Abstract
In this paper, a natural probabilistic model for motif discovery has been used to experimentally test the quality of motif discovery programs. In this model, there are k background sequences, and each character in a background sequence is a random character from an alphabet, Σ. A motif is a string of m characters. In each background sequence is implanted a probabilistically-generated approximate copy of G. For a probabilistically-generated approximate copy of G, every character, , is probabilistically generated, such that the probability for is at most α. We develop two new randomized algorithms and one new deterministic algorithm. They make advancements in the following aspects: (1) The algorithms are much faster than those before. Our algorithms can even run in sublinear time. (2) They can handle any motif pattern. (3) The restriction for the alphabet size is a lower bound of four. This gives them potential applications in practical problems, since gene sequences have an alphabet size of four. (4) All algorithms have rigorous proofs about their performances. The methods developed in this paper have been used in the software implementation. We observed some encouraging results that show improved performance for motif detection compared with other software.
1. Introduction
Motif discovery is an important problem in computational biology and computer science. For instance, it has applications in coding theory [1,2], locating binding sites and conserved regions in unaligned sequences [3,4,5,6], genetic drug target identification [7], designing genetic probes [7] and universal PCR primer design [7,8,9,10].
This paper focuses on the application of motif discovery to find conserved regions in a set of given DNA, RNA or protein sequences. Such conserved regions may represent common biological functions or structures. Many performance measures have been proposed for motif discovery. Let C be a subset of 0–1 sequences of length n. The covering radius of C is the smallest integer, r, such that each vector in is at a distance at most r from a string in C. The decision problem associated with the covering radius for a set of binary sequences is NP-complete [1]. The similar closest string and substring problems were proven to be NP-hard [1,7]. Some approximation algorithms have been proposed. Li et al. [11] gave an approximation scheme for the closest string and substring problems. The related consensus patterns problem is that given n sequences , find a region of length L in each and a string, s, of length L, so that the total Hamming distance from s to these regions is minimized. Approximation algorithms for the consensus patterns problem were reported in [12]. Furthermore, a number of heuristics and programs have been developed [13,14,15,16,17].
In many applications, motifs are faint and may not be apparent when two sequences alone are compared, but may become clearer when more sequences are compared together [18]. For this reason, it has been conjectured that comparing more sequences together can help with identifying faint motifs. This paper is a theoretical approach with a rigorous probabilistic analysis.
We study a natural probabilistic model for motif discovery. In this model, there are k background sequences, and each character in the background sequence is a random character from an alphabet, Σ. A motif is a string of m characters. In each background sequence is implanted a probabilistically-generated approximate copy of G. For a probabilistically-generated approximate copy of G, every character, , is probabilistically generated, such that the probability for , which is called a mutation, is at most α. This model was first proposed in [13] and has been widely used in experimentally testing motif discovery programs [14,15,16,17]. We note that a mutation in our model converts a character, , in the motif into a different character, , without probability restriction. This means that a character, , in the motif may not become any character in with equal probability.
We develop three algorithms for which, under the probabilistic model, one can find the implanted motif with high probability via a tradeoff between computational time and the probability of mutation. Each algorithm has a preprocessing phase and the voting phase. We use a pair of functions, , to describe the computational complexity of the motif detection algorithm, where n is the largest length of the input sequence and k is the number of sequences. Function is the time complexity for the part for preprocessing, and is the time complexity for recovering one character for the motif after preprocessing. The total time is .
(1) There exists a randomized algorithm, such that there are positive constants, and , such that, if the alphabet size is at least four, the number of sequences is at least , the motif length is at least and each character in the motif region has a probability of at most of mutation for some fixed , then the motif can be recovered with a probability of at least in time, where n is the longest length of any input sequences and . The algorithm’s total time is sublinear if the motif length, , is in the range . This is the first sublinear time algorithm with rigorous analysis in this model.
(2) There exists a randomized algorithm, such that there are positive constants, and α, such that if the alphabet size is at least four, the number of sequences is at least , the motif length is at least and each character in the motif region has a probability of at most α of mutation, then the motif can be recovered with a probability of at least in time.
(3) There exists a deterministic algorithm, such that there are positive constants, and α, such that if the alphabet size is at least four, the number of sequences is at least , the motif length is at least and each character in the motif region has a probability of at most α of mutation, then the motif can be recovered with a probability of at least in time.
The research in this model has been reported in [19,20,21]. In [19], Fu et al.. developed an algorithm that needs the alphabet size to be a constant that is much larger than four. In [20], our algorithm cannot handle all possible motif patterns. In [21], Liu et al. designed an algorithm that runs in time and lacks rigorous analysis about its performance. The motif recovery in this natural and simple model has not been fully understood and seems to be a complicated problem.
This paper presents two new randomized algorithms and one new deterministic algorithm. They make advancements in the following aspects: (1) The algorithms are much faster than those before. Our algorithms can even run in sublinear time. (2) They can handle any motif pattern. (3) The restriction for the alphabet size is as small as four, giving them potential applications in practical problems, since gene sequences have an alphabet size of four. (4) All algorithms have rigorous proofs about their performances.
The algorithm for motif detection is named Recover-Motif(.). The entire Recover-Motif(.) is described in Section 4.2. We analyze Algorithm Recover-Motif (.) in Section 6.
2. Notations and the Model of Sequence Generation
For a set, A, denotes the number of elements in A. Σ is an alphabet with . For an integer, , is the set of sequences of a length of n with characters from Σ. For a sequence , denotes the character, , and denotes the substring, , for . denotes the length of the sequence, S. We use ∅ to represent the empty sequence, which has a length of zero.
Let be a fixed sequence of m characters. G is the motif to be discovered by our algorithm. A -sequence has the form , where , each has a probability of to be equal to π for each and has a probability of at most α and not equal to for , where . denotes the motif region of S. A mutation converts a character, , in the motif into an arbitrary, different character, , without probability restriction. This allows a character, , in the motif to change into any character, , in with even a different probability. The motif region of S may start at an arbitrary position in S. Furthermore, a mutation may convert a character, , in the motif into an arbitrary, different character, , only subject to the restriction that will mutate with a probability of at most α.
For two sequences and of the same length, let the relative Hamming distance .
Definition 1.
For two intervals, and , define .
3. Brief Introduction to the Algorithm
Every detection algorithm in this paper has two phases. The first phase is preprocessing, so that the motif regions from multiple sequences can be aligned in the same column region. The second phase is to recover the motif via voting. We use a pair of functions, , to describe the computational complexity of the motif detection algorithm. Function is the time complexity for the preprocessing phase, and is the time complexity for outputting one character for the motif in the voting phase.
The motif, G, is a pattern unknown to algorithm Recover-Motif, and algorithm Recover-Motif will attempt to recover G from a series of -sequences generated by the probabilistic model.
3.1. Algorithm
The algorithm first detects a position that is close to the left motif boundary in a sequence. It finds such a position via sampling and collision between two sequences. After the rough left boundary of a sequence is found, it is used to find the rough boundaries of the rest of the sequences. Similarly, we find those right boundaries of the motif among the input sequences. The exact left boundary of each motif region will be detected in the next phase via voting. Each character of the motif is recovered by voting among all the characters at the same positions in the motif regions of input sequences. For a sequence, S, a sample point is a random position, i, in S. For two sequences, S and , with two sample points, i and j, respectively, a rough motif boundary is detected by the similarity of and for some reasonably large parameter, l.
Descriptions of the Algorithm
Input: , where and are two sets of input sequences.
Output: planted motif in each sequence and consensus string. Start:
Randomly select sample points from each sequence, both in and .
For each pair of sequences selected from and ,
find the rough left and rough right boundaries via the matching at sample points.
Improve the rough boundaries.
If the motif boundaries of each sequence in are not empty,
use the Voting algorithm to get the planted motifs.
End of Algorithm.
3.2. An Example
We provide the following example for the brief idea of our algorithm. Let the following input strings be defined as below. We assume that the original motif is TTTTTAACGATTAGCS. The motif part is displayed with bold font, and the mutated characters in the motif region have been marked by * in their feet.
3.2.1.. Input Sequences
This contains two groups and .
The above two strings are used to detect the initial motif region and use them to deal with the motif in the second group below.
3.2.2.. Select Sample Points
Some sample points of two sequences in are selected randomly and marked with the little dots on the top.
3.2.3.. Collision Detection
In this step, the left and right rough boundaries of two sequences will be marked. The following shows the left collision, which happens nearby the left motif boundary and are marked by two overline and subsequences.
The following shows the right collision, which happens nearby the right motif boundary and is marked by two overline subsequences.
3.2.4.. Improving the Boundaries
In the early phase of the algorithm, we first detect a small piece of the motif in by comparing and . Assume that “” and “” are found in the left and right motif regions of , respectively. The rough motif length will be calculated via the difference of the location of the first character, ‘T’, of the first subsequence and the location of the last character, ‘G’, of the second subsequence. The position marked by “A” is the rough left boundary of the motif, and the position marked by “T” is the rough right boundary of the motif in below.
3.2.5.. Select Sample Points for the Sequences in
Some sample points near the motif boundaries of are selected.
.
Sample points are selected in each sequence in .
3.2.6.. Collision Detection Between with the Sequences in
The rough motif boundaries of the sequences in are detected via the collisions with the subsequences near the motif area of .
.
The rough motif boundaries are marked by the lines over the matched subsequences.
3.2.7.. Improving the Motif Boundaries for the Sequences in
After the collision with the sequences in , we obtain the rough location of the motifs of the sequences in . Their rough motif boundaries for the sequences in are improved to be closer to exact boundaries.
.
The improved motif boundaries of the sequences in are marked below. This phase does not cost much time, as only the positions near the rough motif boundaries are tested.
3.2.8.. Motif Boundaries for the Sequences in
.
Use the pair with and to find the motif boundaries in the sequences of . The rough boundaries of the second group are marked below with underlines. In this phase, the exact motif boundaries for most sequences of can be found.
3.2.9.. Extracting the Motif Regions
The motif regions of the second group will be extracted. The original motif is recovered via voting at each column.
3.2.10.. Recovering Motif via Voting
The original motif, TTTTTAACGATTAGCS, is recovered via voting at all columns. For example, the last S in the motif is recovered via voting among the characters, S, S, S, A, S, in the last column.
3.3. Our Results
We give an algorithm for the case with at most a mutation rate. The performance of the algorithm is stated in Theorem 2. Theorem 2 implies Corollary 3 by selecting with some constant that is large enough.
Theorem 2.
Assume that μ is a fixed number in and the alphabet size, t, is at least four. There exists a randomized algorithm and a constant , such that if the length of the motif, G, is at least , then, given k independent -sequences, the algorithm outputs , such that:
(1) with a probability of at most , ;
(2) for each , with a probability of at most , ; and
(3) with a probability of at most , the algorithm does not stop in time, where n is the longest length of any input sequences, and .
Corollary 3.
There exists a randomized algorithm and positive constants, and μ, such that if the alphabet size is at least four, the number of sequences is at least , the motif length is at least and each character in the motif region has a probability of at most of mutation, then the motif can be recovered with a probability of at least in time, where n is the longest length of any input sequences, and .
We give a randomized algorithm for the case with a mutation rate. The performance of the algorithm is stated in Theorem 4. Theorem 4 implies Corollary 5 by selecting with some constant that is large enough.
Theorem 4.
Assume that the alphabet size, t, is at least four. There exists a randomized algorithm and a constant , such that if the length of the motif G is at least , then, given k independent -sequences, the algorithm outputs , such that:
(1) with a probability of at most , ;
(2) for each , with a probability of at most , ; and
(3) with a probability of at most , the algorithm does not stop in ,
where n is the longest length of any input sequences, and .
Corollary 5.
There exists a randomized algorithm and positive constants, and α, such that if the alphabet size is at least four, the number of sequences is at least , the motif length is at least and each character in the motif region has a probability of at most α of mutation, then the motif can be recovered with a probability of at least in time.
We give a deterministic algorithm for the case with a mutation rate. The performance of the algorithm is stated in Theorem 6. Theorem 6 implies Corollary 7 by selecting with some constant that is large enough.
Theorem 6.
Assume that the alphabet size, t, is at least four. There exists a deterministic algorithm and a constant, , such that if the length of the motif G is at least , then, given k independent -sequences, the algorithm runs in and outputs , such that:
(1) with a probability of at most , ;
(2) for each , with a probability of at most , ; and
(3) with a probability of at most , the algorithm does not stop in time, where nis the longest length of any input sequences, and .
Corollary 7.
There exists a deterministic algorithm and positive constants, and α, such that if the alphabet size is at least four, the number of sequences is at least , the motif length is at least and each character in the motif region has a probability of at most α of mutation, then the motif can be recovered with a probability of at least in time.
4. Algorithm Recover-Motif
In this section, we give a unified approach to describe three algorithms. The performance of the algorithms is stated in Theorems 2, 4 and 6. The description of Algorithm Recover-Motif is given in Section 4.2. The analysis of the algorithm is given at Section 6.
4.1. Some Parameters
Definition 8.
- i.
- Parameter x is selected to be 10. This parameter controls the failure probability of our algorithms to be at most .
- ii.
- The size of the alphabet is .
- iii.
- Select a constant to have inequality (1):
- iv.
- The constant is selected to satisfy:The existence of ϵ follows from inequality (1). The constant ϵ is used to control the mutation in the motif area. It is a part of parameter β defined in item (xiv) of this definition.
- v.
- Let . The constant, c, is used to simplify probabilistic bounds, which are derived from the applications of Chernoff bounds (see Corollary 18).
- vi.
- Define .
- vii.
- Define to be a large constant that, for all :
- viii.
- Select constant , such that:The existence of follows from , which is implied by inequality (2).
- ix.
- Select constant and constant positive integer v that are large enough, such that:
- x.
- Define , and:
- xi.
- Select constant , such that:
- xii.
- The maximal mutation rate, α, for the second algorithm (Theorem 4) and the third algorithm (Theorem 6) are selected as . Since the mutation rate of our sublinear time algorithm is bounded by , the maximal mutation rate α for the first algorithm (Theorem 2) is less than when n is large enough. We always assume that all mutation rates α in our three algorithms are in the range .
- xiii.
- Define . By inequality (10), the definition of and the fact that , we have:Inequality (11) implies . By inequality (6), we have that:
- xiv.
- Let . The parameter, β, controls the similarity of and the original motif, G (see Lemma 27).
- xv.
- Define .
- xvi.
- We define the following .The parameter, , used in Lemma 27 gives an upper bound of the probability that a -sequence, S, whose will not be similar enough to the original motif, G, according to the conditions in Lemma 27.
- xvii.
- Select constant , such that:
- xviii.
- Select constant , such that .
- xix.
- Select number , such that:Since only n is variable, we can make .
- xx.
- For a fixed , define .
4.2. Description of Algorithm Recover-Motif
The algorithms are described in this section. The description combines three algorithms together. The simplest deterministic algorithm is also given in Section 5. Before presenting the algorithm, we define some notions.
Definition 9.
- Two sequences, and , are weakly left matched if: (1) both and are at least ; and (2) for all integers i, , where v is defined in item (ix) in Definition 8.
- Two sequences, and , are left matched if: (1) ; (2) for ; and (3) for all integers i, .
- Two sequences, and , are weakly right matched if and are weakly left matched, where is the inverse sequence of .
- Two sequences, and , are right matched if and are left matched, where is the inverse sequence of .
- Two sequences, and , are matched if and are both left and right matched.
Variable L will be controlled in the range in our algorithm with a high probability. We define the following functions that depend on L.
Definition 10.
Define . Define (see (xx) of Definition 8 for ), where .
We would like to minimize the function . This selection can make the total time complexity sublinear.
Definition 11.
For a sequence, S, define LB(S) to be the left boundary, l, of the motif region in S and RB(S) to be the right boundary, r, of the motif region in S, such that .
4.2.1.. Boundary-Phase of Algorithm Recover-Motif
The first phase of Algorithm Recover-Motif finds the rough motif boundaries of all input sequences. It first detects the rough motif boundaries of one sequence via comparing two input sequences. Then, the rough boundaries of the first sequence are used to find the rough motif boundaries of other input sequences.
Three algorithms share most of the functions. We have a unified approach to describe them. A special variable, “algorithm-type”, selects one of the three algorithms, respectively.
Definition 12.
Let algorithm-type represent one of the three algorithm types, “RANDOMIZED-SUBLINEAR”, “RANDOMIZED-SUBQUADRATIC” and “DETERMINISTIC-SUPERQUADRATIC”.
Definition 13.
Assume that is a set of positions in a sequence, , and is a set of positions in a sequence, . If there are positions and , such that for some position, j, with , is the position of in and is the position of in , then and have a collision at .
In the following function, Collision-Detection, the parameter, , is defined below in the three algorithms.
Function Collision-Detection is used to detect a point, , in the motif area in and another point in the motif area of . The two points, and , are close to the left and right motif boundaries of , respectively. A similar pair of points, and , in is also derived for . See the examples in Section 3.2.3..
Collision-Detection
Input: a pair of -sequences, and ; is a set of locations in for .
Output: the left and right rough boundaries of two sequences.
Let be all subsequences of of a length of with .
Let be all subsequences of of a length of with .
Find two subsequences, and
, such that is the least and .
Find two subsequences, and
, such that is the largest and
.
Find two subsequences, and
, such that is the least and
.
Find two subsequences, and
, such that is the largest and
.
Return .
End of Collision-Detection.
Definition 14.
Let be an interval with two integers boundaries, a and b, and l be a positive integer parameter. Define an l-partition of to be l-, which contains the intervals , such that for and .
For example, the three-partition of the interval is 3-. Function Point-Selection will be defined differently in three different algorithms, where I is an interval of positions in sequence S and L is a positive integer parameter. For randomized algorithms, some random points are selected in L-. For a deterministic algorithm, all points in I are selected. See the examples in Section 3.2.2..
Point-Selection
Input: a pair of -sequences, S, a size parameter, L, of partition and a set of intervals, I, of positions in S.
Output: a set, U, of positions from S, respectively.
Steps:
Let .
If algorithm-type=RANDOMIZED-SUBLINEAR or RANDOMIZED-SUBQUADRATICand
if :
for each interval, , in I, obtain its L-partition in L- and for each interval, J, in L-,
sample (see Definition 10) random positions in J and put them into U.
Else,
put every position of I into .
If algorithm-type=DETERMINISTIC-SUPERQUADRATIC,
put every position of I into U.
Return U.
End of Point-Selection.
The function, Improve-Boundaries, is used to improve the existing rough left and right boundaries, and , of , respectively, and to improve the existing rough left and right boundaries, and , of , respectively. We assume , , and . After calling this function, more accurate approximate boundaries will be derived. From the probabilistic analysis, we have a good chance to get the exact motif boundaries for both and . See the examples in Section 3.2.7..
Improve-Boundaries
Input: a -sequence, , with rough left and right boundaries, and , a -sequences, with rough left and right boundaries, and , and an approximate distance, L, to the nearest motif boundary from those rough boundaries (the parameter, L, usually has the properties of , , and ).
Output: improved rough left and right boundaries for both and .
Steps:
Find two subsequences, and ,
with and , such that and is
the least.
Find two subsequences, and ,
with and , such that and is
the largest.
Find two subsequences, and ,
with and , such that and is
the least.
Find two subsequences, and ,
with and , such that and is
the largest.
Return .
End of Improve-Boundaries.
The function, Initial-Boundaries, detects the motif boundaries for two sequences, and . It first detects rough motif boundaries that are controlled by parameter L. The rough boundaries will be improved to exact motif boundaries via calling Improve-Boundaries. See the examples inSection 3.2.2., Section 3.2.3., Section 3.2.4., Section 3.2.5., Section 3.2.6., Section 3.2.7. and Section 3.2.8..
Initial-Boundaries
Input: a pair of -sequences, and .
Output: rough left boundary of , right boundary of , rough left boundary of and right boundary of .
Steps:
Let .
Let .
Repeat.
Let Point-Selection.
Let Point-Selection.
Let Collision-Detection.
If ( and ),
then go to H.
Else, ,
until .
H: Return Improve-Boundaries.
End of Initial-Boundaries.
If and are the left and right motif boundaries of a sequence, S, then the motif length is . When we have the exact motif boundaries, and , for most sequences, , with high probability, their motif length can be derived via the median in . Therefore, we have the function, Motif-Length-And-Boundaries(), to compute the length of the motif region.
Motif-Length-And-Boundaries()
Input: is a set of independent sequences.
Steps:
For to ,
let =Initial-Boundaries.
Let be the median of .
Return .
End of Motif-Length-And-Boundaries.
4.2.2.. Extract-Phase of Algorithm Recover-Motif
After a set of motif candidates, W, is produced from Boundary-Phase of algorithm Recover-Motif, we use this set to match with another set of input sequences to recover the hidden motif by voting.
Match
Input: a motif left part, (which can be derived from the rough left boundary of an input sequence, S), a motif right part, , and a sequence, , from the group, , with known rough left and right boundaries.
Output: either a rough motif region of or an empty sequence, which means the failure in extracting the motif region, , of .
Steps:
Find a position, a, in with ,
such that and are left matched (see Definition 9).
Find a position, b, in with ,
such that and are right matched (see Definition 9).
If both a and b are found,
then output .
Else, output ∅ (empty string).
End of Match.
If the left, , and right, , motif parts are known, we extract all the motif regions for all sequences in the set, , by the function, Extract).
Extract)
Input and left and right motif parts, and (see function Match).
Steps:
For each with ,
let .
Return .
End of Extract.
The following is Extract-Phase of algorithm Recover-Motif. It extracts the motif regions of another set, , of input sequences. The function is based on the condition that exact motif boundaries can be derived for most sequences. See the examples in Section 3.2.9..
Extract-Phase():
Input is an input sequence with known and for its rough left and right boundaries, respectively, and is a set of input sequences.
Steps:
For each subsequence with
and , with ,
let be the output from Extract).
If the number of empty sequences in is at most ,
then return .
Return ∅ (empty set).
End of Extract-Phase .
4.2.3.. Voting-Phase
The function, Vote, is to generate another sequence, , by voting, where is the most frequent character among . See the examples in Section 3.2.10..
Voting-Phase
Input: sequences, , of the same length, m.
Output: a sequence, , which is derived by voting on every position of the input sequences.
Steps:
For each ,
let be the most frequent character among .
Return .
End of Vote.
4.2.4.. Entire Algorithm Recover-Motif
The entire algorithm is described below. The input has two sets of sequences, and . It detects the motif boundaries for the sequences in via pairwise comparisons and, also, the motif length. The motif regions of the sequences in are detected in the next phase and will be extracted. The original motif is recovered via voting for each column of characters among the extracted motif regions.
We maintain the sizes of and to be roughly equal, which implies:
Algorithm Recover-Motif (Z)
Input: , where and are two sets of input sequences.
Steps:
Preprocessing part:
For each , let (the two boundaries are unknown).
MotifLengthAndBoundaries().
Let .
For to ,
let Point-Selection
Point-Selection.
For to ,
let Point-Selection.
For to ,
for each ,
Let Collision-Detection.
Let =
Improve-Boundaries.
Let be the output from Extract-Phase().
If is not empty,
then go to the Voting part.
Voting part:
Return Voting-Phase().
End of Algorithm Recover-Motif .
5. Deterministic Algorithm
In this section, we give a deterministic algorithm, which is a simplified version of the unified algorithm described before in Section 4.2. It is simpler than the randomized versions. The first phase of Algorithm Recover-Motif (.) finds the rough motif boundaries of all input sequences. It first detects the rough motif boundaries of one sequence via comparing two input sequences. Then, the rough boundaries of the first sequence are used to find the rough motif boundaries of other input sequences. We still let:
Collision-Detection
Input: a pair of -sequences, and ; is a set of locations in for .
Output: the left and right rough boundaries of two sequences.
Let be all subsequences, , of of a length of with .
Let be all subsequences, , of of a length of with .
Find two subsequences, and
, such that is the least and .
Find two subsequences, and
, such that is the largest and
.
Find two subsequences, and
, such that is the least and
.
Find two subsequences, and
, such that is the largest and
.
Return .
End of Collision-Detection.
Function Point-Selection is not used in the deterministic algorithm.
Improve-Boundaries is the same as that in the randomized algorithms.
Initial-Boundaries
Input: a pair of -sequences, and .
Output: rough left boundary of , right boundary of , rough left boundary of and right boundary of .
Steps:
Let .
Let .
Repeat.
Let Collision-Detection.
If ( and ),
then go to H.
Else, ,
until .
H: Return Improve-Boundaries.
End of Initial-Boundaries.
Motif-Length-And-Boundaries() is the same as that before.
Match is the same as that for the randomized algorithm.
Extract) is the same as that for the randomized algorithm.
The following is Extract-Phase of algorithm Recover-Motif. It extracts the motif regions of another set, , of input sequences.
Extract-Phase() is the same as that for the randomized algorithm.
Voting-Phase is the same as that for the randomized algorithm.
The entire deterministic algorithm is described below. We maintain the sizes of and to be roughly equal.
Algorithm Recover-Motif (Z)
Input: , where and are two sets of input sequences.
Steps:
Preprocessing part:
For each , let (the two boundaries are unknown).
MotifLengthAndBoundaries().
Let .
For to ,
for each ,
let Collision-Detection.
Let =
Improve-Boundaries.
Let be the output from Extract-Phase().
If is not empty,
then go to the Voting part.
Voting part:
Return Voting-Phase().
End of Algorithm Recover-Motif .
6. Analysis of the Algorithm
The correctness of the algorithm will be proven via a series of lemmas in Section 6.2 and Section 6.3. Section 6.2 is for Boundary-Phase and Section 6.3 is for Extract-Phase. Furthermore, Section 6.3 gives some lemmas for the two randomized algorithms, and Section 6.4 gives the proof for the deterministic algorithm.
6.1. Review of Some Classical Results in Probability
Some well-known results in classical probability theory are listed. The readers can skip this section if they understand them well. The inclusion of these results make the paper self-contained.
- For a list of events, , .
- For two independent events, A and B, .
- For a random variable, Y, for all positive real numbers, t. This is called Markov inequality.
The analysis of our algorithm employs the Chernoff bound [22] and Corollary 18 below, which can be derived from it (see [11]).
Theorem 15
([22]). Let be n independent random 0–1 variables, where takes one with a probability of . Let , and . Then, for any :
- i.
- and
- ii.
- .
We follow the proof of Theorem 15 to make the following version of the Chernoff bound, so that it can be used in our algorithm analysis.
Theorem 16.
Let be n independent random 0–1 variables, where takes one with a probability of at most p. Let . Then, for any , .
Define . We note that is always strictly less than one for all and is fixed if δ is a constant. This can be verified by checking that the function is decreasing and . This is because , which is less than zero for all .
Theorem 17.
Let be n independent random 0–1 variables, where takes one with a probability of at most p. Let . Then, for any , .
Corollary 18
([11]). Let be n independent random 0–1 variables, and .
i) If takes one with a probability of at most p, then for any , .
ii) If takes one with a probability of at least p, then for any , .
6.2. Analysis of Boundary-Phase of Algorithm Recover-Motif
Lemma 19 shows that with only a small probability, a sequence can match a random sequence. It will be used to prove that when two substrings in two different -sequences are similar, they are unlikely not to coincide with the motif regions in the two -sequences, respectively.
Lemma 19.
Assume that and are two independent sequences of the same length and that every character of is a random character from Σ. Then:
- i.
- if , then the probability that and are matched is (); and
- ii.
- the probability for is at most .
Proof:
The two statements are proven as follows.
Statement i: For every character, , with , the probability is that .
Statement ii: For every character, , with , the probability is for to equal . If , the two sequences, and , are identical in at least positions, but the expected number of positions where the two sequences are identical is . The probability for is at most by Corollary 18, Definition and 9, items (xiv) and xii, equation (8) and inequality (2) in Definition 8. ▮
Lemma 20 shows that with a small probability, an input sequence contains a motif region that has many mutations.
Lemma 20.
- i.
- With a probability of at most , a sequence, S, changes more than characters in its motif region, , with , where c is defined in item (v) in Definition 8.
- ii.
- With a probability of at most , a sequence, S, changes more than characters in its left motif region, , for some t with , where c is defined in item (v) in Definition 8.
Proof:
Statement i: Every character in the region has a probability of at most α to mutate. We know that . By Corollary 18, with a probability of at most , has more than mutations.
Statement ii: We know that . By Corollary 18, with a probability of at most , a sequence, S, in has more than mutations (recall the setting for β in Definition 9) among the first left t characters. The total probability is at most . ▮
Lemma 21 shows that Improve-Boundaries() has a good chance to improve the accuracy of rough motif boundaries. Note that and are the left and right motif boundaries of S, respectively (see Definition 11).
Lemma 21.
Assume that sequence has and for . Then, for =Improve-Boundaries, we have the following two facts:
- i.
- with a probability of at most ; is not in for .
- ii.
- with a probability of at most , is not in for .
- iii.
- Improve-Boundaries runs in time.
Proof:
We need a bound for the following inequality:
Let . Compute the derivative . We also have the closed form for the function , which implies:
Let and . We have .
Statement i. By Lemma 20, with a probability of at most , one of the left motif first y character region of will change characters. Therefore, with a probability of at most , .
For a pair of positions, p, in , and q, in , without loss of generality, assume that p has a larger distance to the left boundary of than q to the left boundary of . Let be the distance from p to the left boundary of .
By Lemma 19, the probability is at most that there will be a match. There are at most cases for q. With a probability of at most , by inequality (20), .
For the cases that one position is in a random region and has a distance more than from the left boundary, the probability is at most by inequality (14).
Therefore, we have a total probability of at most that is not in .
Statement ii. One can also provide a symmetric analogous proof for this statement.
Statement iii. The computation time easily follows from the implementation of Improve-Boundaries . ▮
Lemma 22.
Assume that for each L with , with a probability of at most , for , where Collision-Detection(, Point-Selection and Point-Selection. Then, with a probability of at most , Initial-Boundary returns with or for .
Proof:
It follows from Lemma 21: ▮
Lemma 23.
Assume that with a probability of , each has its rough boundaries, or ; then, with a probability of at most , is not in , where is selected as the median of .
Proof:
If both and , then is in .
If the median of is not in , then there are at least numbers, i, to have or .
On the other hand, the probability is at most p, or . Therefore, this lemma follows from Corollary 18. ▮
For a -sequence S, we often obtain its left rough boundary with . Sometimes, its exact left boundary may be missed in the algorithm.
Definition 24.
- A -sequence, S, misses its left boundary if .
- A -sequence, S, misses its right boundary if .
Definition 25.
- i.
- A -sequence, S, contains a left half stable motif region, , if for all , where , and , as defined in Definition 8 and Section 2, respectively.
- ii.
- A -sequence, S, contains a right half stable motif region, , if for , where and .
- iii.
- A -sequence, S, contains a stable motif region, , satisfying the following conditions: (1) for ; (2) for ; and (3) the S motif region is both left and right half stable, where and .
Definition 26.
Assume that:
- ;
- Scontains both a left half and a right half stable motif region and and (see Definition 8 for and v); and
- for each L with , if has and , then with a probability of at most , for , where Collision-Detection(, Point-Selection Point-Selection and Point-Selection.
- The rough boundaries for all sequences, , are computed via Collision-Detection and = Improve-Boundaries .
Then, with a probability of at most , there are more than sequences in with or .
Proof:
According to the condition of this lemma, with a probability of at most , , where Collision-Detection( and Point-Selection.
For a fixed pattern from S, by Lemma 19, with a probability of at most , it has a distance of more than to the true left boundary. As we need to deal with possible patterns from S, with a probability of at most , .
Similarly, with a probability of at most , . Let .
With a probability of at most , does not contain a left half stable motif region by Lemma 20. Similarly, with a probability of at most , does not contain a right half stable motif region. Let .
Although, S is involved in searching the left boundary with all other sequences. The non-missing condition is to let each sequence not change too many characters in the motif region. Therefore, this is an independent event for each sequence. It is safe to use the Chernoff bound to deal with it.
With a probability of at most , there are more than sequences, , in with or . ▮
6.3. Analysis of Extract-Phase and Voting-Phase of Algorithm Recover-Motif
Lemma 27 shows that with a high probability, the left and last parts of the motif region in a -sequence do not change much.
Lemma 27.
With a probability of at most defined in Equation (13), a -sequence, S, does not contain a stable motif region.
Proof:
The probability is not to satisfy conditions (1) and (2) of item (iii) in Definition 25. Consider condition (3) of item (iii) in Definition 25. Since every character of (notice that ) has a probability of at most α to mutate, by Corollary 18, the probability is at most that . Let , where , as defined in Definition 8. Therefore, the probability is at most that for some . Similarly, we define for the probability on the right-hand side. The probability is at most that for some . The probability that S does not contain a stable motif region is at most . ▮
Definition 28.
Assume that contains , which contains a stable motif region. We fix such a .
- Define to be the left part of the motif region, .
- Define to be the right part of the motif region, .
Lemma 29 shows that with a high probability, Extract-Phase of algorithm Recover-Motif extracts the correct motif regions from the sequences in . It uses to match in another sequences, S. The parameter, R, gives a small probability that the matched region between and S is not in .
Lemma 29.
- i.
- Assume that and are fixed sequences of length . Let S be a -sequence with , and let be the number of characters of M that are not in the region of . Then, the probability is at most R that , where R is defined in (xv) of Definition 8.
- ii.
- The probability is at most that given a -sequence S, .
Proof:
Assume that . Let w be the number of characters outside of on the left of M, and let be the number of characters outside of on the right of M. Clearly, . Since , either or . See Figure 1. Without loss of generality, we assume .

Figure 1.
and M.
Statement i: There are two cases.
Case (a): . By Lemma 19, the probability for this case is at most for a fixed w. The total probability for this case for is at most .
Case (b): . By Lemma 19, the probability is at most for a fixed w. The total probability for is at most .
The probability analysis is similar when . Therefore, the probability for this case is at most for .
Statement ii: By Lemma 27, with a probability of at most , S does not contain a stable motif region. Therefore, we have a probability of at most that given a random -sequence, S, . ▮
Lemma 30 shows that we can use and to extract most of the motif regions for the sequences in if (recall that is defined right after Lemma 27).
Lemma 30.
Assume that and are two sequences of a length of , and for and (recall that each sequence, , is either an empty sequence or a sequence of the length of ).
- i.
- If , and there are no more than () sequences, , with or , then the probability is at most that there are more than sequences, , with .
- ii.
- For arbitrary and , with a probability of at most , , where R is defined in Definition 8.
Proof:
Recall that sequence is selected right after Lemma 27.
Statement i: By Lemma 29, for every , the probability is at most that does not contain a stable motif region, . By Corollary 18, we have a probability of at most that there are more than sequences, , with .
Statement ii: By Lemma 29, the probability is at most R that . By Corollary 18, with a probability of at most , . ▮
Definition 31.
- Given two sequences, and , define:Match.
- For a sequence, S, define to be the , which is the leftmost subsequence of a length of in the motif region of S.
- For a sequence, S, define to be the , which is the rightmost subsequence of a length of in the motif region of S, where ;
Lemma 32.
Assume that we have the following conditions:
- i.
- For each L with , with a probability of at most , or for , where Collision-Detection(, Point-Selection and Point-Selection.
- ii.
- For each L with , if has and , then with a probability of at most , for or , where Collision-Detection(, Point-Selection Point-Selection and Point-Selection .
- iii.
- The inequality holds for some constant , where is defined in Equation (13) and .
- iv.
- The inequality holds, where .
Then, the algorithm generates a set of at most subsequences for voting and votes for a sequence, , such that:
(1) with a probability of at most , ; and
(2) for each , with a probability of at most , .
Before proving Lemma 32, we note that both and are at most for all of the three algorithms. They will be proven by Lemma 47 and Lemma 46 for the case algorithm-type=RANDOMIZED-SUBLINEAR, Lemma 41 and Lemma 40 for the case algorithm-type=RANDOMIZED-SUBQUADRATIC, and Lemma 35 for the case algorithm-type=DETERMINISTIC-SUPERQUADRATIC.
Proof:
By Lemmas 22, with a probability of at most , or .
By Lemma 23, with a probability of at most , the approximate motif length, , is not in the range . Assume in the rest of the proof of this lemma.
By Lemma 27, with a probability of at most , a sequence does not contain a stable motif region. Therefore, with a probability of at most , the following statement is false:
(i) One of for has , and has a stable motif region.
By Lemma 26, with a probability of at most , there are more than sequences with or . In other words, with a probability of at most , the following statement is false:
(ii) There are no more than sequences with or , where .
Assume that Statement (ii) is true. By Lemma 30, with a probability of at most , the following statement is false:
(iii) contains at most empty sequences.
We start from the rough left boundary, , of to match the other left boundaries of for . There are in total at most candidates to consider.
By Lemma 30, if , which consists of matched regions, has at most empty sequences, then it has more than from non-motif regions with a probability of at most. After the pattern is fixed, those events in the matching are considered to be independent of each other. This is why we can apply the Chernoff bound to deal with them. Therefore, the probability is at most ; the following statement is false:
(iv) If contains at most empty sequences, then contains at most elements not from motif regions .
Therefore, with a probability of at most , the sequences are not ready for voting in the next phase, which means the following two conditions are satisfied:
(a) There exists and generated by the algorithm, such that contains at most elements not from motif regions .
(b) For every and that contains at most empty sequences generated by the algorithm, contains at most elements not from motif regions .
Statement (1): For a with at most elements not from motif regions , we still have elements in from motif regions . By the condition (iv) in this lemma, we have . Therefore, is selected to be the length of G in the Voting-Phase().
Statement (2): For a with at most elements not from motif regions , we still have elements in from motif regions . By Corollary 18, with a probability of at most , there are more than characters that are mutated in the same position among all motif regions for the sequences in . We have that by the condition (iv) in this lemma. We let be the most frequent character among in Voting-Phase. Therefore, with a probability of at most , . ▮
We will use multiple variable functions to characterize the computational time for three algorithms. In order to unify the complexity analysis of three algorithms, we introduce the following notation.
Definition 33.
A function, , is nondecreasing if it is nondecreasing on both variables. If for arbitrary positive constants, and , for some positive constant, c, then is slow.
Lemma 34.
Assume that , and are non-decreasing slow functions.Assume that Collision-Detection() returns the result in time and the Point-Selection() selects positions in time. Assume that with a probability of at most , the function Initial-Boundaries() does not stop when , and in the algorithm Recover-Motif is no more than .
Then, with a probability of at most , the entire algorithm Recover-Motif does not stop in the time complexity , where is the largest j, such that and .
Proof:
The function Initial-Boundaries()is executed times. According to the condition that with a probability of at most , the function Initial-Boundaries(.) does not stop when , we have the fact that with a probability of at most , one of those executions of Initial-Boundaries(.) does not stop when .
In the rest of the proof, we assume that all executions of Initial-Boundaries(.) stops when .
When , we detect rough left and right motif boundaries and run Improve-Boundaries(), which takes time by Lemma 21. It takes time to run Initial-Boundaries() one time for one pair () in . It takes time to run Initial-Boundaries() one time for all pairs () in .
It takes time to find the rough boundaries for all sequences in with a fixed sequence, S, from by executing for the loop “For each ” in the algorithm Recover-Motif. It takes time to find the rough boundaries for all sequences in via all sequences, , from through the loop “For each ” in the algorithm Recover-Motif.
Recall that parameters v and are constants, and is . Calling Match() takes time for each . The total times for calling Match() is .
The voting part takes time for executing voting for recovering one character in the motif. ▮
6.4. Deterministic Algorithm for an Mutation Rate
In this section, we give a deterministic algorithm for the case with an mutation rate. The performance of the algorithm is stated in Theorem 6.
Lemma 35.
Let algorithm-type=DETERMINISTIC-SUPERQUADRATIC. Assume that and . Let be a set of intervals of the positions of that satisfies . Let be a set of intervals of the positions of that satisfies . Let Point-Selection, Point-Selection and Collision-Detection. Then:
- i.
- with a probability of at most , the left rough boundary, , has at most distance from and the left rough boundary has at most distance from .
- ii.
- with a probability of at most , the right rough boundary, , has at most distance from and the right boundary of has at most distance from .
Proof:
For two sequences, and , let be the subsequence, , for . By Corollary 18, with a probability of at most (see inequality (8) in Definition 14), there are more than mutations in for .
With a probability of at most , the left boundary position is missed during the matching. We have similar to miss the right boundary.
Assume that and are two positions of and , respectively. If one of two positions is outside the motif region and has more than distance to the motif boundary, with a probability of at most (see inequality (8) in Definition 14), for them to match requires by Lemma 19, where is a subsequence for . ▮
Lemma 36.
For the case algorithm-type=DETERMINISTIC-SUPERQUADRATIC, we have:
- i.
- Collision-Detection() takes time.
- ii.
- Point-Selection() selects positions in time.
- iii.
- in the algorithm Recover-Motif is no more than .
- iv.
- With a probability of at most , the algorithm Recover-Motif does not run in computational complexity .
Proof:
Statement i. The parameter, , is set to be β in Collision-Detection. It follows from the time complexity of the brute force method.
Statement ii. It follows from the implementation of Point-Selection().
Statement iii. It follows from the choice of Point-Selection(.) for the sublinear time algorithm at Recover-Motif(.).
Statement iv. It follows from Lemma 34 and Statements i, ii and iii. ▮
We give the proof for Theorem 6.
Proof:
[Theorem 6] The computational time part of this theorem follows from Lemma 36.
By Lemma 35, we let for the probability bound, , in the condition (i) of Lemma 32.
By Lemma 35, we can let for the probability bound, , in the condition (i) of Lemma 32
By inequality (12), the condition (iii) of Lemma 32 is satisfied.
By inequality (11), we know that the condition (iv) of Lemma 32 can be satisfied.
The failure probability part of this theorem follows from Lemma 21 and Lemma 32 by using the fact that and k are of the same order (see equation (18)). ▮
6.5. Randomized Algorithms for Motif Detection
In this section, we present two randomized algorithms for motif detection. The first one is a sublinear time algorithm that can handle mutation, and the second one is a super-linear time algorithm that can handle mutation. They also share some common functions.
Lemma 37.
Let be a constant in . Assume m and n are two non-negative integers with . Then, for every integer, , with , , where constant as defined in Definition 8.
Proof:
We have the inequalities:
▮
Lemma 38.
Let be a set of n elements with . Assume that are m random elements in S. Then, with a probability of at most , the list, , contains at most different elements from U (in other words, ).
Proof:
For a subset, , with for some integer, , the probability is at most that all elements, , are in . For every subset, , with , there exists another subset, , such that and . We have that . There are subsets of U with a size of . We have the probability of at most that contains at most different elements in U. ▮
Lemma 39.
Let β be a constant in and . Let and for some fixed . Let and be two sets of n elements with and C be a set of the size for some constant . Then, for all large n, with a probability of at most , we have , where and are two sets, which may have multiplicities, of m random elements from and , respectively.
Proof:
In the entire proof of this lemma, we always assume that n is sufficiently large. We are going to give an upper bound about the probability that B does not contain any element in . For each element, , with a probability of at most , is not in . Therefore, the probability is at most that B does not contain any element in .
By Lemma 38, the probability is at most that . We have the inequalities:
The inequality, , which is used from Equation (34) to Equation (35), follows from the fact that . The transition from (35) to (36) follows from the fact that , since , according to the conditions of the lemma.
It is easy to see that for all large n. Thus, (note that as ). Thus, by Lemma 37, . This is why we have the transition from Equation (37) to Equation (38). Therefore, . ▮
6.5.1.. Randomized Algorithm for an Mutation Rate
In this section, we give an algorithm for the case with an mutation rate. The performance of the algorithm is stated in Theorem 4.
Lemma 40.
Let algorithm-type=RANDOMIZED-SUBQUADRATIC. Assume that and . Let be a set of intervals of the positions of that satisfy . Let be a set of intervals of the positions of that satisfy . Let Point-Selection, Point-Selection and Collision-Detection. Then:
- i.
- with a probability of at most , the left rough boundary, , has at most distance from and the left rough boundary, , has at most distance from ;
- ii.
- with a probability of at most , the right rough boundary, , has at most distance from and the right boundary of has at most distance from .
Proof:
The proof is the same as Lemma 35 for the algorithm with algorithm-type=DETERMINISTIC-SUPERQUADRATIC. For two sequences, and , let be the subsequence for . By Corollary 18, with a probability of at most (see inequality (8) in Definition 14), there are more than mutations in for .
With a probability of at most , the left boundary position is missed during the matching. We have similar to miss the right boundary.
Assume that and are two positions of and , respectively. If one of the two positions is outside the motif region and has more than distance to the motif boundary, with a probability of at most (see inequality (8) in Definition 14), for them to match requires by Lemma 19, where is a subsequence for . ▮
Lemma 41.
Let algorithm-type=RANDOMIZED-SUBQUADRATIC. Assume that and . Let be a set of intervals of the positions of that satisfy . Let be a set of intervals of the positions of that satisfy . Let Point-Selection, Point-Selection and Collision-Detection. Then:
- i.
- with a probability of at most , the left rough boundary, , has at most a distance from and the left rough boundary has at most a distance from ;
- ii.
- with a probability of at most , the right rough boundary, , has at most a distance from and the right boundary of has at most a distance from .
Proof:
We prove the following two statements which imply the lemma.
- i.
- With a probability of at most , there are no intervals, from and from , such that: (1) is at least ; (2) the left boundary of has at most a distance from ; (3) the left boundary of has at most a distance from ; and (4) there is a collision between the sampled positions in and .
- ii.
- With a probability pf at most , there are no intervals, from and from , such that: (1) is at least ; (2) the right boundary of has at most a distance from ; (3) the right boundary of has at most a distance from ; and (4) there is a collision between the sampled positions in and .
We only prove statement i. The proof for statement ii is similar.
Select from and from to be the first pair of intervals with . It is easy to see that such a pair exists, and both have a distance from the left boundary with a distance of at most . This is because when a leftmost interval of a length pf L is fully inside the motif region of the first sequence, we can always find the second interval from the second sequence with an intersection of a length of at least .
Replace m by , by (see Definition 10) and n by L to apply Lemma 39. We do not consider any damaged position in this algorithm; therefore, let C be empty.
With a probability of at most , there is a point in . The subsequences of a length of starting at the point from and fail to have the difference bounded by β with a probability of at most by Lemma 20. With a probability of at most , we do not have that the rough boundaries are detected with a distance of at most to exact motif boundaries. ▮
Lemma 42.
For the case algorithm-type=RANDOMIZED-SUBQUADRATIC, we have:
- i.
- Collision-Detection() takes time.
- ii.
- Point-Selection() selects positions in time if .
- iii.
- Point-Selection() selects positions in time if .
- iv.
- in the algorithm Recover-Motif is no more than .
- v.
- With a probability of at most , the algorithm Recover-Motif does not stop in time.
Proof:
Statement i. The parameter, , is set to be β in Collision-Detection. It follows from the time complexity of the brute force method.
Statements ii and iii. They follow from the implementation of Point-Selection().
Statement iv. It follows from the choice of Point-Selection(.) for the sublinear time algorithm at Recover-Motif(.).
Statement iv. It follows from Lemma 40, Lemma 34 and Statements i, ii and iii. ▮
We give the proof for Theorem 6.
Proof:
[Theorem 4] The computational time part of this theorem follows from Lemma 42.
By Lemma 40, we can let for the probability bound, , in the condition (i) of Lemma 32.
By Lemma 41, we can let for the probability bound, , in the condition (ii) of Lemma 32.
By inequality (12), the condition (iii) of Lemma 32 is satisfied.
By inequality (11), we know that the condition (iii) of Lemma 32 can be satisfied.
The failure probability part of this theorem follows from Lemma 21 and Lemma 32 by using the fact that and k are of the same order (see Equation (18)). ▮
6.5.2.. Sublinear Time Algorithm for a Mutation Rate
In this section, we give an algorithm for the case with at most mutation rate. The performance of the algorithm is stated in Theorem 2.
Definition 43.
A position, p, in the motif region, , of an input sequence, S, is damaged if there exists at least one mutation in , where is defined in item (xvii) in Definition 8.
Lemma 44.
Assume that . With a probability of at most , there are more than positions that are from the (see Definition 10 for and ) sampled positions that are damaged in an interval of a length pf L.
Proof:
By Theorem 16, with a probability of at most (let ), there are more than mutations in an interval of a length of L. Therefore, with a probability of at most , there are more than positions that are damaged. Therefore, each random position in an interval of a length of L has at most a probability of to be damaged.
Since and positions are sampled, by Theorem 16, with a probability of at most (let ), the number of damaged positions sampled in an interval of a length of L is more than . Thus, with a total probability of at most , there are more than damaged positions that are from the sampled positions in an interval of a length of L. ▮
Definition 45.
Let A be a set of positions in an input sequence, S, with . Let . In other words, contains all the positions of A in .
Lemma 46. Let
Let algorithm-type=RANDOMIZED-SUBLINEAR. Assume that and L is an integer with . Let be a set of intervals of the positions of that satisfy . Let be a set of intervals of the positions of that satisfy . Let Point-Selection, Point-Selection and Collision-Detection. Then:
- i.
- with a probability of at most , the left rough boundary, , has at most a distance from and the left rough boundary, , has at most a distance from .
- ii.
- with a probability of at most , the right rough boundary, , has at most a distance from and the right boundary of has at most a distance from .
Proof:
For two sequences, and , it is easy to see that there is a common position in both motif regions of the two sequences, such that there is no mutation in the next characters with a high probability. This is because that mutation probability is small.
By Theorem 16, with a probability of at most (let ), there are more than mutated characters in the interval for . Since the mutation probability is , with a probability of at most , there are more than positions to be damaged in .
Similarly, we have a probability of at most that there are more than damaged positions in .
Now, we assume that left side has more than damaged positions and the right side, more than damaged positions in . Since each position in each interval of a length of L is selected in Point-Selection, it is easy to verify the conclusions of this lemma. ▮
Lemma 47.
Let algorithm-type=RANDOMIZED-SUBLINEAR. Assume that and . Let be a set of intervals of the positions of that satisfy . Let be a set of intervals of the positions of that satisfy . Let Point-Selection, Point-Selection and Collision-Detection. Then:
- i.
- with a probability of at most , the left rough boundary, , has at most a distance from and the left rough boundary, , has at most a distance from .
- ii.
- With a probability of at most , the right rough boundary, , has at most a distance from and the right boundary of has at most a distance from .
Proof:
We prove the following two statements, which imply the lemma.
- i.
- With a probability of at most , there are no intervals, from and from , such that: (1) is at least ; (2) the left boundary of has at most a distance from ; (3) the left boundary of has at most a distance from ; and (4) there is a collision between the sampled positions in and .
- ii.
- with a probability of at most , there are no intervals, from and from , such that: (1) is at least ; (2) the right boundary of has at most a distance from ; (3) the right boundary of has at most a distance from ; and (4) there is a collision between the sampled positions in and .
We only prove statement i. The proof for statement ii is similar to that for statement i. Assume that L satisfies the condition of this lemma. Select from and from to be the first pair of intervals of a size of L with . This is because when a leftmost interval of a length of L is fully inside the motif region of the first sequence, we can always find the second interval from the second sequence with an intersection of a length of at least .
Replace m by , by (see Definition 10) and n by L to apply Lemma 39. We also let C be the set of damaged positions in and caused by the mutated positions. With a probability of at most , C has a size of by Lemma 44. With a probability of at most , there is point in . The existence of such a point makes and have a distance of at most to and , respectively. ▮
Lemma 48.
For the case algorithm-type=RANDOMIZED-SUBLINEAR, we have:
- i.
- Collision-Detection() takes time.
- ii.
- Point-Selection() selects positions in time if .
- iii.
- Point-Selection() selects positions in time if .
- iv.
- in the algorithm Recover-Motif is no more than .
- v.
- With a probability of at most , the algorithm Recover-Motif does not stop in time.
Proof:
Statement i. The parameter, , is set to be zero in Collision-Detection. It follows from the time complexity of bucket sorting, which is described in standard algorithm textbooks.
Statements ii and iii. They follow from the implementation of Point-Selection().
Statement iv. It follows from the choice of Point-Selection(.) for the sublinear time algorithm at Recover-Motif(.).
Statement v. It follows from Lemma 46, Lemma 47 and Lemma 34 and Statements i, ii iii and iv. ▮
We give the proof for Theorem 2.
Proof:
[Theorem 2] The computational time part of this theorem follows from Lemma 48.
By Lemma 46, we can let for the probability bound, , in the condition (i) of Lemma 32.
By Lemma 47, we can let for the probability bound, , in the condition (ii) of Lemma 32.
By inequality (12), the condition (iii) of Lemma 32 is satisfied.
By inequality (11), we know that the condition (iv) of Lemma 32 can be satisfied.
The failure probability part of this theorem follows from Lemma 21 and Lemma 32 by using the fact that and k are of the same order (see Equation (18)). ▮
6.6. Experiments on Simulated Datasets
Aiming at solving the motif discovery problem, we implemented our algorithm in Java. Our tests were all done on a laptop with an Intel Dual Core 1.5 G CPU and 3.0 G Memory. In the first experiment, we tested our algorithm on several simulated datasets, which are all generated from our probability model with a small mutation rate. Each input set contains 20 or 15 sequences, which are of a length of 600 or 500 base pairs. Additionally, each bp of all the simulated gene sequences was generated independently with the same occurrence probability. A motif with a fixed length was randomly planted into each input sequence. The minimum Hamming distances between the results and consensus are recorded.
6.7. Experiments
There are many other tools for detecting and analyzing the motifs, like the EMmethod [23], MEME [24], Gibbs [25], Compo [26], MochiView [27], PhyME [28], HeliCis [29] and WebMotif [30], among others. Each of them has their advantages and disadvantages.
Table 1 shows the results on simulated datasets. From the table, we could find that the results of our algorithm for finding the motif on simulated datasets are satisfied. Our algorithm could find all the motifs from each sequence and get the consensus with an accuracy rate of 100%. If the dataset has a high mutation rate, we could increase the number of repetitions, so that the result on the datasets will be more accurate. We also recorded the total time cost for each test, which mainly depends on the number of test repetitions. In the experimental tables, the parameter, N, represents the number of sequences, parameter M represents the length of motifs, parameter M is the maximum length of sequences and R is the number of iterations. GCR1 is a famous DNA binding protein, whose ability to bind DNA is dependent on the CTTCC sequence motif [31]. Several other popular data sets are also used in the experiments of our motif detection and its comparisons to the other methods.

Table 1.
Results on simulated data.
| Set 1 | 20 | 600 | 15 | 60 | 100 | 98 |
| Set 2 | 15 | 600 | 15 | 10 | 100 | 18 |
| Set 3 | 20 | 600 | 12 | 15 | 100 | 22 |
In the second experiment, we tested our algorithm on a real sequence set, which was obtained from SCPD. SCPD contains a large number of gene data and transcription factors of yeast. Sequences in the same set are all regulated by a common motif. We chose 1,000 bp as the length of the input sequences. In order to show the advantages of our algorithm, we also compared the result of our algorithm with the results of several other existing motif finding methods on the same dataset, such as Gibbs, MEME, Info-Gibbs and Consensus. Table 2 shows the details of the data set we used in the experiment.

Table 2.
Number of Sequences and Motif Length.
| N | 6 | 9 | 6 | 15 | 5 |
Table 3 shows the results of the five algorithms on biological data sets. Table 4 and Figure 2 give the average mismatch numbers of each algorithm. We choose four well-known motif-detecting softwares to make the comparisons. From Table 3 and Table 4, we see that the average mismatch numbers of our algorithm on data sets GCN4 and GCR1 are significantly lower than other four well-known methods. On the data sets Bas1,Rap1Ebf1 and HSE-HTSF, our algorithms also shows satisfied performance compared to other methods.
In addition, our algorithm also shows its high speed in computations compared to other four motif finding methods. Because the starting pattern of algorithms are represented by a string, so our algorithm can avoid some extra time consuming computations unlike Gibbs sampling and EM methods, such as computations of likelihoods. According to this feature, we use the consensus string of the voting operation obtained from the last iteration as a new starting pattern in program, and continue doing voting operations repeatedly until there is no further improvement. Experimental results show that if we set the number of iterations to be a large integer, the programs could give more accurate results within a reasonable time.

Table 3.
Total number of mismatch positions.
| Our Algorithm | 10 | 8 | 4 | 45 | 5 |
| Gibbs | 8 | 51 | 5 | 202 | 7 |
| MEME | 8 | 15 | 10 | 32 | 3 |
| InfoGibbs | 9 | 21 | 5 | 46 | 9 |

Table 4.
Average mismatch numbers per sequence.
| Our Algorithm | 1.67 | 0.89 | 0.67 | 3 | 1 |
| Gibbs | 1.33 | 5.6 | 0.83 | 13.46 | 1.4 |
| MEME | 1.33 | 1.67 | 1.67 | 2.13 | 0.6 |
| InfoGibbs | 1.5 | 2.33 | 0.83 | 3.06 | 1.8 |
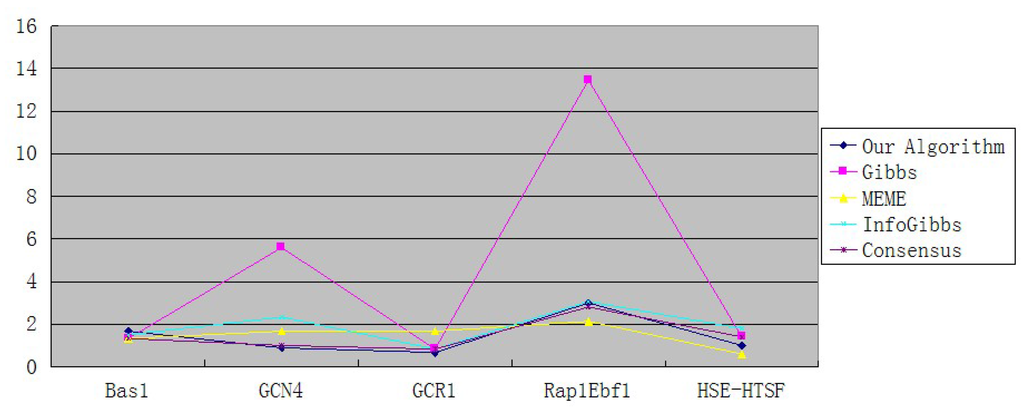
Figure 2.
Average mismatch numbers per sequence.
6.8. Conclusions
We develop algorithms under the probabilistic model. One of them finds the implanted motif with a high probability if the alphabet size is at least four, the motif length is in and each character in the motif region has a probability of at most of mutation. The motif region can be detected, and each motif character can be recovered in sublinear time. A sub-quadratic randomized algorithm is developed to recover the motif with an mutation rate. A quadratic deterministic algorithm is developed to recover the motif with an mutation rate. It is an interesting problem if there is an algorithm to handle the case for an alphabet of a size of three. A more interesting problem is to extend the algorithm to handle larger mutation probability.
6.9. Future Works
Compared with other motif finding methods, our algorithm shows its great advantages. However, there are still some improvements that could be done on this algorithm. For example, though a sequence set has the consensus, the motif in each sequence may have high mutation rates; in addition, the length of each motif could also be different. Therefore, these two factors increase the difficulty for finding motifs, and currently, there is still no effective algorithm that could solve these problems. In the future, we plan to improve the efficiency of our algorithm by combining other motif finding methods, such as MEME; a combination may be made to make our algorithm have better performance in finding motifs from highly mutated sequences.
Acknowledgments
We thank Ming-Yang Kao for introducing us to this topic. We also thank Lusheng Wang and Xiaowen Liu for some discussions. We would like to thank Eugenio De Hayos for his helpful comments. We would like to thank the reviewers, whose comments greatly improved the presentation of this paper.
This research is supported in part by the NSF Early Career Award 0845376, and NSF HRD-1137764.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Frances, M.; Litman, A. On covering problems of codes. Theor. Comput. Sci. 1997, 30, 113–119. [Google Scholar] [CrossRef]
- Ga̧sieniec, L.; Jansson, J.; Lingas, A. Efficient Approximation Algorithms for the Hamming Center Problem. In Proceedings of the 10th Annual ACM-SIAM Symposium on Discrete Algorithms, Baltimore, MD, USA, 17–19 January 1999; pp. S905–S906.
- Stormo, G.; Hartzell, G., III. Identifying protein-binding sites from unaligned DNA fragments. Proc. Natl. Acad. Sci. USA 1991, 88, 5699–5703. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, C.; Reilly, A. An expectation maximization (EM) algorithm for the identification and characterization of common sites in unaligned biopolymer sequences. Proteins 1990, 7, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Hertz, G.; Stormo, G. Identification of Consensus Patterns in Unaligned DNA and Protein Sequences: A Large-Deviation Statistical Basis for Penalizing Gaps. In Proceedings of the 3rd International Conference on Bioinformatics and Genome Research, Tallahassee, USA, 1–4 June 1994; pp. 201–216.
- Stormo, G. Consensus patterns in DNA. In Molecular Evolution: Computer Analysis of Protein and Nucleic Acid Sequences; Doolitle, R.F., Ed.. Methods Enzymol. 1990, 183, 211–221. [Google Scholar]
- Lanctot, J.K.; Li, M.; Ma, B.; Wang, L.; Zhang, L. Distinguishing string selection problems. Inf. Comput. 2003, 185, 41–55. [Google Scholar] [CrossRef]
- Lucas, K.; Busch, M.; Mossinger, S.; Thompson, J. An improved microcomputer program for finding gene- or gene family-specific oligonucleotides suitable as primers for polymerase chain reactions or as probes. Comput. Appl. Biosci. 1991, 7, 525–529. [Google Scholar] [CrossRef] [PubMed]
- Dopazo, J.; Rodríguez, A.; Sáiz, J.C.; Sobrino, F. Design of primers for PCR amplification of highly variable genomes. Comput. Appl. Biosci. 1993, 9, 123–125. [Google Scholar] [PubMed]
- Proutski, V.; Holme, E.C. Primer master: A new program for the design and analysis of PCR primers. Comput. Appl. Biosci. 1996, 12, 253–255. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Ma, B.; Wang, L. On The Closest String and Substring Problems. J. ACM 2002, 49, 157–171. [Google Scholar] [CrossRef]
- Li, M.; Ma, B.; Wang, L. Finding Similar Regions in Many Strings. In Proceedings of the 31st Annual ACM Symposium on Theory of Computing, Atlanta, GA, USA, 1–4 May 1999; pp. 473–482.
- Pevzner, P.; Sze, S. Combinatorial Approaches to Finding Subtle Signals in DNA Sequences. In Proceedings of the 8th International Conference on Intelligent Systems for Molecular Biology, Toronto, ON, Canada, 19–23 July 2000; pp. 269–278.
- Keich, U.; Pevzner, P. Finding motifs in the twilight zone. Bioinformatics 2002, 18, 1374–1381. [Google Scholar] [CrossRef] [PubMed]
- Keich, U.; Pevzner, P. Subtle motifs: Defining the limits of motif finding algorithms. Bioinformatics 2002, 18, 1382–1390. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Dong, L. Randomized algorithms for motif detection. J. Bioinform. Comput. Biol. 2005, 3, 1039–1052. [Google Scholar] [CrossRef] [PubMed]
- Chin, F.; Leung, H. Voting Algorithms for Discovering Long Motifs. In Proceedings of the 3rd Asia-Pacific Bioinformatics Conference, Singapore, 17–21 January 2005; pp. 261–272.
- Gusfield, D. Algorithms on Strings, Trees, and Sequences; Cambridge University Press: Cambridge, UK, 1997. [Google Scholar]
- Fu, B.; Kao, M.Y.; Wang, L. Probabilistic analysis of a motif discovery algorithm for multiple sequences. SIAM J. Discret. Math. 2009, 23, 1715–173. [Google Scholar] [CrossRef]
- Fu, B.; Kao, M.Y.; Wang, L. Discovering almost any hidden motif from multiple sequences. ACM Transactions on Algorithms 2011, 7(2), 26. [Google Scholar] [CrossRef]
- Liu, X.; Ma, B.; Wang, L. Voting Algorithms for the Motif Problem. In Proceedings of Computational Systems Bioinformatics Conference, (CSB’08), Stanford, CA, USA, 26–29 August 2008; pp. 37–47.
- Motwani, R.; Raghavan, P. Randomized Algorithms; Cambridge University Press: Cambridge, UK, 2000. [Google Scholar]
- Dempster, A.P.; Laird, N.M.; Rubin, D.B. Maximum likelihood from complete data vis the EM algorithm. J. R. Stat. Soc. 1977, 39, 1–38. [Google Scholar]
- D’haesler, P. How does DNA sequence motif discovery work? Nat. Biotechnol. 2006, 24, 959–961. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, C.; Altschul, S.; Boguski, M.; Liu, J.; Neuwald, A.; Wootton, J. Detecting subtle sequence signals: A gibbs sampling strategy for multiple alignment. Science 1993, 262, 262–5131. [Google Scholar] [CrossRef]
- Sandve, G.K.K.; Abul, O.; Drabløs, F. Compo: Composite motif discovery using discrete models. BMC Bioinform. 2008, 9. [Google Scholar] [CrossRef] [PubMed]
- Homann, O.; Johnson, A. MochiView: Versatile software for genome browsing and DNA motif analysis. BMC Biol. 2010, 8. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.; Blanchette, M.; Tompa, M. PhyME: A probabilistic algorithm for finding motifs in sets of orthologous sequences. BMC Bioinform. 2004, 5. [Google Scholar] [CrossRef]
- Larsson, E.; Lindahl, P.; Mostad, P. HeliCis: A DNA motif discovery tool for colocalized motif pairs with periodic spacing. BMC Bioinform. 2007, 8. [Google Scholar] [CrossRef] [PubMed]
- Romer, K.; Kayombya, G.R.; Fraenkel, E. WebMOTIFS: Automated discovery, filtering and scoring of DNA sequence motifs using multiple programs and Bayesian approaches. Nucleic Acids Res. 2007, 35, W217–W220. [Google Scholar] [CrossRef] [PubMed]
- Baker, H. GCR1 of Saccharomyces cerevisiae encodes a DNA binding protein whose binding is abolished by mutations in the CTTCC sequence motif. Proc. Natl. Acad. Sci. USA 1991, 88, 9443–9447. [Google Scholar] [CrossRef] [PubMed]
© 2013 by the authors licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).