signal Transduct. Target. Ther. 2022, 7, 8. [Google Scholar] [CrossRef]
- Yao, H.; Song, Y.; Chen, Y.; Wu, N.; Xu, J.; Sun, C.; Zhang, J.; Weng, T.; Zhang, Z.; Wu, Z.; et al. Molecular Architecture of the SARS-CoV-2 Virus. Cell 2020, 183, 730–738.e13. [Google Scholar] [CrossRef]
- Milanetti, E.; Miotto, M.; Di Rienzo, L.; Nagaraj, M.; Monti, M.; Golbek, T.W.; Gosti, G.; Roeters, S.J.; Weidner, T.; Otzen, D.E.; et al. In-Silico Evidence for a Two Receptor Based Strategy of SARS-CoV-2. Front. Mol. Biosci. 2021, 8, 690655. [Google Scholar] [CrossRef] [PubMed]
- Burckhardt, C.J.; Greber, U.F. Virus Movements on the Plasma Membrane Support Infection and Transmission between Cells. PLoS Pathog. 2009, 5, e1000621. [Google Scholar] [CrossRef] [PubMed]
- Caldas, L.A.; Carneiro, F.A.; Higa, L.M.; Monteiro, F.L.; da Silva, G.P.; da Costa, L.J.; Durigon, E.L.; Tanuri, A.; de Souza, W. Ultrastructural analysis of SARS-CoV-2 interactions with the host cell via high resolution scanning electron microscopy. Sci. Rep. 2020, 10, 16099. [Google Scholar] [CrossRef] [PubMed]
- Seyran, M.; Takayama, K.; Uversky, V.N.; Lundstrom, K.; Palù, G.; Sherchan, S.P.; Attrish, D.; Rezaei, N.; Aljabali, A.A.A.; Ghosh, S.; et al. The structural basis of accelerated host cell entry by SARS-CoV-2. FEBS J. 2021, 288, 5010–5020. [Google Scholar] [CrossRef] [PubMed]
- Hao, W.; Ma, B.; Li, Z.; Wang, X.; Gao, X.; Li, Y.; Qin, B.; Shang, S.; Cui, S.; Tan, Z. Binding of the SARS-CoV-2 Spike Protein to Glycans. Sci. Bull. 2021, 66, 1205–1214. [Google Scholar] [CrossRef]
- Baker, A.N.; Richards, S.-J.; Guy, C.S.; Congdon, T.R.; Hasan, M.; Zwetsloot, A.J.; Gallo, A.; Lewandowski, J.R.; Stansfeld, P.J.; Straube, A.; et al. The SARS-COV-2 Spike Protein Binds Sialic Acids and Enables Rapid Detection in a Lateral Flow Point of Care Diagnostic Device. ACS Cent. Sci. 2020, 6, 2046–2052. [Google Scholar] [CrossRef]
- Yang, Q.; Hughes, T.A.; Kelkar, A.; Yu, X.; Cheng, K.; Park, S.; Huang, W.-C.; Lovell, J.F.; Neelamegham, S. Inhibition of SARS-CoV-2 viral entry upon blocking N- and O-glycan elaboration. eLife 2020, 9, e61552. [Google Scholar] [CrossRef]
- Chu, H.; Hu, B.; Huang, X.; Chai, Y.; Zhou, D.; Wang, Y.; Shuai, H.; Yang, D.; Hou, Y.; Zhang, X.; et al. Host and viral determinants for efficient SARS-CoV-2 infection of the human lung. Nat. Commun. 2021, 12, 134. [Google Scholar] [CrossRef]
- Sun, X.-L. The role of cell surface sialic acids for SARS-CoV-2 infection. Glycobiology 2021, 31, 1245–1253. [Google Scholar] [CrossRef]
- Nguyen, L.; McCord, K.A.; Bui, D.T.; Bouwman, K.M.; Kitova, E.N.; Elaish, M.; Kumawat, D.; Daskhan, G.C.; Tomris, I.; Han, L.; et al. Sialic Acid-Containing Glycolipids Mediate Binding and Viral Entry of SARS-CoV-2. Nat Chem Biol 2022, 18, 81–90. [Google Scholar] [CrossRef]
- Fantini, J.; Di Scala, C.; Chahinian, H.; Yahi, N. Structural and molecular modelling studies reveal a new mechanism of action of chloroquine and hydroxychloroquine against SARS-CoV-2 infection. Int. J. Antimicrob. Agents 2020, 55, 105960. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, I.S.; Boulware, D.R.; Lee, T.C. Hydroxychloroquine for COVID19: The curtains close on a comedy of errors. Lancet Reg. Health Am. 2022, 11, 100268. [Google Scholar] [CrossRef] [PubMed]
- Cagno; Tseligka; Jones; Tapparel Heparan Sulfate Proteoglycans and Viral Attachment: True Receptors or Adaptation Bias? Viruses 2019, 11, 596. [CrossRef] [PubMed]
- Clausen, T.M.; Sandoval, D.R.; Spliid, C.B.; Pihl, J.; Perrett, H.R.; Painter, C.D.; Narayanan, A.; Majowicz, S.A.; Kwong, E.M.; McVicar, R.N.; et al. SARS-CoV-2 Infection Depends on Cellular Heparan Sulfate and ACE2. Cell 2020, 183, 1043–1057.e15. [Google Scholar] [CrossRef] [PubMed]
- Kearns, F.L.; Sandoval, D.R.; Casalino, L.; Clausen, T.M.; Rosenfeld, M.A.; Spliid, C.B.; Amaro, R.E.; Esko, J.D. Spike-heparan sulfate interactions in SARS-CoV-2 infection. Curr. Opin. Struct. Biol. 2022, 76, 102439. [Google Scholar] [CrossRef]
- Liu, L.; Chopra, P.; Li, X.; Bouwman, K.M.; Tompkins, S.M.; Wolfert, M.A.; de Vries, R.P.; Boons, G.-J. Heparan Sulfate Proteoglycans as Attachment Factor for SARS-CoV-2. ACS Cent. Sci. 2021, 7, 1009–1018. [Google Scholar] [CrossRef]
- Milewska, A.; Zarebski, M.; Nowak, P.; Stozek, K.; Potempa, J.; Pyrc, K. Human Coronavirus NL63 Utilizes Heparan Sulfate Proteoglycans for Attachment to Target Cells. J. Virol. 2014, 88, 13221–13230. [Google Scholar] [CrossRef]
- Lang, J.; Yang, N.; Deng, J.; Liu, K.; Yang, P.; Zhang, G.; Jiang, C. Inhibition of SARS Pseudovirus Cell Entry by Lactoferrin Binding to Heparan Sulfate Proteoglycans. PLoS ONE 2011, 6, e23710. [Google Scholar] [CrossRef]
- Hu, Y.; Meng, X.; Zhang, F.; Xiang, Y.; Wang, J. The in vitro antiviral activity of lactoferrin against common human coronaviruses and SARS-CoV-2 is mediated by targeting the heparan sulfate co-receptor. Emerg. Microbes Infect. 2021, 10, 317–330. [Google Scholar] [CrossRef]
- Zheng, Y.; Zhao, J.; Li, J.; Guo, Z.; Sheng, J.; Ye, X.; Jin, G.; Wang, C.; Chai, W.; Yan, J.; et al. SARS-CoV-2 spike protein causes blood coagulation and thrombosis by competitive binding to heparan sulfate. Int. J. Biol. Macromol. 2021, 193, 1124–1129. [Google Scholar] [CrossRef]
- Ng, A.; Xavier, R.J. Leucine-rich repeat (LRR) proteins: Integrators of pattern recognition and signaling in immunity. Autophagy 2011, 7, 1082–1084. [Google Scholar] [CrossRef] [PubMed]
- Ray, U.; Pathoulas, C.L.; Thirusangu, P.; Purcell, J.W.; Kannan, N.; Shridhar, V. Exploiting LRRC15 as a Novel Therapeutic Target in Cancer. Cancer Res. 2022, 82, 1675–1681. [Google Scholar] [CrossRef] [PubMed]
- O’Prey, J.; Wilkinson, S.; Ryan, K.M. Tumor Antigen LRRC15 Impedes Adenoviral Infection: Implications for Virus-Based Cancer Therapy. J. Virol. 2008, 82, 5933–5939. [Google Scholar] [CrossRef] [PubMed]
- Shilts, J.; Crozier, T.W.M.; Teixeira-Silva, A.; Gabaev, I.; Greenwood, E.J.D.; Watson, S.J.; Ortmann, B.M.; Gawden-Bone, C.M.; Pauzaite, T.; Hoffmann, M.; et al. LRRC15 mediates an accessory interaction with the SARS-CoV-2 spike protein. bioRxiv 2021. [Google Scholar]
- Loo, L.; Waller, M.A.; Cole, A.J.; Stella, A.O.; Moreno, C.L.; Denes, C.E.; Hamoudi, Z.; Chung, F.; Aggarwal, A.; Low, J.K.K.; et al. LRRC15 suppresses SARS-CoV-2 infection and controls collagen production. bioRxiv 2021. [Google Scholar]
- Satoh, K.; Hata, M.; Yokota, H. A Novel Member of the Leucine-Rich Repeat Superfamily Induced in Rat Astrocytes by β-Amyloid. Biochem. Biophys. Res. Commun. 2002, 290, 756–762. [Google Scholar] [CrossRef]
- Song, J.; Chow, R.D.; Pena-Hernandez, M.; Zhang, L.; Loeb, S.A.; So, E.-Y.; Liang, O.D.; Wilen, C.B.; Lee, S. LRRC15 is an inhibitory receptor blocking SARS-CoV-2 spike-mediated entry in trans. bioRxiv 2021. [Google Scholar]
- Gudowska-Sawczuk, M.; Mroczko, B. The Role of Neuropilin-1 (NRP-1) in SARS-CoV-2 Infection: Review. J. Clin. Med. 2021, 10, 2772. [Google Scholar] [CrossRef]
- Cantuti-Castelvetri, L.; Ojha, R.; Pedro, L.D.; Djannatian, M.; Franz, J.; Kuivanen, S.; van der Meer, F.; Kallio, K.; Kaya, T.; Anastasina, M.; et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science 2020, 370, 856–860. [Google Scholar] [CrossRef]
- Daly, J.L.; Simonetti, B.; Klein, K.; Chen, K.-E.; Williamson, M.K.; Antón-Plágaro, C.; Shoemark, D.K.; Simón-Gracia, L.; Bauer, M.; Hollandi, R.; et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science 2020, 370, 861–865. [Google Scholar] [CrossRef]
- Peacock, T.P.; Goldhill, D.H.; Zhou, J.; Baillon, L.; Frise, R.; Swann, O.C.; Kugathasan, R.; Penn, R.; Brown, J.C.; Sanchez-David, R.Y.; et al. The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets. Nat. Microbiol. 2021, 6, 899–909. [Google Scholar] [CrossRef] [PubMed]
- Colomer-Lluch, M.; Ruiz, A.; Moris, A.; Prado, J.G. Restriction Factors: From Intrinsic Viral Restriction to Shaping Cellular Immunity Against HIV-1. Front. Immunol. 2018, 9, 2876. [Google Scholar] [CrossRef] [PubMed]
- Winstone, H.; Lista, M.J.; Reid, A.C.; Bouton, C.; Pickering, S.; Galao, R.P.; Kerridge, C.; Doores, K.J.; Swanson, C.M.; Neil, S.J.D. The Polybasic Cleavage Site in SARS-CoV-2 Spike Modulates Viral Sensitivity to Type I Interferon and IFITM2. J. Virol. 2021, 95, e02422-20. [Google Scholar] [CrossRef]
- Meng, B.; Abdullahi, A.; Ferreira, I.A.T.M.; Goonawardane, N.; Saito, A.; Kimura, I.; Yamasoba, D.; Gerber, P.P.; Fatihi, S.; Rathore, S.; et al. Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity. Nature 2022, 603, 706–714. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, A.; Akerman, A.; Milogiannakis, V.; Silva, M.R.; Walker, G.; Stella, A.O.; Kindinger, A.; Angelovich, T.; Waring, E.; Amatayakul-Chantler, S.; et al. SARS-CoV-2 Omicron BA.5: Evolving tropism and evasion of potent humoral responses and resistance to clinical immunotherapeutics relative to viral variants of concern. eBioMedicine 2022, 84, 104270. [Google Scholar] [CrossRef] [PubMed]
- Varki, A. Sialic acids in human health and disease. Trends Mol. Med. 2008, 14, 351–360. [Google Scholar] [CrossRef]
- Yu, R.K.; Tsai, Y.-T.; Ariga, T.; Yanagisawa, M. Structures, Biosynthesis, and Functions of Gangliosides-an Overview. J. Oleo Sci. 2011, 60, 537–544. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. SARS-CoV-2 Variant Classifications and Definitions 2022; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2022.
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.-L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef]
- Yan, Y.; Zhang, D.; Zhou, P.; Li, B.; Huang, S.-Y. HDOCK: A web server for protein–protein and protein–DNA/RNA docking based on a hybrid strategy. Nucleic Acids Res. 2017, 45, W365–W373. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).