Abstract
Myotonic dystrophy type 1 (DM1) is an autosomal dominant hereditary disease caused by abnormal expansion of unstable CTG repeats in the 3′ untranslated region of the myotonic dystrophy protein kinase (DMPK) gene. This disease mainly affects skeletal muscle, resulting in myotonia, progressive distal muscle weakness, and atrophy, but also affects other tissues and systems, such as the heart and central nervous system. Despite some studies reporting therapeutic strategies for DM1, many issues remain unsolved, such as the contribution of metabolic and mitochondrial dysfunctions to DM1 pathogenesis. Therefore, it is crucial to identify molecular target candidates associated with metabolic processes for DM1. In this study, resorting to a bibliometric analysis, articles combining DM1, and metabolic/metabolism terms were identified and further analyzed using an unbiased strategy of automatic text mining with VOSviewer software. A list of candidate molecular targets for DM1 associated with metabolic/metabolism was generated and compared with genes previously associated with DM1 in the DisGeNET database. Furthermore, g:Profiler was used to perform a functional enrichment analysis using the Gene Ontology (GO) and REAC databases. Enriched signaling pathways were identified using integrated bioinformatics enrichment analyses. The results revealed that only 15 of the genes identified in the bibliometric analysis were previously associated with DM1 in the DisGeNET database. Of note, we identified 71 genes not previously associated with DM1, which are of particular interest and should be further explored. The functional enrichment analysis of these genes revealed that regulation of cellular metabolic and metabolic processes were the most associated biological processes. Additionally, a number of signaling pathways were found to be enriched, e.g., signaling by receptor tyrosine kinases, signaling by NRTK1 (TRKA), TRKA activation by NGF, PI3K-AKT activation, prolonged ERK activation events, and axon guidance. Overall, several valuable target candidates related to metabolic processes for DM1 were identified, such as NGF, NTRK1, RhoA, ROCK1, ROCK2, DAG, ACTA, ID1, ID2 MYOD, and MYOG. Therefore, our study strengthens the hypothesis that metabolic dysfunctions contribute to DM1 pathogenesis, and the exploitation of metabolic dysfunction targets is crucial for the development of future therapeutic interventions for DM1.
1. Introduction
Myotonic dystrophy type 1 (DM1), also known as Steinert’s disease, is the most common type of muscular dystrophy in adults, with a prevalence of approximately 5 to 20 per 100,000 individuals worldwide [1,2]. DM1 is a multisystemic neuromuscular disorder mainly characterized by myotonia, muscle weakness, and atrophy. Other features observed in patients with DM1 include alterations in the central nervous system (CNS), gonadal atrophy, insulin resistance, dyslipidemia, cardiac conduction deficiencies, and breathing difficulties [3]. This disorder is caused by a trinucleotide expansion of unstable repetitions of CTG in the 32,032 untranslated region of the myotonic dystrophy protein kinase (DMPK) gene located at chromosome 19 q13.3 [4,5]. Fewer than 38 CTG repeats are found in normal individuals, whereas DM1 patients have at least 50 CTG repeats [6]. Interestingly, the age of onset seems to be inversely correlated with CTG repeat length, influencing the severity of clinical symptoms [7]. Despite research efforts in the field, there is currently no cure for DM1, and diagnosis is also challenging to halt or slow down DM1 progression.
Metabolic disorders are known to result from metabolism problems, which affect neurodegeneration, longevity, and aging [8,9]. Various metabolic defects, including glucose resistance, hyperinsulinemia, and diabetes mellitus, are found in DM1 patients [10]. Moreover, whereas DM1 diagnosis using genetic tests and magnetic resonance, electromyography, and skeletal muscle histopathology are standard methodologies [11], creatine kinase is also known to be the only reasonable biochemical marker in DM1 patients, although not a disease-specific marker [12,13]. Few studies have been conducted regarding the advance of non-invasive biomarkers for DM1, but promising results have been reported on the use of microRNAs (miRNAs) [14]. miR-1, miR-27b, miR-133a, miR-133b, miR-140-3p, miR-206, miR-454, and miR-574 have been reported to be deregulated in DM1 patients compared to healthy individuals [15,16,17]. Additionally, promising treatments for DM1 have been proposed, which might be able to suppress or eliminate its associated molecular effects, including MBNL1 (Muscleblind Like Splicing Regulator 1) expression enhancers, and toxic RNA degradation [18]. It was also recently reported that treatment of DM1 patient-derived fibroblasts with metformin reversed metabolic and mitochondrial defects, including impaired proliferation [19]. Metformin has the potential to delay aging at both the cellular and organismal level, and metformin treatment may show efficiency in a preclinical setting [20]. Therefore, an in-depth study of the molecular mechanisms and therapeutic targets in DM1 has become a key research topic. However, the abovementioned approaches are not currently treatment options for DM1. Therefore, approaches to understand the contribution of metabolism and mitochondrial dysfunction in DM1 seems to be crucial to unraveling DM1 pathogenesis. However, only few studies have been carried out to identify novel putative metabolic and therapeutic targets for DM1. In the present work, we aimed to identify novel metabolism-associated molecular targets, resorting to bibliometric and visualization-based analyses, together with bioinformatics, to map the biological processes and associated signaling pathways. These potential novel molecular targets will hopefully highlight a new perspective to address research gaps in DM1 pathogenesis.
2. Material and Methods
2.1. Search Approach
Literature searches were performed in the Web of Science (WOS) and Scopus databases. The Scopus database was chosen over the WOS because more results were retrieved, and information is updated on a daily basis [21,22]. The first search query included the following terms: “myotonic and dystrophy and type 1” and “metabolic or syndrome”. The second query included the following terms: “myotonic and dystrophy and type 1” and “metabolic or metabolism or skeletal or muscle or metabolism”. Both queries aimed to identify scientific articles related to metabolism and DM1. These search queries included studies from the 1960s onwards, and the generic terms “review”, “animals”, and “non-English” were excluded from the search tab.
2.2. Automatic Text-Mining Analysis Using VOSviewer
VOSviewer is a software tool used to reveal scientific landscapes and to show associated terms from selected scientific articles through a network [23]. To extract the keywords related to DM1, we used VOSviewer (v. 1.6.16), whereby the layout was established in a context for mapping and clustering. To create the bibliometric networks, a co-occurrence analysis of all closely related terms was carried out to indicate how closely keywords were associated by computing and extracting keywords, in addition to numerical evaluation of the content of a group of documents. Usually, if keywords are grouped into the same cluster, they likely to reflect similar topics. Thereafter, these results can be analyzed to build a conceptual structure of the desired field [24,25]. We aimed to determine the co-occurrence of keywords between different clusters. All the metadata from the articles were uploaded by combining the keywords included in queries 1 and 2. These include “myotonic and dystrophy and type 1” and “metabolic or syndrome or metabolism or skeletal or muscle”. Generic terms such as ‘follow-up’, ‘article’, ‘review’, ‘male’, ‘female’, and many other irrelevant terms were removed by manual curation in VOSviewer. Thereafter, networks were created based on the relation strength of the search terms and keywords from the different clusters, which were displayed in different colors. Possible molecular associations were depicted if a specific keyword had a simultaneous association with DM1 or any identical keywords by setting a minimum number of occurrences of a keyword to 63. Based on this analysis, we anticipated that we would be able to identify popular research topics in DM1 research.
Furthermore, to retrieve a gene list from VOSviewer, we combined the total 8 keywords from both queries using Scopus search. The resulting files were uploaded, and we manually ticked the genes/proteins present in this list in VOSviewer. We manually curated duplicates and plural names of genes/proteins from the final list to reduce redundancy. The UniProt Knowledgebase (UniProtKB) website [26] was used to review entries and obtain the gene/protein identifiers. Only the reviewed (Swiss-Prot) entries that contained manually annotated records were chosen. Each identifier was recorded manually for further analyses. However, some of the genes downloaded from Scopus had no UniProt identifier because the gene was broad. Therefore, the nearest gene/protein was chosen to explore the possibility of any similarities.
2.3. DisGeNET Search
To complement the VOSviewer results (Section 2.2) and identify the genes previously associated with DM1, the DisGeNET database (version 7.0) was searched. DisGeNET is an open-access management program that combines information on genes and gene variants related to human diseases from numerous resources with data obtained by text mining of scientific information [27,28]. We searched for “Myotonic dystrophy type 1” (C3250443 code) in the query box and opted for a summary of gene–disease associations, keeping the default query focused on diseases. The jvenn plug-in tool [29] was used to compare the lists of results obtained from DisGeNET and VOSviewer.
2.4. Protein–Protein Interaction Network Construction
Protein–protein interaction (PPI) network analysis was performed using the STRING web tool (version 11.5) [30] to illustrate the physical and functional association between the genes retrieved in the VOSviewer analysis and that were not previously associated with DM1. The interaction score was kept at the highest confidence (0.900) to easily view the strength connection between the genes.
2.5. Gene Ontology Functional and Pathway Enrichment Analyses
A functional enrichment analysis was performed using the g:Profiler tool (version e106_eg53_p16_65fcd97), which maps genes to functional data sources and identifies statistically notable gene ontology (GO) terms [31]. GO enrichment analysis (GO biological process) and REAC signaling pathway analysis were performed. The default parameters of the enrichment analysis were as follows: human was chosen as a specific organism, and the user threshold was 0.05.
3. Results
Considering the aim of the present manuscript, a literature search was performed, followed by automatic text mining and data functional enrichment analysis to identify relevant associations between metabolism/metabolic processes and DM1. A summary of the research strategy used in the present manuscript is represented in Figure 1.
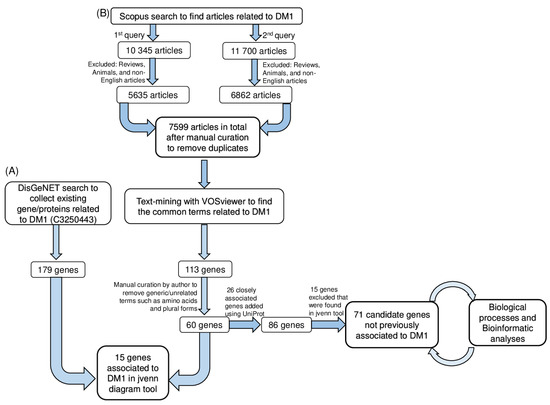
Figure 1.
Workflow of the strategy used for literature search and bibliometric analysis to unravel a functional association between myotonic dystrophy type 1 and metabolism. (A) DisGeNET-based collection of previously reported molecular associations with DM1. Terminologies corresponding to genes only were selected and compared with DisGeNET data. This analysis yielded a set of 15 genes previously associated with DM1 in DisGeNET. (B) Workflow used to obtain the common terms related to metabolism/metabolic and DM1 using VOSviewer software. Following the literature search, text-mining with VOSviewer software was used to uncover the most appropriate terminologies from scientific articles. We also focused on the 71 genes identified in VOSviewer analysis but that were not previously associated with DM1 in DisGeNET. These genes emerged as potential DM1 targets. DM1, myotonic dystrophy type 1.
3.1. Literature Search and Automatic Text-Mining Analysis to Unveil Novel Metabolism Molecular Targets in DM1
The Scopus literature search yielded 12 497 papers using both queries after excluding articles on animals, as well as non-English and review articles. After manual curation by the author to remove duplicates from both queries, a total of 7599 papers were obtained (Figure 1). VOSviewer software was used to perform a text-mining analysis of the article data by construction and visualization of the co-occurrence networks of relevant terms (Figure 1).
Using 45 co-occurrences as a threshold in VOSviewer software settings, 863 terms were obtained and grouped into 4 clusters represented by different colors (Figure 2). The size of nodes reflects the frequency of appearance of the keywords, and the distance between two nodes represents their correlation. Cluster 1 (red) is related to metabolism and includes keywords such as gene expression, protein binding, and RNA-binding protein (Figure 2A), with the second highest total link strength (Table 1) after myotonic dystrophy. Cluster 2 (green) is related to myotonic dystrophy and includes keywords such as myotonia, muscular dystrophy, myotonic dystrophy type 1, and myopathy (Figure 2A). Cluster 3 (blue) is related to trinucleotide repeat, nucleotide repeat, gene mutation, genetic analysis, and pathogenesis (Figure 2A). Cluster 4 (yellow) includes keywords such as skeletal muscle, muscle fibers, skeletal, muscle cell, and calcium (Figure 2A). Subnetworks around the terms “Metabolism” (Figure 2B) and “Myotonic Dystrophy” (Figure 2C) were created to analyze the associated terms. Overall, it is clear that the keywords metabolism and myotonic dystrophy are represented as large nodes with a high total link strength, suggesting the significance of these keywords, followed by gene expression, skeletal muscle, muscle skeletal, and RNA-binding proteins.
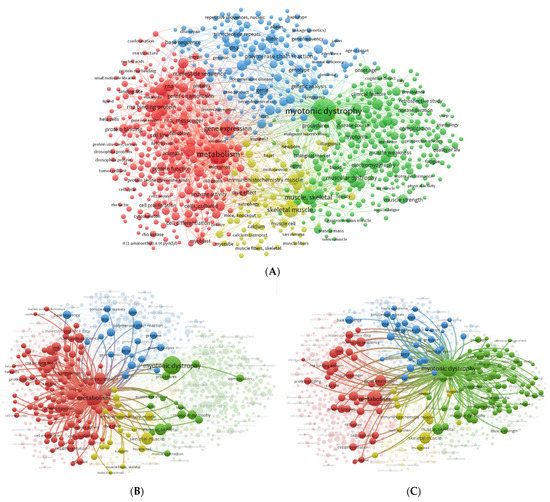
Figure 2.
Keyword network visualization map. (A) Global network of the co-occurrence of all keywords co-. (B) Metabolism co-occurrence network. (C) Myotonic dystrophy co-occurrence network. The co-occurrence network visualization map was created using VOSviewer (https://www.vosviewer.com/ (accessed on 21 December 2022). The node size reflects the occurrence of the term, and the edges point out co-occurring terms. The lines demonstrate keywords occurring together. The total link strength shows the number of total publications in which two keywords appear. The size of the nodes reflects the co-occurrence of the keywords, and the distance between two nodes represents the association between the keywords. Terms are grouped into clusters according to research interests, which are represented by different colors: cluster 1, red; cluster 2, green; cluster 3, blue; and cluster 4, yellow.

Table 1.
Top six keywords from the global network visualization map. In the network visualization map, a link expresses a co-occurrence association between keywords. These links have a strength identified by a positive number; a higher number indicates a stronger link. The total link strength indicates the total number of publications in which two keywords appear.
Table 1 summarizes the top six keywords according to their total link strength relative to the global network of all keyword co-occurrences (Figure 2A). Table 1 also indicates that the keyword myotonic dystrophy has the highest link strength, and RNA-binding proteins has the lowest strength.
In order to identify the genes related to DM1 in VOSviewer, using the default settings, 39,565 terms were found to meet the threshold by setting the minimum number of occurrences to 1. By default, 5 clusters with a total of 113 genes (Figure S1 and Table S1). Among the 113 genes, ‘amino acids, drosophila, duplicates, plurals and generic terms’ were manually curated from the list, and a total of 60 genes were attained. Furthermore, to obtain relevant information about these 60 genes, closely associated genes were further retrieved from the UniProt database, and 26 genes were added (Table S4). Thereafter, a total of 86 genes was considered for further analysis (Figure 1).
3.2. Comparative Analysis of the Novel Identified Metabolism-Associated Molecular Targets in DM1 Using VOSviewer with Molecular Associations Previously Described in DisGeNET
In order to compare the novel identified metabolism targets associated with DM1 (Table S1) with the genes/proteins already associated with DM1 in the literature, a search of the DisGeNET database was performed. A total of 179 genes associated with DM1 were retrieved (Table S2). A comparative analysis using the jvenn tool revealed that 15 common genes: DMPK, MBNL, CELF1, INSR, IL6, IGF1, DMD, PRKCA, ACTB, ATP2A1, ATP2A2, RYR1, CASP3, TGFB1, and CKM (Figure 3 and Table 2).
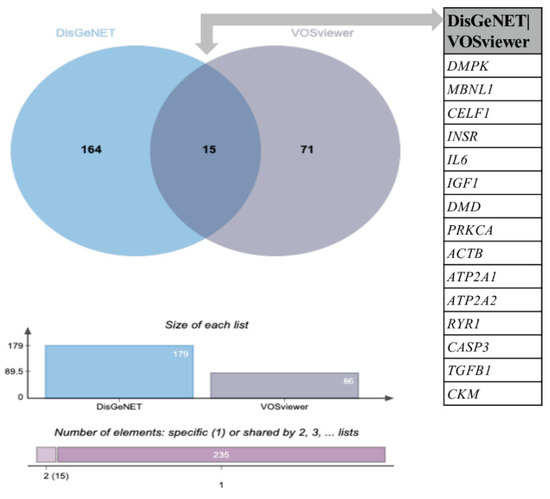
Figure 3.
Venn diagram analysis of the DisGeNET and VOSviewer lists of genes. The Venn diagram shows an intersection between the DisGeNET and VOSviewer lists of genes. Each circle corresponds to an entry/gene list. Fifteen (15) common genes were found between the DisGeNET and VOSviewer lists; 164 are exclusive to the DisGeNET list, and 71 are exclusive to the VOSviewer list. The chart presented below the Venn diagram shows the list size and intersection size repartition. A comparative analysis was performed with jvenn software (http://jvenn.toulouse.inra.fr/app/example.html (accessed on 28 November 2022)).

Table 2.
Detailed list of common genes identified in DisGeNET and VOSviewer analysis. Proteomics DB (https://www.proteomicsdb.org/(accessed on 21 December 2022) and UniProt (https://www.uniprot.org/ (accessed on 21 December 2022) were used to investigate the biological processes in which these genes participate. More information on their subcellular localization and molecular function are presented in Table S3.
For further analysis, we excluded these 15 common genes between VOSviewer and DisGeNET, and a list of 71 candidate genes associated with metabolism was obtained.
3.3. Characterization and Functional Enrichment Analysis of the Novel Identified Metabolism Associated Molecular Targets in DM1
Given that the main goal of the present manuscript is the identification of novel putative metabolism-associated targets for DM1, we next focused our analysis on the 71 genes exclusively identified using the VOSviewer analysis (Figure 1). First, we gathered information regarding its subcellular localization, molecular function, and biological process using ProteomicsDB and UniProt (Table 3 and Table S5). More importantly, their dysfunction in DM1, if already reported, or gene information retrieved through the NCBI (National Center for Biotechnology Information) (Table 3) was searched, as these genes may reveal a great potential as metabolism-associated targets for DM1.

Table 3.
Summary of the 71 potential novel molecular targets for DM1 that were not previously associated with the disease in DisGeNET and the correspondent VOSviewer occurrence score. ProteomicsDB and UniProt were used to investigate biological processes in which these 71 genes participate.
Furthermore, we analyzed the protein–protein interactions (PPI) of these candidate targets using the STRING database. The resulting network outlines the arrangement of anticipated associations for the 71 genes considering all possible protein interactions, which comprised 69 nodes (genes) interacting with each other through 60 edges, showing a significant PPI enrichment p-value: 1.53 × 10−14; therefore, these genes are possibly biologically connected (Figure 4). The most interactions were observed for INS and NTRK1 proteins. AKT1, CDC42, and LAMA1 proteins also presented a higher number of interactions.
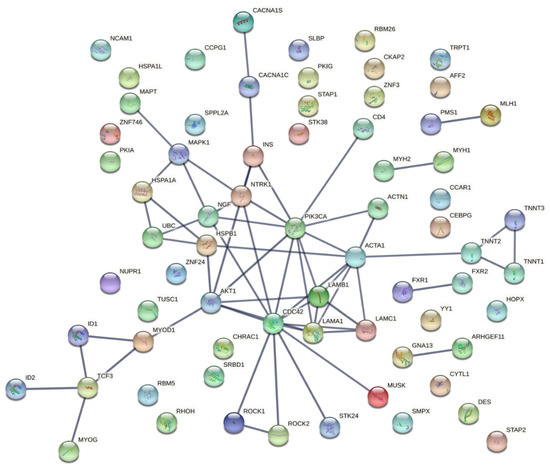
Figure 4.
Protein–protein interaction network of the 71 candidate targets for myotonic dystrophy type 1 generated using the STRING database. Network analysis of the 71 genes considering all the possible interactions. The thickness of the lines indicates the degree of confidence of the interaction (wider lines indicate stronger evidence of interactions).
Moreover, using g:Profiler, we performed a functional enrichment analysis in terms of biological processes (BPs) and pathways of the 71 candidate molecular targets for DM1 (Figure 5). A total of 187 GO terms were found to be enriched in the BP category (Table S6). The candidate molecular targets for DM1 were mainly associated with the regulation of primary metabolic processes, regulation of cellular metabolic processes, regulation of nitrogen compound metabolic processes, gene expression, and cell differentiation (Figure 5A). Furthermore, several signaling pathways were found to be associated with the candidate molecular targets for DM1 using the REAC database (Table S7), from which signaling by receptor tyrosine kinases, axon guidance, and NTRK1 (TRKA) presented a higher number of associated genes/proteins (Figure 5B).
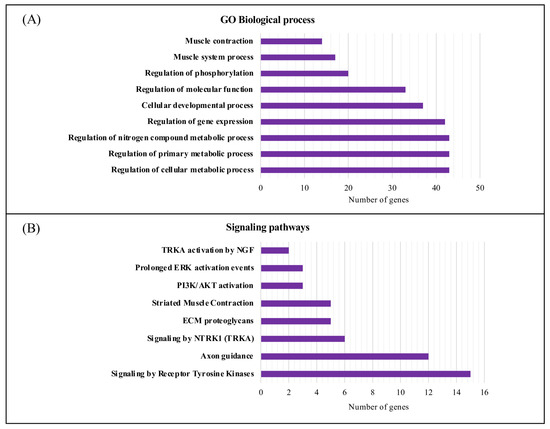
Figure 5.
Biological process and signaling pathway functional enrichment analysis of the candidate molecular targets for DM1. (A) The top 10 biological processes associated with the candidate molecular targets for DM1. (B) REAC signaling pathway enrichment analysis of the eight overrepresented signaling pathways using g:Profiler (https://biit.cs.ut.ee/gprofiler/gost (accessed on 29 August 2022). The purple bars represent the number of genes annotated in the category. GO, gene ontology; NGF, nerve growth factor; ERK, extracellular signal-regulated kinase; PI3K/AKT, phosphatidylinositol-3-kinase/ protein kinase B; ECM, extracellular matrix; NTRK1 (TRKA), neurotrophic tyrosine kinase receptor type 1.
Based on the previous analysis, it was evident that biological processes associated with metabolic processes are overrepresented. Therefore, we compiled a list of the genes associated with metabolic processes, which comprises 44 genes/proteins. The PPI network comprising the 44 identified proteins is presented in Supplementary Figure S2. Five clusters were constructed using the STRING database by applying the Markov clustering (MCL) algorithm after summarizing the complied list of genes associated with metabolic processes (Figure 6). Cluster 1 includes the RHOH, AKT1, ACTA1, ACTN1, HSPB1, HSPA1A, and MAPT genes. Two chaperones (HSPB1 and HSPA1A) were present in this cluster. Cluster 2 includes the MYOD1, ID1, ID2, TCF3, and MYOG genes. Cluster 3 comprises the NGF, NTRK1, PIK3CA, and INS genes. It was possible to verify that cluster 3 comprises genes known to be important signaling pathways, such as PI3K/AKT activation and signaling by NTRK1. Lastly, cluster 4 includes the FXR1, FXR2, and MAPK1 genes, whereas cluster 5 includes the ROCK1 and ROCK2 genes. As anticipated, this cluster-based functional characterization of the metabolic gene list unveiled specific physiological roles/signaling pathways for several subgroups of interacting genes (metabolic processes and myogenesis/muscle contraction) (Figure 6).
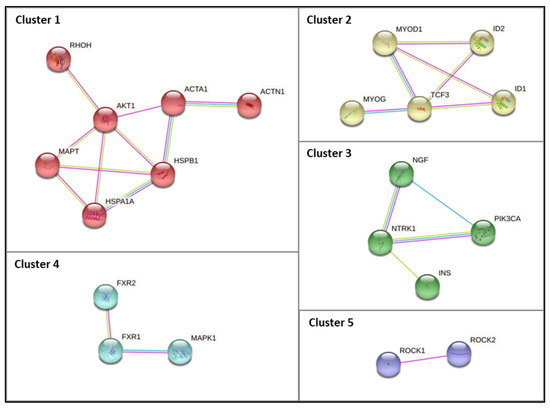
Figure 6.
PPI subnetworks of clusters (1 to 5) comprising the genes identified in VOSviewer analysis associated with GO biological processes related to protein metabolism. The PPI subnetwork was constructed using the STRING database by applying the Markov clustering (MCL) algorithm (inflation parameter of 1.8). The edges indicate that the directly linked proteins are part of the same physical complex, although in large complexes, this may not signify that they are directly bound to each other. The edge color denotes the interaction source; blue edges denote known interactions from curated databases, pink edges denote experimentally determined known interactions, and green edges denote interactions inferred by text mining.
4. Discussion
DM1 is the most prevalent form of muscular dystrophy among adults, causing multisystemic symptoms in patients and with no treatment currently available. In the present work, we offer a comprehensive analysis based on a bibliometric approach to identify novel candidate targets for DM1 related to metabolism. Upon a systematic literature search, the work was divided into four steps: analysis of keyword co-occurrences, selection of candidate gene targets, analysis of the shortlisted targets, and analysis of biological and signaling pathways associated with the obtained gene list. Interestingly, the term metabolism frequently co-occurs with terms such as gene expression, protein binding, protein phosphorylation, and protein function (Figure 2B). This is not unexpected, given that in previous work, our described that metabolic and mitochondria dysfunction contribute to DM1 pathophysiology [57]. In fact, there are several pieces of evidence of metabolic defects in DM1 patients, such as insulin resistance, hyperinsulinemia, and diabetes mellitus [34]. Furthermore, previous work by our team associated these metabolic alterations with lipins; phosphatidate phosphatase enzymes, which regulate the lipid signaling pathways; DAG levels; triacylglycerol levels; and phospholipids [57].
Myotonic dystrophy also co-occurs with keywords that are associated with main clinical features observed in patients with DM1, such as muscle weakness, myopathy, myotonia, and muscle contraction (Figure 2C), which is in line with other previous studies [102,103].
Further analysis of automatic text mining carried out in VOSviewer in combination with DisGeNET data showed an intersection of 15 genes that were common between the two software programs, of which DMPK, MBNL, CELF1 are important players in DM1 pathology and are the most studied in DM1 [104,105,106]. Other common genes include INSR, IL6, IGF1, DMD, PRKCA, ACTB, ATP2A1, ATP2A2, RYR1, CASP3, TGFB1, and CKM (Table 2). Remarkably, with this analysis, we identified 71 genes not previously associated with DM1. Among these, several valuable candidate targets for DM1 related to metabolic process emerged (Figure 6, Table 3).
The last part of our work focused on performing a biological process and pathway enrichment analysis of the 71 genes not previously associated with DM1. It was evident that more genes participate in regulation of cellular metabolic process, regulation of primary metabolic process, and regulation of nitrogen compound metabolic processes (Figure 5). Furthermore, cluster-based functional characterization of the metabolic gene list grouped the genes in five different and highly relevant clusters (Figure 6), which are discussed below. Additionally, a novel automated string-based approach for cluster analysis is a possible alternative and an interesting approach to that used in the present study [107].
4.1. Dysregulation of Biological Processes and Signaling Pathways Associated with DM1
4.1.1. Muscle Function and Associated Pathways
Cluster 1 comprises genes important for skeletal muscle (ACTA1) and muscle function (ACTN1) and includes a crucial gene (AKT1) known to regulate insulin signaling, cell survival, and tumor progression (Table 3), as well as two chaperones: HSPA1A and HSPB1 (heat shock proteins (HSPs)). HSPs are known to be expressed in skeletal muscle and play key roles in muscle growth and development [108]. Both HSPA1A and HSPA1L are known as HSP70 or commonly as HSP72 [109]. HSP70 is stimulated in response to non-damaging and damaging stress stimuli. Hence, its overexpression leads to the maintenance of muscle fiber cohesion and enables muscle reconstruction and repair. However, HSP70 expression is reduced during muscle aging, and the loss of HSP70 as a vital mechanism may lead to contractile dysfunction and muscle atrophy [110]. HSP70 upregulation in skeletal muscle has been recognized in rodent models of muscle injury, muscle atrophy, and muscular dystrophy, emphasizing the role of HSP70 as an important therapeutic target for the treatment of different disorders that have a negative impact on skeletal muscle function [111]. Likewise, glycogen synthase kinase 3 beta (GSK3β) is a familiar conserved serine/threonine kinase with roles in pathways regulating myogenesis, inflammation, neurogenesis, metabolism, and cellular processes in skeletal muscle [112]. Although evidence shows a vital role for GSK3β in skeletal muscle metabolism, it is also involved in the development of clinical conditions. Notably, increased activity of GSK3 is described in DM1 [113]. In fact, it was shown that GSK3β inhibition improved defects in myotonia, myogenesis, and muscle strength in a DM1 mouse model [114]. It was also shown that normalization of CUGBP1 activity with GSK3 inhibitors had a positive effect on reducing skeletal muscle and CNS morphology in DM1 mouse models [115]. Overall, GSK3β can be pointed to as a therapeutic target [114] for the treatment skeletal muscle wasting induced by aging and a number of chronic diseases [116].
4.1.2. NGF and TRK Signaling Pathways
Moreover, in the PPI analysis network (Figure 4), a strong degree of interaction was revealed between INS and NTRK1, which was further evidenced by cluster analysis (cluster 3, Figure 6). NTRK genes comprise NTRK1, NTRK2, and NTRK3, encoding the proteins of tropomyosin receptor kinase (TRK) family TRKA, TRKB, and TRKC, respectively, which are transmembrane tyrosine kinases receptors. The latter receptors have been associated with several functions, including precursor cell survival and proliferation, differentiation, metabolism, sensory neuron function, synaptic strength, and plasticity [117]. These tyrosine kinase receptors have received some attention from the clinical point of view, given that NTRK gene fusions including NTRK1, NTRK2, and NTRK3 are identified as oncogenic drivers in various types of tumors, including an increase in cancer in DM1 [118]. The NTRK1 gene encodes the TRKA protein, which binds to the nerve growth factor (NGF), inducing tyrosine phosphorylation and tyrosine kinase activity of TRKA [119]. Furthermore, the ligand binding to TRK receptors causes TRK receptor dimerization and activates three main intracellular signaling pathways, namely phospholipase C-γ (PLCγ), PI3 kinase (PI3K), and mitogen-activated protein kinase (MAPK/ERK) pathways (Figure 7) [120]. The ROCK pathway can also be activated both via the Rac-RhoA or the Raf-MAPK-RSK signaling pathway [121,122,123,124]. Together, these pathways play important and distinct roles in cell functioning. The MAPK/ERK pathway is involved in cell growth and proliferation, whereas the PLCγ pathway regulates neuronal differentiation, survival, and metabolism. The PI3K pathway is responsible for metabolism, survival, and apoptosis prevention [125]. Interestingly, crosstalk between these signaling pathways occurs to coregulate biological functions mediated by NTRK genes. The appropriate activation of TRK receptors is critical to nervous system development and cell survival.
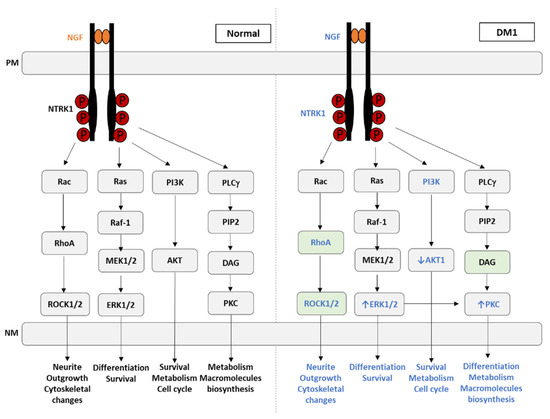
Figure 7.
NGF-mediated TRK receptor dimerization and activation of three main intracellular signaling pathways under normal conditions and its deregulation in myotonic dystrophy type 1. The eight genes found in our analysis associated with the TRK signaling pathways are identified in blue, as well as the biological processes affected in DM1. The green boxes indicate the genes never associated with DM1. PM, plasma membrane; NM, nuclear membrane; NGF, nerve growth factor; NTRK1, neurotrophic receptor tyrosine kinase 1; RAC, Ras-related C3 botulinum toxin substrate 1; RAS, rat sarcoma; PI3K, phosphoinositide 3-kinase; PLC-γ, phospholipase C gamma; RhoA, Ras homolog family member A; Raf1, Raf-1 proto-oncogene, serine/threonine kinase; PIP2, phosphatidylinositol 4,5-bisphosphate; MEK1/2, MAP kinase kinases MEK-1 and MEK-2; AKT, protein kinase B; DAG, diacylglycerol; ROCK1/2, Rho-associated protein kinase 1/2; ERK1/2, extracellular signal-regulated protein kinase; PKC, protein kinase C.
In the present work, eight genes associated with the TRK signaling pathways were identified as being related to DM1 (Figure 7, indicated in blue). Among these, only four were previously reported as altered in DM1, namely ERK1/2, PI3K, AKT1, and PKC (Figure 7). The MEK/ERK pathway mainly promotes and regulates cellular proliferation [126], and in DM1, it is abnormally active in the early stages of myoblast differentiation [127]. Additionally, the aberrant expression of other cell proliferation stimulators observed in DM1, such as protein kinase R (PKR), protein kinase R-like ER kinase (PERK), and pyruvate kinase M2 (PKM2), is also observed in cancer, which may explain why DM1 patients present with increased cancer susceptibility [128,129,130].
AKT and mTOR, which are important stimulators of anabolic pathways such as glucose uptake, glycogen storage, and protein synthesis did not respond to insulin stimulation and were therefore found to be decreased in DM1. In addition, the MEK/ERK pathway is responsible for the growth-promoting effects of insulin [131]. Together, they regulate several biological processes, such as transcription, protein synthesis, cell growth, and differentiation [132]. ERK1/2 was also found to be decreased in DM1 upon insulin stimulation [133].
We hypothesized that NGF signaling and downstream pathways are highly relevant to the pathophysiological mechanism of DM1, i.e., NGF, NTRK1, RhoA, ROCK1/2, and DAG novel putative DM1 targets. The latter is a second messenger, and its levels are correlated with PKC and PKD levels, given that these kinases are DAG sensors. Aberrant activation of PKC and PKD contributes to the development of metabolic diseases. Given that PKC is upregulated in DM1, we hypothesized that DAG is also increased in DM1, accumulating in many organs and leading to metabolic homeostasis disruption. The follow-up of DAG levels in DM1 is of paramount importance, given that it could be used as DM1 biomarker and as a disease progression biomarker.
The role of downstream TRK receptor signaling pathways has been intensively studied with respect to neuronal survival. The truncated TRK receptors, which are most abundant in non-neuronal tissues, seem to be important for skeletal muscle, as their presence is well documented during late developmental, early postnatal, and adult stages. However, TRK levels during myoblasts division, differentiation, and myotube formation seems very low. Conversely, p75NTR seems to be the most abundant neurotrophic receptor during early development, and receptor complexes involving both p75NTR and TRK might occur only in the late stages of development [134].
Furthermore, the impact of neurotrophins on muscle cell differentiation has been investigated both in vitro and in vivo. Essentially, an effect on differentiation and a trophic antiapoptotic effect of NGF in early myotubes have been suggested [134]. It was demonstrated that NGF signaling, through its low-affinity p75NTR receptor, is mediated by RhoA in muscle cells and is required for physiological myoblast fusion and to maintain a functional cytoskeletal organization of myotubes [135]. These results are particularly relevant in vivo when fibers are damaged, given that the activation of myogenic precursors is needed, as well as their fusion with existing myofibers, contributing for the formation of fibers with a functional contractile unit, allowing for efficient repair of damaged areas of skeletal muscle [135].
4.1.3. Insulin Signaling Pathways
As mentioned above, there is crosstalk between insulin-mediated signaling and NGF signaling. In our PPI analysis, we observed interactions of insulin with NGF, NTRK1, PI3K, and AKT1 (Figure 4). This is particularly interesting, given that insulin exerts its biological functions by binding to insulin binding receptors such as insulin receptor (IR) and insulin growth factor (IGF). Besides insulin, ligands structurally similar to insulin are able to activate insulin receptors, such as insulin growth factors. The same happens when insulin activates the downstream effectors by binding to insulin growth factors. Upon ligand–receptor interaction, signal transduction occurs through kinase domains of IR, IGF1/IR, and IGFR receptors. The main phosphorylation targets are the insulin response elements (IRSs), which further transduce the signal to different signaling cascades, exerting the various effects of insulin and IGFs [34]. These could be metabolic or mitogenic and include the PI3K/SREBP pathway (lipid synthesis), the PI3K/AKT/GSK3/eIF2B pathway (glycogen and protein synthesis and apoptosis regulation), and the RAS/MAPK/ERKS/RSK/ELK1 pathway (gene transcription, protein regulation, cell proliferation, and synaptic plasticity) [34]. Overall, both insulin and NGF are able to transduce signals that culminate with the activation of the MEK/ERK pathway. As described above, in DM1, there is a deregulation of this pathway (increase in ERK1/ERK2 and decrease upon insulin stimulation).
4.1.4. Myogenesis- and Signaling-Associated Pathways
The majority of genes included in cluster 2 (Figure 6), in particular MYOG, ID1, ID2, and MYOD1, play a significant role in skeletal muscle, as mentioned in Table 3. During embryogenesis, mononucleated myoblasts differentiate into myocytes, which fuse to originate multinucleated myotubes to be maturated into multinucleated myofibers that are grouped into highly oriented bundles, forming a single long cylinder (Figure 8). The reorganization of diverse upstream regulators of muscle progenitor differentiation and commitment to the myogenic fate requires the expression of the early myogenic regulatory factors MYF5, MRF4, and MYOD, as well as the late differentiation marker MYOG [136].
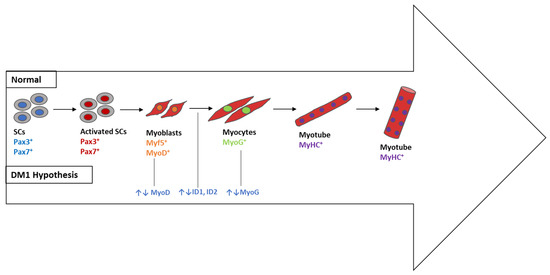
Figure 8.
Skeletal muscle differentiation in vitro and its deregulation in DM1 (DM1 hypothesis). The differentiation process starts when Pax3+ and/or Pax7+ progenitors begin to express Myf5 or MyoD as committed myoblasts. The latter gradually express myogenin (MyoG) and form single-nucleated nascent myotubes with myosin heavy chain (MHC+). The last step consists of myotube fusion to form multinucleated myotubes. SCs, satellite cells; MYF5, myogenic factor 5; MyHC, myosin heavy chain; Pax3/Pax7, paired box transcription factor 3/paired box transcription factor 7; MyoG, myogenin; MyoD, myogenic differentiation 1; ID1, inhibitor of DNA binding 1; ID2, inhibitor of DNA binding 2.
During development, the balance between proliferation and differentiation of myogenic progenitors is tightly regulated to allow for muscle growth while maintaining a pool of undifferentiated progenitors. Several signaling pathways play a role in the switch between proliferation and differentiation of skeletal muscle progenitor cells during development [137]. The major regulator of the muscle progenitor pool is the NOTCH signaling pathway, inhibiting myoblast differentiation in various organisms. Consistent with this idea, NOTCH signaling seems to be prevalent in muscle progenitors and declines throughout differentiation [138]. The NOTCH downstream target split-1 (HES1) is expressed in an oscillatory manner in developing and adult muscle stem cells [139]. Cyclic HES1 expression regulates the downstream target, MYOD, the levels of which also oscillate, allowing for the maintenance of the undifferentiated and proliferative state of the stem cells [139]. Additional signaling pathways are also involved in maintaining muscle progenitors in a proliferative state, such as BMP signaling [140]. Inhibitors of DNA-binding (ID) proteins are negative regulators of basic helix–loop–helix transcription factors and generally stimulate cell proliferation and inhibit differentiation. In addition, the BMP target gene inhibitor of DNA-binding protein 1 (ID1) is required to maintain satellite cells (SCs) in a proliferative and undifferentiated state [141,142]. In fact, both ID1 and ID2 are considered genes that inhibit differentiation through promotion of cell proliferation.
As mentioned above, the regulation of skeletal muscle formation (myogenesis) involves several different signaling pathways that are tightly regulated to control cell differentiation and proliferation [126,143,144]. In DM1, the activity of pathways that promote differentiation is decreased, whereas the activity of pathways that promote proliferation is increased, which significantly impairs and delays myogenesis [69]. Based on our results, one can hypothesize that ID1 and ID2 are deregulated in DM1, favoring cell proliferation instead of differentiation and leading to delayed myogenesis, which is consistent with the muscle immaturity observed in patients with DM1. Conversely, MYOG expression could be somehow deregulated, impairing the transition of myoblasts to myocytes and decreasing the initiation of the differentiation process. Therefore, we propose four genes as novel target candidates highly relevant to the pathophysiology of DM1 that should be explored in future studies, namely MYOD, MYOG, ID1, and ID2.
Based on the results of this study, it is clear that the genes identified in the cluster analysis are related to either metabolism and/or skeletal muscle. With regard to DM1, we suggest that genes in clusters 1, 2, and 3 are crucial for the pathophysiological mechanisms of this neuromuscular disease. Analyses have provided insights into the identification of novel metabolic process targets for DM1, as well as potential candidate genes and pathways that might be altered in this disorder and that should be further studied.
5. Conclusions
To the best of our knowledge, this is the first study using an automatic text-mining analysis to explore novel metabolism-related targets for DM1. The number of articles in this area has drastically increased in recent years, meaning considerable efforts are being applied to discover the pathophysiological mechanisms underlying DM1. The identified gene candidates could connect aspects of the disease that were previously considered unrelated and could be important to further explore new therapeutic approaches for DM1. However, it needs to be noted that the results reported herein were obtained exclusively using a bioinformatic approach. Therefore, although the novel identified metabolic process targets are a valuable point of reference for potential studies, they need to be further validated on molecular and cellular levels. Overall, the most relevant putative targets identified in the present study are NGF, NTRK1, ROCK1, ROCK2, DAG, ACTA1, ID1, ID2, MyoD, and MyoG. We strongly believe that some of these targets could be druggable for DM1. However, the next step will be the evaluation of these relevant putative targets in DM1 samples (cells, as well as muscle and biofluids) to confirm whether their expression and/or activity is dysregulated in DM1. Subsequently, the modulation of their levels is mandatory to understand whether the disease phenotype can be improved. The genes that improve disease phenotype can be used as novel therapeutic targets for DM1. This work is expected to promote further research and development with respect to the study of the pathophysiological mechanisms of DM1 and to provide guidance for the treatment of DM1 in patients.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijerph20032283/s1, Table S1: Genes obtained from VOSviewer; Table S2: Genes corresponding to the terms resulting from the DisGeNET search (disease: C3250443). Table S3: Gene terms common to DisGeNET and VOSviewer; Table S4: Additional genes/proteins corresponding to the terms resulting from the closely related generic terms in VOSviewer; Table S5: Subcellular localization and molecular function of the list of 71 putative genes; Table S6: g:Profiler: GO: Biological Process (BP); Table S7: g:Profiler: REAC signaling pathways; Table S8: VOSviewer occurrences Score; Figure S1: Network map of occurrence distribution of the similar genes found both in Scopus and DisGeNET; Figure S2: PPI network comprising the 44 genes.
Author Contributions
S.R. and R.V. contributed to the design of the study. D.H.K. and F.M. conducted the experiments. D.H.K., F.M., R.V. and S.R. analyzed the data. D.H.K. drafted the first version of the manuscript. D.H.K., F.M., R.V. and S.R. revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Fundação para a Ciência e a Tecnologia (FCT) through the Institute of Biomedicine (iBiMED) (UIDB/BIM/04501/2020/UIDP/04501/2020) and by the MEDISIS project (CENTRO-01-0246-FEDER-000018).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data used to support the findings of this study are included within the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Siciliano, G.; Manca, M.L.; Gennarelli, M.; Angelini, C.; Rocchi, A.; Iudice, A.; Miorin, M.; Mostacciuolo, M.L. Epidemiology of Myotonic Dystrophy in Italy: Re-Apprisal after Genetic Diagnosis. Clin. Genet. 2001, 59, 344–349. [Google Scholar] [CrossRef] [PubMed]
- Johnson, N.E.; Butterfield, R.J.; Mayne, K.; Newcomb, T.; Imburgia, C.; Dunn, D.; Duval, B.; Feldkamp, M.L.; Weiss, R.B. Population-Based Prevalence of Myotonic Dystrophy Type 1 Using Genetic Analysis of Statewide Blood Screening Program. Neurology 2021, 96, e1045–e1053. [Google Scholar] [CrossRef] [PubMed]
- Johnson, N.E.; Aldana, E.Z.; Angeard, N.; Ashizawa, T.; Berggren, K.N.; Marini-Bettolo, C.; Duong, T.; Ekström, A.B.; Sansone, V.; Tian, C.; et al. Consensus-Based Care Recommendations for Congenital and Childhood-Onset Myotonic Dystrophy Type 1. Neurol. Clin. Pract. 2019, 9, 443–454. [Google Scholar] [CrossRef] [PubMed]
- Aslanidis, C.; Jansen, G.; Amemiya, C.; Shutler, G.; Mahadevan, M.; Tsilfidis, C.; Chen, C.; Alleman, J.; Wormskamp, N.G.M.; Vooijs, M.; et al. Cloning of the Essential Myotonic Dystrophy Region and Mapping of the Putative Defect. Nature 1992, 355, 548–551. [Google Scholar] [CrossRef]
- Fu, Y.H.; Pizzuti, A.; Fenwick, R.G.; King, J.; Rajnarayan, S.; Dunne, P.W.; Dubel, J.; Nasser, G.A.; Ashizawa, T.; De Jong, P.; et al. An Unstable Triplet Repeat in a Gene Related to Myotonic Muscular Dystrophy. Science 1992, 255, 1256–1258. [Google Scholar] [CrossRef]
- Yum, K.; Wang, E.T.; Kalsotra, A. Myotonic Dystrophy: Disease Repeat Range, Penetrance, Age of Onset, and Relationship between Repeat Size and Phenotypes. Curr. Opin. Genet. Dev. 2017, 44, 30–37. [Google Scholar] [CrossRef]
- De Antonio, M.; Dogan, C.; Hamroun, D.; Mati, M.; Zerrouki, S.; Eymard, B.; Katsahian, S.; Bassez, G. Unravelling the Myotonic Dystrophy Type 1 Clinical Spectrum: A Systematic Registry-Based Study with Implications for Disease Classification. Rev. Neurol. 2016, 172, 572–580. [Google Scholar] [CrossRef]
- López-Otín, C.; Galluzzi, L.; Freije, J.M.P.; Madeo, F.; Kroemer, G. Metabolic Control of Longevity. Cell 2016, 166, 802–821. [Google Scholar] [CrossRef]
- Mattson, M.P.; Arumugam, T.V. Hallmarks of Brain Aging: Adaptive and Pathological Modification by Metabolic States. Cell Metab. 2018, 27, 1176–1199. [Google Scholar] [CrossRef]
- Mateos-Aierdi, A.J.; Goicoechea, M.; Aiastui, A.; Fernández-Torrón, R.; Garcia-Puga, M.; Matheu, A.; de Munain, A. Muscle Wasting in Myotonic Dystrophies: A Model of Premature Aging. Front. Aging Neurosci. 2015, 7, 125. [Google Scholar] [CrossRef]
- Turner, C.; Hilton-Jones, D. Myotonic Dystrophy: Diagnosis, Management and New Therapies. Curr. Opin. Neurol. 2014, 27, 599–606. [Google Scholar] [CrossRef]
- Lee, J.E.; Cooper, T.A. Pathogenic Mechanisms of Myotonic Dystrophy. Biochem. Soc. Trans. 2009, 37, 1281–1286. [Google Scholar] [CrossRef] [PubMed]
- Udd, B.; Krahe, R. The Myotonic Dystrophies: Molecular, Clinical, and Therapeutic Challenges. Lancet Neurol. 2012, 11, 891–905. [Google Scholar] [CrossRef] [PubMed]
- Koutsoulidou, A.; Koutalianos, D.; Georgiou, K.; Kakouri, A.C.; Oulas, A.; Tomazou, M.; Kyriakides, T.C.; Roos, A.; Papadimas, G.K.; Papadopoulos, C.; et al. Serum MiRNAs as Biomarkers for the Rare Types of Muscular Dystrophy. Neuromuscul. Disord. 2022, 32, 332–346. [Google Scholar] [CrossRef] [PubMed]
- Koutsoulidou, A.; Photiades, M.; Kyriakides, T.C.; Georgiou, K.; Prokopi, M.; Kapnisis, K.; Lusakowska, A.; Nearchou, M.; Christou, Y.; Papadimas, G.K.; et al. Identification of Exosomal Muscle-Specific MiRNAs in Serum of Myotonic Dystrophy Patients Relating to Muscle Disease Progress. Hum. Mol. Genet. 2017, 26, 3285–3302. [Google Scholar] [CrossRef]
- Koehorst, E.; Ballester-lopez, A.; Arechavala-gomeza, V.; Martínez-piñeiro, A.; Nogales-gadea, G. The Biomarker Potential of Mirnas in Myotonic Dystrophy Type I. J. Clin. Med. 2020, 9, 3939. [Google Scholar] [CrossRef]
- Pegoraro, V.; Cudia, P.; Baba, A.; Angelini, C. MyomiRNAs and Myostatin as Physical Rehabilitation Biomarkers for Myotonic Dystrophy. Neurol. Sci. 2020, 41, 2953–2960. [Google Scholar] [CrossRef]
- Espinosa-Espinosa, J.; González-Barriga, A.; López-Castel, A.; Artero, R. Deciphering the Complex Molecular Pathogenesis of Myotonic Dystrophy Type 1 through Omics Studies. Int. J. Mol. Sci. 2022, 23, 1441. [Google Scholar] [CrossRef]
- García-Puga, M.; Saenz-Antoñanzas, A.; Fernández-Torrón, R.; de Munain, A.L.; Matheu, A. Myotonic Dystrophy Type 1 Cells Display Impaired Metabolism and Mitochondrial Dysfunction That Are Reversed by Metformin. Aging 2020, 12, 6260–6275. [Google Scholar] [CrossRef]
- García-Puga, M.; Saenz-Antoñanzas, A.; Matheu, A.; de Munain, A. Targeting Myotonic Dystrophy Type 1 with Metformin. Int. J. Mol. Sci. 2022, 23, 2901. [Google Scholar] [CrossRef]
- Burnham, J.F. Scopus Database: A Review. Biomed. Digit. Libr. 2006, 3, 1. [Google Scholar] [CrossRef] [PubMed]
- Falagas, M.E.; Pitsouni, E.I.; Malietzis, G.A.; Pappas, G. Comparison of PubMed, Scopus, Web of Science, and Google Scholar: Strengths and Weaknesses. FASEB J. 2008, 22, 338–342. [Google Scholar] [CrossRef] [PubMed]
- Van Eck, N.J.; Waltman, L. Software Survey: VOSviewer, a Computer Program for Bibliometric Mapping. Scientometrics 2010, 84, 523–538. [Google Scholar] [CrossRef] [PubMed]
- Callon, M.; Courtial, J.P.; Turner, W.A.; Bauin, S. From Translations to Problematic Networks: An Introduction to Co-Word Analysis. Soc. Sci. Inf. 1983, 22, 191–235. [Google Scholar] [CrossRef]
- Börner, K.; Chen, C.; Boyack, K.W. Visualizing Knowledge Domains. Annu. Rev. Inf. Sci. Technol. 2003, 37, 179–255. [Google Scholar] [CrossRef]
- Bateman, A. UniProt: A Worldwide Hub of Protein Knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef]
- Piñero, J.; Ramírez-Anguita, J.M.; Saüch-Pitarch, J.; Ronzano, F.; Centeno, E.; Sanz, F.; Furlong, L.I. The DisGeNET Knowledge Platform for Disease Genomics: 2019 Update. Nucleic Acids Res. 2020, 48, D845–D855. [Google Scholar] [CrossRef]
- Piñero, J.; Saüch, J.; Sanz, F.; Furlong, L.I. The DisGeNET Cytoscape App: Exploring and Visualizing Disease Genomics Data. Comput. Struct. Biotechnol. J. 2021, 19, 2960–2967. [Google Scholar] [CrossRef]
- Bardou, P.; Mariette, J.; Escudié, F.; Djemiel, C.; Klopp, C. Jvenn: An Interactive Venn Diagram Viewer. BMC Bioinform. 2014, 15, 293. [Google Scholar] [CrossRef]
- Snel, B.; Lehmann, G.; Bork, P.; Huynen, M.A. String: A Web-Server to Retrieve and Display the Repeatedly Occurring Neighbourhood of a Gene. Nucleic Acids Res. 2000, 28, 3442–3444. [Google Scholar] [CrossRef]
- Reimand, J.; Arak, T.; Adler, P.; Kolberg, L.; Reisberg, S.; Peterson, H.; Vilo, J. G:Profiler—A Web Server for Functional Interpretation of Gene Lists (2016 Update). Nucleic Acids Res. 2016, 44, W83–W89. [Google Scholar] [CrossRef] [PubMed]
- Silla, T.; Schmid, M.; Dou, Y.; Garland, W.; Milek, M.; Imami, K.; Johnsen, D.; Polak, P.; Andersen, J.S.; Selbach, M.; et al. The Human ZC3H3 and RBM26/27 Proteins Are Critical for PAXT-Mediated Nuclear RNA Decay. Nucleic Acids Res. 2020, 48, 2518–2530. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Tan, L.; Yin, Z.; Zhu, W.; Tao, K.; Wang, G.; Shi, W.; Gao, J. MIR17HG-MiR-18a/19a Axis, Regulated by Interferon Regulatory Factor-1, Promotes Gastric Cancer Metastasis via Wnt/β-Catenin Signalling. Cell Death Dis. 2019, 10, 454. [Google Scholar] [CrossRef] [PubMed]
- Nieuwenhuis, S.; Okkersen, K.; Widomska, J.; Blom, P.; ’t Hoen, P.A.C.; van Engelen, B.; Glennon, J.C. Insulin Signaling as a Key Moderator in Myotonic Dystrophy Type 1. Front. Neurol. 2019, 10, 1229. [Google Scholar] [CrossRef]
- Vihola, A.; Bachinski, L.L.; Sirito, M.; Olufemi, S.-E.; Hajibashi, S.; Baggerly, K.A.; Raheem, O.; Haapasalo, H.; Suominen, T.; Holmlund-Hampf, J.; et al. Differences in Aberrant Expression and Splicing of Sarcomeric Proteins in the Myotonic Dystrophies DM1 and DM2. Acta Neuropathol. 2010, 119, 465–479. [Google Scholar] [CrossRef]
- Pantic, B.; Trevisan, E.; Citta, A.; Rigobello, M.P.; Marin, O.; Bernardi, P.; Salvatori, S.; Rasola, A. Myotonic Dystrophy Protein Kinase (DMPK) Prevents ROS-Induced Cell Death by Assembling a Hexokinase II-Src Complex on the Mitochondrial Surface. Cell Death Dis. 2013, 4, e858. [Google Scholar] [CrossRef]
- Shi, D.-L.; Grifone, R. RNA-Binding Proteins in the Post-Transcriptional Control of Skeletal Muscle Development, Regeneration and Disease. Front. Cell Dev. Biol. 2021, 9, 738978. [Google Scholar] [CrossRef]
- Laing, N.G.; Dye, D.E.; Wallgren-Pettersson, C.; Richard, G.; Monnier, N.; Lillis, S.; Winder, T.L.; Lochmüller, H.; Graziano, C.; Mitrani-Rosenbaum, S.; et al. Mutations and Polymorphisms of the Skeletal Muscle α-Actin Gene (ACTA1). Hum. Mutat. 2009, 30, 1267–1277. [Google Scholar] [CrossRef]
- Velcicky, J.; Bodendorf, U.; Rigollier, P.; Epple, R.; Beisner, D.R.; Guerini, D.; Smith, P.; Liu, B.; Feifel, R.; Wipfli, P.; et al. Discovery of the First Potent, Selective, and Orally Bioavailable Signal Peptide Peptidase-Like 2a (SPPL2a) Inhibitor Displaying Pronounced Immunomodulatory Effects In Vivo. J. Med. Chem. 2018, 61, 865–880. [Google Scholar] [CrossRef]
- Zhang, T.; Cheng, G.; Deng, L.; Yang, Y.; Sun, L.; Chen, P.; He, X.; Su, D.; Bi, N.; Qiu, B. Silence of S1 RNA Binding Domain 1 Represses Cell Growth and Promotes Apoptosis in Human Non-Small Cell Lung Cancer Cells. Transl. Lung Cancer Res. 2019, 8, 760. [Google Scholar] [CrossRef]
- Hintze, S.; Knaier, L.; Limmer, S.; Schoser, B.; Meinke, P. Nuclear Envelope Transmembrane Proteins in Myotonic Dystrophy Type. Front. Physiol. 2018, 9, 1532. [Google Scholar] [CrossRef]
- Rübben, A.; Wahl, R.U.; Eggermann, T.; Dahl, E.; Ortiz-Brüchle, N.; Cacchi, C. Mutation Analysis of Multiple Pilomatricomas in a Patient with Myotonic Dystrophy Type 1 Suggests a DM1-Associated Hypermutation Phenotype. PLoS ONE 2020, 15, e0230003. [Google Scholar] [CrossRef]
- Shi, J.; Wu, X.; Surma, M.; Vemula, S.; Zhang, L.; Yang, Y.; Kapur, R.; Wei, L. Distinct Roles for ROCK1 and ROCK2 in the Regulation of Cell Detachment. Cell Death Dis. 2013, 4, e483. [Google Scholar] [CrossRef]
- Sun, L.; Trausch-Azar, J.S.; Ciechanover, A.; Schwartz, A.L. Ubiquitin-Proteasome-Mediated Degradation, Intracellular Localization, and Protein Synthesis of MyoD and Id1 during Muscle Differentiation. J. Biol. Chem. 2005, 280, 26448–26456. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, H.; Goedert, M. Phosphorylation of Microtubule-Associated Protein Tau by AMPK-Related Kinases. J. Neurochem. 2012, 120, 165–176. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.S.; Bedrin, N.G.; Ades, P.A.; Palmer, B.M.; Toth, M.J. Molecular Determinants of Force Production in Human Skeletal Muscle Fibers: Effects of Myosin Isoform Expression and Cross-Sectional Area. Am. J. Physiol. Cell Physiol. 2015, 308, C473–C484. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.A.; Curry, E.G.; Blue, R.E.; Roden, C.; Dundon, S.E.R.; Rodríguez-Vargas, A.; Jordan, D.C.; Chen, X.; Lyons, S.M.; Crutchley, J.; et al. FXR1 Splicing Is Important for Muscle Development and Biomolecular Condensates in Muscle Cells. J. Cell Biol. 2020, 219, e201911129. [Google Scholar] [CrossRef]
- Shelton, D.L.; Reichardt, L.F. Studies on the Expression of the Beta Nerve Growth Factor (NGF) Gene in the Central Nervous System: Level and Regional Distribution of NGF MRNA Suggest That NGF Functions as a Trophic Factor for Several Distinct Populations of Neurons. Proc. Natl. Acad. Sci. USA 1986, 83, 2714–2718. [Google Scholar] [CrossRef]
- Harding, H.P.; Lackey, J.G.; Hsu, H.-C.; Zhang, Y.; Deng, J.; Xu, R.-M.; Damha, M.J.; Ron, D. An Intact Unfolded Protein Response in Trpt1 Knockout Mice Reveals Phylogenic Divergence in Pathways for RNA Ligation. RNA 2008, 14, 225–232. [Google Scholar] [CrossRef]
- Wei, X.W.; Liu, X.Q.; Zhang, Y.C.; Qin, C.M.; Lin, Y.; Tian, F.J. TCF3 Regulates Human Endometrial Stromal Cell Proliferation and Migration in RPL. Reproduction 2022, 163, 281–291. [Google Scholar] [CrossRef]
- Xu, F.; Na, L.; Li, Y.; Chen, L. RETRACTED ARTICLE: Roles of the PI3K/AKT/MTOR Signalling Pathways in Neurodegenerative Diseases and Tumours. Cell Biosci. 2020, 10, 54. [Google Scholar] [CrossRef]
- Bourque, J.; Kousnetsov, R.; Hawiger, D. Roles of Hopx in the Differentiation and Functions of Immune Cells. Eur. J. Cell Biol. 2022, 101, 151242. [Google Scholar] [CrossRef] [PubMed]
- Xia, Z.; Zhang, X.; Liu, P.; Zhang, R.; Huang, Z.; Li, D.; Xiao, X.; Wu, M.; Ning, N.; Zhang, Q.; et al. GNA13 Regulates BCL2 Expression and the Sensitivity of GCB-DLBCL Cells to BCL2 Inhibitors in a Palmitoylation-Dependent Manner. Cell Death Dis. 2021, 12, 54. [Google Scholar] [CrossRef]
- Huang, X.; Liu, N.; Xiong, X. ZNF24 Is Upregulated in Prostate Cancer and Facilitates the Epithelial-to-mesenchymal Transition through the Regulation of Twist1. Oncol. Lett. 2020, 19, 3593–3601. [Google Scholar] [CrossRef] [PubMed]
- Aavani, P.; Allen, L.J.S. The Role of CD4 T Cells in Immune System Activation and Viral Reproduction in a Simple Model for HIV Infection. Appl. Math. Model. 2019, 75, 210–222. [Google Scholar] [CrossRef]
- Gobbi, G.; Galli, D.; Carubbi, C.; Neri, L.M.; Masselli, E.; Pozzi, G.; Vitale, M.; Mirandola, P. PKC Proteins and Muscular Dystrophy. J. Funct. Morphol. Kinesiol. 2018, 3, 12. [Google Scholar] [CrossRef]
- Mateus, T.; Martins, F.; Nunes, A.; Herdeiro, M.T.; Rebelo, S. Metabolic Alterations in Myotonic Dystrophy Type 1 and Their Correlation with Lipin. Int. J. Environ. Res. Public. Health 2021, 18, 1794. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.D.; Harley, M.E.; Kemp, A.J.; Wills, J.; Lee, M.; Arends, M.; von Kriegsheim, A.; Behrends, C.; Wilkinson, S. CCPG1 Is a Non-Canonical Autophagy Cargo Receptor Essential for ER-Phagy and Pancreatic ER Proteostasis. Dev. Cell 2018, 44, 217–232. [Google Scholar] [CrossRef]
- Huang, Y.; Xing, Y.; Li, H. Heterozygous Desmin Gene (DES) Mutation Contributes to Familial Dilated Cardiomyopathy. J. Int. Med. Res. 2021, 49, 03000605211006598. [Google Scholar] [CrossRef]
- Mosaddeghzadeh, N.; Ahmadian, M.R. The RHO Family GTPases: Mechanisms of Regulation and Signaling. Cells 2021, 10, 1831. [Google Scholar] [CrossRef]
- Zhao, Z.; Manser, E. Myotonic Dystrophy Kinase-Related Cdc42-Binding Kinases (MRCK), the ROCK-like Effectors of Cdc42 and Rac1. Small GTPases 2015, 6, 81–88. [Google Scholar] [CrossRef]
- Li, Y.; Gong, H.; Wang, P.; Zhu, Y.; Peng, H.; Cui, Y.; Li, H.; Liu, J.; Wang, Z. The Emerging Role of ISWI Chromatin Remodeling Complexes in Cancer. J. Exp. Clin. Cancer Res. 2021, 40, 346. [Google Scholar] [CrossRef]
- Blum, R.; Dynlacht, B.D. The Role of MyoD1 and Histone Modifications in the Activation of Muscle Enhancers. Epigenetics 2013, 8, 778–784. [Google Scholar] [CrossRef] [PubMed]
- Shan, Z.; Shakoori, A.; Bodaghi, S.; Goldsmith, P.; Jin, J.; Wiest, J.S. TUSC1, a Putative Tumor Suppressor Gene, Reduces Tumor Cell Growth In Vitro and Tumor Growth In Vivo. PLoS ONE 2013, 8, e66114. [Google Scholar] [CrossRef] [PubMed]
- Hasan, N.; Ray, T.A.; Gregg, R.G. CACNA1S Expression in Mouse Retina: Novel Isoforms and Antibody Cross-Reactivity with GPR179. Vis. Neurosci. 2016, 33, E009. [Google Scholar] [CrossRef] [PubMed]
- Faralli, H.; Dilworth, F.J. Turning on Myogenin in Muscle: A Paradigm for Understanding Mechanisms of Tissue-Specific Gene Expression. Comp. Funct. Genomics 2012, 2012, 836374. [Google Scholar] [CrossRef] [PubMed]
- Bedford, L.; Lowe, J.; Dick, L.R.; Mayer, R.J.; Brownell, J.E. Ubiquitin-like Protein Conjugation and the Ubiquitin–Proteasome System as Drug Targets. Nat. Rev. Drug Discov. 2011, 10, 29–46. [Google Scholar] [CrossRef]
- Kästle, M.; Grune, T. Chapter 4—Interactions of the Proteasomal System with Chaperones: Protein Triage and Protein Quality Control. In The Proteasomal System in Aging and Disease; Grune, T., Ed.; Progress in Molecular Biology and Translational Science; Academic Press: Cambridge, MA, USA, 2012; Volume 109, pp. 113–160. [Google Scholar]
- André, L.M.; Ausems, C.R.M.; Wansink, D.G.; Wieringa, B. Abnormalities in Skeletal Muscle Myogenesis, Growth, and Regeneration in Myotonic Dystrophy. Front. Neurol. 2018, 9, 368. [Google Scholar] [CrossRef]
- Zhu, S.; Kuek, V.; Bennett, S.; Xu, H.; Rosen, V.; Xu, J. Protein Cytl1: Its Role in Chondrogenesis, Cartilage Homeostasis, and Disease. Cell. Mol. Life Sci. 2019, 76, 3515–3523. [Google Scholar] [CrossRef]
- Simoncini, C.; Spadoni, G.; Lai, E.; Santoni, L.; Angelini, C.; Ricci, G.; Siciliano, G. Central Nervous System Involvement as Outcome Measure for Clinical Trials Efficacy in Myotonic Dystrophy Type 1. Front. Neurol. 2020, 11, 624. [Google Scholar] [CrossRef]
- Sen, S.; Dong, M.; Kumar, S. Isoform-Specific Contributions of α-Actinin to Glioma Cell Mechanobiology. PLoS ONE 2009, 4, e8427. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Zhu, Z.; Xu, L.; Chen, X.; Li, X.; Lan, T.; Li, W.; Yuan, K.; Zeng, Y. ARHGEF11 Promotes Proliferation and Epithelial-Mesenchymal Transition of Hepatocellular Carcinoma through Activation of β-Catenin Pathway. Aging 2020, 12, 20235–20253. [Google Scholar] [CrossRef]
- Fusco, N.; Malapelle, U.; Fassan, M.; Marchiò, C.; Buglioni, S.; Zupo, S.; Criscitiello, C.; Vigneri, P.; Dei Tos, A.P.; Maiorano, E.; et al. PIK3CA Mutations as a Molecular Target for Hormone Receptor-Positive, HER2-Negative Metastatic Breast Cancer. Front. Oncol. 2021, 11, 644737. [Google Scholar] [CrossRef]
- Jiang, Y.; Wu, S.-Y.; Chen, Y.-L.; Zhang, Z.-M.; Tao, Y.-F.; Xie, Y.; Liao, X.-M.; Li, X.-L.; Li, G.; Wu, D.; et al. CEBPG Promotes Acute Myeloid Leukemia Progression by Enhancing EIF4EBP1. Cancer Cell Int. 2021, 21, 598. [Google Scholar] [CrossRef] [PubMed]
- Kunitomi, H.; Kobayashi, Y.; Wu, R.-C.; Takeda, T.; Tominaga, E.; Banno, K.; Aoki, D. LAMC1 Is a Prognostic Factor and a Potential Therapeutic Target in Endometrial Cancer. J. Gynecol. Oncol. 2020, 31, e11. [Google Scholar] [CrossRef] [PubMed]
- Zou, D.; Qin, B.; Wang, J.; Shi, Y.; Zhou, P.; Yi, Y.; Liao, J.; Lu, X. AFF2 Is Associated With X-Linked Partial (Focal) Epilepsy with Antecedent Febrile Seizures. Front. Mol. Neurosci. 2022, 15, 795840. [Google Scholar] [CrossRef]
- Souidi, A.; Zmojdzian, M.; Jagla, K. Dissecting Pathogenetic Mechanisms and Therapeutic Strategies in Drosophila Models of Myotonic Dystrophy Type 1. Int. J. Mol. Sci. 2018, 19, 4104. [Google Scholar] [CrossRef]
- Kim, J.H.; Yang, C.K.; Heo, K.; Roeder, R.G.; An, W.; Stallcup, M.R. CCAR1, a Key Regulator of Mediator Complex Recruitment to Nuclear Receptor Transcription Complexes. Mol. Cell 2008, 31, 510–519. [Google Scholar] [CrossRef]
- McAlear, T.S.; Bechstedt, S. The Mitotic Spindle Protein CKAP2 Potently Increases Formation and Stability of Microtubules. eLife 2022, 11, e72202. [Google Scholar] [CrossRef]
- Zhang, Y.; O’Connor, J.P.; Siomi, M.C.; Srinivasan, S.; Dutra, A.; Nussbaum, R.L.; Dreyfuss, G. The Fragile X Mental Retardation Syndrome Protein Interacts with Novel Homologs FXR1 and FXR2. EMBO J. 1995, 14, 5358–5366. [Google Scholar] [CrossRef]
- Takahashi, S.; Andreoletti, G.; Chen, R.; Munehira, Y.; Batra, A.; Afzal, N.A.; Beattie, R.M.; Bernstein, J.A.; Ennis, S.; Snyder, M. De Novo and Rare Mutations in the HSPA1L Heat Shock Gene Associated with Inflammatory Bowel Disease. Genome Med. 2017, 9, 8. [Google Scholar] [CrossRef] [PubMed]
- Geuens, T.; De Winter, V.; Rajan, N.; Achsel, T.; Mateiu, L.; Almeida-Souza, L.; Asselbergh, B.; Bouhy, D.; Auer-Grumbach, M.; Bagni, C.; et al. Mutant HSPB1 Causes Loss of Translational Repression by Binding to PCBP1, an RNA Binding Protein with a Possible Role in Neurodegenerative Disease. Acta Neuropathol. Commun. 2017, 5, 5. [Google Scholar] [CrossRef] [PubMed]
- Navarro, M.; Valentinis, B.; Belletti, B.; Romano, G.; Reiss, K.; Baserga, R. Regulation of Id2 Gene Expression by the Type 1 IGF Receptor and the Insulin Receptor Substrate-1. Endocrinology 2001, 142, 5149–5157. [Google Scholar] [CrossRef] [PubMed]
- Elmas, M.; Gogus, B.; Solak, M. Understanding What You Have Found: A Family with a Mutation in the LAMA1 Gene with Literature Review. Clin. Med. Insights Case Rep. 2020, 13, 1179547620948666. [Google Scholar] [CrossRef]
- Ran, T.; Chen, Z.; Zhao, L.; Ran, W.; Fan, J.; Hong, S.; Yang, Z. LAMB1 Is Related to the T Stage and Indicates Poor Prognosis in Gastric Cancer. Technol. Cancer Res. Treat. 2021, 20, 15330338211004944. [Google Scholar] [CrossRef]
- Campbell, C.S.; Hombauer, H.; Srivatsan, A.; Bowen, N.; Gries, K.; Desai, A.; Putnam, C.D.; Kolodner, R.D. Mlh2 Is an Accessory Factor for DNA Mismatch Repair in Saccharomyces Cerevisiae. PLoS Genet. 2014, 10, e1004327. [Google Scholar] [CrossRef]
- Franc, D.T.; Muetzel, R.L.; Robinson, P.R.; Rodriguez, C.P.; Dalton, J.C.; Naughton, C.E.; Mueller, B.A.; Wozniak, J.R.; Lim, K.O.; Day, J.W. Cerebral and Muscle MRI Abnormalities in Myotonic Dystrophy. Neuromuscul. Disord. 2012, 22, 483–491. [Google Scholar] [CrossRef]
- Telese, R.; Pagliarani, S.; Lerario, A.; Ciscato, P.; Fagiolari, G.; Cassandrini, D.; Grimoldi, N.; Conte, G.; Cinnante, C.; Santorelli, F.M.; et al. MYH2 Myopathy, a New Case Expands the Clinical and Pathological Spectrum of the Recessive Form. Mol. Genet. Genom. Med. 2020, 8, e1320. [Google Scholar] [CrossRef] [PubMed]
- Amatu, A.; Sartore-Bianchi, A.; Bencardino, K.; Pizzutilo, E.G.; Tosi, F.; Siena, S. Tropomyosin Receptor Kinase (TRK) Biology and the Role of NTRK Gene Fusions in Cancer. Ann. Oncol. 2019, 30, viii5. [Google Scholar] [CrossRef]
- Xu, Y.; Su, Z.; Li, J.; Wang, Q.; Meng, G.; Zhang, Y.; Yang, W.; Zhang, J.; Gao, P. Role of RNA-binding Protein 5 in the Diagnosis and Chemotherapeutic Response of Lung Cancer (Review). Oncol. Lett. 2019, 17, 2013–2019. [Google Scholar] [CrossRef]
- Zhao, X.; McKillop-Smith, S.; Müller, B. The Human Histone Gene Expression Regulator HBP/SLBP Is Required for Histone and DNA Synthesis, Cell Cycle Progression and Cell Proliferation in Mitotic Cells. J. Cell Sci. 2004, 117, 6043–6051. [Google Scholar] [CrossRef] [PubMed]
- Loaiza, N.; Hartgers, M.L.; Reeskamp, L.F.; Balder, J.-W.; Rimbert, A.; Bazioti, V.; Wolters, J.C.; Winkelmeijer, M.; Jansen, H.P.G.; Dallinga-Thie, G.M.; et al. Taking One Step Back in Familial Hypercholesterolemia. Arterioscler. Thromb. Vasc. Biol. 2020, 40, 973–985. [Google Scholar] [CrossRef] [PubMed]
- Kitai, Y.; Iwakami, M.; Saitoh, K.; Togi, S.; Isayama, S.; Sekine, Y.; Muromoto, R.; Kashiwakura, J.; Yoshimura, A.; Oritani, K.; et al. STAP-2 Protein Promotes Prostate Cancer Growth by Enhancing Epidermal Growth Factor Receptor Stabilization. J. Biol. Chem. 2017, 292, 19392–19399. [Google Scholar] [CrossRef]
- Bisikirska, B.C.; Adam, S.J.; Alvarez, M.J.; Rajbhandari, P.; Cox, R.; Lefebvre, C.; Wang, K.; Rieckhof, G.E.; Felsher, D.W.; Califano, A. STK38 Is a Critical Upstream Regulator of MYC’s Oncogenic Activity in Human B-Cell Lymphoma. Oncogene 2013, 32, 5283–5291. [Google Scholar] [CrossRef] [PubMed]
- Xu-Monette, Z.Y.; Medeiros, L.J.; Li, Y.; Orlowski, R.Z.; Andreeff, M.; Bueso-Ramos, C.E.; Greiner, T.C.; McDonnell, T.J.; Young, K.H. Dysfunction of the TP53 Tumor Suppressor Gene in Lymphoid Malignancies. Blood 2012, 119, 3668–3683. [Google Scholar] [CrossRef]
- Rasmussen, M.; Jin, J.-P. Troponin Variants as Markers of Skeletal Muscle Health and Diseases. Front. Physiol. 2021, 12, 1623. [Google Scholar] [CrossRef]
- Bosè, F.; Renna, L.V.; Fossati, B.; Arpa, G.; Labate, V.; Milani, V.; Botta, A.; Micaglio, E.; Meola, G.; Cardani, R. TNNT2 Missplicing in Skeletal Muscle as a Cardiac Biomarker in Myotonic Dystrophy Type 1 but Not in Myotonic Dystrophy Type 2. Front. Neurol. 2019, 10, 992. [Google Scholar] [CrossRef]
- Rhodes, J.D.; Lott, M.C.; Russell, S.L.; Moulton, V.; Sanderson, J.; Wormstone, I.M.; Broadway, D.C. Activation of the Innate Immune Response and Interferon Signalling in Myotonic Dystrophy Type 1 and Type 2 Cataracts. Hum. Mol. Genet. 2012, 21, 852–862. [Google Scholar] [CrossRef]
- Hale, M.A.; Richardson, J.I.; Day, R.C.; McConnell, O.L.; Arboleda, J.; Wang, E.T.; Berglund, J.A. An Engineered RNA Binding Protein with Improved Splicing Regulation. Nucleic Acids Res. 2018, 46, 3152–3168. [Google Scholar] [CrossRef]
- Bae, J.-H.; Jeong, H.-J.; Kim, H.; Leem, Y.-E.; Ryu, D.; Park, S.C.; Lee, Y.-I.; Cho, S.C.; Kang, J.-S. ZNF746/PARIS Overexpression Induces Cellular Senescence through FoxO1/P21 Axis Activation in Myoblasts. Cell Death Dis. 2020, 11, 359. [Google Scholar] [CrossRef]
- Esposito, F.; Cè, E.; Rampichini, S.; Monti, E.; Limonta, E.; Fossati, B.; Meola, G. Electromechanical Delays during a Fatiguing Exercise and Recovery in Patients with Myotonic Dystrophy Type 1. Eur. J. Appl. Physiol. 2017, 117, 551–566. [Google Scholar] [CrossRef]
- Johnson, N.E. Myotonic Muscular Dystrophies. Contin. Lifelong Learn. Neurol. 2019, 25, 1682–1695. [Google Scholar] [CrossRef] [PubMed]
- Kalsotra, A.; Xiao, X.; Ward, A.J.; Castle, J.C.; Johnson, J.M.; Burge, C.B.; Cooper, T.A. A Postnatal Switch of CELF and MBNL Proteins Reprograms Alternative Splicing in the Developing Heart. Proc. Natl. Acad. Sci. USA 2008, 105, 20333–20338. [Google Scholar] [CrossRef] [PubMed]
- Overby, S.J.; Cerro-Herreros, E.; Llamusi, B.; Artero, R. RNA-Mediated Therapies in Myotonic Dystrophy. Drug Discov. Today 2018, 23, 2013–2022. [Google Scholar] [CrossRef]
- Ozimski, L.L.; Sabater-Arcis, M.; Bargiela, A.; Artero, R. The Hallmarks of Myotonic Dystrophy Type 1 Muscle Dysfunction. Biol. Rev. 2021, 96, 716–730. [Google Scholar] [CrossRef] [PubMed]
- Cauteruccio, F.; Stamile, C.; Terracina, G.; Ursino, D.; Sappey-Marinier, D. An Automated String-Based Approach to Extracting and Characterizing White Matter Fiber-Bundles. Comput. Biol. Med. 2016, 77, 64–75. [Google Scholar] [CrossRef]
- Thakur, S.S.; Swiderski, K.; Ryall, J.G.; Lynch, G.S. Therapeutic Potential of Heat Shock Protein Induction for Muscular Dystrophy and Other Muscle Wasting Conditions. Philos. Trans. R. Soc. B Biol. Sci. 2017, 373, 20160528. [Google Scholar] [CrossRef]
- Kampinga, H.H.; Hageman, J.; Vos, M.J.; Kubota, H.; Tanguay, R.M.; Bruford, E.A.; Cheetham, M.E.; Chen, B.; Hightower, L.E. Guidelines for the Nomenclature of the Human Heat Shock Proteins. Cell Stress Chaperones 2009, 14, 105–111. [Google Scholar] [CrossRef]
- Senf, S.M.; Dodd, S.L.; Judge, A.R. FOXO Signaling Is Required for Disuse Muscle Atrophy and Is Directly Regulated by Hsp70. Am. J. Physiol. Cell Physiol. 2010, 298, C38–C45. [Google Scholar] [CrossRef]
- Senf, S. Skeletal Muscle Heat Shock Protein 70: Diverse Functions and Therapeutic Potential for Wasting Disorders. Front. Physiol. 2013, 4, 330. [Google Scholar] [CrossRef]
- Wang, S.-B.; Venkatraman, V.; Crowgey, E.L.; Liu, T.; Fu, Z.; Holewinski, R.; Ranek, M.; Kass, D.A.; O’Rourke, B.; Van Eyk, J.E. Protein S-Nitrosylation Controls Glycogen Synthase Kinase 3β Function Independent of Its Phosphorylation State. Circ. Res. 2018, 122, 1517–1531. [Google Scholar] [CrossRef]
- Wei, C.; Stock, L.; Valanejad, L.; Zalewski, Z.A.; Karns, R.; Puymirat, J.; Nelson, D.; Witte, D.; Woodgett, J.; Timchenko, N.A.; et al. Correction of GSK3ß at Young Age Prevents Muscle Pathology in Mice with Myotonic Dystrophy Type 1. FASEB J. 2018, 32, 2073–2085. [Google Scholar] [CrossRef] [PubMed]
- Grande, V.; Hathazi, D.; O’Connor, E.; Marteau, T.; Schara-Schmidt, U.; Hentschel, A.; Gourdon, G.; Nikolenko, N.; Lochmüller, H.; Roos, A. Dysregulation of GSK3β-Target Proteins in Skin Fibroblasts of Myotonic Dystrophy Type 1 (DM1) Patients. J. Neuromuscul. Dis. 2021, 8, 603–619. [Google Scholar] [CrossRef] [PubMed]
- Mei, W.; Wen-Chin, W.; Lauren, S.; Diana, L.; Ana, M.; Genevieve, G.; Nikolai, T.; Mike, S.; Lubov, T. Correction of Glycogen Synthase Kinase 3β in Myotonic Dystrophy 1 Reduces the Mutant RNA and Improves Postnatal Survival of DMSXL Mice. Mol. Cell. Biol. 2019, 39, e00155-19. [Google Scholar] [CrossRef]
- Mirzoev, T.M.; Sharlo, K.A.; Shenkman, B.S. The Role of GSK-3β in the Regulation of Protein Turnover, Myosin Phenotype, and Oxidative Capacity in Skeletal Muscle under Disuse Conditions. Int. J. Mol. Sci. 2021, 22, 5081. [Google Scholar] [CrossRef] [PubMed]
- Huang, E.J.; Reichardt, L.F. Neurotrophins: Roles in Neuronal Development and Function. Annu. Rev. Neurosci. 2001, 24, 677–736. [Google Scholar] [CrossRef] [PubMed]
- Westphalen, C.B.; Krebs, M.G.; Le Tourneau, C.; Sokol, E.S.; Maund, S.L.; Wilson, T.R.; Jin, D.X.; Newberg, J.Y.; Fabrizio, D.; Veronese, L.; et al. Genomic Context of NTRK1/2/3 Fusion-Positive Tumours from a Large Real-World Population. NPJ Precis. Oncol. 2021, 5, 69. [Google Scholar] [CrossRef]
- Kaplan, D.R.; Martin-Zanca, D.; Parada, L.F. Tyrosine Phosphorylation and Tyrosine Kinase Activity of the Trk Proto-Oncogene Product Induced by NGF. Nature 1991, 350, 158–160. [Google Scholar] [CrossRef]
- Arévalo, J.C.; Wu, S.H. Neurotrophin Signaling: Many Exciting Surprises! Cell. Mol. Life Sci. CMLS 2006, 63, 1523–1537. [Google Scholar] [CrossRef]
- Fujisawa, K.; Fujita, A.; Ishizaki, T.; Saito, Y.; Narumiya, S. Identification of the Rho-Binding Domain of P160ROCK, a Rho-Associated Coiled-Coil Containing Protein Kinase. J. Biol. Chem. 1996, 271, 23022–23028. [Google Scholar] [CrossRef]
- Sahai, E.; Ishizaki, T.; Narumiya, S.; Treisman, R. Transformation Mediated by RhoA Requires Activity of ROCK Kinases. Curr. Biol. 1999, 9, 136–145. [Google Scholar] [CrossRef] [PubMed]
- Sahai, E.; Olson, M.F.; Marshall, C.J. Cross-Talk between Ras and Rho Signalling Pathways in Transformation Favours Proliferation and Increased Motility. EMBO J. 2001, 20, 755–766. [Google Scholar] [CrossRef] [PubMed]
- Ehrenreiter, K.; Piazzolla, D.; Velamoor, V.; Sobczak, I.; Small, J.V.; Takeda, J.; Leung, T.; Baccarini, M. Raf-1 Regulates Rho Signaling and Cell Migration. J. Cell Biol. 2005, 168, 955–964. [Google Scholar] [CrossRef] [PubMed]
- Amatu, A.; Sartore-Bianchi, A.; Siena, S. NTRK Gene Fusions as Novel Targets of Cancer Therapy across Multiple Tumour Types. ESMO Open 2016, 1, e000023. [Google Scholar] [CrossRef]
- Zou, J.; Lei, T.; Guo, P.; Yu, J.; Xu, Q.; Luo, Y.; Ke, R.; Huang, D. Mechanisms Shaping the Role of ERK1/2 in Cellular Senescence (Review). Mol. Med. Rep. 2018, 19, 759–770. [Google Scholar] [CrossRef]
- Beffy, P.; Del Carratore, R.; Masini, M.; Furling, D.; Puymirat, J.; Masiello, P.; Simili, M. Altered Signal Transduction Pathways and Induction of Autophagy in Human Myotonic Dystrophy Type 1 Myoblasts. Int. J. Biochem. Cell Biol. 2010, 42, 1973–1983. [Google Scholar] [CrossRef]
- Gadalla, S.M.; Lund, M.; Pfeiffer, R.M.; Gørtz, S.; Mueller, C.M.; Moxley, R.T.; Kristinsson, S.Y.; Björkholm, M.; Shebl, F.M.; Hilbert, J.E.; et al. Cancer Risk Among Patients with Myotonic Muscular Dystrophy. JAMA 2011, 306, 2480–2486. [Google Scholar] [CrossRef]
- Win, A.K.; Perattur, P.G.; Pulido, J.S.; Pulido, C.M.; Lindor, N.M. Increased Cancer Risks in Myotonic Dystrophy. Mayo Clin. Proc. 2012, 87, 130–135. [Google Scholar] [CrossRef] [PubMed]
- Alsaggaf, R.; St. George, D.M.M.; Zhan, M.; Pfeiffer, R.M.; Wang, Y.; Wagner, K.R.; Greene, M.H.; Amr, S.; Gadalla, S.M. Cancer Risk in Myotonic Dystrophy Type I: Evidence of a Role for Disease Severity. JNCI Cancer Spectr. 2018, 2, pky052. [Google Scholar] [CrossRef]
- Roskoski, R. ERK1/2 MAP Kinases: Structure, Function, and Regulation. Pharmacol. Res. 2012, 66, 105–143. [Google Scholar] [CrossRef]
- Mao, Z.; Zhang, W. Role of MTOR in Glucose and Lipid Metabolism. Int. J. Mol. Sci. 2018, 19, 2043. [Google Scholar] [CrossRef] [PubMed]
- Renna, L.V.; Bosè, F.; Brigonzi, E.; Fossati, B.; Meola, G.; Cardani, R. Aberrant Insulin Receptor Expression Is Associated with Insulin Resistance and Skeletal Muscle Atrophy in Myotonic Dystrophies. PLoS ONE 2019, 14, e0214254. [Google Scholar] [CrossRef] [PubMed]
- Chevrel, G.; Hohlfeld, R.; Sendtner, M. The Role of Neurotrophins in Muscle under Physiological and Pathological Conditions. Muscle Nerve 2006, 33, 462–476. [Google Scholar] [CrossRef]
- Deponti, D.; Buono, R.; Catanzaro, G.; De Palma, C.; Longhi, R.; Meneveri, R.; Bresolin, N.; Bassi, M.T.; Cossu, G.; Clementi, E.; et al. The Low-Affinity Receptor for Neurotrophins P75NTR Plays a Key Role for Satellite Cell Function in Muscle Repair Acting via RhoA. Mol. Biol. Cell 2009, 20, 3620–3627. [Google Scholar] [CrossRef] [PubMed]
- Romagnoli, C.; Iantomasi, T.; Brandi, M.L. Available In Vitro Models for Human Satellite Cells from Skeletal Muscle. Int. J. Mol. Sci. 2021, 22, 13221. [Google Scholar] [CrossRef]
- Esteves de Lima, J.; Relaix, F. Master Regulators of Skeletal Muscle Lineage Development and Pluripotent Stem Cells Differentiation. Cell Regen. 2021, 10, 31. [Google Scholar] [CrossRef] [PubMed]
- Mourikis, P.; Sambasivan, R.; Castel, D.; Rocheteau, P.; Bizzarro, V.; Tajbakhsh, S. A Critical Requirement for Notch Signaling in Maintenance of the Quiescent Skeletal Muscle Stem Cell State. Stem Cells 2012, 30, 243–252. [Google Scholar] [CrossRef]
- Lahmann, I.; Bröhl, D.; Zyrianova, T.; Isomura, A.; Czajkowski, M.T.; Kapoor, V.; Griger, J.; Ruffault, P.-L.; Mademtzoglou, D.; Zammit, P.S.; et al. Oscillations of MyoD and Hes1 Proteins Regulate the Maintenance of Activated Muscle Stem Cells. Genes Dev. 2019, 33, 524–535. [Google Scholar] [CrossRef]
- Borok, M.J.; Mademtzoglou, D.; Relaix, F. Bu-M-P-Ing Iron: How BMP Signaling Regulates Muscle Growth and Regeneration. J. Dev. Biol. 2020, 8, 4. [Google Scholar] [CrossRef]
- Friedrichs, M.; Wirsdöerfer, F.; Flohé, S.B.; Schneider, S.; Wuelling, M.; Vortkamp, A. BMP Signaling Balances Proliferation and Differentiation of Muscle Satellite Cell Descendants. BMC Cell Biol. 2011, 12, 26. [Google Scholar] [CrossRef]
- Ono, Y.; Calhabeu, F.; Morgan, J.E.; Katagiri, T.; Amthor, H.; Zammit, P.S. BMP Signalling Permits Population Expansion by Preventing Premature Myogenic Differentiation in Muscle Satellite Cells. Cell Death Differ. 2010, 18, 222–234. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.S.L.; Cui, W. Proliferation, Survival and Metabolism: The Role of PI3K/AKT/MTOR Signalling in Pluripotency and Cell Fate Determination. Development 2016, 143, 3050–3060. [Google Scholar] [CrossRef] [PubMed]
- Huichalaf, C.H.; Sakai, K.; Wang, G.-L.; Timchenko, N.A.; Timchenko, L. Regulation of the Promoter of CUG Triplet Repeat Binding Protein, Cugbp1, during Myogenesis. Gene 2007, 396, 391–402. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).