Abstract
Molnupiravir is a β-d-N4-hydroxycytidine-5′-isopropyl ester (NHC) compound that exerts antiviral activity against various RNA viruses such as influenza, SARS, and Ebola viruses. Thus, the repurposing of Molnupiravir has gained significant attention for combatting infection with SARS-CoV-2, the etiological agent of COVID-19. Recently, Molnupiravir was granted authorization for the treatment of mild-to-moderate COVID-19 in adults. Findings from in vitro experiments, in vivo studies and clinical trials reveal that Molnupiravir is effective against SARS-CoV-2 by inducing viral RNA mutagenesis, thereby giving rise to mutated complementary RNA strands that generate non-functional viruses. To date, the data collectively suggest that Molnupiravir possesses promising antiviral activity as well as favorable prophylactic efficacy, attributed to its effective mutagenic property of disrupting viral replication. This review discusses the mechanisms of action of Molnupiravir and highlights its clinical utility by disabling SARS-CoV-2 replication, thereby ameliorating COVID-19 severity. Despite relatively few short-term adverse effects thus far, further detailed clinical studies and long-term pharmacovigilance are needed in view of its mutagenic effects.
Keywords:
COVID-19; SARS-CoV-2; repurposing; repositioning; antiviral; Molnupiravir; NHC; N4-hydroxycytidine 1. Introduction
The recent COVID-19 pandemic has exerted a significant impact on global health and the economy. The etiological agent causing this disastrous pandemic is known as the Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2). The term “coronavirus” is no stranger to many as the emergence of coronaviruses causing SARS and Middle East Respiratory Syndrome (MERS) have left an indelible impression over the past two decades. Afflicting more than 530 million individuals accompanied by 6.3 million deaths worldwide, SARS-CoV-2 infection continues to progress at a very rapid rate on a global scale [1]. The zoonotic origin of SARS-CoV-2 has been postulated to be bats, similar to its previous counterpart, SARS-CoV [2]. Despite exacting a lower estimated case fatality rate (3.4%) compared to its predecessors (9.6% for SARS-CoV, and 40% for MERS-CoV), SARS-CoV-2 shows greater transmissibility along with the rapid emergence of new variants [3]. To facilitate public health and research activities, WHO has reclassified the variants of SARS-CoV-2—there are variants of concern (VOC), variants of interest (VOI), and variants under monitoring (VUM), which are named after the Greek alphabets [4]. The more recent VOCs are delta and omicron with Pango lineages B.1.617.2 from India, and B.1.1.529 first detected in South Africa, respectively [4]. On the other hand, VOIs fall under the “previous circulating” category, encompassing the epsilon, zeta, iota, eta, theta, kappa, lambda, and mu variants [4].
It is noteworthy that COVID-19 may lead to certain critical complications, such as pneumonia, septic shock, multiple organ failure, and the more pronounced acute respiratory distress syndrome or ARDS [5]. In such a time of urgent need, drug repositioning attempts represent a suitable strategy to develop antiviral therapy against COVID-19. Drug repositioning—also known as drug repurposing, drug recycling, or drug reprofiling—is described as an alternative approach to find new uses for a previously established drug to treat disease(s) other than its initially intended one [6]. The benefits of drug repositioning undoubtedly include lower costs since the drug has already undergone rigorous safety and pharmacokinetic profiling, with a shorter drug development time for its new repositioned target [7]. Currently, there is an approved nucleoside analog-based antiviral agent against COVID-19, i.e., Remdesivir [8]. A number of existing drug candidates have been proposed and/or evaluated as antivirals against SARS-CoV-2, e.g., Lopinavir-Ritonavir, Ivermectin, and others in the pipeline [9].
Lopinavir-Ritonavir is one of the first co-formulated HIV-1 protease inhibitors [10]. It was reported to have brought substantial improvements over the previous standard therapy with nucleoside reverse transcriptase inhibitors (NRTIs) [11]. The combination of Lopinavir-Ritonavir has been investigated for its efficacy for COVID-19 treatment, in view of its formation of stable complexes with viral 3-chymotrypsin-like protease (3CLpro) or main protease (Mpro) which regulate the proteolytic activity for viral replication [12]. Ivermectin, an anti-parasitic drug, has gained attention as a potential repurposed drug against COVID-19. It was first developed and commercialized as a veterinary drug in 1981 due to its promising nematicidal, acaricidal, and insecticidal activities [13]. Ivermectin is an FDA-approved drug for treating onchocerciasis and Strongyloides infection in humans [14,15]. The proposed modes of SARS-CoV-2 inhibition by Ivermectin include: the disruption of host importin heterodimer complex (IMPα/β1), inhibition of viral entry via the host angiotensin-converting enzyme 2 (ACE2) receptor, and disruption of the viral 3CLpro enzyme—thereby reducing the efficiency of viral replication [15].
Notably, RNA-dependent RNA polymerase (RdRp) represents a preferred target for developing drug inhibitors against RNA viruses, and SARS-CoV-2 is no exception. This is attributed to the highly conserved nature of RdRp domains within the Coronaviridae family, especially with respect to the predecessor SARS-CoV. Comparative genomic analysis reveals high homology of RdRp domains between SARS-CoV and SARS-CoV-2 with 96.3%, 98.8%, and 97.5% similarity in NSP12, NSP7, and NSP8, respectively—rendering RdRp a highly suitable target for drug repositioning [16,17,18]. In particular, Remdesivir, a broad-spectrum nucleoside analog inhibitor of RdRp, was the first intravenous antiviral drug against COVID-19 approved by the US Food and Drug Administration (FDA) [19]. Although initially developed against Ebola virus, Remdesivir also exhibits positive antiviral activity against multiple viruses, such as filoviruses, paramyxoviruses, coronaviruses (e.g., SARS-CoV), and others [20,21]. Given the favorable clinical trial outcomes coupled with the close RdRp similarity between SARS-CoV-2 and SARS-CoV, this therapeutic agent has been approved for the management of severe COVID-19 patients.
More recently, Molnupiravir, a newly emerged repositioned synthetic nucleoside-derived RdRp inhibitor, has also gained attention for the treatment of COVID-19. This drug is also known as β-d-N4-hydroxycytidine-5′-isopropyl ester (NHC) or Emory Institute of Drug Development-2801 (EIDD-2801) [22]. The first synthesis of Molnupiravir was reported by the Drug Innovation Ventures at Emory University back in 2018—later acquired by Ridgeback Biotherapeutics and partnered with Merck for further development [23,24]. Molnupiravir has been proven effective in multiple antiviral treatments against influenza A virus (IAV), Venezuelan equine encephalitis virus (VEEV), Ebola virus, SARS-CoV, and most recently SARS-CoV-2 [25,26]. Remdesivir acts as a nucleoside analog that interferes directly with RdRp activity. However, the mechanism of Molnupiravir is via its interactions with the RNA building blocks instead. Molnupiravir works primarily as a mutagenesis agent that induces RNA mutations by incorporating the incorrect nucleo-base into the viral RNA genome leading to catastrophic errors [27]. The RdRp then replicates and generates new virions with the errors brought forward, thereby effectively hampering viral replication capacity of the new virions [27].
The UK Medicines and Healthcare Products Regulatory Agency recently approved Molnupiravir as an oral antiviral drug against COVID-19 for adults in emergency and urgent settings [28]. This agent has the potential to ameliorate disease severity and contribute to lower morbidity and mortality among COVID-19 patients. Hence, this review aims to highlight consolidated knowledge on the molecular mechanisms underpinning the inhibition of SARS-CoV-2 replication of by Molnupiravir.
3. The General Genomic Organization of SARS-CoV-2
The general genomic configuration of SARS-CoV-2 closely resembles its beta-coronavirus counterparts, SARS-CoV and MERS-CoV [43]. SARS-CoV-2 is an enveloped, non-segmented, positive-sense RNA virus with a size of 65–125 nm in diameter [44]. Within the envelope resides a 29.9-kb RNA genome, with two-thirds containing the open reading frame 1a and 1b (ORF1ab) replicase that also encodes various non-structural proteins (NSP1–16) [45]. The remaining one-third encodes different structural proteins, namely the spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins [45]. In addition, there are multiple ORFs at the 3′-end, which encode several accessory proteins (Figure 1). The typical organization of the SARS-CoV-2 genome can be denoted as: 5′-leader-UTR-replicase-S-E-M-N-3′-UTR-poly(A) tail—with the accessory genes scattered between the structural genes (S-E-M-N) at the 3′ end [46].
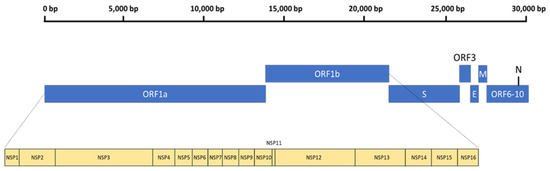
Figure 1.
The general genomic organization of SARS-CoV-2.
While the structural proteins play important roles in the integrity of new virions, the NSPs are vital for viral replication. Thus, NSP3 and NSP5 impede innate immunity, causing aberrant cytokine expression and viral polyprotein cleavages [47]. Notably, NSP12 is critical for viral replication, and the RdRp complex (RNA replicase) plays a vital role in genome replication and viral transcription [48]. The positive-ssRNA genome of SARS-CoV-2 can function as mRNA for direct protein translation, or serve as template for the production of the negative-strand RNA via RdRp [49]. The SARS-CoV-2 RdRp structure is a complex that comprises NSP12, NSP7, and NSP8, similar to SARS-CoV. Within the NSP12 catalytic subunits are three right-handed structures, namely the fingers (residues 366–581 and 621–679), palm (residues 582–620 and 680–815), and thumb (residues 816–920) subdomains, with polymerase motifs A to G spanning across the RdRp domain (Figure 2). NSP7 and NSP8 are predicted to further stabilize the conformation of NSP12, thereby enhancing the binding and processivity of the RdRp complex [50]. The general viral replication process initiates via nucleotide triphosphate (NTP) binding. This is then followed by a conformational change of the active site, leading to phosphatidyl transfer and subsequent formation of the phosphodiester bond with the existing nucleotide chain. This process is aided by magnesium (Mg2+) ions. Finally, the translocation of the newly bound NTP and chain elongation occur [51].
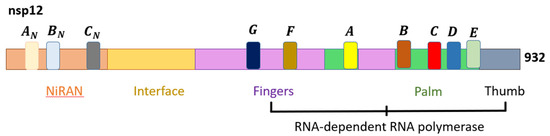
Figure 2.
The domain structure of SARS-CoV-2 NSP12 that comprises the nidovirus RdRp-associated nucleotidyltransferase (NiRAN), interface, and RdRp domains [47].
Given the highly conserved nature of RdRP within the coronavirus family, RdRp constitutes an attractive target for developing drug therapies against COVID-19 [16]. With the success of other antiviral interventions against RdRp (such as Remdesivir), it is pertinent to elucidate the mechanisms of Molnupiravir, an N4-hydroxycytidine that inhibits SARS-CoV-2 viral replication.
4. The Molecular Mechanisms of Molnupiravir on SARS-CoV-2
As mentioned, the RdRp is well-conserved among coronaviruses, in which it plays a pivotal role in the replication of the SARS-CoV-2 genome. RdRp catalyzes viral RNA replication from the original template, where RdRp synthesizes the complementary negative-sense RNA genome (minus-gRNA) from the positive-strand template. The minus-gRNA serves as a new template for further replication of the positive-sense RNA genome or plus-gRNA [52]. Hence, RdRp is a promising target for treatment strategies against COVID-19. To date, there are a limited number of FDA-approved RdRp inhibitors against SARS-CoV-2, including Remdesivir [53].
The main mechanism underpinning NHC is the inhibition of RdRp by acting as a ribonucleoside analog for RNA polymerase. Unlike Remdesivir which attenuates RNA synthesis, NHC primarily functions as a mutagen by increasing the frequency of transition mutations (G-to-A and C-to-U) [25,54]. A two-step model is deduced for NHC-induced RNA mutagenesis. As NHC enters the cell, it is cleaved in the plasma, followed by phosphorylation by host kinases into its active form, NHC-5′-triphosphate (NHC-TP) [55]. Subsequently, NHC-TP is incorporated into the synthesized minus-gRNA and sub-genomic RNA by RdRp, instead of C or U (when referring to the plus-gRNA template). Notably, NHC-TP predominantly competes with C for incorporation as compared to U. As a result, the NHC-TP-containing minus-gRNA is used as a template for the synthesis of plus-gRNA and positive-strand sub-genomic mRNA, culminating in mutations in the positive-strand RNA products and the formation of non-functional viruses [56].
With the incorporation of NHC-TP in the template, NHC-TP can form a base-pair with either G or A, since NHC exists as two tautomers. The amino or hydroxylamine form of NHC mimics C to base-pair with G via three hydrogen bonds, while the imino or oxime form mimics U to base-pair with A via two hydrogen bonds [57,58]. Although the incorporation of G (NHC-TP: G) in the product-RNA strand will hinder RNA synthesis, the increase in G concentrations will overcome this inhibitory effect with no mutation formed. In contrast, the incorporation of A will result in G-to-A transition mutation due to the [G: NHC-TP: A] base-pairing. The higher intracellular concentrations of A than G may also explain the higher frequency of [G: NHC-TP: A] formation [58]. Furthermore, the presence of C in the positive-strand viral RNA template will result in C-to-U transition mutation due to [C: G: NHC-TP: A: U] base-pairings (Figure 3). Thus, tautomerization and the resultant preference of NHC-TP to function as a C analog promotes the formation of [G: NHC-TP: A] and [C: G: NHC-TP: A: U] base-pairings, which are necessary for NHC-induced mutagenesis [59]. Other than inducing mutagenesis, studies suggest that NHC may disrupt the secondary structure of viral RNA or hinder the release of virions [55,60]. Akin to Remdesivir, NHC possesses the ability to escape viral RNA proofreading by viral exonuclease due to the stability of [NHC-TP: G] and [NHC-TP: A] base-pairings, in which the backtracking of RdRp is not induced [58,61]. The lack of interruption in RNA synthesis may bypass the proofreading ability as well [62].
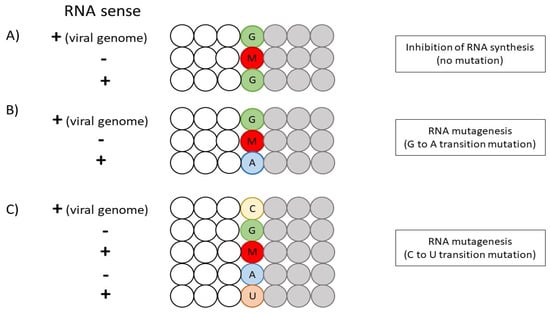
Figure 3.
An overview of NHC-induced mutagenesis of SARS-CoV-2 RNA. (A) The inhibition of RNA synthesis via [G: NHC-TP: G] base-pairing. (B) NHC-induced RNA mutagenesis via G-A transition mutation. (C) NHC-induced RNA mutagenesis via C-U transition mutation. The letters G (green), C (yellow), A (blue) and U (orange) represent the ribonucleotide bases, while the letter M (red) refers to Molnupiravir (NHC-TP). The gray circles illustrate the incorporation of NTP after M (NHC-TP).
5. Clinical and Preclinical Studies on the Efficacy of Molnupiravir
Since the discovery of SARS-CoV-2 [63], there have been a few studies on the efficacy and safety of NHC. Based on a randomized, double-blinded Phase I clinical trial on safety, tolerability, and pharmacokinetics, NHC illustrated good potential for SARS-CoV-2 inhibition [64]. The participants (18–60 years old, predominantly Caucasian males with a mean body mass index of 24.4–25.4 kg/m2) were subjected to either single-ascending dosage (50–1600 mg NHC or placebo) or multiple-ascending dosage (50–800 mg NHC or placebo, b.i.d.) for 5.5 days. The NHC pharmacokinetic profile revealed relatively good plasma absorption and safety across doses ranging from 50 to 1600 mg. The study also suggested that food intake does not influence the therapeutic efficacy of NHC. Overall, the reported adverse effects for single-dose and multiple-dose NHC include headache (12.5%), diarrhea (7.1%), and rashes. There were minimal negative effects on vital functions, electrocardiogram data, and hematological parameters, i.e., NHC is generally safe within the stipulated dosage and study timeframe.
Interim data from a phase III clinical trial of 775 patients released by Merck Sharp and Dohme (MSD) concluded that NHC is capable of reducing the risk of hospital admission and death by 50%. The data reported that 7.3% (28 of 385) of NHC-treated patients were admitted to the hospital or died as compared to 14.1% (53 of 377) of placebo-treated patients over the course of 29 days. No further deaths were reported in NHC-treated groups after day 29, while eight deaths were observed in the placebo-treated group. Although the incidence of adverse events was similar in both groups (35% NHC versus 40% placebo), there were fewer participants from the NHC group who discontinued treatment due to adverse events from the placebo group (1.3% versus 3.4%) [65,66].
The antiviral activity of NHC has also been demonstrated by in vivo and in vitro preclinical experiments. For instance, the numbers of infectious particles were successfully reduced by 4.4 logs (at 24 h) and 1.5 logs (at 48 h) in NHC-treated immunodeficient mice with implanted human lung tissue [67]. NHC administration at 12 h prior to SARS-CoV-2 infection, and at every 12 h thereafter, reduced virus titers by 100,000-fold. These studies suggest that NHC clearly exerts a prophylactic effect, and can be a strong candidate to treat SARS-CoV-2 infection, although early administration is preferred for superior outcomes [67]. Similarly, Cox et al. (2021) documented a significant reduction of virus shedding at 12 h and 36 h (peak shedding) post-infection in NHC-treated ferret models (fed b.i.d.). Infectious particles were undetectable within 24 h of treatment, with only traces of SARS-CoV-2 RNA detected in nasal tissues. Notably, the close proximity of NHC-treated infected ferrets with two untreated ferrets for 3 days revealed no infectious particles and SARS-CoV-2 RNA in the nasal lavages and intestinal tissue samples from the untreated contacts, further suggesting the efficacy of NHC to inhibit SARS-CoV-2 replication [68].
Furthermore, one study showed that 10 μM of NHC successfully inhibited virus production in primary human airway epithelial cells with a maximal titer reduction of >5 logs of MERS-CoV and >3 logs of SARS-CoV. Upon NHC administration at 10 μM dosage, the mutation rate of NHC-treated MERS-CoV RNA was significantly elevated by up to ten-fold. With NHC treatment, potent dose-dependent virus reduction was observed, i.e., 1.5 × 102 PFU/mL with 10 μM NHC versus 2.96 × 10⁴ PFU/mL with 1 μM NHC, compared with 3.96 × 10⁶ PFU/mL for vehicle control. Overall, error rates of 3-fold and 6-fold were accompanied by 138-fold and 26,000-fold reductions in virus titer during treatment with 1 μM and 10 μM of NHC, respectively [40]. Additionally, a significant decrease in lung hemorrhage was also observed in infected mice administered with 500 mg/kg of NHC [40]. Collectively, these experiments suggest that NHC confers a prophylactic effect, and that earlier administration of NHC could significantly reduce SARS-CoV-2 replication and lung hemorrhage, thereby improving the survival rate of COVID-19 patients. There are multiple clinical trials (NCT04575584, NCT04939428, NCT04405739, NCT04746183) to evaluate the efficacy and safety of NHC against SARS-CoV-2 infection [69,70,71,72].
Despite the encouraging outcomes, the mutagenic effect of NHC has raised concerns on its potential host cell mutagenesis. One study demonstrated that there is no accumulation of host ISG15 mutations at high NHC concentrations (up to 500 mg/kg), accompanied by efficient removal of ribonucleotides from host DNA [40,73]. Mutagenicity assays conducted on high concentrations of NHC displayed no significant differences in mutation rates between NHC-treated and untreated animals. NHC was also shown to be safe from inducing chromosomal damage in micronucleus in vitro and in vivo [74]. There was also no increase in mutational load in SARS-CoV-2-infected and NHC-treated golden hamster lung biopsies [75]. Nevertheless, there are contradictory reports that NHC may potentially cause mutagenesis in human host cells. One report stated that NHC could be metabolized to 2′-deoxyribonucleotide form (dNHC) by host ribonucleotide reductase, leading to the incorporation of DNA and subsequent host mutagenesis. The same study also illustrated that the ribonucleoside form of NHC (rNHC) was mutagenic to Chinese hamster ovary (CHO-K1) cells by converting to dNHC in a dose-dependent manner, resulting in the loss of HPRT gene function via missense substitutions and frameshift mutations. This phenomenon was further supported by rNHC-treated cells which induced resistance to the toxic base analog 6-TG (which functional HPRT genes are sensitive to) at concentrations that do not affect cell viability [76]. Animal reproduction studies have also alluded that Molnupiravir may cause fetal harm when administered to pregnant women. Hence, the drug is not recommended for use during pregnancy [77]. Despite the limited reports on host cell mutagenesis, future detailed studies and long-term observations are essential for patients administered with NHC to further evaluate its safety profile in the longer term.
7. Conclusions
The rapid progression and sustained global persistence of COVID-19 has placed enormous pressure to develop and harness therapeutic agents against SARS-CoV-2. In such critical times, drug repositioning can play a pivotal role to fulfil this urgent need. Molnupiravir (NHC) possesses antiviral activity against RNA viruses such as IAV, VEEV, Ebola virus and SARS-CoV, and shows promise as an antiviral drug against SARS-CoV-2. The main mode of antiviral action of Molnupiravir involves the inhibition of RdRp by acting as a ribonucleoside analog for viral RNA polymerase. NHC mainly functions as a mutagen by increasing the frequency of transition mutations (G-to-A and C-to-U) in the viral genes. As a consequence, the negative-gRNA strand containing NHC-TP leads to mutations in the complementary positive-strand RNA, thereby generating non-functional viruses. Owing to its antiviral and prophylactic effects, Molnupiravir may be deployed as one therapeutic solution to ameliorate SARS-CoV-2 infection, by reducing disease severity and fatality. Finally, more comprehensive clinical trial data and long-term surveillance are urgently warranted to support the clinical utility and safety of Molnupiravir against COVID-19.
Author Contributions
A.J.W.Y. and Z.Y.L. Low contributed equally to the preparation and drafting of the manuscript. V.T.K.C. and S.K.L. critically revised the article. All authors have read and agreed to the published version of the manuscript.
Funding
This publication was supported by internal funds to Sunil K. Lal from the School of Science, Monash University Malaysia (SoS Strategic Grant 2020), and to Vincent T. K. Chow from the National University of Singapore.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- COVID Live—Coronavirus Statistics—Worldometer. Available online: https://www.worldometers.info/coronavirus/ (accessed on 7 June 2022).
- Low, Z.Y.; Wen Yip, A.J.; Chow, V.T.K.; Lal, S.K. The Suppressor of Cytokine Signalling Family of Proteins and Their Potential Impact on COVID-19 Disease Progression. Rev. Med. Virol. 2022, 32, e2300. [Google Scholar] [CrossRef] [PubMed]
- Low, Z.Y.; Yip, A.J.W.; Sharma, A.; Lal, S.K. SARS Coronavirus Outbreaks Past and Present—A Comparative Analysis of SARS-CoV-2 and Its Predecessors. Virus Genes 2021, 57, 307–317. [Google Scholar] [CrossRef]
- World Health Organization. Tracking SARS-CoV-2 Variants. Available online: https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/ (accessed on 29 March 2022).
- Wehbe, Z.; Hammoud, S.; Soudani, N.; Zaraket, H.; El-Yazbi, A.; Eid, A.H. Molecular Insights into SARS COV-2 Interaction With Cardiovascular Disease: Role of RAAS and MAPK Signaling. Front. Pharmacol. 2020, 11, 836. [Google Scholar] [CrossRef] [PubMed]
- Low, Z.Y.; Farouk, I.A.; Lal, S.K. Drug Repositioning: New Approaches and Future Prospects for Life-Debilitating Diseases and the COVID-19 Pandemic Outbreak. Viruses 2020, 12, 1058. [Google Scholar] [CrossRef]
- Jin, G.; Wong, S.T.C. Toward Better Drug Repositioning: Prioritising and Integrating Existing Methods into Efficient Pipelines. Drug Discov. Today 2014, 19, 637–644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- U.S. Food & Drug Administration. Remdesivir (Veklury) Approval for the Treatment of COVID-19—The Evidence for Safety and Efficacy. Available online: https://www.fda.gov/drugs/news-events-human-drugs/remdesivir-veklury-approval-treatment-covid-19-evidence-safety-and-efficacy (accessed on 29 March 2022).
- Sharma, A.; Ahmad Farouk, I.; Lal, S.K. COVID-19: A Review on the Novel Coronavirus Disease Evolution, Transmission, Detection, Control and Prevention. Viruses 2021, 13, 202. [Google Scholar] [CrossRef] [PubMed]
- Shuter, J. Lopinavir/Ritonavir in the Treatment of HIV-1 Infection: A Review. Ther. Clin. Risk Manag. 2008, 4, 1023–1033. [Google Scholar] [CrossRef] [Green Version]
- Sham, H.L.; Kempf, D.J.; Molla, A.; Marsh, K.C.; Kumar, G.N.; Chen, C.-M.; Kati, W.; Stewart, K.; Lal, R.; Hsu, A.; et al. ABT-378, a Highly Potent Inhibitor of the Human Immunodeficiency Virus Protease. Antimicrob. Agents Chemother. 1998, 42, 3218–3224. [Google Scholar] [CrossRef] [Green Version]
- Bolcato, G.; Bissaro, M.; Pavan, M.; Sturlese, M.; Moro, S. Targeting the Coronavirus SARS-CoV-2: Computational Insights into the Mechanism of Action of the Protease Inhibitors Lopinavir, Ritonavir and Nelfinavir. Sci. Rep. 2020, 10, 20927. [Google Scholar] [CrossRef]
- Õmura, S.; Crump, A. The Life and Times of Ivermectin—A Success Story. Nat. Rev. Microbiol. 2004, 2, 984–989. [Google Scholar] [CrossRef]
- Muñoz-Muñoz, L.; Shoen, C.; Sweet, G.; Vitoria, A.; Bull, T.J.; Cynamon, M.; Thompson, C.J.; Ramón-García, S. Repurposing Avermectins and Milbemycins against Mycobacteroides Abscessus and Other Nontuberculous Mycobacteria. Antibiotics 2021, 10, 381. [Google Scholar] [CrossRef] [PubMed]
- Low, Z.Y.; Yip, A.J.W.; Lal, S.K. Repositioning Ivermectin for Covid-19 Treatment: Molecular Mechanisms of Action against SARS-CoV-2 Replication. Biochim. Biophys. Acta BBA Mol. Basis Dis. 2022, 1868, 166294. [Google Scholar] [CrossRef]
- Amin, S.A.; Jha, T. Fight against Novel Coronavirus: A Perspective of Medicinal Chemists. Eur. J. Med. Chem. 2020, 201, 112559. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Wu, J.; Wang, H.; Gao, Y.; Liu, Q.; Mu, A.; Ji, W.; Yan, L.; Zhu, Y.; Zhu, C.; et al. Structural Basis for RNA Replication by the SARS-CoV-2 Polymerase. Cell 2020, 182, 417–428.e13. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.L.; Tan, K.S.W.; Chu, J.J.H.; Chow, V.T. Combination Treatment with Remdesivir and Ivermectin Exerts Highly Synergistic and Potent Antiviral Activity Against Murine Coronavirus Infection. Front. Cell. Infect. Microbiol. 2021, 11, 700502. [Google Scholar] [CrossRef] [PubMed]
- U.S. Food & Drug Administration. FDA Approves First Treatment for COVID-19. Available online: https://www.fda.gov/news-events/press-announcements/fda-approves-first-treatment-covid-19 (accessed on 29 March 2022).
- Warren, T.K.; Jordan, R.; Lo, M.K.; Ray, A.S.; Mackman, R.L.; Soloveva, V.; Siegel, D.; Perron, M.; Bannister, R.; Hui, H.C.; et al. Therapeutic Efficacy of the Small Molecule GS-5734 against Ebola Virus in Rhesus Monkeys. Nature 2016, 531, 381–385. [Google Scholar] [CrossRef]
- Malin, J.J.; Suárez, I.; Priesner, V.; Fätkenheuer, G.; Rybniker, J. Remdesivir against COVID-19 and Other Viral Diseases. Clin. Microbiol. Rev. 2020, 34, e00162-20. [Google Scholar] [CrossRef]
- Lee, C.-C.; Hsieh, C.-C.; Ko, W.-C. Molnupiravir—A Novel Oral Anti-SARS-CoV-2 Agent. Antibiotics 2021, 10, 1294. [Google Scholar] [CrossRef]
- Singh, A.K.; Singh, A.; Singh, R.; Misra, A. Molnupiravir in COVID-19: A Systematic Review of Literature. Diabetes Metab. Syndr. Clin. Res. Rev. 2021, 15, 102329. [Google Scholar] [CrossRef]
- Ridgebackbio. Lagevrio (Molnupiravir). Available online: https://ridgebackbio.com/pipeline/lagevrio/ (accessed on 29 May 2022).
- Agostini, M.L.; Pruijssers, A.J.; Chappell, J.D.; Gribble, J.; Lu, X.; Andres, E.L.; Bluemling, G.R.; Lockwood, M.A.; Sheahan, T.P.; Sims, A.C.; et al. Small-Molecule Antiviral β-d-N4-Hydroxycytidine Inhibits a Proofreading-Intact Coronavirus with a High Genetic Barrier to Resistance. J. Virol. 2019, 93, e01348-19. [Google Scholar] [CrossRef] [Green Version]
- Reynard, O.; Nguyen, X.-N.; Alazard-Dany, N.; Barateau, V.; Cimarelli, A.; Volchkov, V. Identification of a New Ribonucleoside Inhibitor of Ebola Virus Replication. Viruses 2015, 7, 6233–6240. [Google Scholar] [CrossRef] [PubMed]
- Abdelnabi, R.; Foo, C.S.; De Jonghe, S.; Maes, P.; Weynand, B.; Neyts, J. Molnupiravir Inhibits Replication of the Emerging SARS-CoV-2 Variants of Concern in a Hamster Infection Model. J. Infect. Dis. 2021, 224, 749–753. [Google Scholar] [CrossRef] [PubMed]
- Merck. Merck and Ridgeback’s Molnupiravir, an Oral COVID-19 Antiviral Medicine, Receives First Authorization in the World. Available online: https://www.merck.com/news/merck-and-ridgebacks-molnupiravir-an-oral-covid-19-antiviral-medicine-receives-first-authorization-in-the-world/ (accessed on 29 March 2022).
- Eisfeld, A.J.; Neumann, G.; Kawaoka, Y. At the Centre: Influenza A Virus Ribonucleoproteins. Nat. Rev. Microbiol. 2015, 13, 28–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fodor, E.; Te Velthuis, A.J.W. Structure and Function of the Influenza Virus Transcription and Replication Machinery. Cold Spring Harb. Perspect. Med. 2020, 10, a038398. [Google Scholar] [CrossRef] [Green Version]
- Toots, M.; Plemper, R.K. Next-Generation Direct-Acting Influenza Therapeutics. Transl. Res. 2020, 220, 33–42. [Google Scholar] [CrossRef] [Green Version]
- Toots, M.; Yoon, J.-J.; Cox, R.M.; Hart, M.; Sticher, Z.M.; Makhsous, N.; Plesker, R.; Barrena, A.H.; Reddy, P.G.; Mitchell, D.G.; et al. Characterisation of Orally Efficacious Influenza Drug with High Resistance Barrier in Ferrets and Human Airway Epithelia. Sci. Transl. Med. 2019, 11, eaax5866. [Google Scholar] [CrossRef]
- Yoon, J.-J.; Toots, M.; Lee, S.; Lee, M.-E.; Ludeke, B.; Luczo, J.M.; Ganti, K.; Cox, R.M.; Sticher, Z.M.; Edpuganti, V.; et al. Orally Efficacious Broad-Spectrum Ribonucleoside Analog Inhibitor of Influenza and Respiratory Syncytial Viruses. Antimicrob. Agents Chemother. 2018, 62, e00766-18. [Google Scholar] [CrossRef] [Green Version]
- Ma, B.; Huang, C.; Ma, J.; Xiang, Y.; Zhang, X. Structure of Venezuelan Equine Encephalitis Virus with Its Receptor LDLRAD3. Nature 2021, 598, 677–681. [Google Scholar] [CrossRef]
- Sharma, A.; Knollmann-Ritschel, B. Current Understanding of the Molecular Basis of Venezuelan Equine Encephalitis Virus Pathogenesis and Vaccine Development. Viruses 2019, 11, 164. [Google Scholar] [CrossRef] [Green Version]
- Urakova, N.; Kuznetsova, V.; Crossman, D.K.; Sokratian, A.; Guthrie, D.B.; Kolykhalov, A.A.; Lockwood, M.A.; Natchus, M.G.; Crowley, M.R.; Painter, G.R.; et al. β-d-N4-Hydroxycytidine Is a Potent Anti-Alphavirus Compound That Induces a High Level of Mutations in the Viral Genome. J. Virol. 2018, 92, e01965-17. [Google Scholar] [CrossRef] [Green Version]
- Painter, G.R.; Bowen, R.A.; Bluemling, G.R.; DeBergh, J.; Edpuganti, V.; Gruddanti, P.R.; Guthrie, D.B.; Hager, M.; Kuiper, D.L.; Lockwood, M.A.; et al. The Prophylactic and Therapeutic Activity of a Broadly Active Ribonucleoside Analog in a Murine Model of Intranasal Venezuelan Equine Encephalitis Virus Infection. Antiviral Res. 2019, 171, 104597. [Google Scholar] [CrossRef] [PubMed]
- Bartlam, M.; Yang, H.; Rao, Z. Structural Insights into SARS Coronavirus Proteins. Curr. Opin. Struct. Biol. 2005, 15, 664–672. [Google Scholar] [CrossRef] [PubMed]
- Yoshimoto, F.K. The Proteins of Severe Acute Respiratory Syndrome Coronavirus-2 (SARS CoV-2 or n-COV19), the Cause of COVID-19. Protein J. 2020, 39, 198–216. [Google Scholar] [CrossRef] [PubMed]
- Sheahan, T.P.; Sims, A.C.; Zhou, S.; Graham, R.L.; Pruijssers, A.J.; Agostini, M.L.; Leist, S.R.; Schafer, A.; Dinnon III, K.H.; Stevens, L.J.; et al. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl. Med. 2020, 12, eabb5883. [Google Scholar] [CrossRef] [Green Version]
- Pruijssers, A.J.; Denison, M.R. Nucleoside Analogues for the Treatment of Coronavirus Infections. Curr. Opin. Virol. 2019, 35, 57–62. [Google Scholar] [CrossRef]
- Barnard, D.L.; Hubbard, V.D.; Burton, J.; Smee, D.F.; Morrey, J.D.; Otto, M.J.; Sidwell, R.W. Inhibition of Severe Acute Respiratory Syndrome-Associated Coronavirus (SARSCoV) by Calpain Inhibitors and β-D-N4-Hydroxycytidine. Antivir. Chem. Chemother. 2004, 15, 15–22. [Google Scholar] [CrossRef] [Green Version]
- van der Hoek, L.; Pyrc, K.; Jebbink, M.F.; Vermeulen-Oost, W.; Berkhout, R.J.M.; Wolthers, K.C.; Wertheim-van Dillen, P.M.E.; Kaandorp, J.; Spaargaren, J.; Berkhout, B. Identification of a New Human Coronavirus. Nat. Med. 2004, 10, 368–373. [Google Scholar] [CrossRef]
- Shereen, M.A.; Khan, S.; Kazmi, A.; Bashir, N.; Siddique, R. COVID-19 Infection: Emergence, Transmission, and Characteristics of Human Coronaviruses. J. Adv. Res. 2020, 24, 91–98. [Google Scholar] [CrossRef]
- Zhu, Z.; Lian, X.; Su, X.; Wu, W.; Marraro, G.A.; Zeng, Y. From SARS and MERS to COVID-19: A Brief Summary and Comparison of Severe Acute Respiratory Infections Caused by Three Highly Pathogenic Human Coronaviruses. Respir. Res. 2020, 21, 224. [Google Scholar] [CrossRef]
- Neuman, B.W.; Adair, B.D.; Yoshioka, C.; Quispe, J.D.; Orca, G.; Kuhn, P.; Milligan, R.A.; Yeager, M.; Buchmeier, M.J. Supramolecular Architecture of Severe Acute Respiratory Syndrome Coronavirus Revealed by Electron Cryomicroscopy. J. Virol. 2006, 80, 7918–7928. [Google Scholar] [CrossRef] [Green Version]
- Low, Z.Y.; Yip, A.J.W.; Lal, S.K. Repositioning anticancer drugs as novel COVID-19 antivirals: Targeting structural and functional similarities between viral proteins and cancer. Expert Rev. Mol. Med. 2022, 24, e20. [Google Scholar] [CrossRef] [PubMed]
- Hillen, H.S.; Kokic, G.; Farnung, L.; Dienemann, C.; Tegunov, D.; Cramer, P. Structure of Replicating SARS-CoV-2 Polymerase. Nature 2020, 584, 154–156. [Google Scholar] [CrossRef] [PubMed]
- Machitani, M.; Yasukawa, M.; Nakashima, J.; Furuichi, Y.; Masutomi, K. RNA-Dependent RNA Polymerase, RdRP, a Promising Therapeutic Target for Cancer and Potentially COVID-19. Cancer Sci. 2020, 111, 3976–3984. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Yan, L.; Huang, Y.; Liu, F.; Zhao, Y.; Cao, L.; Wang, T.; Sun, Q.; Ming, Z.; Zhang, L.; et al. Structure of the RNA-Dependent RNA Polymerase from COVID-19 Virus. Science 2020, 368, 779–782. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Gong, P. Visualising the Nucleotide Addition Cycle of Viral RNA-Dependent RNA Polymerase. Viruses 2018, 10, 24. [Google Scholar] [CrossRef] [Green Version]
- Venkataraman, S.; Prasad, B.; Selvarajan, R. RNA Dependent RNA Polymerases: Insights from Structure, Function and Evolution. Viruses 2018, 10, 76. [Google Scholar] [CrossRef] [Green Version]
- Pirzada, R.H.; Haseeb, M.; Batool, M.; Kim, M.; Choi, S. Remdesivir and Ledipasvir among the FDA-Approved Antiviral Drugs Have Potential to Inhibit SARS-CoV-2 Replication. Cells 2021, 10, 1052. [Google Scholar] [CrossRef]
- Imran, M.; Kumar Arora, M.; Asdaq, S.M.B.; Khan, S.A.; Alaqel, S.I.; Alshammari, M.K.; Alshehri, M.M.; Alshrari, A.S.; Mateq Ali, A.; Al-shammeri, A.M.; et al. Discovery, Development, and Patent Trends on Molnupiravir: A Prospective Oral Treatment for COVID-19. Molecules 2021, 26, 5795. [Google Scholar] [CrossRef]
- Ehteshami, M.; Tao, S.; Zandi, K.; Hsiao, H.-M.; Jiang, Y.; Hammond, E.; Amblard, F.; Russell, O.O.; Merits, A.; Schinazi, R.F. Characterization of β-d-N4-Hydroxycytidine as a Novel Inhibitor of Chikungunya Virus. Antimicrob. Agents Chemother. 2017, 61, e02395-16. [Google Scholar] [CrossRef] [Green Version]
- Kabinger, F.; Stiller, C.; Schmitzová, J.; Dienemann, C.; Kokic, G.; Hillen, H.S.; Höbartner, C.; Cramer, P. Mechanism of Molnupiravir-Induced SARS-CoV-2 Mutagenesis. Nat. Struct. Mol. Biol. 2021, 28, 740–746. [Google Scholar] [CrossRef]
- Jena, N.R. Role of Different Tautomers in the Base-Pairing Abilities of Some of the Vital Antiviral Drugs Used against COVID-19. Phys. Chem. Chem. Phys. 2020, 22, 28115–28122. [Google Scholar] [CrossRef] [PubMed]
- Traut, T.W. Physiological Concentrations of Purines and Pyrimidines. Mol. Cell. Biochem. 1994, 140, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Gordon, C.J.; Tchesnokov, E.P.; Schinazi, R.F.; Götte, M. Molnupiravir Promotes SARS-CoV-2 Mutagenesis via the RNA Template. J. Biol. Chem. 2021, 297, 100770. [Google Scholar] [CrossRef] [PubMed]
- Stuyver, L.J.; Whitaker, T.; McBrayer, T.R.; Hernandez-Santiago, B.I.; Lostia, S.; Tharnish, P.M.; Ramesh, M.; Chu, C.K.; Jordan, R.; Shi, J.; et al. Ribonucleoside Analogue That Blocks Replication of Bovine Viral Diarrhea and Hepatitis C Viruses in Culture. Antimicrob. Agents Chemother. 2003, 47, 244–254. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Malone, B.; Llewellyn, E.; Grasso, M.; Shelton, P.M.M.; Olinares, P.D.B.; Maruthi, K.; Eng, E.T.; Vatandaslar, H.; Chait, B.T.; et al. Structural Basis for Helicase-Polymerase Coupling in the SARS-CoV-2 Replication-Transcription Complex. Cell 2020, 182, 1560–1573.e13. [Google Scholar] [CrossRef]
- Malone, B.; Campbell, E.A. Molnupiravir: Coding for Catastrophe. Nat. Struct. Mol. Biol. 2021, 28, 706–708. [Google Scholar] [CrossRef]
- Han, K.; Blair, R.V.; Iwanaga, N.; Liu, F.; Russell-Lodrigue, K.E.; Qin, Z.; Midkiff, C.C.; Golden, N.A.; Doyle-Meyers, L.A.; Kabir, M.E.; et al. Lung Expression of Human Angiotensin-Converting Enzyme 2 Sensitises the Mouse to SARS-CoV-2 Infection. Am. J. Respir. Cell Mol. Biol. 2021, 64, 79–88. [Google Scholar] [CrossRef]
- Painter, W.P.; Holman, W.; Bush, J.A.; Almazedi, F.; Malik, H.; Eraut, N.C.J.E.; Morin, M.J.; Szewczyk, L.J.; Painter, G.R. Human Safety, Tolerability, and Pharmacokinetics of Molnupiravir, a Novel Broad-Spectrum Oral Antiviral Agent with Activity against SARS-CoV-2. Antimicrob. Agents Chemother. 2021, 65, e02428-20. [Google Scholar] [CrossRef]
- Merck. Merck and Ridgeback’s Investigational Oral Antiviral Molnupiravir Reduced the Risk of Hospitalisation or Death by Approximately 50 Percent Compared to Placebo for Patients with Mild or Moderate COVID-19 in Positive Interim Analysis of Phase 3 Study. Available online: https://www.merck.com/news/merck-and-ridgebacks-investigational-oral-antiviral-molnupiravir-reduced-the-risk-of-hospitalization-or-death-by-approximately-50-percent-compared-to-placebo-for-patients-with-mild-or-moderat/ (accessed on 29 March 2022).
- Mahase, E. COVID-19: Molnupiravir Reduces Risk of Hospital Admission or Death by 50% in Patients at Risk, MSD Reports. BMJ 2021, 375, n2422. [Google Scholar] [CrossRef]
- Wahl, A.; Gralinski, L.E.; Johnson, C.E.; Yao, W.; Kovarova, M.; Dinnon, K.H.; Liu, H.; Madden, V.J.; Krzystek, H.M.; De, C.; et al. SARS-CoV-2 Infection Is Effectively Treated and Prevented by EIDD-2801. Nature 2021, 591, 451–457. [Google Scholar] [CrossRef]
- Cox, R.M.; Wolf, J.D.; Plemper, R.K. Therapeutically Administered Ribonucleoside Analogue MK-4482/EIDD-2801 Blocks SARS-CoV-2 Transmission in Ferrets. Nat. Microbiol. 2021, 6, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Merck Sharp & Dohme. A Phase 3, Multicenter, Randomized, Double-Blind, Placebo-Controlled Study to Evaluate the Efficacy and Safety of MK-4482 for the Prevention of COVID-19 (Laboratory-Confirmed SARS-CoV-2 Infection with Symptoms) in Adults Residing with a Person with COVID-19. Available online: https://clinicaltrials.gov/ct2/show/NCT04939428 (accessed on 29 March 2022).
- Ridgeback Biotherapeutics. The Safety of EIDD-2801 and Its Effect on Viral Shedding of SARS-CoV-2 (END-COVID). Available online: https://clinicaltrials.gov/ct2/show/NCT04405739 (accessed on 29 March 2022).
- University of Liverpool. AGILE (Early Phase Platform Trial for COVID-19). Available online: https://clinicaltrials.gov/ct2/show/NCT04746183 (accessed on 29 March 2022).
- Merck Sharp & Dohme. Efficacy and Safety of Molnupiravir (MK-4482) in Hospitalized Adult Participants With COVID-19) in Adults (MK-4482-001). Available online: https://clinicaltrials.gov/ct2/show/NCT04575584 (accessed on 29 March 2022).
- Masyeni, S.; Iqhrammullah, M.; Frediansyah, A.; Nainu, F.; Tallei, T.; Emran, T.B.; Ophinni, Y.; Dhama, K.; Harapan, H. Molnupiravir: A lethal mutagenic drug against rapidly mutating severe acute respiratory syndrome coronavirus 2 - A narrative review. J. Med. Virol. 2022, 94, 3006–3016. [Google Scholar] [CrossRef] [PubMed]
- Painter, G.R.; Natchus, M.G.; Cohen, O.; Holman, W.; Painter, W.P. Developing a direct acting, orally available antiviral agent in a pandemic: The evolution of molnupiravir as a potential treatment for COVID-19. Curr. Opin. Virol. 2021, 50, 17–22. [Google Scholar] [CrossRef]
- Githaka, J.M. Molnupiravir Does Not Induce Mutagenesis in Host Lung Cells during SARS-CoV-2 Treatment. Bioinform. Biol. Insights 2022, 16, 11779322221085077. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Hill, C.S.; Sarkar, S.; Tse, L.V.; Woodburn, B.M.D.; Schinazi, R.F.; Sheahan, T.P.; Baric, R.S.; Heise, M.T.; Swanstrom, R. β-D-N4-hydroxycytidine Inhibits SARS-CoV-2 Through Lethal Mutagenesis but Is Also Mutagenic to Mammalian Cells. J. Infect. Dis. 2021, 223, 415–419. [Google Scholar] [CrossRef]
- Merck. Important Safety Information Regarding Use of LAGEVRIO™ (molnupiravir) in Pregnancy and Individuals of Childbearing Potential. Available online: https://fda.gov/media/155101/download (accessed on 7 June 2022).
- Ji, W.; Luo, G. Zika Virus NS5 Nuclear Accumulation Is Protective of Protein Degradation and Is Required for Viral RNA Replication. Virology 2020, 541, 124–135. [Google Scholar] [CrossRef]
- Yip, T.-F.; Selim, A.S.M.; Lian, I.; Lee, S.M.-Y. Advancements in Host-Based Interventions for Influenza Treatment. Front. Immunol. 2018, 9, 1547. [Google Scholar] [CrossRef]
- Miorin, L.; Kehrer, T.; Sanchez-Aparicio, M.T.; Zhang, K.; Cohen, P.; Patel, R.S.; Cupic, A.; Makio, T.; Mei, M.; Moreno, E.; et al. SARS-CoV-2 Orf6 Hijacks Nup98 to Block STAT Nuclear Import and Antagonise Interferon Signaling. Proc. Natl. Acad. Sci. USA 2020, 117, 28344–28354. [Google Scholar] [CrossRef]
- Yang, S.; Atkinson, S.; Fraser, J.; Wang, C.; Maher, B.; Roman, N.; Forwood, J.; Wagstaff, K.; Borg, N.; Jans, D. Novel Flavivirus Antiviral That Targets the Host Nuclear Transport Importin α/Β1 Heterodimer. Cells 2019, 8, 281. [Google Scholar] [CrossRef] [Green Version]
- Jans, D.A.; Martin, A.J.; Wagstaff, K.M. Inhibitors of Nuclear Transport. Curr. Opin. Cell Biol. 2019, 58, 50–60. [Google Scholar] [CrossRef]
- Saha, J.K.; Raihan, M.J. The Binding Mechanism of Ivermectin and Levosalbutamol with Spike Protein of SARS-CoV-2. Struct. Chem. 2021, 32, 1985–1992. [Google Scholar] [CrossRef] [PubMed]
- Mody, V.; Ho, J.; Wills, S.; Mawri, A.; Lawson, L.; Ebert, M.C.C.J.C.; Fortin, G.M.; Rayalam, S.; Taval, S. Identification of 3-Chymotrypsin like Protease (3CLPro) Inhibitors as Potential Anti-SARS-CoV-2 Agents. Commun. Biol. 2021, 4, 93. [Google Scholar] [CrossRef] [PubMed]
- Anand, K.; Ziebuhr, J.; Wadhwani, P.; Mesters, J.R.; Hilgenfeld, R. Coronavirus Main Proteinase (3CL pro) Structure: Basis for Design of Anti-SARS Drugs. Science 2003, 300, 1763–1767. [Google Scholar] [CrossRef] [Green Version]
- Camprubí, D.; Almuedo-Riera, A.; Martí-Soler, H.; Soriano, A.; Hurtado, J.C.; Subirà, C.; Grau-Pujol, B.; Krolewiecki, A.; Muñoz, J. Lack of Efficacy of Standard Doses of Ivermectin in Severe COVID-19 Patients. PLoS ONE 2020, 15, e0242184. [Google Scholar] [CrossRef] [PubMed]
- Krolewiecki, A.; Lifschitz, A.; Moragas, M.; Travacio, M.; Valentini, R.; Alonso, D.F.; Solari, R.; Tinelli, M.A.; Cimino, R.O.; Álvarez, L.; et al. Antiviral Effect of High-Dose Ivermectin in Adults with COVID-19: A Proof-of-Concept Randomised Trial. EClinicalMedicine 2021, 37, 100959. [Google Scholar] [CrossRef] [PubMed]
- López-Medina, E.; López, P.; Hurtado, I.C.; Dávalos, D.M.; Ramirez, O.; Martínez, E.; Díazgranados, J.A.; Oñate, J.M.; Chavarriaga, H.; Herrera, S.; et al. Effect of Ivermectin on Time to Resolution of Symptoms Among Adults with Mild COVID-19: A Randomized Clinical Trial. JAMA 2021, 325, 1426. [Google Scholar] [CrossRef] [PubMed]
- Ramírez, C.; Herrera-Paz, E.F.; Peralta, G.; Rodríguez, G.; Durón, R.M. Is Ivermectin Ready to Be Part of a Public Health Policy for COVID-19 Prophylaxis? EClinicalMedicine 2021, 32, 100744. [Google Scholar] [CrossRef]
- Kokic, G.; Hillen, H.S.; Tegunov, D.; Dienemann, C.; Seitz, F.; Schmitzova, J.; Farnung, L.; Siewert, A.; Höbartner, C.; Cramer, P. Mechanism of SARS-CoV-2 Polymerase Stalling by Remdesivir. Nat. Commun. 2021, 12, 279. [Google Scholar] [CrossRef]
- Gordon, C.J.; Tchesnokov, E.P.; Woolner, E.; Perry, J.K.; Feng, J.Y.; Porter, D.P.; Götte, M. Remdesivir Is a Direct-Acting Antiviral That Inhibits RNA-Dependent RNA Polymerase from Severe Acute Respiratory Syndrome Coronavirus 2 with High Potency. J. Biol. Chem. 2020, 295, 6785–6797. [Google Scholar] [CrossRef] [Green Version]
- Tchesnokov, E.P.; Gordon, C.J.; Woolner, E.; Kocinkova, D.; Perry, J.K.; Feng, J.Y.; Porter, D.P.; Götte, M. Template-Dependent Inhibition of Coronavirus RNA-Dependent RNA Polymerase by Remdesivir Reveals a Second Mechanism of Action. J. Biol. Chem. 2020, 295, 16156–16165. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, D.; Du, G.; Du, R.; Zhao, J.; Jin, Y.; Fu, S.; Gao, L.; Cheng, Z.; Lu, Q.; et al. Remdesivir in Adults with Severe COVID-19: A Randomised, Double-Blind, Placebo-Controlled, Multicentre Trial. Lancet 2020, 395, 1569–1578. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).