Identifying Endogenous Cellular Proteins Destabilizing the Propagation of Swi1 Prion upon Overproduction
Abstract
1. Introduction
2. Materials and Methods
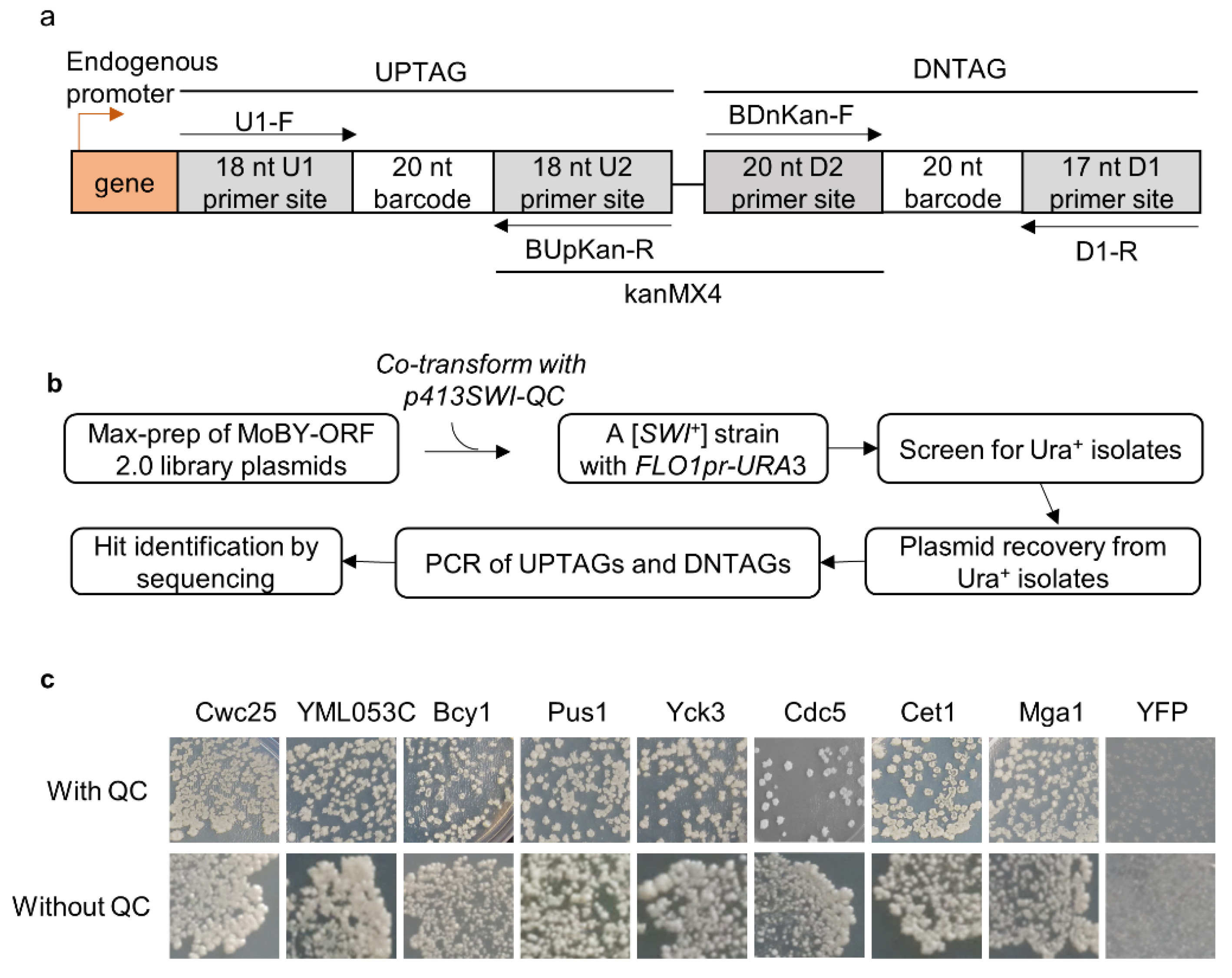
2.1. Yeast Strains, Media, Plasmids, Primers, and Library
2.2. Primary Screen and Candidate Identification
2.3. Prioritizing Hits and Performing Alternative Anti-[SWI+] Activity Test Assays
2.4. Colony Visualization Assays to Test the Activities against Other Yeast Prions
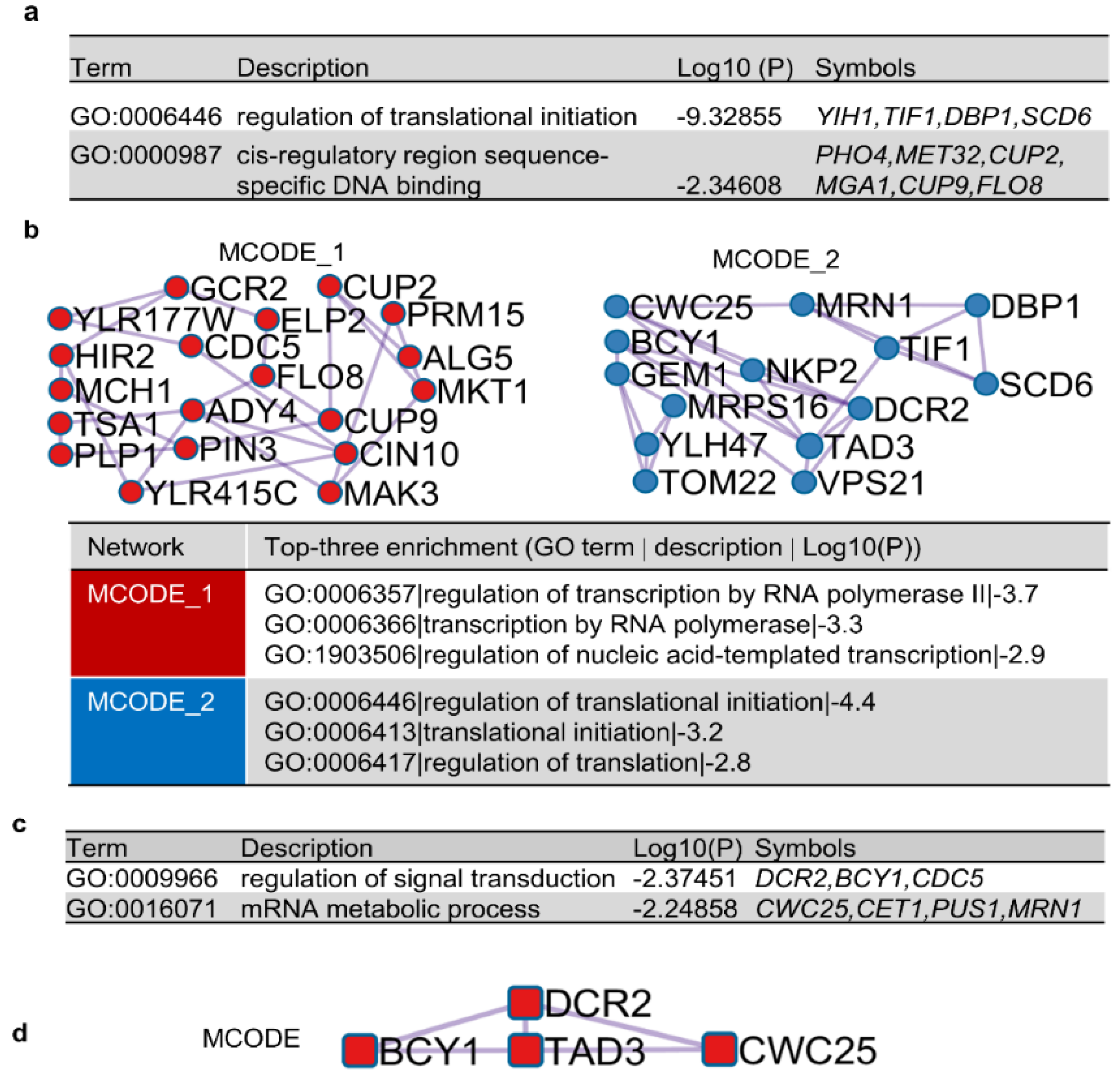
2.5. Bioinformatics Analyses
3. Results
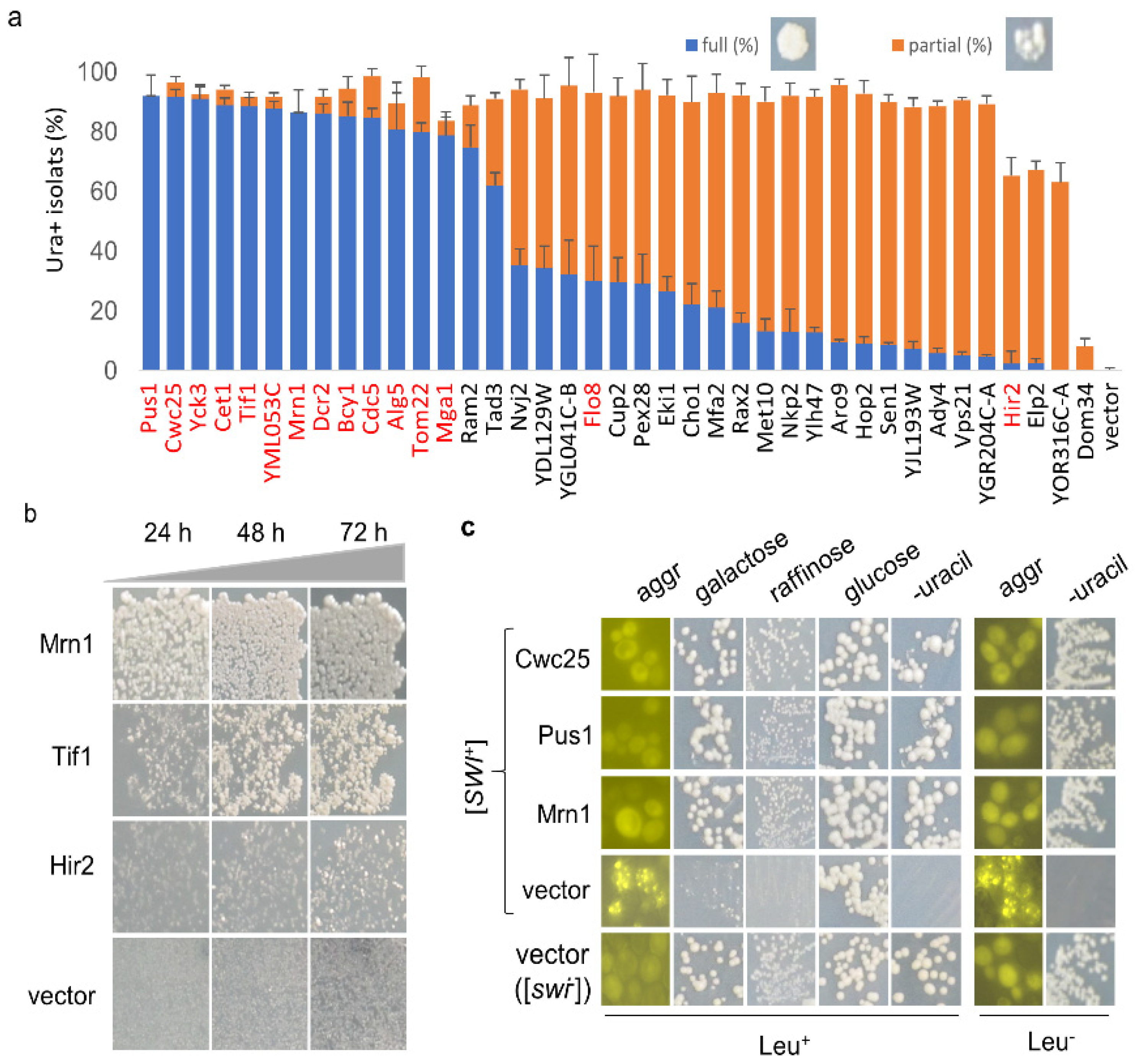
3.1. Eighty-Two Different Cellular Proteins Showing Suppression Activities against [SWI+]-Conferred Ura-Phenotype upon Overproduction
3.2. Prioritizing the Identified [SWI+] Inhibitors
3.3. The Nature of the [SWI+] Phenotypic Suppression
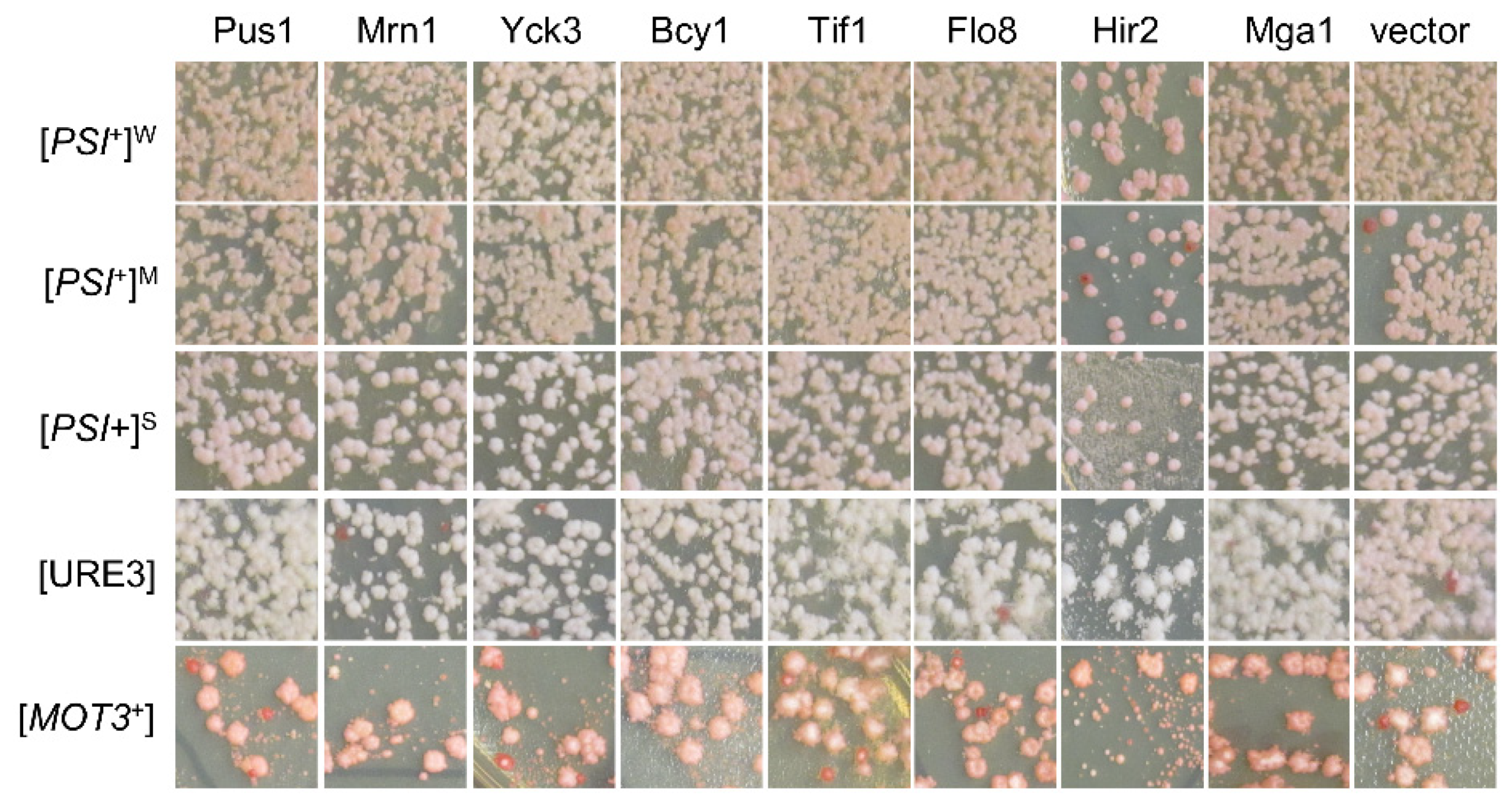
3.4. The [SWI+]-Specific Feature of the Identified Prion Destabilizers
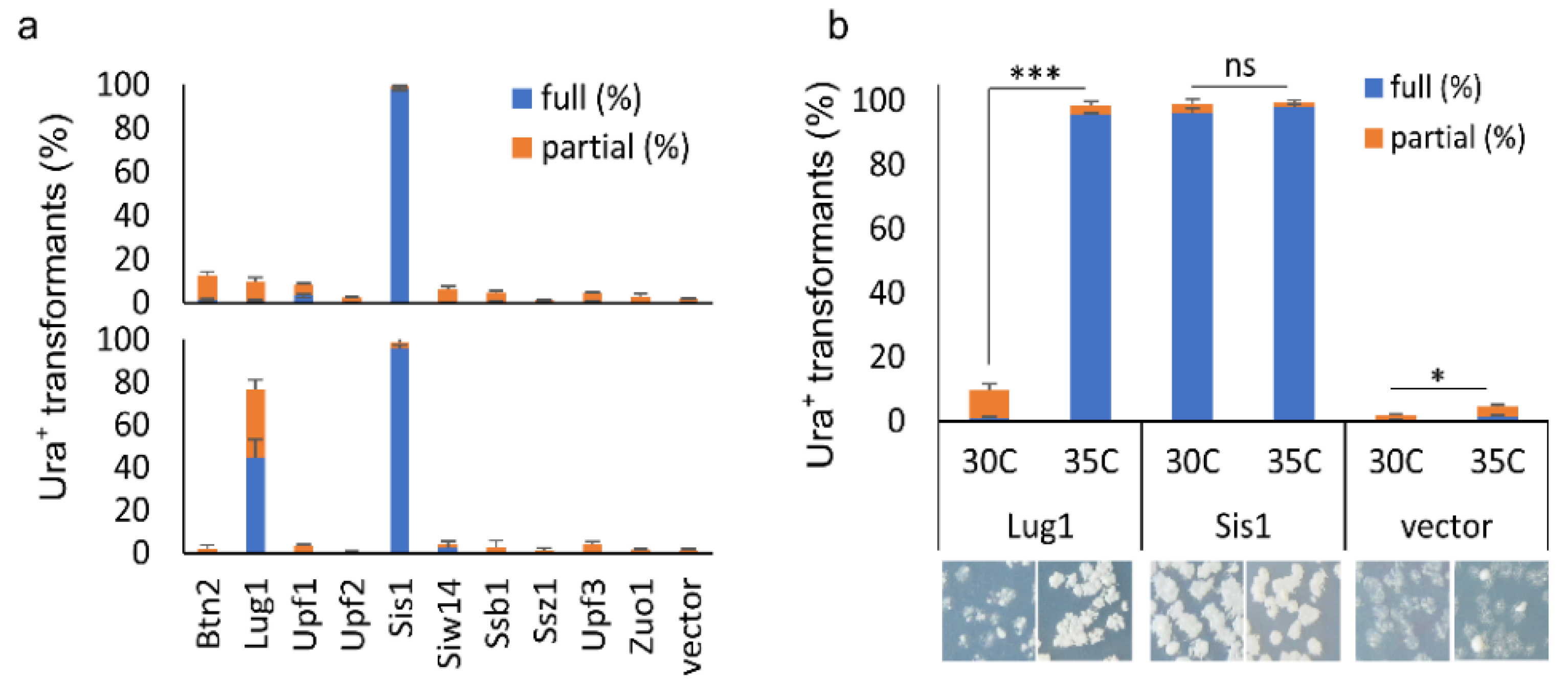
3.5. The Effects of a Selected Group of Documented Prion Destabilizers on [SWI+]
3.6. Possible Mechanisms of [SWI+] Destabilization by Identified Suppressors
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Prusiner, S.B. Novel proteinaceous infectious particles cause scrapie. Science 1982, 216, 136–144. [Google Scholar] [CrossRef] [PubMed]
- Jaunmuktane, Z.; Mead, S.; Ellis, M.; Wadsworth, J.D.; Nicoll, A.J.; Kenny, J.; Launchbury, F.; Linehan, J.; Richard-Loendt, A.; Walker, A.S.; et al. Evidence for human transmission of amyloid-beta pathology and cerebral amyloid angiopathy. Nature 2015, 525, 247–250. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, A.; Morales-Scheihing, D.; Salvadores, N.; Moreno-Gonzalez, I.; Gonzalez, C.; Taylor-Presse, K.; Mendez, N.; Shahnawaz, M.; Gaber, A.O.; Sabek, O.M.; et al. Induction of IAPP amyloid deposition and associated diabetic abnormalities by a prion-like mechanism. J. Exp. Med. 2017, 214, 2591–2610. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kwon, S.H.; Kam, T.I.; Panicker, N.; Karuppagounder, S.S.; Lee, S.; Lee, J.H.; Kim, W.R.; Kook, M.; Foss, C.A.; et al. Transneuronal Propagation of Pathologic alpha-Synuclein from the Gut to the Brain Models Parkinson’s Disease. Neuron 2019, 103, 627–641.e7. [Google Scholar] [CrossRef]
- Sampson, T.R.; Challis, C.; Jain, N.; Moiseyenko, A.; Ladinsky, M.S.; Shastri, G.G.; Thron, T.; Needham, B.D.; Horvath, I.; Debelius, J.W.; et al. A gut bacterial amyloid promotes alpha-synuclein aggregation and motor impairment in mice. eLife 2020, 9, e53111. [Google Scholar] [CrossRef]
- Wickner, R.B.; Edskes, H.K.; Son, M.; Wu, S.; Niznikiewicz, M. How Do Yeast Cells Contend with Prions? Int. J. Mol. Sci. 2020, 21, 4742. [Google Scholar] [CrossRef]
- Crow, E.T.; Li, L. Newly identified prions in budding yeast, and their possible functions. Semin. Cell Dev. Biol. 2011, 22, 452–459. [Google Scholar] [CrossRef][Green Version]
- Liebman, S.W.; Chernoff, Y.O. Prions in yeast. Genetics 2012, 191, 1041–1072. [Google Scholar] [CrossRef]
- Chernova, T.A.; Wilkinson, K.D.; Chernoff, Y.O. Prions, Chaperones, and Proteostasis in Yeast. Cold Spring Harb. Perspect. Biol. 2017, 9, a023663. [Google Scholar] [CrossRef]
- Chakrabortee, S.; Byers, J.S.; Jones, S.; Garcia, D.M.; Bhullar, B.; Chang, A.; She, R.; Lee, L.; Fremin, B.; Lindquist, S.; et al. Intrinsically Disordered Proteins Drive Emergence and Inheritance of Biological Traits. Cell 2016, 167, 369–381.e12. [Google Scholar] [CrossRef]
- Chakravarty, A.K.; Smejkal, T.; Itakura, A.K.; Garcia, D.M.; Jarosz, D.F. A Non-amyloid Prion Particle that Activates a Heritable Gene Expression Program. Mol. Cell 2020, 77, 251–265.e9. [Google Scholar] [CrossRef] [PubMed]
- Harvey, Z.H.; Chakravarty, A.K.; Futia, R.A.; Jarosz, D.F. A Prion Epigenetic Switch Establishes an Active Chromatin State. Cell 2020, 180, 928–940.e14. [Google Scholar] [CrossRef] [PubMed]
- Halfmann, R.; Wright, J.R.; Alberti, S.; Lindquist, S.; Rexach, M. Prion formation by a yeast GLFG nucleoporin. Prion 2012, 6, 391–399. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Park, K.W.; Yu, H.; Fan, Q.; Li, L. Newly identified prion linked to the chromatin-remodeling factor Swi1 in Saccharomyces cerevisiae. Nat. Genet. 2008, 40, 460–465. [Google Scholar] [CrossRef] [PubMed]
- Dutta, A.; Gogol, M.; Kim, J.H.; Smolle, M.; Venkatesh, S.; Gilmore, J.; Florens, L.; Washburn, M.P.; Workman, J.L. Swi/Snf dynamics on stress-responsive genes is governed by competitive bromodomain interactions. Genes Dev. 2014, 28, 2314–2330. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Regan, J.; Bartom, E.; Wu, W.S.; Zhang, L.; Goncharoff, D.K.; Li, L. Elucidating the regulatory mechanism of Swi1 prion in global transcription and stress responses. Sci. Rep. 2020, 10, 21838. [Google Scholar] [CrossRef] [PubMed]
- Malovichko, Y.V.; Antonets, K.S.; Maslova, A.R.; Andreeva, E.A.; Inge-Vechtomov, S.G.; Nizhnikov, A.A. RNA Sequencing Reveals Specific TranscriptomicSignatures Distinguishing Effects of the [SWI(+)] Prion and SWI1 Deletion in Yeast Saccharomyces cerevisiae. Genes 2019, 10, 212. [Google Scholar] [CrossRef]
- Du, Z.; Zhang, Y.; Li, L. The Yeast Prion [SWI(+)] Abolishes Multicellular Growth by Triggering Conformational Changes of Multiple Regulators Required for Flocculin Gene Expression. Cell Rep. 2015, 13, 2865–2878. [Google Scholar] [CrossRef]
- Du, Z.; Crow, E.T.; Kang, H.S.; Li, L. Distinct subregions of Swi1 manifest striking differences in prion transmission and SWI/SNF function. Mol. Cell. Biol. 2010, 30, 4644–4655. [Google Scholar] [CrossRef]
- Du, Z. The complexity and implications of yeast prion domains. Prion 2011, 5, 311–316. [Google Scholar] [CrossRef]
- Tuite, M.F. Yeast prions and their prion-forming domain. Science 2000, 287, 562–563. [Google Scholar] [CrossRef]
- Crow, E.T.; Du, Z.; Li, L. A small, glutamine-free domain propagates the [SWI(+)] prion in budding yeast. Mol. Cell. Biol. 2011, 31, 3436–3444. [Google Scholar] [CrossRef] [PubMed]
- Valtierra, S.; Du, Z.; Li, L. Analysis of Small Critical Regions of Swi1 Conferring Prion Formation, Maintenance, and Transmission. Mol. Cell. Biol. 2017, 37, e00206-17. [Google Scholar] [CrossRef] [PubMed]
- Goncharoff, D.K.; Cabral, R.; Applebey, S.V.; Pagadala, M.; Du, Z.; Li, L. Defining Key Residues of the Swi1 Prion Domain in Prion Formation and Maintenance. Mol. Cell. Biol. 2021, 41, e0004421. [Google Scholar] [CrossRef]
- Hines, J.K.; Li, X.; Du, Z.; Higurashi, T.; Li, L.; Craig, E.A. [SWI], the prion formed by the chromatin remodeling factor Swi1, is highly sensitive to alterations in Hsp70 chaperone system activity. PLoS Genet. 2011, 7, e1001309. [Google Scholar] [CrossRef]
- Du, Z.; Goncharoff, D.K.; Cheng, X.; Li, L. Analysis of [SWI+] formation and propagation events. Mol. Microbiol. 2017, 104, 105–124. [Google Scholar] [CrossRef]
- Du, Z.; Valtierra, S.; Li, L. An insight into the complex prion-prion interaction network in the budding yeast Saccharomyces cerevisiae. Prion 2014, 8, 387–392. [Google Scholar] [CrossRef][Green Version]
- Du, Z.; Li, L. Investigating the interactions of yeast prions: [SWI+], [PSI+], and [PIN+]. Genetics 2014, 197, 685–700. [Google Scholar] [CrossRef]
- Nizhnikov, A.A.; Ryzhova, T.A.; Volkov, K.V.; Zadorsky, S.P.; Sopova, J.V.; Inge-Vechtomov, S.G.; Galkin, A.P. Interaction of Prions Causes Heritable Traits in Saccharomyces cerevisiae. PLoS Genet. 2016, 12, e1006504. [Google Scholar] [CrossRef]
- Du, Z.; Valtierra, S.; Cardona, L.R.; Dunne, S.F.; Luan, C.H.; Li, L. Identifying Anti-prion Chemical Compounds Using a Newly Established Yeast High-Throughput Screening System. Cell Chem. Biol. 2019, 26, 1664–1680. [Google Scholar] [CrossRef]
- Chernoff, Y.O.; Kiktev, D.A. Dual role of ribosome-associated chaperones in prion formation and propagation. Curr. Genet. 2016, 62, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Killian, A.N.; Miller, S.C.; Hines, J.K. Impact of Amyloid Polymorphism on Prion-Chaperone Interactions in Yeast. Viruses 2019, 11, 349. [Google Scholar] [CrossRef] [PubMed]
- Matveenko, A.G.; Barbitoff, Y.A.; Jay-Garcia, L.M.; Chernoff, Y.O.; Zhouravleva, G.A. Differential effects of chaperones on yeast prions: CURrent view. Curr. Genet. 2018, 64, 317–325. [Google Scholar] [CrossRef] [PubMed]
- Alexandrov, I.M.; Ter-Avanesyan, M.D.; Kushnirov, V.V.; Chernova, T.A. Pleiotropic effects of Ubp6 loss on drug sensitivities and yeast prion are due to depletion of the free ubiquitin pool. J. Biol. Chem. 2003, 278, 49636–49643. [Google Scholar]
- Allen, K.D.; Chernova, T.A.; Tennant, E.P.; Wilkinson, K.D.; Chernoff, Y.O. Effects of ubiquitin system alterations on the formation and loss of a yeast prion. J. Biol. Chem. 2007, 282, 3004–3013. [Google Scholar] [CrossRef]
- Ali, M.; Chernova, T.A.; Newnam, G.P.; Yin, L.; Shanks, J.; Karpova, T.S.; Lee, A.; Laur, O.; Subramanian, S.; Kim, D.; et al. Stress-dependent proteolytic processing of the actin assembly protein Lsb1 modulates a yeast prion. J. Biol. Chem. 2014, 289, 27625–27639. [Google Scholar] [CrossRef]
- Magtanong, L.; Ho, C.H.; Barker, S.L.; Jiao, W.; Baryshnikova, A.; Bahr, S.; Smith, A.M.; Heisler, L.E.; Choy, J.S.; Kuzmin, E.; et al. Dosage suppression genetic interaction networks enhance functional wiring diagrams of the cell. Nat. Biotechnol. 2011, 29, 505–511. [Google Scholar] [CrossRef]
- Tautz, D.; Renz, M. An optimized freeze-squeeze method for the recovery of DNA fragments from agarose gels. Anal. Biochem. 1983, 132, 14–19. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Chan, K.M.; Liu, Y.T.; Ma, C.H.; Jayaram, M.; Sau, S. The 2 micron plasmid of Saccharomyces cerevisiae: A miniaturized selfish genome with optimized functional competence. Plasmid 2013, 70, 2–17. [Google Scholar] [CrossRef] [PubMed]
- Wickner, R.B. Anti-prion systems in yeast. J. Biol. Chem. 2019, 294, 1729–1738. [Google Scholar] [CrossRef] [PubMed]
- Greene, L.E.; Saba, F.; Silberman, R.E.; Zhao, X. Mechanisms for Curing Yeast Prions. Int. J. Mol. Sci. 2020, 21, 6536. [Google Scholar] [CrossRef] [PubMed]
- Higurashi, T.; Hines, J.K.; Sahi, C.; Aron, R.; Craig, E.A. Specificity of the J-protein Sis1 in the propagation of 3 yeast prions. Proc. Natl. Acad. Sci. USA 2008, 105, 16596–16601. [Google Scholar] [CrossRef]
- Tipton, K.A.; Verges, K.J.; Weissman, J.S. In vivo monitoring of the prion replication cycle reveals a critical role for Sis1 in delivering substrates to Hsp104. Mol. Cell 2008, 32, 584–591. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Du, Z.; Cho, B.; Li, L. Identifying Endogenous Cellular Proteins Destabilizing the Propagation of Swi1 Prion upon Overproduction. Viruses 2022, 14, 1366. https://doi.org/10.3390/v14071366
Du Z, Cho B, Li L. Identifying Endogenous Cellular Proteins Destabilizing the Propagation of Swi1 Prion upon Overproduction. Viruses. 2022; 14(7):1366. https://doi.org/10.3390/v14071366
Chicago/Turabian StyleDu, Zhiqiang, Brandon Cho, and Liming Li. 2022. "Identifying Endogenous Cellular Proteins Destabilizing the Propagation of Swi1 Prion upon Overproduction" Viruses 14, no. 7: 1366. https://doi.org/10.3390/v14071366
APA StyleDu, Z., Cho, B., & Li, L. (2022). Identifying Endogenous Cellular Proteins Destabilizing the Propagation of Swi1 Prion upon Overproduction. Viruses, 14(7), 1366. https://doi.org/10.3390/v14071366




