Molecular Cloning and Functional Characterization of Bisabolene Synthetase (SaBS) Promoter from Santalum album
Abstract
1. Introduction
2. Materials and Methods
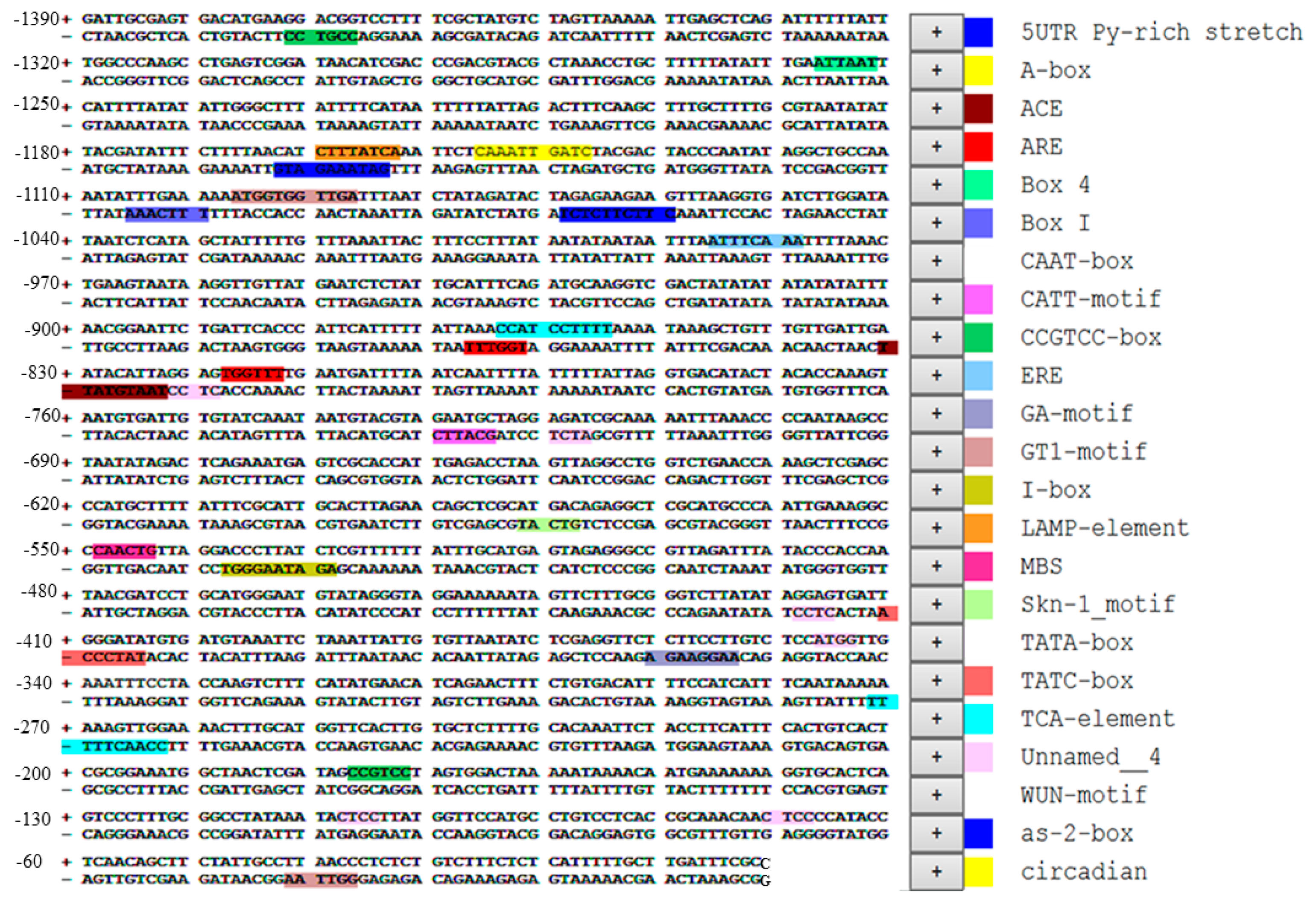
2.1. Isolation and Sequence Analysis of the SaBS Promoter
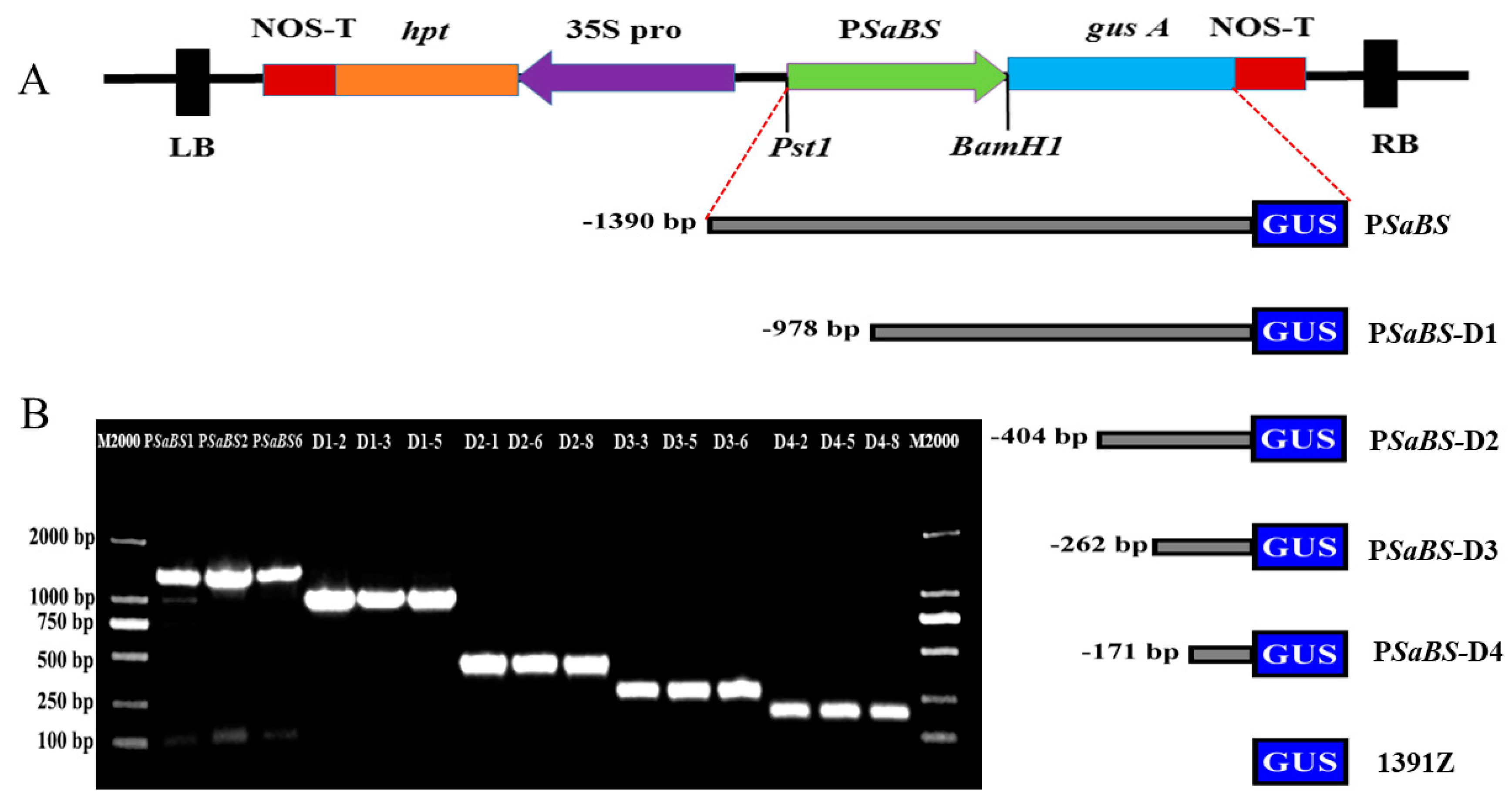
2.2. Construction of GUS Plasmid and Deletion Segements
2.3. Arabidopsis Transformation and Screening Positive Transgenic Lines
2.4. Histochemical GUS Staining and GUS Fluorometric Assays
2.5. Statistical Analyses
3. Results
3.1. Isolation and Sequence Analysis of the SaBS Promoter (PSaBS)
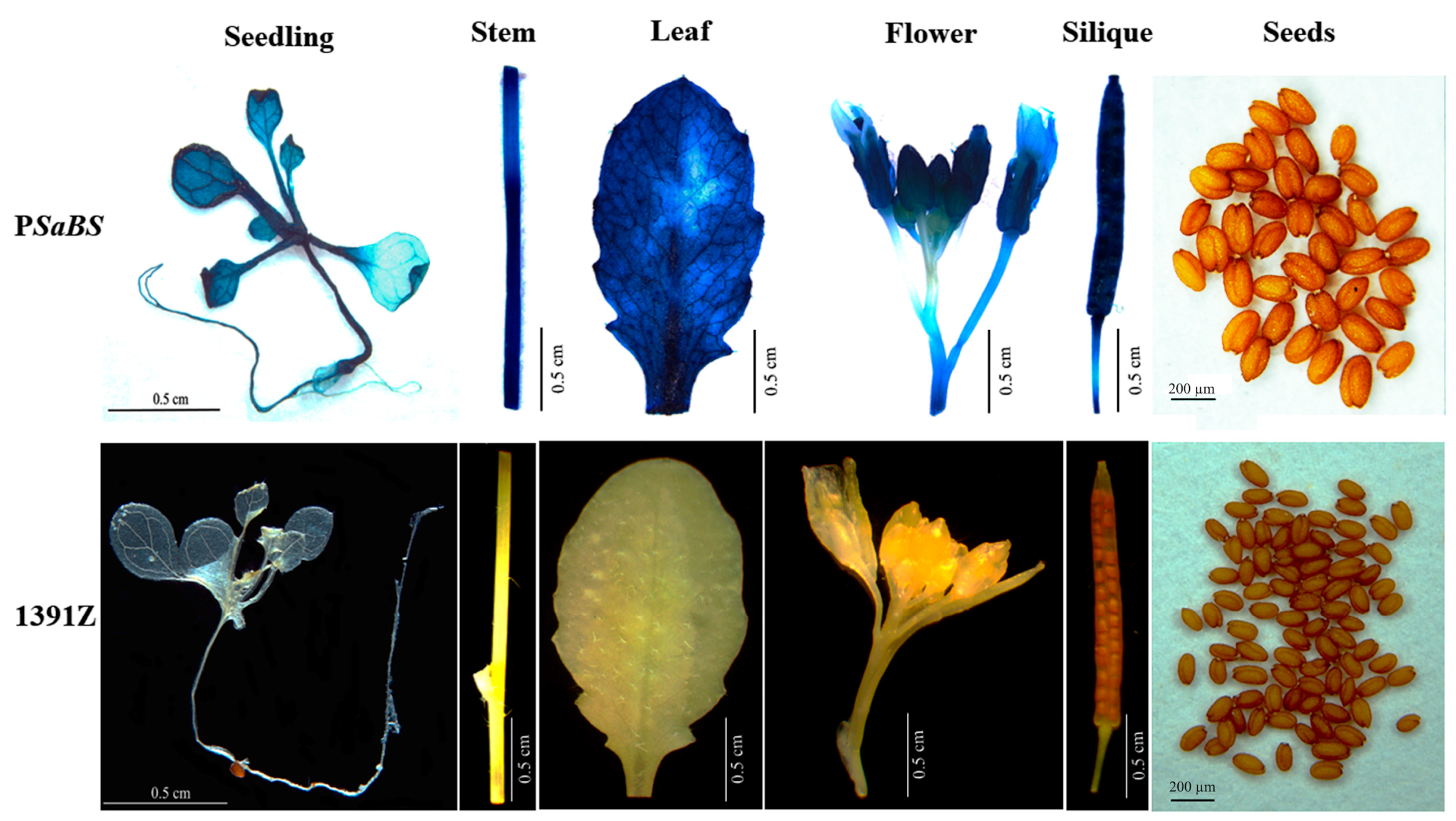
3.2. Spatiotemporal and Tissue-Specific Expression Patterns of PSaBS
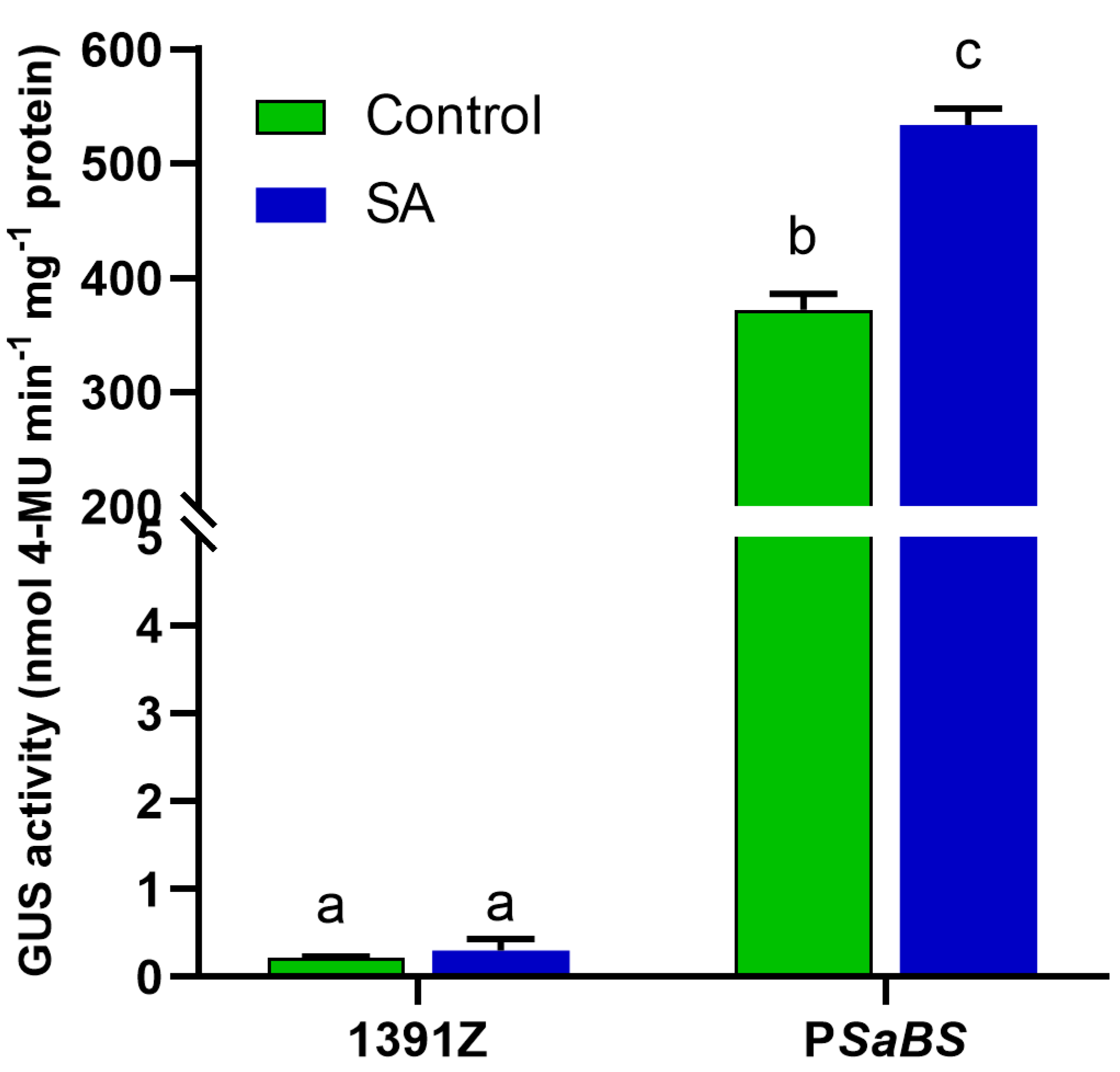
3.3. SA Enhanced GUS Activity Directed by PSaBS
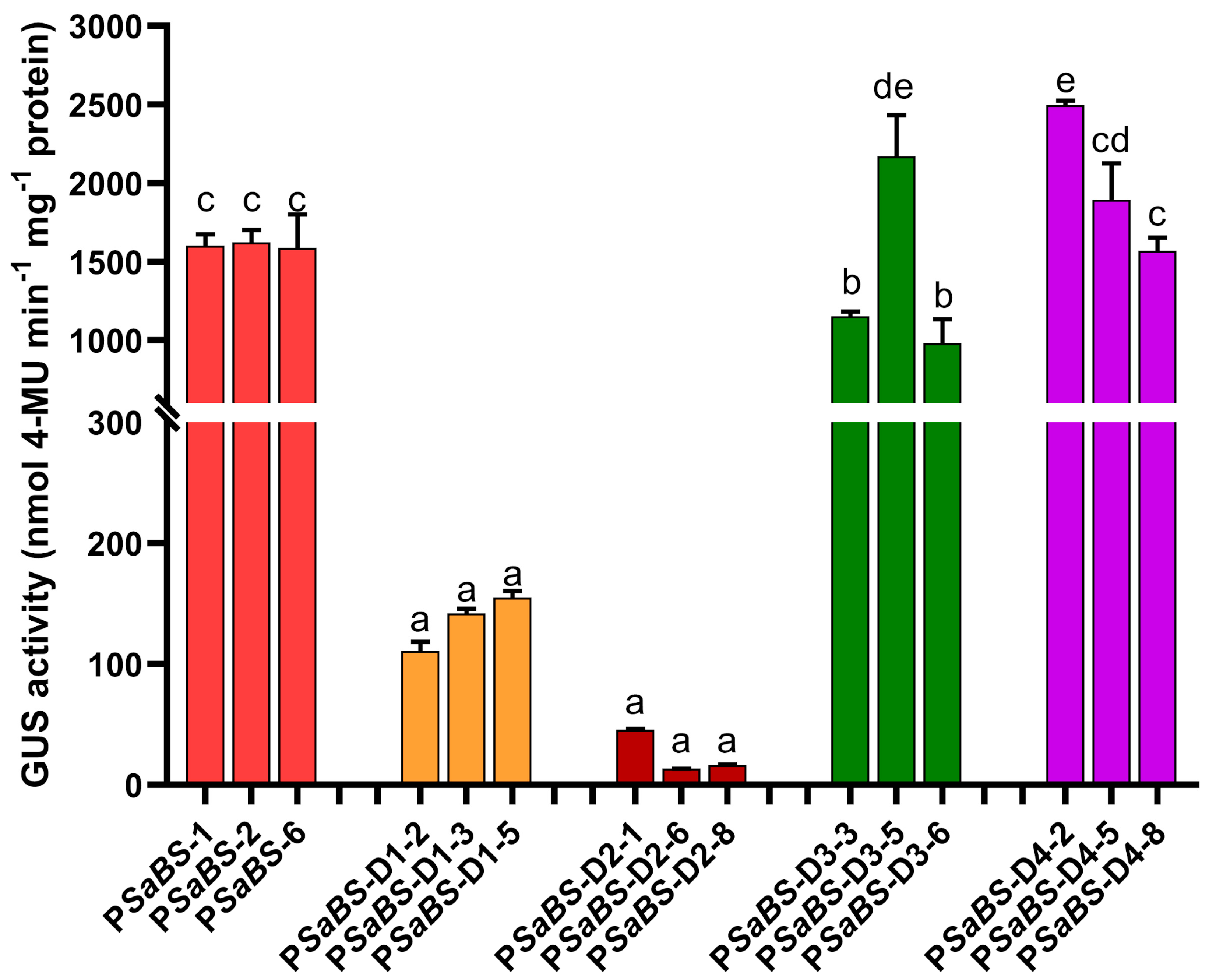
3.4. Deletion Analysis of PSaBS in Transgenic Arabidopsis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Orellana-Paucar, A.M.; Serruys, A.S.; Afrikanova, T.; Maes, J.; de Borggraeve, W.; Alen, J.; Leon-Tamariz, F.; Wilches-Arizabala, I.M.; Crawford, A.D.; de Witte, P.A.; et al. Anticonvulsant activity of bisabolene sesquiterpenoids of Curcuma longa in zebrafish and mouse seizure models. Epilepsy Behav. 2012, 24, 14–22. [Google Scholar] [CrossRef]
- Yu, Z.-L.; Peng, Y.-L.; Wang, C.; Cao, F.; Huo, X.-K.; Tian, X.-G.; Feng, L.; Ning, J.; Zhang, B.-J.; Sun, C.-P.; et al. Alismanoid A, an unprecedented 1,2-seco bisabolene from Alisma orientale, and its protective activity against H2O2-induced damage in SH-SY5Y cells. New J. Chem. 2017, 41, 12664–12670. [Google Scholar] [CrossRef]
- Jou, Y.J.; Hua, C.H.; Lin, C.S.; Wang, C.Y.; Wan, L.; Lin, Y.J.; Huang, S.H.; Lin, C.W. Anticancer activity of γ-bisabolene in human neuroblastoma cells via induction of p53-mediated mitochondrial apoptosis. Molecules 2016, 21, 601. [Google Scholar] [CrossRef] [PubMed]
- Yeo, S.K.; Ali, A.Y.; Hayward, O.A.; Turnham, D.; Jackson, T.; Bowen, I.D.; Clarkson, R. β-Bisabolene, a sesquiterpene from the essential oil extract of opoponax (Commiphora guidottii), exhibits cytotoxicity in breast cancer cell lines. Phytother. Res. 2016, 30, 418–425. [Google Scholar] [CrossRef] [PubMed]
- Raharivelomanana, P.; Cambon, A.; Azzaro, M.; Bianchini, J.-P.; Faure, R. β-Bisabolenol and β-bisabolenal, two new bisabolene sesquiterpenes from Neocallitropsis pancheri. J. Nat. Prod. 1993, 56, 272–274. [Google Scholar] [CrossRef]
- Xi, F.M.; Ma, S.G.; Liu, Y.B.; Li, L.; Yu, S.S. Artaboterpenoids A and B, bisabolene-derived sesquiterpenoids from Artabotrys hexapetalus. Org. Lett. 2016, 18, 3374–3377. [Google Scholar] [CrossRef]
- Raiju, K.; Hitora, Y.; Kato, H.; Ise, Y.; Angkouw, E.D.; Mangindaan, R.E.P.; Tsukamoto, S. Halichonic acid, a new rearranged bisabolene-type sesquiterpene from a marine sponge Halichondria sp. Tetrahedron Lett. 2019, 60, 1079–1081. [Google Scholar] [CrossRef]
- Baldovini, N.; Delasalle, C.; Joulain, D. Phytochemistry of the heartwood from fragrant Santalum species: A review. Flavour. Frag. J. 2011, 26, 7–26. [Google Scholar] [CrossRef]
- Jones, C.G.; Keeling, C.I.; Ghisalberti, E.L.; Barbour, E.L.; Plummer, J.A.; Bohlmann, J. Isolation of cDNAs and functional characterisation of two multi-product terpene synthase enzymes from sandalwood, Santalum album L. Arch. Biochem. Biophys. 2008, 477, 121–130. [Google Scholar] [CrossRef]
- Srivastava, P.L.; Daramwar, P.P.; Krithika, R.; Pandreka, A.; Shankar, S.S.; Thulasiram, H.V. Functional characterization of novel sesquiterpene synthases from Indian sandalwood, Santalum album. Sci. Rep. 2015, 5, 10095. [Google Scholar] [CrossRef]
- Hernandez-Garcia, C.M.; Finer, J.J. Identification and validation of promoters and cis-acting regulatory elements. Plant Sci. 2014, 217–218, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.Y.; Wang, J.P.; Yang, H.F. Identification and functional characterization of the NAC gene promoter from Populus euphratica. Planta 2016, 244, 417–427. [Google Scholar] [CrossRef] [PubMed]
- Dao, L.T.M.; Spicuglia, S. Transcriptional regulation by promoters with enhancer function. Transcription 2018, 9, 307–314. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Chen, P.; Guo, D.; Li, H.; Wang, Y.; Dai, H.; Mei, W.; Peng, S. Identification and functional characterization of the DcF3’H promoter from Dracaena cambodiana. Trop. Plant Biol. 2018, 11, 192–198. [Google Scholar] [CrossRef]
- Duraisamy, G.S.; Mishra, A.K.; Kocabek, T.; Matoušek, J. Identification and characterization of promoters and cis-regulatory elements of genes involved in secondary metabolites production in hop (Humulus lupulus L.). Comput. Biol. Chem. 2016, 64, 346–352. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, Y.; Wang, W.; Cao, Z.; Fu, Q.; Bao, M.; He, Y. Functional analysis of the marigold (Tagetes erecta) lycopene ε-cyclase (TeLCYe) promoter in transgenic tobacco. Mol. Biotechnol. 2019, 61, 703–713. [Google Scholar] [CrossRef]
- Liu, Y.-G.; Mitsukawa, N.; Oosumi, T.; Whittier, R.F. Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 1995, 8, 457–463. [Google Scholar] [CrossRef]
- Zhang, X.; Henriques, R.; Lin, S.S.; Niu, Q.W.; Chua, N.H. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 2006, 1, 641–646. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Jefferson, R.A.; Kavanagh, T.A.; Bevan, M.W. GUS fusion: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987, 6, 3901–3907. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Zou, C.; Sun, K.; Mackaluso, J.D.; Seddon, A.E.; Jin, R.; Thomashow, M.F.; Shiu, S.H. Cis-regulatory code of stress-responsive transcription in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2011, 108, 14992–14997. [Google Scholar] [CrossRef] [PubMed]
- Shahmuradov, I.A.; Gammerman, A.J.; Hancock, J.M.; Bramley, P.M.; Solovyev, V.V. PlantProm: A database of plant promoter sequences. Nucleic Acids Res. 2003, 31, 114–117. [Google Scholar] [CrossRef] [PubMed]
- Cvetesic, N.; Lenhard, B. Core promoters across the genome. Nat. Biotechnol. 2017, 35, 123–124. [Google Scholar] [CrossRef] [PubMed]
- Haberle, V.; Stark, A. Eukaryotic core promoters and the functional basis of transcription initiation. Nat. Rev. Mol. Cell Biol. 2018, 19, 621–637. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Xuan, L.; Liu, B.; Hu, T.; Wang, C.; Wang, X. Effects of heterologous expression of Populus euphratica brassinosteroids biosynthetic enzyme genes CPD (PeCPD) and DWF4 (PeDWF4) on tissue dedifferentiation and growth of Arabidopsis thaliana seedlings. Plant Cell Tissue Org. Cult. 2017, 132, 111–121. [Google Scholar] [CrossRef]
- Grishin, D.V.; Samoilenko, V.A.; Gladilina, Y.A.; Zhdanov, D.D.; Pokrovskaya, M.V.; Aleksandrova, S.S.; Pokrovsky, V.S.; Sokolov, N.N. Effect of heterologous expression of chemotaxis proteins from genus Thermotoga on the growth kinetics of Escherichia coli cells. Bull. Exp. Biol. Med. 2019, 167, 375–379. [Google Scholar] [CrossRef]
- Yu, H.; Khalid, M.H.B.; Lu, F.; Sun, F.; Qu, J.; Liu, B.; Li, W.; Fu, F. Isolation and identification of a vegetative organ-specific promoter from maize. Physiol. Mol. Biol. Plants 2019, 25, 277–287. [Google Scholar] [CrossRef]
- Lai, G.; Song, S.; Liu, Y.; Fu, P.; Xiang, J.; Lu, J. RPW8 promoter is involved in pathogen- and stress-inducible expression from Vitis pseudoreticulata. J. Phytopathol. 2019, 167, 65–74. [Google Scholar] [CrossRef]
- Zhang, M.; Liu, J.; Li, K.; Yu, D. Identification and characterization of a novel monoterpene synthase from soybean restricted to neryl diphosphate precursor. PLoS ONE 2013, 8, e75972. [Google Scholar] [CrossRef]
- De Costa, F.; Yendo, A.C.; Fleck, J.D.; Gosmann, G.; Fett-Neto, A.G. Accumulation of a bioactive triterpene saponin fraction of Quillaja brasiliensis leaves is associated with abiotic and biotic stresses. Plant Physiol. Biochem. 2013, 66, 56–62. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.J.; Sukweenadhi, J.; Khorolragchaa, A.; Mathiyalagan, R.; Subramaniyam, S.; Kim, Y.J.; Kim, H.B.; Kim, M.J.; Kim, Y.J.; Yang, D.C. Overexpression of Panax ginseng sesquiterpene synthase gene confers tolerance against Pseudomonas syringae pv. tomato in Arabidopsis thaliana. Physiol. Mol. Biol. Plants 2016, 22, 485–495. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Sun, Y.; Yang, Q.; Kang, J.; Zhang, T.; Gruber, M.Y.; Fang, F. Cloning and function analysis of an alfalfa (Medicago sativa L.) zinc finger protein promoter MsZPP. Mol. Biol. Rep. 2012, 39, 8559–8569. [Google Scholar] [CrossRef] [PubMed]
- Lou, H.; Li, H.; Ho, K.J.; Cai, L.L.; Huang, A.S.; Shank, T.R.; Verneris, M.R.; Nickerson, M.L.; Dean, M.; Anderson, S.K. The human TET2 gene contains three distinct promoter regions with differing tissue and developmental specificities. Front Cell Dev. Biol. 2019, 7, 99. [Google Scholar] [CrossRef]
- Iqbal, A.; Tang, W.; Wang, T.; Wang, H. Identification and functional characterization of the promoter driving “xyloglucan endotransglucosylase/hydrolase gene (XET)” gene for root growth in the desert Populus euphratica. S. Afr. J. Bot. 2017, 112, 437–446. [Google Scholar] [CrossRef]
- Han, Y.J.; Kim, Y.M.; Hwang, O.J.; Kim, J.I. Characterization of a small constitutive promoter from Arabidopsis translationally controlled tumor protein (AtTCTP) gene for plant transformation. Plant Cell Rep. 2015, 34, 265–275. [Google Scholar] [CrossRef]
- Wang, H.-J.; Jiang, Y.-H.; Qi, Y.-W.; Dai, J.-Y.; Liu, Y.-L.; Zhu, X.-B.; Liu, C.-H.; LÜ, Y.-R.; Ren, X.-L. Identification and functional characterization of the MdHB-1 gene promoter sequence from Malus×domestica. J. Integr. Agric. 2017, 16, 1730–1741. [Google Scholar] [CrossRef]


| Primer Name | Sequence (5′–3′) |
|---|---|
| AD1 | NTCGASTWTSGWGTT |
| AD2 | NGTCGASWGANAWGAA |
| AD3 | WGTGNAGWANCANAGA |
| AD4 | TGWGNAGWANCASAGA |
| AD5 | AGWGNAGWANCAWAGG |
| SaBStail1 | GCTTCTACGACCAACCTGCC |
| SaBStail2 | TTCTTCGGGCACTCATACGC |
| SaBStail3 | CCTTTTCCTTGCCCTTGTCGTG |
| PSaBSF | AACTGCAGGATTGCGAGTGACATGAAG |
| PSaBSR | CGGGATCCGGCGAAATCAAGCAAAAAT |
| PSaBS-D1F | AACTGCAGTTTTAAACTGAAGTAATAA |
| PSaBS-D2F | AACTGCAGTGTGATGTAAATTCTAAAT |
| PSaBS-D3F | AACTGCAGAAAACTTTGCATGGTTCAC |
| PSaBS-D4F | AACTGCAGTAGTGGACTAAAAATAAAA |
| Element | Sequence | Element Number | Function |
|---|---|---|---|
| A-box | CCGTCC | 2 | Regulatory element |
| ACE | CTAACGTATT | 1 | Element involved in light responsiveness |
| ARE | AAACCA | 2 | Essential for anaerobic induction |
| Box 4 | ATTAAT | 1 | Light responsiveness |
| Box I | TTTCAAA | 2 | Light-responsive element |
| CAAT-box | CAAT | 37 | Enhancer regions |
| CATT-motif | GCATCC | 1 | Part of a light-responsive element |
| CCGTCC-box | CCGTCC | 2 | Meristem-specific activation |
| ERE | ATTTCAAA | 1 | Ethylene-responsive element |
| GA-motif | AAGGAAGA | 1 | Part of a light-responsive element |
| GT1-motif | ATGGTGGTTGG GGTTAA | 2 | Light-responsive element |
| I-box | GATAAGGGT AGATAAGG | 2 | Part of a light-responsive element |
| LAMP-element | CTTTATCA | 1 | Part of a light-responsive element |
| MBS | CAACTG | 1 | MYB binding site involved in drought-inducibility |
| Skn-1 motif | GTCAT | 1 | Required for endosperm expression |
| TATC-box | TATCCCA | 1 | Gibberellin responsiveness |
| TCA-element | CCATCTTTTT | 2 | Salicylic acid responsiveness |
| WUN-motif | AAATTTCCT | 1 | Wound-responsive element |
| as-2 box | GATAATGATG | 1 | Shoot-specific expression and light responsiveness |
| Circadian | CAANNNNATC | 1 | Circadian control |
| TATA-box | TATA | 75 | Core promoter element around -30 of transcription start |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, H.; Xiong, Y.; Teixeira da Silva, J.A.; Pang, J.; Zhang, T.; Yu, X.; Zhang, X.; Niu, M.; Ma, G. Molecular Cloning and Functional Characterization of Bisabolene Synthetase (SaBS) Promoter from Santalum album. Forests 2020, 11, 85. https://doi.org/10.3390/f11010085
Yan H, Xiong Y, Teixeira da Silva JA, Pang J, Zhang T, Yu X, Zhang X, Niu M, Ma G. Molecular Cloning and Functional Characterization of Bisabolene Synthetase (SaBS) Promoter from Santalum album. Forests. 2020; 11(1):85. https://doi.org/10.3390/f11010085
Chicago/Turabian StyleYan, Haifeng, Yuping Xiong, Jaime A. Teixeira da Silva, Jinhui Pang, Ting Zhang, Xincheng Yu, Xinhua Zhang, Meiyun Niu, and Guohua Ma. 2020. "Molecular Cloning and Functional Characterization of Bisabolene Synthetase (SaBS) Promoter from Santalum album" Forests 11, no. 1: 85. https://doi.org/10.3390/f11010085
APA StyleYan, H., Xiong, Y., Teixeira da Silva, J. A., Pang, J., Zhang, T., Yu, X., Zhang, X., Niu, M., & Ma, G. (2020). Molecular Cloning and Functional Characterization of Bisabolene Synthetase (SaBS) Promoter from Santalum album. Forests, 11(1), 85. https://doi.org/10.3390/f11010085





