Abstract
Hepatocarcinogenesis is a complex process that remains still partly understood. That might be explained by the multiplicity of etiologic factors, the genetic/epigenetic heterogeneity of tumors bulks and the ignorance of the liver cell types that give rise to tumorigenic cells that have stem cell-like properties. The DNA stress induced by hepatocyte turnover, inflammation and maybe early oncogenic pathway activation and sometimes viral factors, leads to DNA damage response which activates the key tumor suppressive checkpoints p53/p21Cip1 and p16INK4a/pRb responsible of cell cycle arrest and cellular senescence as reflected by the cirrhosis stage. Still obscure mechanisms, but maybe involving the Wnt signaling and Twist proteins, would allow pre-senescent hepatocytes to bypass senescence, acquire immortality by telomerase reactivation and get the last genetic/epigenetic hits necessary for cancerous transformation. Among some of the oncogenic pathways that might play key driving roles in hepatocarcinogenesis, c-myc and the Wnt/β-catenin signaling seem of particular interest. Finally, antiproliferative and apoptosis deficiencies involving TGF-β, Akt/PTEN, IGF2 pathways for instance are prerequisite for cancerous transformation. Of evidence, not only the transformed liver cell per se but the facilitating microenvironment is of fundamental importance for tumor bulk growth and metastasis.
1. Introduction
Hepatocellular carcinoma (HCC) is one of the most prevalent cancers worldwide, developing mainly in cirrhosis. Hepatitis B (HBV) or C virus (HCV) chronic infections account for 75% of HCCs whereas nonviral etiologies as alcohol, genetic or metabolic disorders represent less than 25% of cases. Furthermore, western countries suffer from a substantial and constant increase of HCC incidence due to HCV infection. Dramatically, HCC is a poor prognosis tumor, and is the first cause of death in cirrhotic patients. Current therapies are rather inefficient, mainly due to usually late diagnosis and high recurrence rates within the remaining cirrhotic liver after surgical resection [1-3].
Hepatocarcinogenesis is tightly linked to chronic liver damage, and rarely develops in healthy liver. That might be due to the possible requirement of chronic inflammation and cell divisions in a context of cellular stress which lead towards the step-wise acquisition of genetic and epigenetic hits necessary for cellular transformation. In addition, the virus persistence per se can trigger deregulation of the cellular machinery. By contrast to HCV, HBV can integrate into the host genome, leading to genomic instability, rearrangements and more rarely cis- or trans- activation of proto-oncogenes. Although the direct involvement of viral proteins in hepatocarcinogenesis is not clear, it seems that HBx and Pre-S2 for HBV as well as core and others for HCV can interact with and deregulate cellular machinery. However, data was obtained from in vitro transfection assays or in vivo transgenic mouse models. Nevertheless, these models support supra-natural viral protein expression levels, and nothing is known about these interplays in a context of natural viral infection [4-6].
Herein we will aim to describe the general mechanisms which could be involved in hepatocarcinogenesis independently of etiologic factors underlying the chronic liver disease.
2. Tumor Bulk and Cancer Stem Cell Concept
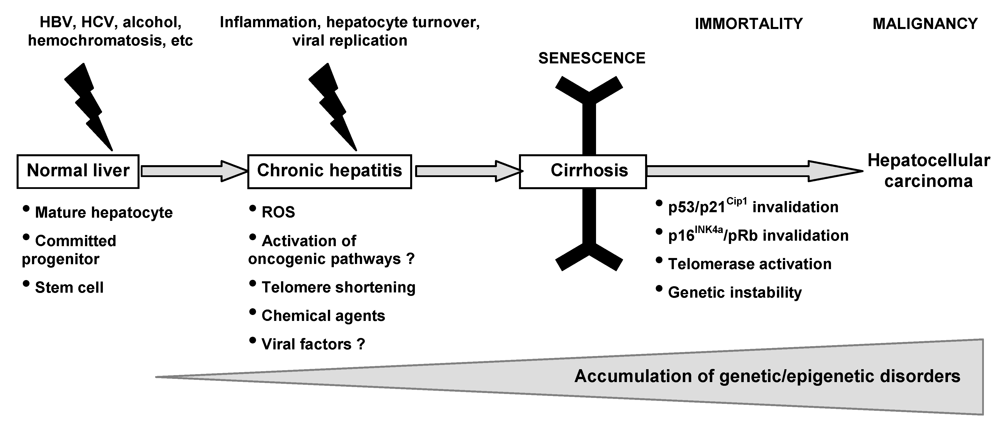
The most common and unifying condition associated with hepatocarcinogenesis is cirrhosis, which develops after long latencies (20-40 years) of chronic liver disease. HCC risk remains low during chronic liver disease but dramatically increases at the cirrhotic stage. Hepatocarcinogenesis remains partly obscure. Initially, a variety of genetic and epigenetic alterations have been detected in human and experimental HCCs. Later on, DNA microarray analysis has led to an extensive integrative approach, leading to identification of clusters of HCCs that allow comparison between phenotypes in experimental and human HCCs, and may predict outcome of patients. However, none of the identified genes is universally expressed by tumor cells that are heterogeneous in their morphology, clinical behaviour, and molecular profiles in the tumor bulk [7-9]. These observations lead to the suspicion that the current studies might have focused only on the heterogeneous “end products” – i.e. “adult” tumor cells within the tumor bulk - but not the “root” cell (Figure 1) [10-13]. More interestingly, molecular signatures from cirrhotic nonneoplastic tissues can predict occurrence/reccurrence of HCC [14], letting hypothesize that although “root” cells exist within the heterogeneous “end product” tumor bulk, they also might be present in cirrhotic tissues prone to develop HCC.
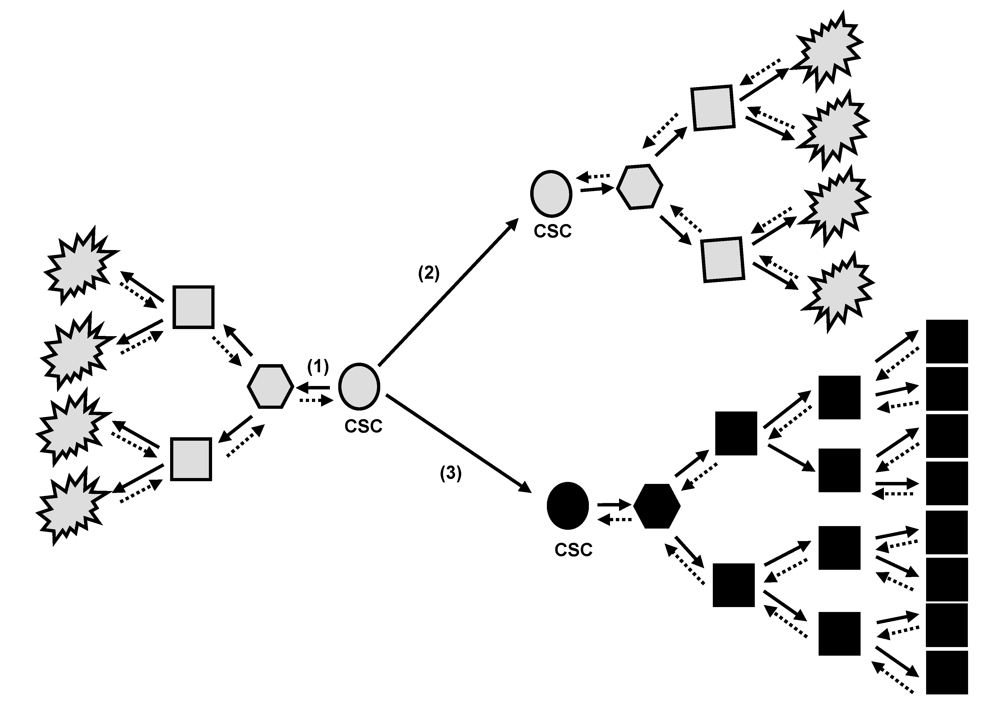
Figure 1.
The cancer stem cells (CSC) model. Proposed hierarchical organization of a malignant clone. CSC have self-renew and tumorigenic capacities. They can give rise to more and more differentiated cancerous cells which the last ones in the hierarchical organization may have low or absent proliferative capabilities (1). During asymetric division at self-renew, the original CSC can give birth to the identical CSC (2) that will lead to the same tumor clone (grey), or can give birth to a genetically/epigenetically modified CSC (3) that will lead to a more aggressive tumor clone (black) with differentiation blockage and higher proliferative capabilities. To date, it is not clear whether the more differentiated cells can revert back and regain a more stem cell properties. As appreciated from the definition of a CSC, this cell is not necessarily derived from a normal tissue stem cell. Alternatively, hypothetical genetic/epigenetic changes caused in more committed and differentiated cancerous cells might be very well involved in mechanisms allowing these cells to acquire cancer stem cell capabilities.
Tumors originate from normal cells as a result of accumulated genetic/epigenetic changes. It was initially believed that cancers arose exclusively by de-differentiation of mature cells, and tumor cell heterogeneity could be explained by the clonal evolution model [15]. More recent findings suggest that heterogeneity may come from derivation of endogenous progenitor/stem cells or de-differentiation of a transformed cell [16-17]. This hypothesis supports an early proposal that cancers represent “blocked ontogeny” and a derivative that cancers are transformed stem cells (Figure 1). The normal liver has tissue-determined stem cells and ductal bipotent committed progenitors that are scarce and not brought into play except with the occurrence of both liver injury and inhibition of proliferation of mature hepatocytes. These latter are not terminally differentiated and can respond to injury by highly regulated proliferation. Although the cell type giving rise to HCC has been shown as dependent on many factors in experimental hepatocarcinogenesis, few is known in humans. One can speculate that “root” cells might come from the neoplastic transformation of different normal liver cell types: periductal stem cells, bipolar ductal committed progenitor cells, or differentiated hepatocytes. Thus, it would not be surprising to find HCC as arising differentially from one (or several) of them, depending on extrinsic factors such as viral infection, or deregulation of intrinsic key pathways [18-23]. The maturation arrest of cells at various stages of differentiation in a hierarchical cell lineage may best explain the various types of human liver cancer. From analysis of established HCCs, we might speculate that HCCs contain cancer stem cells (CSC) – i.e. cells with stem-cell-like properties of immortality, resistance to therapy, and transplantability. As hepatocarcinogenesis is likely a dynamic process leading from a normal cell towards an initiated root cell and thereafter a hugely heterogeneous tumor bulk, abnormalities useful for initiation of root cells are not mandatory found in all cancerous cells forming the tumor bulk. In addition, abnormalities found in tumor bulks might evolve with time and/or under pressure of anti-cancer therapies, thus strengthening the need of cautious when interpreting a molecular profile. However, some fundamental events have been described as key steps in cellular transformation and very likely necessary and sufficient to allow each cell to get and keep the cancerous phenotype.
3. Oncogenic Stress and Cellular Behaviour
Cancer cells contain multiple genetic/epigenetic alterations, and chromosomal aberrations. It has been rationalized that a long period of time is required for any individual cell to accumulate the right combination of alterations that promote the cancer cell phenotype. Alterations consistently found in cancer cells are selected because they confer a growth advantage by either activating growth promoting pathways, inactivating growth inhibitory cascades or allowing alterations to accumulate [24]. Over the life span any individual cell can acquire multiple alterations with oncogenic potential, yet only a fraction of them will experience cancer transformation. This fact suggests that organisms evolved mechanisms to prevent oncogenic transformation, the so called anti-oncogenes or tumor suppressor genes. Tumor suppressors may avert cancer by preventing alterations, inducing cell death or a program of cell division arrest known as cellular senescence. This senescence has been experimentally modeled by enforcing the expression of oncogenes in primary cells. The astonishing outcome of these manipulations is that oncogenes trigger antiproliferative responses preventing progression to malignant transformation. These responses bring to an end-proliferation due to cell death or a permanent cell cycle arrest called senescence (Figure 2). On a general point of view, and although it remains to be validated in hepatocarcinogenesis, these results imply mechanisms of DNA damage in cells expressing oncogenes, that may be secondary to reactive oxygen species and/or some form of “oncogenic stress” that affect normal DNA replication. Interestingly, DNA damage signals persist in cells that escape from senescence and go ahead towards cancer transformation [25].
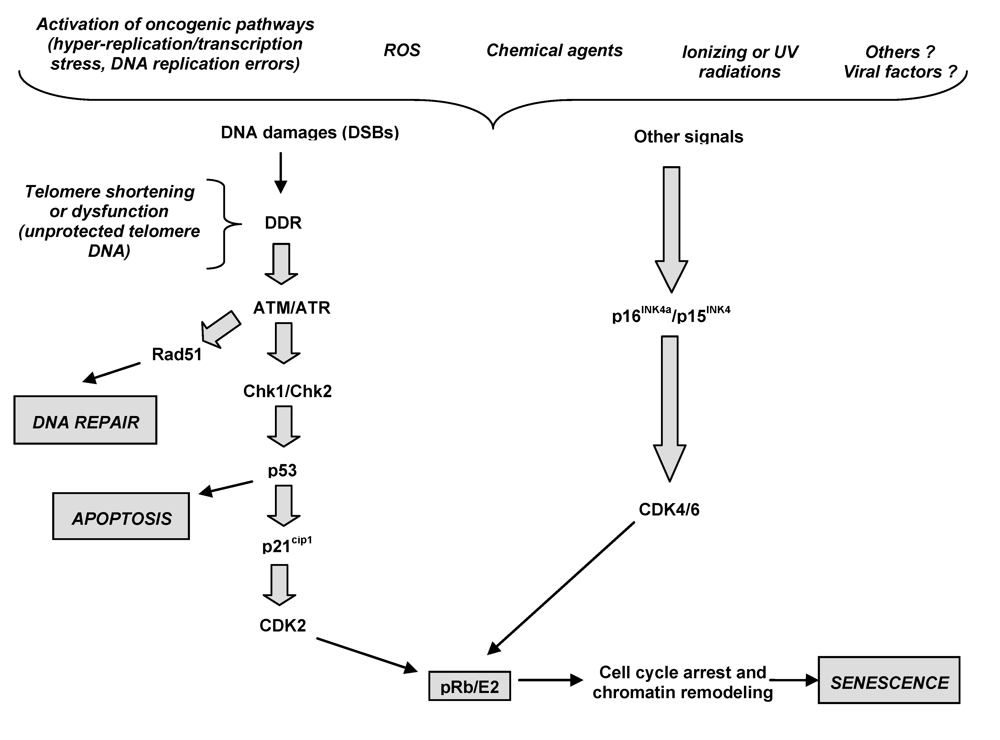
Figure 2.
The proposed molecular mechanisms of cellular senescence. Oncogenic stress result in DNA damages as double strand breaks (DSBs) that lead to activation of ATM/ATR kinases. This leads to stabilization of the Rad51 repair protein needed for DNA DSB repair. Besides DNA repair induction, cooperative action of ATM/ATR, ARF and Chk1/Chk2 and p53 is crucial for the induction of either apoptosis or cell cycle arrest and induction of senescence. Another possibility is that other factors such as reactive oxygen species (ROS) or other physical, chemical of viral agents could induce the accumulation of DNA damages or activate, by alternative mechanisms, the p15INK4b/p16INK4a cascade. All known senescence pathways converge at the level of activation of CDKIs (p15INK4b, p16INK4a and p21Cip1) that keep the pRb protein under its active form. The pRb inhibits E2F and prevents the expression of growth-promoting genes for cell cycle exit. Furthermore, pRb recruits growth-promoting genes into a facultative chromatin structure for permanent silencing and growth arrest.
3.1. Senescence pathways as tumor suppressor mechanisms in chronic liver disease
In healthy adult livers, hepatocytes are quiescent cells, being renewed slowly, approximately once a year. However, the liver has an extremely powerful regenerative capacity, as demonstrated experimentally in rodents, and as observed in patients with chronic liver diseases [26]. This regenerative capacity is due mostly to the ability of mature hepatocytes to proliferate in response to a diminution of the total liver mass either experimentally, or following exposure to viral and nonviral hepatotoxic agents. In addition, the adult liver seems to harbor hepatocyte-progenitor cells (< 0.1% of total hepatocyte mass) that are able to restore liver hepatocyte populations [27]. However, hepatocytes do not have unlimited replicative capacity, due to the lack of telomerase activity that is needed to avoid telomere shortening during successive cell divisions. This is best exemplified by decreased hepatocyte proliferation in the cirrhosis stage of chronic liver diseases, providing in vivo evidence for the exhaustion of hepatocyte proliferation capacity [28].
Senescence mechanisms in hepatocytes and in liver tissue are not well known. However, a limited number of in vitro studies with hepatocytes, as well as numerous descriptive in vivo studies in liver tissue provide sufficient evidence that hepatocytes can undergo senescence type changes. Limited proliferative capacity of somatic cells is controlled by replicative senescence. By contrast to primary hepatocytes which do not proliferate in culture, fetal hepatocytes display better proliferation capacity and can enter replicative senescence [29]. This is accompanied by progressive shortening of telomeres in a context of telomerase-free activity. In contrast to in vitro studies, in vivo senescence of human hepatocytes is better known. Replicative senescence displays a gradual increase from 10% in normal liver, to more than 80% in cirrhosis, being detected in 60% HCCs [30]. Telomere shortening during aging is slow and stabilizes at mid age in healthy liver, so that the loss of telomeric DNA does not reach a level to induce telomere dysfunction and DNA damage response (DDR). On the other hand, telomere loss is accelerated in chronic liver disease to reach lowest levels in the cirrhotic liver. Therefore, one plausible mechanism involved in cirrhosis is probably telomere-dependent senescence, the so-called replicative senescence [31].
Hepatocyte senescence that is observed in severe chronic liver diseases may also be induced by telomere-independent mechanisms, such as ROS-induced senescence (RIS) and oncogene-induced senescence (OIS). RIS and OIS are rare events under normal physiological conditions, but could more commonly occur in context of ROS overproduction and/or oncogenic signals activation, both leading to DDR and activation of the ATM/Chk/p53 pathway and, by alternative mechanisms, the p16INK4a/pRb pathway. OIS is a DNA damage response triggered by DNA hyper-replication [32], and several oncogenic pathways – i.e. activated Ras, c-myc or Wnt/β-catenin - have been shown as potentially involved in OIS in mammary cells or fibroblasts, but nothing is known about liver cell types [33-34]. As witness of DDR occurrence in chronic liver diseases, upregulation of DNA repair enzymes which may reflect increased DNA damages, has been reported in cirrhosis [35]. ROS production can be favored by the context of chronic liver injury with inflammation, cell death, oxidative stress as well as some of the etiological factors such as HCV and alcohol induce mitochondrial dysfunction [36]. Later on, we will describe the main oncogenic pathways found as activated in HCC and that might lead to OIS although this phenomenon of senescence-induction/senescence-escape has not been clearly demonstrated so far in liver cells during hepatocarcinogenesis.
4. Activation of Oncogenic Pathways
Hepatocarcinogenesis is multifactorial and various oncogenes, tumor-suppressor genes, growth factor genes and virologic factors are implicated [53]. Activation of oncogenic pathways in human HCC appears to be more heterogeneous compared with other cancer types. It remains largely unknown the exact sequence of the oncogenic pathways that could be activated during the different steps of hepatocarcinogenesis (Figure 3): i) at the pre-senescent step which leads from normal cells to senescent ones in cirrhosis; and/or ii) at the senescent steps which might allow the cell to bypass senescence, re-enter cell cycle progression and accumulate the next genetic/epigenetic hits required for acquisition of the cancerous phenotype; and/or iii) at the neoplastic step which allow cancerous cells to keep their cancerous phenotype which can evolve with time to aggressiveness, invasiveness and metastatic properties. Herein, we will focus on three major oncogenic pathways that might play a driving role in hepatocarcinogenesis - i.e. Ras, c-myc and Wnt/β-catenin. These pathways have been shown as giving oncogenic stress in vitro in different models of carcinogenesis. Although they have been found activated in human HCC tisssues and/or their matched nontumorous counterparts by comparison to normal livers, experimental data are almost missing regarding their transforming properties on normal nontransformed liver cell types.
4.1. Ras
Proto-oncogenes of the ras family (H-ras, K-ras et N-ras) play a key role in transduction of the mitogenic signal linked to mitogen-activated protein kinases (MAPK). In humans, they can be activated by point mutation at codons 12, 13 and 61. In hepatocarcinogenesis, these mutations are uncommon and, when present, found at codon 12 for K-ras and H-ras, and at codon 61 for K-ras and N-ras [54]. In contrast, the p21-Ras protein is frequently expressed in cirrhosis and HCCs [55]. In vitro, ectopic expression of mutated ras is a strong oncogenic stress factor and leads frequently to cancerous transformation when the p53/p21Cip1 and p16INK4a/pRb checkpoints are inhibited and telomerase reactivated in mammary cell models [33]. However, few are known concerning liver cells.
4.2. c-Myc
The c-myc proto-oncogene stimulates a pattern of cellular gene expression by its regulatory elements and is involved in gene expression during cell growth and differentiation [56]. It has been shown to be important and acts as a driver oncogene in the process of experimental hepatocarcinogenesis since it is able to initiate and promote hepatocarcinogenesis in transgenic mice by giving rise sequentially to liver dysplasia and HCC [5,57-59]. However, its role in human hepatocarcinogenesis remains unclear. Previous studies have shown that c-myc is barely expressed in normal liver tissues [60]. By contrast, c-myc has been found overexpressed in most of the human hepatoma cell lines, and its repression by ribozyme or antisense therapy induces differentiation and growth inhibition of these cells [61-62]. Also, in vivo studies have shown that c-myc expression may gradually increase from normal liver to chronic hepatitis, cirrhosis and HCC [63].
Mechanisms of c-myc overexpression during human hepatocarcinogenesis are poorly understood, but might be related to amplification of the gene (40%-60% of HCCs, but rare in liver dysplasia) or hypomethylation of its regulatory sequences [64-65]. Speculation of c-myc overexpression as being an early event in the premalignant steps of human hepatocarcinogenesis may be of major importance since it has been found in more than 50% of human HCCs and their corresponding adjacent nontumor liver. Recent studies have shown that activation of c-myc is strongly associated with the malignant conversion of preneoplastic high grade dysplastic liver nodules in HCC [66]. A cytogenetic tumor progression model constructed by Poon and colleagues have determined that gains of 8q22-24 (bearing the c-myc allele) are among the earliest genomic events associated with HCC development [67]. Furthermore, it has experimentally been shown that c-myc is able to induce oncogenic stress in mouse embryonic fibroblasts [34], although it remains to be determined in normal human liver cells. Several observations have initially let hypothesize that, although c-myc triggering cellular growth may play a role in the premalignant steps during the process of malignant transformation (as reflected by its high mRNA steady state level in peritumor liver), it might not play a significant role in sustaining the growth of the tumor cells (as reflected by its lower mRNA steady state level in the corresponding tumor tissue) [68-69]. However, more recent data strengthen the role of c-MYC in maintenance of the cancerous phenotype since, although c-myc gene mRNA steady state level might not change or decrease in HCCs by comparison to the matched adjacent preneoplastic tissues, c-MYC activity can be enhanced by posttranscriptional modifications affecting the half-life of the protein. Phosphorylation of the Thr-58 residue in the Myc box I domain targets MYC for ubiquitination and consequent proteasosome-mediated degradation [70]. Although mutations of this region are frequently observed in Burkitt lymphomas and lead to stabilization of MYC, as well as to disruption of its pro-apoptotic function, these mutations have not been found in human HCC. A member of the COP9 signalosome – i.e. CSN5 - located at 8q13 locus, regulates activity of the ubiquitin ligase complex. It has recently been shown that CSN5 overexpression occurs at the early stages of hepatocarcinogenesis and shows a significant association with the presence of the c-MYC-regulated expression signature. These results are consistent with the notion that CSN5 plays an important role in liver cancer progression by a mechanism involving stabilization of the c-MYC protein and enhancement of its activity [66]. Another mechanism of increased c-MYC-activity could be its stabilizing interaction with HIF (hypoxia inducible factors) [71]. Finally, that transcriptional activity of c-MYC itself could be regulated by multiple pathways, including RAS/RAF/MAPK, JAK/STAT, and Wnt/β-catenin signaling, which may result in a significant overlap between the c-MYC and other oncogenic pathways [72-75]. Thus it appears that c-MYC could be a central regulator of malignant transformation in early hepatocarcinogenesis.
4.3. Wnt/β-catenin
The Wnt/β-catenin pathway is regulated tightly during early liver development [76]. The β-catenin molecule is an important multifactorial protein which is involved in cell-cell adhesion by strengthening the linkage between cadherin and α-catenin to the actin cytoskeleton. Its soluble form can translocate from cytosol to nucleus where it transactivates genes involved in cell fate during physiological homeostasis as well as for the setting of cancer properties. The Wnt/Frizzled signaling network controls activation of the canonical Wnt/β-catenin signalling cascade and/or the noncanonical c-Jun N-terminal kinase (JNK) and protein kinase C (PKC) [77].
Although evidence is accumulating that alterations of the Wnt/β-catenin pathway, due to or unrelated to β-catenin gene mutation, is a common event in hepatocarcinogenesis [78-84], little is known concerning the noncanonical elements. The previous finding of inappropriate activation of the Wnt/β-catenin pathway in hepatocarcinogenesis resulting from CTNNB1 gene mutations has provided clues toward understanding this process [78]. Different cohort studies of human HCC tissues have shown that activating mutations hitting the Wnt/β-catenin pathways have been observed within the heterogeneous “end product” tumor bulk : the CTNNB1 gene encoding for the β-catenin protein in 10% to 30% of HCCs, the AXIN-1 gene in 7% to 9%, whereas APC gene mutations are exceptional [85-87]. However, the meaning of these mutations is questionable due to their absence in liver dysplasia, cirrhotic nodules or chronic hepatitis tissues, and due to their heterogeneity in HCC bulks since they concern only a fraction of tumor cells. Additionally, it appears that aberrant activation of the Wnt/β-catenin pathway as manifested by cellular and nuclear accumulation of the β-catenin protein occurs in a higher percentage of HCCs - i.e. 35% to 85% - while the Wnt/β-catenin pathway gene mutations are absent [81-83]. More recently, it has been shown that, in absence of Wnt/β-catenin pathway gene mutation, activation of the Wnt/β-catenin pathway can occur by enhancement of the Wnt/Frizzled-mediated signalling. Indeed, it has been shown that binding of the WNT3 ligand on the FZD7 receptor can activate the canonical Wnt/β-catenin pathway in human and rodent HCCs, enhancing the cancerous phenotype of cancerous human hepatoma cell lines. Interestingly, WNT3 and/or FZD7 are overexpressed not only in 60% to 90% of human HCCs but also in 35-60% of the surrounding preneoplastic liver tissues, letting hypothesize that activation of the WNT3/FZD7-mediated signalling might be an early event in hepatocarcinogenesis [79,80,88-90]. Additionally, down-regulation of the Wnt/Frizzled-mediated signaling per se has recently been shown as being an early key signal for senescence of primary human cells, suggesting at opposite that activation of Wnt/Frizzled-mediated signaling might force the presenescent cell to bypass the senescence program [91]. Thus it remains to be confirmed that activation of the Wnt/Frizzled-mediated signaling might be involved in the early steps of hepatocarcinogeneis through induction of cellular oncogenic stress and allowing to bypass senescence.
5. Liver Inflammation and Hepatocarcinogenesis
HCC arises most frequently in the setting of chronic liver inflammation due to viral infection, metabolic injury, toxic insults or autoimmune reactions. Liver cirrhosis itself is considered as the result of persistent liver damage and chronic inflammation. Cirrhosis also changes the microenvironment, which impacts on tumor formation. One of the hallmarks of cirrhosis is the activation of stellate cells, resulting in increased production of extracellular matrix proteins, cytokines, growth factors, and products of oxidative stress [92]. During recent years evidence has been accumulating to show that inflammation has an important role in initiation, promotion and progression of tumours, and that NF-κB signalling is at the heart of the issue [93].
NF-κB might be activated by the concerted action of cytokines or interleukins, such as TNF-α and IL-6, chemokines and viral proteins, which likely will promote cell survival of pre-cancerous hepatocytes [94]. Furthermore, cellular pathways such as EFGR-mediated cascade can activate NF-κB signalling leading to inhibition of c-Myc-induced apoptosis [95]. In the same view, NF-κB signalling can activate pro-survival factors such as Bcl-Xl and the XIAP caspase inhibitor [96-97]. Finally, the generation of pro-inflammatory cytokines and growth factors produced by tumour infiltrating macrophages, lymphocytes and other cell types in the tumour microenvironment provokes activation of NF-κB, protects against pro-apoptotic host immune defence mechanisms, influences cell differentiation and exert proangiogenic effects which stimulate the growth of cancer cells, tumour invasiveness and metastasis [98]. As a paradigm, the influence of inflammatory signalling on hepatocarcinogenesis can be context-dependent. Indeed, deletion of NF-κB-dependent inflammatory responses can enhance HCC formation in carcinogen-treated mice [99]. Similarly, deletion of NF-κB-essential modulator/IκB kinase (NEMO/IKK), an activator of NF-κB, induces steatohepatitis and HCC in mice [100]. In contrast, inhibition of NF-κB impairs HCC progression in a mouse model of cholestatic hepatitis [101].
Conversely, IL-6 production has been shown as occurring through MyD88-mediated Toll-like receptor (TLR) stimulation in rodent models, demonstrating the implication of the innate immune response in the hepatocarcinogenic process [102]. TLRs can be activated by nucleic acids in a context of viral infection, or by the damage-associated molecular patterns such as products of liver cell necrosis [103]. TLRs can thus act as finely tuned sensors of tissue damage fuelling inflammation and tissue reorganization following injury. Along these lines, the deletion of the suppressor of cytokine signalling-3 (SOCS3), a negative regulator of interleukin-6, promotes hepatitis-induced hepatocarcinogenesis in mice [104].
6. Antiproliferative and Apoptosis Deficiency and Liver Cell Transformation
The frequent inactivation of p53 in human HCC indicates that abrogation of p53-dependent apoptosis could promote hepatocarcinogenesis. In addition, p53-independent pathways can induce apoptosis in response to telomere dysfunction [47], although the role of impairments of p53-independent apoptosis for hepatocarcinogenesis remains to be defined. It was shown that Hint2 (an apoptosis sensitizer acting at the mitochondria) is down-regulated in human HCC, correlating with poor prognosis [105]. The transforming growth factor-β (TGF-β) pathway is frequently activated at the cirrhosis stage and induces apoptosis by activating Smad3-mediated BCL2 repression [106]. Apoptosis resistance also could involve insulin-receptor signalling and activation of the Akt pathway [107].
The insulin-like growth factor 2 receptor (IGF2R) impairs cell proliferation by promoting degradation of the IGF2 mitogen and by activation of TGF-β signalling [108]. Loss of heterozygosity in IGF2R locus is a frequent and early event in human hepatocarcinogenesis since occurring in more than 60% of dysplastic nodules and HCC [109]. Loss of IGF2R could cooperate with the IGF2 growth factor overexpression, which is a common feature in human HCCs. In addition, an abrogation of IGF2R could represent a selective advantage at the cirrhosis stage by impairing antiproliferative and pro-apoptotic signals induced by TGF-β, which is overexpressed in cirrhosis. Interestingly, IGF2R is located in the subtelomeric region of chromosome 6 in humans, indicating that telomere shortening could influence recombination rates and loss of heterozygosity of this locus [110].
Activation of the Akt signalling and impaired expression of phosphatase and tensin homolog (PTEN) (a negative regulator of Akt) have been reported in 40% to 60% of human HCC. Activation of the Akt pathway suppresses TGF-β-induced apoptosis and growth-inhibitory activity of CCAAT/enhancer binding protein α. Both effects could promote tumor formation at the cirrhosis stage. Activation of the Akt pathway has been linked to an activation of β-catenin signalling in intestinal stem cells [111-112].
7. Conclusion
HCC is a major problem of public health, and a better understanding of hepatocarcinogenesis will help to identify pertinent molecular targets for innovative therapies. Although high output microarray analysis from tumor bulks have allowed to classify HCCs to predict outcome of patients, huge efforts are being done to identify liver cell types the most permissive to support the different genetic/epigenetic hits needed for cancerous transformation, and to accurately determine the sequence of these events. It has been clearly shown that cancer stem cells are indispensable for tumorigenicity, and much is being done to understand whether they come from transformation of normal liver stem cells or from de-differentiation of mature heptocytes that would have re-acquired stem cell properties. At preneoplastic steps, senescence has been shown as being a powerful anti-oncogenic mechanism in different cellular models supporting DNA stress secondary to oncogenic pathway activation, ROS production and/or telomere shortening for instance. Few is known in hepatocarcinogenesis at preneoplastic steps, but c-myc and Wnt/β-catenin oncogenic pathways might be of relevance as well as ROS production and maybe viral factors. Cirrhosis could represent the senescent step where cell cycle arrest of hepatocytes is controlled by key checkpoints such as p53/p21Cip1 and p16INK4a/pRb. Their inactivation is a prerequisite for the pre-senescent cell to bypass senescence and to re-enter cell cycle progression, mechanisms that might be controlled by the Wnt signalling and the activity of Twist proteins, although it remains to be determined in hepatocarcinogenesis. Finally, abrogation of anti-proliferative signals and of apoptosis, allow the post-senescent cell to become cancerous. However, one should keep in mind that tumors arise in a context of facilitating microenvironment (mesenchymal cells, immune response), and genetic/epigenetic hits necessary for cancerous phenotype in one single liver cell is not enough per se to allow tumor bulk to develop. All this angle of hepatocarcinogenesis has not been tackled in this paper. Additionally, of interest will be the chapter on interplays between HBV and HCV factors in hepatocarcinogenesis.
References
- Parkin, D.M.; Bray, F.; Ferlay, J.; Pisani, P. Estimating the world cancer burden: Globocan 2000. Int. J. Cancer 2001, 94, 153–156. [Google Scholar] [CrossRef] [PubMed]
- Llovet, J.M.; Burroughs, A.; Bruix, J. Hepatocellular carcinoma. Lancet 2003, 362, 1907–1917. [Google Scholar] [CrossRef] [PubMed]
- El-Serag, H.; Davila, J.A.; Petersen, N.J.; McGlynn, K.A. The continuing increase in the incidence of hepatocellular carcinoma in the United States: an update. Ann. Intern. Med. 2003, 139, 817–823. [Google Scholar] [PubMed]
- Wang, J.; Chenivesse, X.; Henglein, B.; Brechot, C. Hepatitis B virus integration in a cyclin A gene in a hepatocellular carcinoma. Nature 1990, 343, 555–557. [Google Scholar] [CrossRef] [PubMed]
- Etiemble, J.; Degott, C.; Renard, C.A.; Fourel, G.; Shamoon, B.; Vitvitski-Trépo, L.; Hsu, T.Y.; Tiollais, P.; Babinet, C.; Buendia, M.A. Liver-specific expression and high oncogenic efficiency of a c-myc transgene activated by woodchuck hepatitis virus insertion. Oncogene 1994, 9, 727–737. [Google Scholar] [PubMed]
- Moriya, K.; Fujie, H.; Shintani, Y.; Yotsuyanagi, H.; Tsutsumi, T.; Ishibashi, K.; Matsuura, Y.; Kimura, S.; Miyamura, T.; Koike, K. The core protein of hepatitis C virus induces hepatocellular carcinoma in transgenic mice. Nat. Med. 1998, 4, 1065–1067. [Google Scholar] [CrossRef]
- Heppner, G.H. Tumor heterogeneity. Cancer Res. 1984, 44, 2259–2265. [Google Scholar] [PubMed]
- Thorgeirsson, S.S.; Grisham, J.W. Molecular pathogenesis of human hepatocellular carcinoma. Nat. Genet. 2002, 31, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Heo, J.; Libbrecht, L.; Chu, I.S.; Kaposi-Novak, P.; Calvisi, D.F.; Mikaelyan, A.; Roberts, L.R.; Demetris, A.J.; Sun, Z.; Nevens, F.; Roskams, T.; Thorgeirsson, S.S. A novel prognostic subtype of human hepatocellular carcinoma derived from hepatic progenitor cells. Nat. Med. 2006, 12, 410–416. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Thorgeirsson, S.S. Comparative and integrative functional genomics of HCC. Oncogene 2006, 25, 3801–3809. [Google Scholar] [CrossRef] [PubMed]
- Boyault, S.; Rickman, D.S.; de Reyniès, A.; Balabaud, C.; Rebouissou, S.; Jeannot, E.; Hérault, A.; Saric, J.; Belghiti, J.; Franco, D.; Bioulac-Sage, P.; Laurent-Puig, P.; Zucman-Rossi, J. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology 2007, 45, 42–52. [Google Scholar] [CrossRef] [PubMed]
- Al-Hajj, M.; Wicha, M.S.; Benito-Hernandez, A.; Morrison, S.J.; Clarke, M.F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA 2003, 100, 3983–3988. [Google Scholar] [CrossRef]
- O’Brien, C.A.; Pollett, A.; Gallinger, S.; Dick, J.E. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature 2007, 445, 106–110. [Google Scholar] [CrossRef] [PubMed]
- Hoshida, Y.; Villanueva, A.; Kobayashi, M.; Peix, J.; Chiang, D.Y.; Camargo, A.; Gupta, S.; Moore, J.; Wrobel, M.J.; Lerner, J.; Reich, M.; Chan, J.A.; Glickman, J.N.; Ikeda, K.; Hashimoto, M.; Watanabe, G.; Daidone, M.G.; Roayaie, S.; Schwartz, M.; Thung, S.; Salvesen, H.B.; Gabriel, S.; Mazzaferro, V.; Bruix, J.; Friedman, S.L.; Kumada, H.; Llovet, J.M.; Golub, T.R. Gene expression in fixed tissues and outcome in hepatocellular carcinoma. N. Engl. J. Med. 2008, 359, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef] [PubMed]
- Clarke, M.F.; Dick, J.E.; Dirks, P.B.; Eaves, C.J.; Jamieson, C.H.; Jones, D.L.; Visvader, J.; Weissman, I.L.; Wahl, G.M. Cancer stem cells – perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer Res. 2006, 66, 9339–9344. [Google Scholar] [CrossRef] [PubMed]
- Vermeulen, L.; Sprick, M.R.; Kemper, K.; Stassi, G.; Medema, J.P. Cancer stem cells - old concepts, new insights. Cell. Death Diff. 2008, 15, 947–958. [Google Scholar] [CrossRef]
- Sell, S.; Pierce, G.B. Maturation arrest of stem cell differentiation is a common pathway for the cellular origin of teratocarcinomas and epithelial cancers. Lab. Invest. 1994, 70, 6–22. [Google Scholar] [PubMed]
- Sell, S. Heterogeneity and plasticity of hepatocyte lineage cells. Hepatology 2001, 33, 738–750. [Google Scholar] [CrossRef] [PubMed]
- Sell, S. Cellular origin of hepatocellular carcinoma. Cell. Develop. Biology 2002, 13, 419–424. [Google Scholar] [CrossRef]
- Sell, S. Stem cell origin of cancer and differentiation therapy. Crit. Rev. Oncology Hematology 2004, 52, 1–28. [Google Scholar] [CrossRef]
- Sell, S.; Leffert, H.L. Liver cancer stem cells. J. Clin. Oncol. 2008, 26, 2800–2805. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, T.; Ji, J.; Budhu, A.; Forgues, M.; Yang, W.; Wang, H.Y.; Jia, H.; Ye, Q.; Qin, L.X.; Wauthier, E.; Reid, L.M.; Minato, H.; Honda, M.; Kaneko, S.; Tang, Z.Y.; Wang, X.W. EpCAM-positive hepatocellular carcinoma cells are tumor-initiating cells with stem/progenitor cell features. Gastroenterology 2009, 136, 1012–1024. [Google Scholar] [CrossRef] [PubMed]
- Vogelstein, B.; Kinzler, K.W. Cancer genes and the pathways they control. Nat. Med. 2004, 10, 789–799. [Google Scholar] [CrossRef] [PubMed]
- Mallette, F.A.; Ferbeyre, G. The DNA damage signaling pathway connects oncogenic stress to cellular senescence. Cell Cycle 2007, 6, 1831–1836. [Google Scholar] [CrossRef] [PubMed]
- Michalopoulos, G.K. Liver regeneration. J. Cell. Physiol. 2007, 213, 286–300. [Google Scholar] [CrossRef] [PubMed]
- Utoh, R.; Tateno, C.; Yamasaki, C.; Hiraga, N.; Kataoka, M.; Shimada, T.; Chayama, K.; Yoshizato, K. Susceptibility of chimeric mice with livers repopulated by serially subcultured human hepatocytes to hepatitis B virus. Hepatology 2008, 47, 435–446. [Google Scholar] [CrossRef] [PubMed]
- Delhaye, M.; Louis, H.; Degraef, C.; Le Moine, O.; Devière, J.; Gulbis, B.; Jacobovitz, D.; Adler, M.; Galand, P. Relationship between hepatocyte proliferative activity and liver functional reserve in human cirrhosis. Hepatology 1996, 23, 1003–1011. [Google Scholar] [CrossRef] [PubMed]
- Wege, H.; Le, H.T.; Chui, MS.; Lui, L.; Wu, J.; Giri, G.; Malhi, H.; Sappal, B.S.; Kumaran, V.; Gupta, S.; Zern, M.A. Telomerase reconstitution immortalized human fetal hepatocytes without disrupting their differentiation potential. Gastroenterology 2003, 124, 432–444. [Google Scholar] [CrossRef] [PubMed]
- Paradis, V.; Youssef, N.; Dargere, D.; Ba, N.; Bonvoust, F.; Deschatrette, J.; Bedossa, P. Replicative senescence in normal liver, chronic hepatitis C, and hepatocellular carcinomas. Hum. Pathol. 2001, 32, 327–332. [Google Scholar] [CrossRef] [PubMed]
- Aikata, H.; Takaishi, H.; Kawakami, Y.; Takahashi, S.; Kitamoto, M.; Nakanishi, T.; Nakamura, Y.; Shimamoto, F.; Kajiyama, G.; Ide, T. Telomere reduction in human liver tissues with age and chronic inflammation. Exp. Cell. Res. 2000, 256, 578–582. [Google Scholar] [CrossRef] [PubMed]
- Di Micco, R.; Fumagalli, M.; Cicalese, A.; Piccinin, S.; Gasparini, P.; Luise, C.; Schurra, C.; Garre, M.; Nuciforo, P.G.; Bensimon, A.; Maestro, R.; Pelicci, P.G.; d’Adda di Fagagna, F. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature 2006, 444, 638–642. [Google Scholar] [CrossRef] [PubMed]
- Sarkisian, C.J.; Keister, B.A.; Stairs, D.B.; Boxer, R.B.; Moody, S.E.; Chodosh, L.A. Dose-dependent oncogene-induced senescence in vivo and its evasion during mammary tumorigenesis. Nat. Cell. Biol. 2007, 9, 493–505. [Google Scholar] [CrossRef]
- Pauklin, S.; Kristjuhan, A.; Maimets, T.; Jaks, V. ARF and ATM/ATR cooperate in p53-mediated apoptosis upon oncogenic stress. Biochem. Biophys. Res. Com. 2005, 334, 386–394. [Google Scholar] [CrossRef]
- Zindy, P.; Andrieux, L.; Bonnier, D.; Musso, O.; Langouët, S.; Campion, J.P.; Turlin, B.; Clément, B.; Théret, N. Upregulation of DNA repair genes in active cirrhosis associated with hepatocellular carcinoma. FEBS Lett. 2005, 579, 95–99. [Google Scholar] [CrossRef] [PubMed]
- Shimoda, R.; Nagashima, M.; Sakamoto, M.; Yamaguchi, N.; Hirohashi, S.; Yokota, J.; Kasai, H. Increased formation of oxidative DNA damage, 8-hydroxydeoxyguanosine, in human livers with chronic hepatitis. Cancer Res. 1994, 54, 3171–3172. [Google Scholar] [PubMed]
- Ohtani, N.; Mann, D.; Hara, E. Cellular senescence: its role in tumor suppression and aging. Cancer Sci. 2009, 5, 792–797. [Google Scholar] [CrossRef]
- Oishi, N.; Shilagardi, K.; Nakamoto, Y.; Honda, M.; Kaneko, S.; Murakami, S. Hepatitis B virus X protein overcomes oncogenic RAS-induced senescence in human immortalized cells. Cancer Sci. 2007, 98, 1540–1548. [Google Scholar] [CrossRef] [PubMed]
- Higashitsuji, H.; Higashitsuji, H.; Itoh, K.; Sakurai, T.; Nagao, T.; Sumitomo, Y.; Masuda, T.; Dawson, S.; Shimada, Y.; Mayer, R.J.; Fujita, J. The oncoprotein gankyrin binds to MDM2/HDM2, enhancing ubiquitylation and degradation of p53. Cancer Cell 2005, 8, 75–87. [Google Scholar] [CrossRef]
- Plentz, R.R.; Park, Y.N.; Lechel, A.; Kim, H.; Nellessen, F.; Langkopf, B.H.; Wilkens, L.; Destro, A.; Fiamengo, B.; Manns, M.P.; Roncalli, M.; Rudolph, K.L. Telomere shortening and inactivation of cell cycle checkpoints characterize human hepatocarcinogenesis. Hepatology 2007, 45, 968–976. [Google Scholar] [CrossRef] [PubMed]
- Roncalli, M.; Bianchi, P.; Bruni, B.; Laghi, L.; Destro, A.; Di Gioia, S.; Gennari, L.; Tommasini, M.; Malesci, A.; Coggi, G. Methylation framework of cell cycle gene inhibitors in cirrhosis and associated hepatocellular carcinoma. Hepatology 2002, 36, 427–432. [Google Scholar] [CrossRef] [PubMed]
- Kalinichenko, V.V.; Major, M.L.; Wang, X.; Petrovic, V.; Kuechle, J.; Yoder, H.M.; Dennewitz, M.B.; Shin, B.; Datta, A.; Raychaudhuri, P.; Costa, R.H. Foxm1b transcription factor is essential for development of hepatocellular carcinomas and is negatively regulated by the p19ARF tumor suppressor. Genes Dev. 2004, 18, 830–850. [Google Scholar] [CrossRef] [PubMed]
- Azechi, H.; Nishida, N.; Fukuda, Y.; Nishimura, T.; Minata, M.; Katsuma, H.; Kuno, M.; Ito, T.; Komeda, T.; Kita, R.; Takahashi, R.; Nakao, K. Disruption of the p16/cyclin D1/retinoblastoma protein pathway in the majority of human hepatocellular carcinomas. Oncology 2001, 60, 346–354. [Google Scholar] [CrossRef] [PubMed]
- Llovet, J.M.; Chen, Y.; Wurmbach, E.; Roayaie, S.; Fiel, M.I.; Schwartz, M.; Thung, S.N.; Khitrov, G.; Zhang, W.; Villanueva, A.; Battiston, C.; Mazzaferro, V.; Bruix, J.; Waxman, S.; Friedman, S.L. A molecular signature to discriminate dysplastic nodules from early hepatocellular carcinoma in HCV cirrhosis. Gastroenterology 2006, 131, 1758–1767. [Google Scholar] [CrossRef] [PubMed]
- Farazi, P.A.; Glickman, J.; Jiang, S.; Yu, A.; Rudolph, K.L.; DePinho, R.A. Differential impact of telomere dysfunction on initiation and progression of hepatocellular carcinoma. Cancer Res. 2003, 63, 5021–5027. [Google Scholar] [PubMed]
- Djojosubroto, M.W.; Chin, A.C.; Go, N.; Schaetzlein, S.; Manns, M.P.; Gryaznov, S.; Harley, C.B.; Rudolph, K.L. Telomerase antagonists GRN163 and GRN163L inhibit tumor growth and increase chemosensitivity of human hepatoma. Hepatology 2005, 42, 1127–1136. [Google Scholar] [CrossRef] [PubMed]
- Lechel, A.; Holstege, H.; Begus, Y.; Schienke, A.; Kamino, K.; Lehmann, U.; Kubicka, S.; Schirmacher, P.; Jonkers, J.; Rudolph, K.L. Telomerase deletion limits progression of p53-mutant hepatocellular carcinoma with short telomeres in chronic liver disease. Gastroenterology 2007, 132, 1465–1475. [Google Scholar] [CrossRef] [PubMed]
- Murakami, Y.; Saigo, K.; Takashima, H.; Minami, M.; Okanoue, T.; Brechot, C.; Paterlini-Brechot, P. Large scaled analysis of hepatitis B virus (HBV) DNA integration in HBV related hepatocellular carcinomas. Gut 2005, 54, 1162–1168. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Luan, F.; Ju, Y.; Shen, H.; Gao, L.; Wang, X.; Liu, S.; Zhang, L.; Sun, W.; Ma, C. In vitro transfection of the hepatitis B virus PreS2 gene into the human hepatocarcinoma cell line HepG2 induces upregulation of human telomerase reverse transcriptase. Biochem. Biophys. Res. Commun. 2007, 355, 379–384. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Yu, Q.; Subrahmanyam, R.; Difilippantonio, M.J.; Ried, T.; Sen, J.M. β-Catenin expression results in p53-independent DNA damage and oncogene-induced senescence in prelymphomagenic thymocytes in vivo. Mol. Cell. Biol. 2008, 28, 1713–1723. [Google Scholar] [CrossRef] [PubMed]
- Ansieau, S.; Bastid, J.; Doreau, A.; Morel, A.P.; Bouchet, B.P.; Thomas, C.; Fauvet, F.; Puisieux, I.; Doglioni, C.; Piccinin, S.; Maestro, R.; Voeltzel, T.; Selmi, A.; Valsesia-Wittmann, S.; Caron de Fromentel, C.; Puisieux, A. Induction of EMT by Twist proteins as a collateral effect of tumor-promoting inactivation of premature senescence. Cancer Cell 2008, 14, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Weinberg, R.A. Twisted epithelial–mesenchymal transition blocks senescence. Nat. Cell. Biol. 2008, 10, 1021–1023. [Google Scholar] [CrossRef] [PubMed]
- Buendia, M.A. Genetics of hepatocellular carcinoma. Semin. Cancer Biol. 2000, 10, 185–200. [Google Scholar] [CrossRef] [PubMed]
- Challen, C.; Guo, K.; Collier, J.D.; Cavanagh, D.; Bassendine, M.F. Infrequent point mutations in codons 12 and 61 of ras oncogenes in human hepatocellular carcinomas. J. Hepatol. 1992, 14, 342–346. [Google Scholar] [CrossRef] [PubMed]
- Jadirgar, J.; Nonomura, A.; Patil, J.; Thor, A.; Paronetto, F. Ras oncoprotein p21 expression in hepatocellular carcinoma. J. Exp. Pathol. 1989, 4, 37–46. [Google Scholar] [PubMed]
- Evan, G.I.; Littlewood, T.D. The role of c-myc in cell growth. Curr. Opin. Genet. Dev. 1993, 3, 44–49. [Google Scholar] [CrossRef]
- Hsu, T.; Moroy, T.; Etiemble, J.; Louise, A.; Trepo, C.; Tiollais, P.; Buendia, M.A. Activation of c-myc by woodchuck hepatitis virus insertion in hepatocellular carcinoma. Cell 1988, 55, 627–635. [Google Scholar] [CrossRef] [PubMed]
- Merle, P.; Chevallier, M.; Levy, R.; Maisonnas, M.; Terradillos, O.; Si Ahmed, S.N.; Trepo, C.; Buendia, M.A.; Vitvitski-Trepo, L. Preliminary results of interferon-alpha therapy on woodchuck hepatitis virus-induced hepatocarcinogenesis: possible benefit in female transgenic mice. J. Hepatol. 2001, 34, 562–569. [Google Scholar] [CrossRef] [PubMed]
- Merle, P.; Barraud, L.; Lefrancois, L.; Chevallier, M.; Guerret, S.; Maisonnas, M.; Bordes, I.; Savre-Train, I.; Trepo, C.; Vitvitski-Trepo, L. Long-term high-dose interferon-alpha therapy delays Hepadnavirus-related hepatocarcinogenesis in X/myc transgenic mice. Oncogene 2003, 22, 2762–2771. [Google Scholar] [CrossRef] [PubMed]
- Voravud, N.; Foster, C.S.; Gilbertson, J.A.; Sikora, K.; Waxman, J. Oncogene expression in cholangiocarcinoma and in normal hepatic development. Hum. Pathol. 1989, 12, 1163–1168. [Google Scholar] [PubMed]
- Ebinuma, H.; Saito, H.; Saito, Y.; Wakabayashi, K.; Nakamura, M.; Kurose, I.; Ishii, H. Antisense oligodeoxynucleotide against c-myc mRNA induces differentiation of human hepatocellular carcinoma cells. Int. J. Oncol. 1999, 5, 991–999. [Google Scholar] [PubMed]
- Cheng, J.; Luo, J.; Zhang, X.; Hu, J.; Hui, H.; Wang, C.; Stern, A. Inhibition of cell proliferation in HCC-9204 hepatoma cells by a c-myc specific ribozyme. Cancer Gene Ther. 2000, 3, 407–412. [Google Scholar] [PubMed]
- Himeno, Y.; Fukuda, Y.; Hatanaka, M.; Imura, H. Expression of oncogenes in human liver disease. Liver 1988, 4, 208–212. [Google Scholar] [CrossRef]
- Shen, L.; Fang, J.; Qiu, D.; Zhang, T.; Yang, J.; Chen, S.; Xiao, S. Correlation between DNA methylation and pathological changes in human hepatocellular carcinoma. Hepatogastroenterology 1998, 23, 1753–1759. [Google Scholar]
- Kawate, S.; Kukusato, T.; Ohwada, S.; Watanuki, A.; Morishita, Y. Amplification of c-myc in hepatocellular carcinoma: correlation with clinicopathologic features, proliferative activity and p53 overexpression. Oncology 1999, 57, 157–163. [Google Scholar] [CrossRef] [PubMed]
- Kaposi-Novak, P.; Libbrecht, L.; Woo, Y.G.; Lee, Y.H.; Sears, N.C.; Conner, E.A.; Factor, V.M.; Roskams, T.; Thorgeirsson, S.S. Central role of c-myc during malignant conversion in human hepatocarcinogenesis. Cancer Res. 2009, 69, 2775–2782. [Google Scholar] [CrossRef] [PubMed]
- Poon, T.C.; Wong, N.; Lai, P.B.; Rattray, M.; Johnson, P.J.; Sung, J.J. A tumor progression model for hepatocellular carcinoma: bioinformatic analysis of genomic data. Gastroenterology 2006, 131, 1262–1270. [Google Scholar] [CrossRef] [PubMed]
- Su, T.S.; Lin, L.H.; Lui, W.Y.; Chang, C.M.; Chou, C.K.; Ting, L.P.; Hu, C.P.; Han, S.H.; P’eng, F.K. Biochem. Biophys. Res. Commun. 1985, 1, 264–268. [Google Scholar] [CrossRef]
- Yuen, M.F.; Wu, P.C.; Lai, V.C.H.; Lau, J.Y.N.; Lai, C.L. Expression of c-Myc, c-Fos, and c-jun in hepatocellular carcinoma. Cancer 2001, 91, 106–112. [Google Scholar] [CrossRef] [PubMed]
- Wei, N.; Deng, X.W. The COP9 signalosome. Annu. Rev. Cell. Dev. Biol. 2003, 19, 261–286. [Google Scholar] [CrossRef] [PubMed]
- Dang, C.V.; Kim, J.W.; Gao, P.; Yustein, J. The interplay between MYC and HIF in cancer. Nat. Rev. 2008, 8, 51–56. [Google Scholar] [CrossRef]
- Sears, R.; Leone, G.; DeGregori, J.; Nevins, J.R. Ras enhances Myc protein stability. Mol. Cell. 1999, 3, 169–179. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Levens, D. Making myc. Curr. Top. Microbiol. Immunol. 2006, 302, 1–32. [Google Scholar] [PubMed]
- He, T.C.; Sparks, A.B.; Rago, C.; Hermeking, H.; Zawel, L.; da Costa, L.T.; Morin, P.J.; Vogelstein, B.; Kinzler, K.W. Identification of c-MYC as a target of the APC pathway. Science 1998, 281, 1509–1512. [Google Scholar] [CrossRef] [PubMed]
- Kolligs, F.T.; Bommer, G.; Goke, B. Wnt/β-catenin/Tcf signaling: a critical pathway in gastrointestinal tumorigenesis. Digestion 2002, 66, 131–134. [Google Scholar] [CrossRef] [PubMed]
- Micsenyi, A.; Tan, X.; Sneddon, T.; Luo, J.H.; Michalopoulos, G.K.; Monga, S.P. β-catenin is temporally regulated during normal liver development. Gastroenterology 2004, 126, 1134–1146. [Google Scholar] [CrossRef] [PubMed]
- Katoh, M.; Katoh, M. WNT signaling pathway and stem cell signaling network. Clin. Cancer Res. 2007, 13, 4042–4045. [Google Scholar] [CrossRef]
- de La Coste, A.; Romagnolo, B.; Billuart, P.; Renard, C.A.; Buendia, M.A.; Soubrane, O.; Fabre, M.; Chelly, J.; Beldjord, C.; Kahn, A.; Perret, C. Somatic mutations of the β-catenin gene are frequent in mouse and human hepatocellular carcinoma. Proc Natl Acad Sci USA 1998, 95, 8847–8851. [Google Scholar] [CrossRef] [PubMed]
- Merle, P.; de la Monte, S.; Kim, M.; Herrmann, M.; Tanaka, S.; Von Dem Bussche, A.; Kew, M.C.; Trepo, C.; Wands, J.R. Functional consequences of Frizzled-7 receptor overexpression in human hepatocellular carcinoma. Gastroenterology 2004, 127, 1110–1122. [Google Scholar] [CrossRef] [PubMed]
- Bengochea, A.; de Souza, M.M.; Lefrancois, L.; Le Roux, E.; Galy, O.; Chemin, I.; Kim, M.; Wands, J.R.; Trepo, C.; Hainaut, P.; Scoazec, J.Y.; Vitvitski, L.; Merle, P. Common dysregulation of Wnt/Frizzled receptor elements in human hepatocellular carcinoma. Br J Cancer 2008, 99, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Hsu, H.C.; Jeng, Y.M.; Mao, T.L.; Chu, J.S.; Lai, P.L.; Peng, S.Y. β-catenin mutations are associated with a subset of low-stage hepatocellular carcinoma negative for hepatitis B virus and with favorable prognosis. Am J Pathol 2000, 157, 763–770. [Google Scholar] [PubMed]
- Devereux, T.R.; Stern, M.C.; Flake, G.P.; Yu, M.C.; Zhang, Z.Q.; London, S.J.; Taylor, J.A. CTNNB1 mutations and beta-catenin protein accumulation in human hepatocellular carcinomas associated with high exposure to aflatoxin B1. Mol. Carcinog. 2001, 31, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.M.; Fan, S.T.; Ng, I.O. β-catenin mutation and overexpression in hepatocellular carcinoma: clinicopathologic and prognostic significance. Cancer 2001, 92, 136–145. [Google Scholar] [CrossRef] [PubMed]
- Inagawa, S.; Itabashi, M.; Adachi, S.; Kawamoto, T.; Hori, M.; Shimazaki, J.; Yoshimi, F.; Fukao, K. Expression and prognostic roles of beta-catenin in hepatocellular carcinoma: correlation with tumor progression and postoperative survival. Chin. Cancer Res. 2002, 8, 450–456. [Google Scholar]
- Laurent-Puig, P.; Legoix, P.; Bluteau, O.; Belghiti, J.; Franco, D.; Binot, F.; Mones, G.; Thomas, G.; Bioulac-Sage, P.; Zucman-Rossi, J. Genetic alterations associated with hepatocellular carcinomas define distinct pathways of hepatocarcinogenesis. Gastroenterology 2001, 120, 1763–1773. [Google Scholar] [CrossRef] [PubMed]
- Satoh, S.; Daigo, Y.; Furukawa, Y.; Kato, T.; Miwa, N.; Nishiwaki, T.; Kawasoe, T.; Ishiguro, H.; Fujita, M.; Tokino, T.; Sasaki, Y.; Imaoka, S.; Murata, M.; Shimano, T.; Yamaoka, Y.; Nakamura, Y. AXIN1 mutations in hepatocellular carcinomas, and growth suppression in cancer cells by virus-mediated transfer of AXIN1. Nat. Genet. 2000, 24, 245–250. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Fuji, H.; Sankila, A.; Mahler-Araujo, B.M.; Matsuda, M.; Cathomas, G.; Ohgaki, H. β-catenin mutations are frequent in human hepatocellular carcinomas associated with hepatitis C virus infection. Am. J. Pathol. 1999, 155, 1795–1801. [Google Scholar] [PubMed]
- Merle, P.; Kim, M.; Herrmann, M.; Gupte, A.; Lefrançois, L.; Califano, S.; Trepo, C.; Tanaka, S.; Vitvitski, L.; de la Monte, S.; Wands, J.R. Oncogenic role of the Frizzled-7/β-catenin pathway in hepatocellular carcinoma. J. Hepatol. 2005, 43, 854–862. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Lee, H.C.; Tsedensodnom, O.; Hartley, R.; Lim, Y.S.; Yu, E.; Merle, P.; Wands, J.R. Functional interaction between Wnt3 and Frizzled-7 leads to activation of the Wnt/β-catenin signaling pathway in hepatocellular carcinoma cells. J. Hepatol. 2008, 48, 780–791. [Google Scholar] [CrossRef] [PubMed]
- Longato, L.; de la Monte, S.; Kuzushita, N.; Horimoto, M.; Rogers, A.B.; Slagle, B.L.; Wands, J.R. Overexpression of insulin receptor substrate-1 and hepatitis Bx genes causes premalignant alterations in the liver. Hepatology 2009, 49, 1935–1943. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Zerlanko, B.; Kennedy, A.; Banumathy, G.; Zhang, R.; Adams, P.D. Downregulation of Wnt signaling is an early signal for formation of facultative heterochromatin and onset of cell senescence in primary human cells. Mol. Cell. 2007, 27, 183–196. [Google Scholar] [CrossRef] [PubMed]
- Bataller, R.; Brenner, D.A. Liver fibrosis. J. Clin. Invest. 2005, 115, 209–218. [Google Scholar] [PubMed]
- Naugler, W.E.; Karin, M. NF-kappaB and cancer-identifying targets and mechanisms. Curr. Opin. Genet. Dev. 2008, 18, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Arsura, M.; Cavin, L.G. Nuclear factor-kappaB and liver carcinogenesis. Cancer Lett. 2005, 229, 157–169. [Google Scholar] [CrossRef] [PubMed]
- Qiao, L.; Zhang, H.; Yu, J.; Francisco, R.; Dent, P.; Ebert, MP.; Rocken, C.; Farrell, G. Constitutive activation of NF kappaB in human hepatocellular carcinoma: evidence of a cytoprotective role. Hum. Gene Ther. 2006, 17, 280–290. [Google Scholar] [CrossRef] [PubMed]
- Kaur, S.; Wang, F.; Venkatraman, M.; Arsura, M. X-linked inhibitor of apoptosis (XIAP) inhibits c-Jun N-terminal kinase 1 (JNK1) activation by transforming growth factor beta1 (TGF-beta1) through ubiquitin-mediated proteosomal degradation of the TGF-beta1-activated kinase 1 (TAK1). J. Biol. Chem. 2005, 280, 38599–38608. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Karin, M. NF-kappaB signaling, liver disease and hepatoprotective agents. Oncogene 2008, 27, 6228–6244. [Google Scholar] [CrossRef] [PubMed]
- Karin, M. Nuclear factor-kappa B in cancer development and progression. Nature 2006, 441, 431–436. [Google Scholar] [CrossRef] [PubMed]
- Sakurai, T.; Maeda, S.; Chang, L.; Karin, M. Loss of hepatic NF kappa B activity enhances chemical hepatocarcinogenesis through sustained c-Jun N-terminal kinase 1 activation. Proc. Natl. Acad. Sci. USA, 2006, 103, 10544–10551. [Google Scholar] [CrossRef]
- Luedde, T.; Beraza, N.; Kotsikoris, V.; van Loo, G.; Nenci, A.; De Vos, R.; Roskams, T.; Trautwein, C.; Pasparakis, M. Deletion of NEMO/IKKgamma in liver parenchymal cells causes steatohepatitis and hepatocellular carcinoma. Cancer Cell 2007, 11, 119–132. [Google Scholar] [CrossRef] [PubMed]
- Pikarsky, E.; Porat, R.M.; Stein, I.; Abramovitch, R.; Amit, S.; Kasem, S.; Gutkovich-Pyest, E.; Urieli-Shoval, S.; Galun, E.; Ben-Neriah, Y. NF-kappaB functions as a tumour promoter in inflammation-associated cancer. Nature 2004, 431, 461–466. [Google Scholar] [CrossRef] [PubMed]
- Naugler, W.E.; Sakurai, T.; Kim, S.; Maeda, S.; Kim, K.; Elsharkawy, A.M.; Karin, M. Gender disparity in liver cancer due to sex differences in MyD88-dependent IL-6 production. Science 2007, 317, 121–124. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Schluesener, H.J. Mammalian toll-like receptors: from endogenous ligands to tissue regeneration. Cell. Mol. Life Sci. 2006, 63, 2901–2907. [Google Scholar] [CrossRef] [PubMed]
- Ogata, H.; Kobayashi, T.; Chinen, T.; Takaki, H.; Sanada, T.; Minoda, Y.; Koga, K.; Takaesu, G.; Maehara, Y.; Iida, M.; Yoshimura, A. Deletion of the SOCS3 gene in liver parenchymal cells promotes hepatitis-induced hepatocarcinogenesis. Gastroenterology 2006, 131, 179–193. [Google Scholar] [CrossRef] [PubMed]
- Martin, J.; Magnino, F.; Schmidt, K.; Piguet, A.C.; Lee, J.S.; Semela, D.; St-Pierre, M.V.; Ziemiecki, A.; Cassio, D.; Brenner, C.; Thorgeirsson, S.S.; Dufour, J.F. Hint2, a mitochondrial apoptotic sensitizer down-regulated in hepatocellular carcinoma. Gastroenterology 2006, 130, 2179–2188. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.A.; Zhang, G.M.; Feigenbaum, L.; Zhang, Y.E. Smad3 reduces susceptibility to hepatocarcinoma by sensitizing hepatocytes to apoptosis through downregulation of Bcl-2. Cancer Cell 2006, 9, 445–457. [Google Scholar] [CrossRef]
- Chen, R.H.; Su, Y.H.; Chuang, R.L.; Chang, T.Y. Suppression of transforming growth factor-beta-induced apoptosis through a phosphatidylinositol 3-kinase/Akt-dependent pathway. Oncogene 1998, 17, 1959–1968. [Google Scholar] [PubMed]
- Dennis, P.A.; Rifkin, D.B. Cellular activation of latent transforming growth factor beta requires binding to the cation-independent mannose 6-phosphate/insulin-like growth factor type II receptor. Proc. Natl. Acad. Sci. USA 1991, 88, 580–584. [Google Scholar] [CrossRef]
- Yamada, T.; de Souza, A.T.; Finkelstein, S.; Jirtle, R.L. Loss of the gene encoding mannose 6-phosphate/insulin-like growth factor II receptor is an early event in liver carcinogenesis. Proc. Natl. Acad. Sci. USA 1997, 94, 10351–10355. [Google Scholar] [CrossRef]
- Acquati, F.; Malgaretti, N.; Hauptschein, R.; Rao, P.; Gaidano, G.; Taramelli, R. A 2-Mb YAC contig linking the plasminogen-apoprotein(a) gene family to the insulin-like growth factor 2 receptor (IGF2R) gene on the telomeric region of chromosome 6 (6q26-q27). Genomics 1994, 22, 664–666. [Google Scholar] [CrossRef] [PubMed]
- Hu, T.H.; Huang, C.C.; Lin, P.R.; Chang, H.W.; Ger, L.P.; Lin, Y.W.; Changchien, C.S.; Lee, C.M.; Tai, M.H. Expression and prognostic role of tumor suppressor gene PTEN/MMAC1/TEP1 in hepatocellular carcinoma. Cancer 2003, 97, 1929–1940. [Google Scholar] [CrossRef] [PubMed]
- He, X.C.; Yin, T.; Grindley, J.C.; Tian, Q.; Sato, T.; Tao, W.A.; Dirisina, R.; Porter-Westpfahl, K.S.; Hembree, M.; Johnson, T.; Wiedemann, L.M.; Barrett, T.A.; Hood, L.; Wu, H.; Li, L. PTEN-deficient intestinal stem cells initiate intestinal polyposis. Nat. Genet. 2007, 39, 189–198. [Google Scholar] [CrossRef] [PubMed]
© 2009 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.