MiRNA: Biological Regulator in Host-Parasite Interaction during Malaria Infection
Abstract
:1. Introduction
2. MicroRNAs
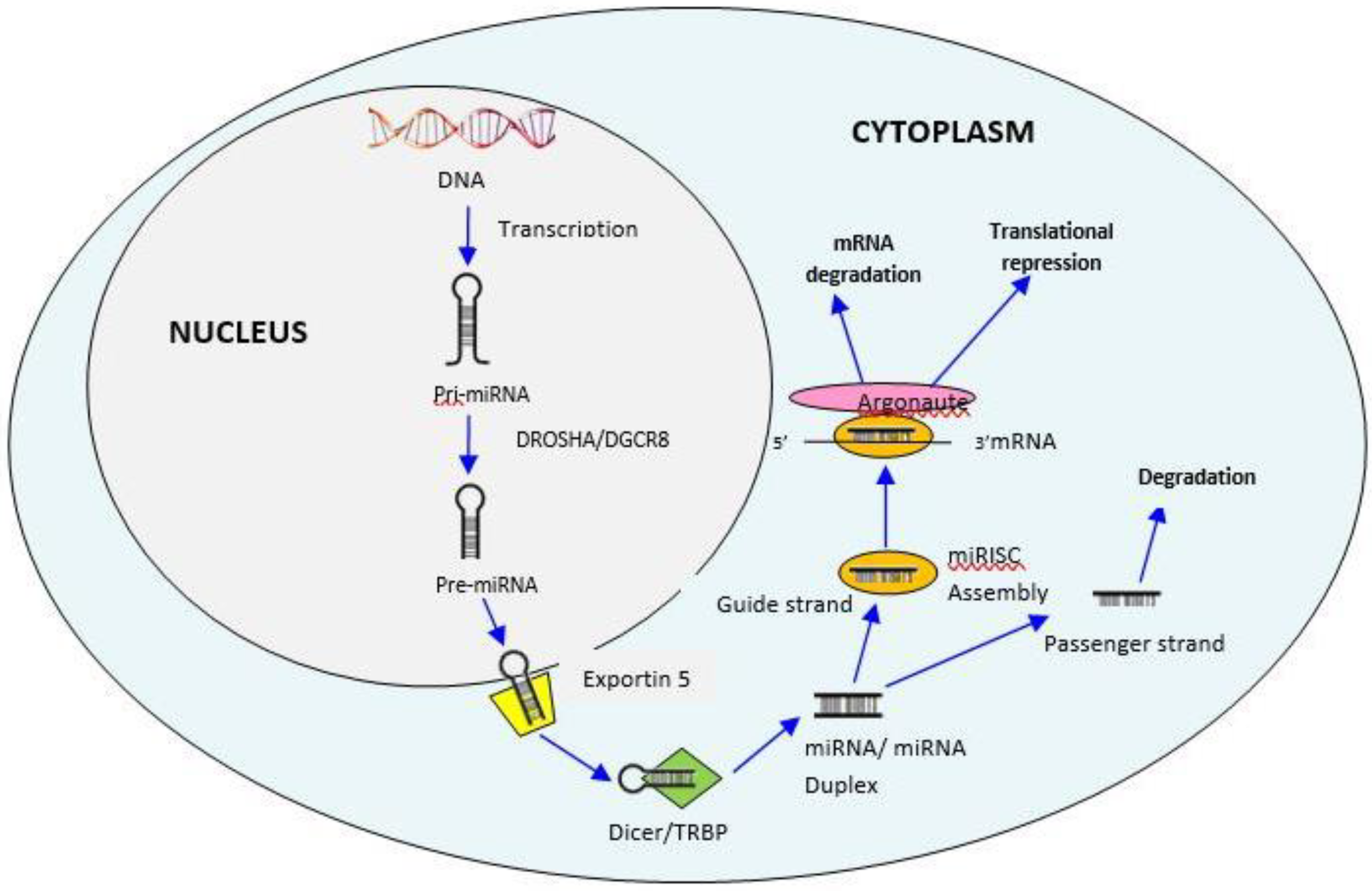
3. Biogenesis of MiRNAs
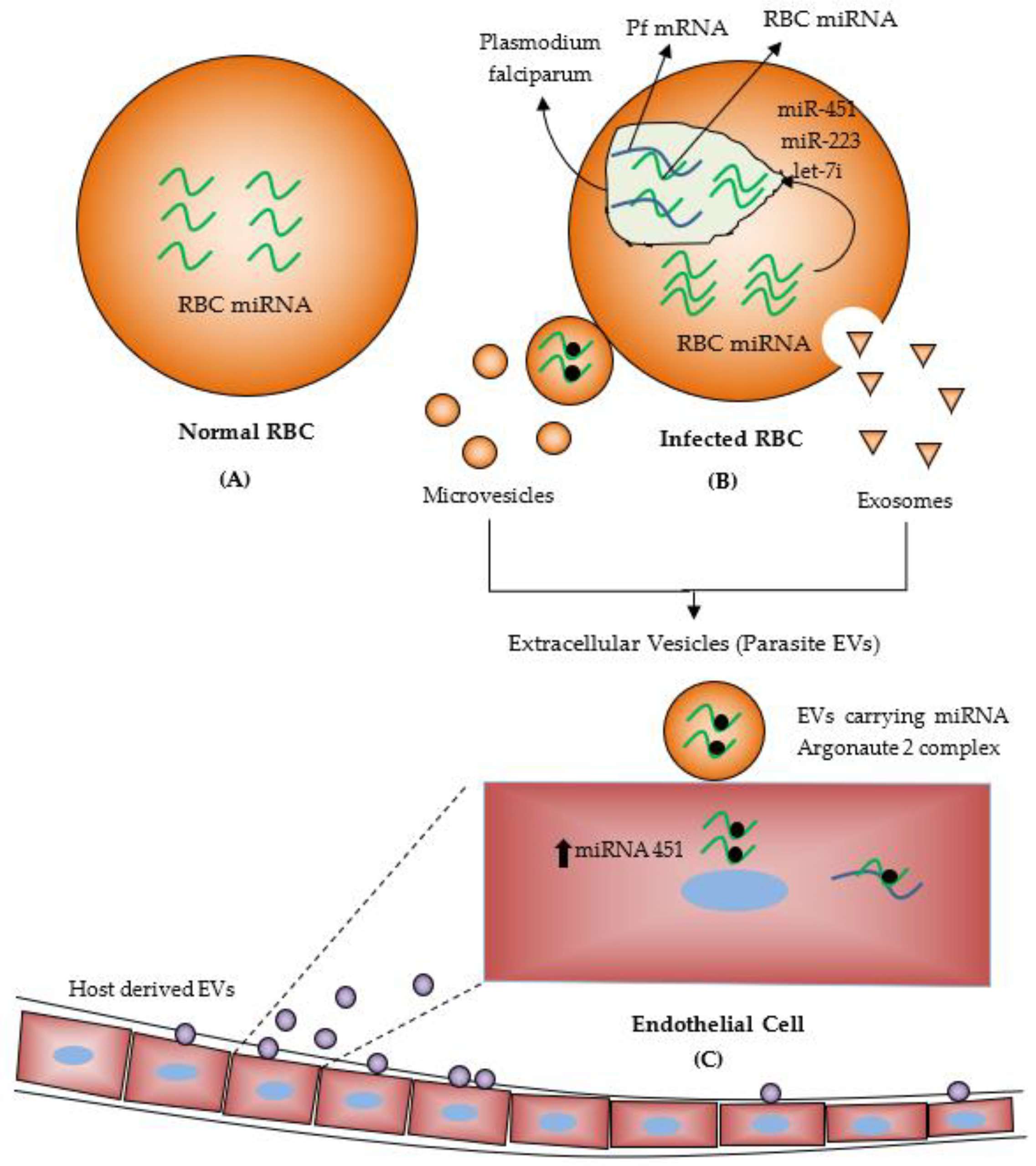
4. MiRNAs during Malaria Infection
5. MiRNAs as Diagnostic Tools for Malaria and Other Diseases
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rubio, M.; Bassat, Q.; Estivill, X.; Mayor, A. Tying malaria and microRNAs: From the biology to future diagnostic perspectives. Malar. J. 2016, 15, 167. [Google Scholar] [CrossRef] [Green Version]
- Tangpukdee, N.; Duangdee, C.; Wilairatana, P.; Krudsood, S. Malaria diagnosis: A brief review. Korean J. Parasitol. 2009, 47, 93. [Google Scholar] [CrossRef] [Green Version]
- Chamnanchanunt, S.; Kuroki, C.; Desakorn, V.; Enomoto, M.; Thanachartwet, V.; Sahassananda, D.; Sattabongkot, J.; Jenwithisuk, R.; Fucharoen, S.; Svasti, S. Downregulation of plasma miR-451 and miR-16 in Plasmodium vivax infection. Exp. Parasitol. 2015, 155, 19–25. [Google Scholar] [CrossRef]
- Ardekani, A.M.; Naeini, M.M. The role of microRNAs in human diseases. Avicenna J. Med. Biotechnol. 2010, 2, 161. [Google Scholar]
- Lee, R.C.; Ambros, V. An extensive class of small RNAs in Caenorhabditis elegans. Science 2001, 294, 862–864. [Google Scholar] [CrossRef] [Green Version]
- Ecsedi, M.; Großhans, H. LIN-41/TRIM71: Emancipation of a miRNA target. Genes Dev. 2013, 27, 581–589. [Google Scholar] [CrossRef] [Green Version]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [Green Version]
- Condrat, C.E.; Thompson, D.C.; Barbu, M.G.; Bugnar, O.L.; Boboc, A.; Cretoiu, D.; Suciu, N.; Cretoiu, S.M.; Voinea, S.C. miRNAs as biomarkers in disease: Latest findings regarding their role in diagnosis and prognosis. Cells 2020, 9, 276. [Google Scholar] [CrossRef] [Green Version]
- Kim, V.N. MicroRNA biogenesis: Coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 2005, 6, 376–385. [Google Scholar] [CrossRef]
- Paul, S.; Ruiz-Manriquez, L.M.; Serrano-Cano, F.I.; Estrada-Meza, C.; Solorio-Diaz, K.A.; Srivastava, A. Human microRNAs in host–parasite interaction: A review. 3 Biotech 2020, 10, 510. [Google Scholar] [CrossRef]
- Hammond, S.M. An overview of microRNAs. Adv. Drug Deliv. Rev. 2015, 87, 3–14. [Google Scholar] [CrossRef] [Green Version]
- Lodde, V.; Floris, M.; Muroni, M.R.; Cucca, F.; Idda, M.L. Non-coding RNAs in malaria infection. Wiley Interdiscip. Rev. RNA 2021, e1697. [Google Scholar] [CrossRef]
- Jopling, C. Liver-specific microRNA-122: Biogenesis and function. RNA Biol. 2012, 9, 137–142. [Google Scholar] [CrossRef] [Green Version]
- Ha, M.; Pang, M.; Agarwal, V.; Chen, Z.J. Interspecies regulation of microRNAs and their targets. Biochim. Biophys. Acta (BBA) Gene Regul. Mech. 2008, 1779, 735–742. [Google Scholar] [CrossRef] [Green Version]
- Xue, X.; Zhang, Q.; Huang, Y.; Feng, L.; Pan, W. No miRNA were found in Plasmodium and the ones identified in erythrocytes could not be correlated with infection. Malar. J. 2008, 7, 47. [Google Scholar] [CrossRef] [Green Version]
- Dandewad, V.; Vindu, A.; Joseph, J.; Seshadri, V. Import of human miRNA-RISC complex into Plasmodium falciparum and regulation of the parasite gene expression. J. Biosci. 2019, 44, 50. [Google Scholar] [CrossRef]
- Wilde, M.-L.; Triglia, T.; Marapana, D.; Thompson, J.K.; Kouzmitchev, A.A.; Bullen, H.E.; Gilson, P.R.; Cowman, A.F.; Tonkin, C.J. Protein kinase A is essential for invasion of Plasmodium falciparum into human erythrocytes. MBio 2019, 10, e01972-19. [Google Scholar] [CrossRef] [Green Version]
- Bandje, K.; Naissant, B.; Bigey, P.; Lohezic, M.; Vayssières, M.; Blaud, M.; Kermasson, L.; Lopez-Rubio, J.-J.; Langsley, G.; Lavazec, C. Characterization of an A-kinase anchoring protein-like suggests an alternative way of PKA anchoring in Plasmodium falciparum. Malar. J. 2016, 15, 248. [Google Scholar] [CrossRef] [Green Version]
- LaMonte, G.; Philip, N.; Reardon, J.; Lacsina, J.R.; Majoros, W.; Chapman, L.; Thornburg, C.D.; Telen, M.J.; Ohler, U.; Nicchitta, C.V. Translocation of sickle cell erythrocyte microRNAs into Plasmodium falciparum inhibits parasite translation and contributes to malaria resistance. Cell Host Microbe 2012, 12, 187–199. [Google Scholar] [CrossRef] [Green Version]
- Rathjen, T.; Nicol, C.; McConkey, G.; Dalmay, T. Analysis of short RNAs in the malaria parasite and its red blood cell host. FEBS Lett. 2006, 580, 5185–5188. [Google Scholar] [CrossRef]
- Chandan, K.; Gupta, M.; Sarwat, M. Role of host and pathogen-derived microRNAs in immune regulation during infectious and inflammatory diseases. Front. Immunol. 2020, 3081. [Google Scholar] [CrossRef] [Green Version]
- Verma, P.; Pandey, R.K.; Prajapati, P.; Prajapati, V.K. Circulating MicroRNAs: Potential and emerging biomarkers for diagnosis of human infectious diseases. Front. Microbiol. 2016, 7, 1274. [Google Scholar] [CrossRef]
- Chakrabarti, M.; Garg, S.; Rajagopal, A.; Pati, S.; Singh, S. Targeted repression of Plasmodium apicortin by host microRNA impairs malaria parasite growth and invasion. Dis. Model. Mech. 2020, 13, dmm042820. [Google Scholar] [CrossRef]
- Reid, G.; Kirschner, M.B.; van Zandwijk, N. Circulating microRNAs: Association with disease and potential use as biomarkers. Crit. Rev. Oncol. Hematol. 2011, 80, 193–208. [Google Scholar] [CrossRef]
- Wang, Z.; Xi, J.; Hao, X.; Deng, W.; Liu, J.; Wei, C.; Gao, Y.; Zhang, L.; Wang, H. Red blood cells release microparticles containing human argonaute 2 and miRNAs to target genes of Plasmodium falciparum: MPs and hAgo2-miRNAs targeting P. falciparum. Emerg. Microbes Infect. 2017, 6, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Sampaio, N.G.; Cheng, L.; Eriksson, E.M. The role of extracellular vesicles in malaria biology and pathogenesis. Malar. J. 2017, 16, 245. [Google Scholar] [CrossRef] [Green Version]
- Ketprasit, N.; Cheng, I.S.; Deutsch, F.; Tran, N.; Imwong, M.; Combes, V.; Palasuwan, D. The characterization of extracellular vesicles-derived microRNAs in Thai malaria patients. Malar. J. 2020, 19, 285. [Google Scholar] [CrossRef]
- Kirschner, M.B.; Kao, S.C.; Edelman, J.J.; Armstrong, N.J.; Vallely, M.P.; van Zandwijk, N.; Reid, G. Haemolysis during sample preparation alters microRNA content of plasma. PLoS ONE 2011, 6, e24145. [Google Scholar] [CrossRef]
- Chapman, L.M.; Ture, S.K.; Field, D.J.; Morrell, C.N. miR-451 limits CD4+ T cell proliferative responses to infection in mice. Immunol. Res. 2017, 65, 828–840. [Google Scholar] [CrossRef]
- Bayer-Santos, E.; Marini, M.M.; da Silveira, J.F. Non-coding RNAs in host–pathogen interactions: Subversion of mammalian cell functions by protozoan parasites. Front. Microbiol. 2017, 8, 474. [Google Scholar] [CrossRef]
- Tkach, M.; Théry, C. Communication by extracellular vesicles: Where we are and where we need to go. Cell 2016, 164, 1226–1232. [Google Scholar] [CrossRef] [Green Version]
- El-Assaad, F.; Hempel, C.; Combes, V.; Mitchell, A.J.; Ball, H.J.; Kurtzhals, J.A.; Hunt, N.H.; Mathys, J.-M.; Grau, G.E. Differential microRNA expression in experimental cerebral and noncerebral malaria. Infect. Immun. 2011, 79, 2379–2384. [Google Scholar] [CrossRef] [Green Version]
- Cohen, A.; Zinger, A.; Tiberti, N.; Grau, G.E.; Combes, V. Differential plasma microvesicle and brain profiles of microRNA in experimental cerebral malaria. Malar. J. 2018, 17, 285. [Google Scholar] [CrossRef] [Green Version]
- Martin-Alonso, A.; Cohen, A.; Quispe-Ricalde, M.A.; Foronda, P.; Benito, A.; Berzosa, P.; Valladares, B.; Grau, G.E. Differentially expressed microRNAs in experimental cerebral malaria and their involvement in endocytosis, adherens junctions, FoxO and TGF-β signalling pathways. Sci. Rep. 2018, 8, 11277. [Google Scholar] [CrossRef]
- van Loon, W.; Gai, P.P.; Hamann, L.; Bedu-Addo, G.; Mockenhaupt, F.P. MiRNA-146a polymorphism increases the odds of malaria in pregnancy. Malar. J. 2019, 18, 7. [Google Scholar] [CrossRef] [Green Version]
- Sharma, S.; Kumar, S.; Ahmed, M.; Bhardwaj, N.; Singh, J.; Kumari, S.; Savargaonkar, D.; Anvikar, A.R.; Das, J. Advanced Multiplex Loop Mediated Isothermal Amplification (mLAMP) Combined with Lateral Flow Detection (LFD) for Rapid Detection of Two Prevalent Malaria Species in India and Melting Curve Analysis. Diagnostics 2022, 12, 32. [Google Scholar] [CrossRef]
- Poti, K.E.; Sullivan, D.J.; Dondorp, A.M.; Woodrow, C.J. HRP2: Transforming malaria diagnosis, but with caveats. Trends Parasitol. 2020, 36, 112–126. [Google Scholar] [CrossRef] [Green Version]
- Berzosa, P.; de Lucio, A.; Romay-Barja, M.; Herrador, Z.; González, V.; García, L.; Fernández-Martínez, A.; Santana-Morales, M.; Ncogo, P.; Valladares, B. Comparison of three diagnostic methods (microscopy, RDT, and PCR) for the detection of malaria parasites in representative samples from Equatorial Guinea. Malar. J. 2018, 17, 333. [Google Scholar] [CrossRef]
- Tribolet, L.; Kerr, E.; Cowled, C.; Bean, A.G.; Stewart, C.R.; Dearnley, M.; Farr, R.J. MicroRNA biomarkers for infectious diseases: From basic research to biosensing. Front. Microbiol. 2020, 11, 1197. [Google Scholar] [CrossRef]
- De Guire, V.; Robitaille, R.; Tetreault, N.; Guerin, R.; Menard, C.; Bambace, N.; Sapieha, P. Circulating miRNAs as sensitive and specific biomarkers for the diagnosis and monitoring of human diseases: Promises and challenges. Clin. Biochem. 2013, 46, 846–860. [Google Scholar] [CrossRef]
- Lawrie, C.H.; Gal, S.; Dunlop, H.M.; Pushkaran, B.; Liggins, A.P.; Pulford, K.; Banham, A.H.; Pezzella, F.; Boultwood, J.; Wainscoat, J.S. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br. J. Haematol. 2008, 141, 672–675. [Google Scholar] [CrossRef]
- Meng, W.; McElroy, J.P.; Volinia, S.; Palatini, J.; Warner, S.; Ayers, L.W.; Palanichamy, K.; Chakravarti, A.; Lautenschlaeger, T. Comparison of microRNA deep sequencing of matched formalin-fixed paraffin-embedded and fresh frozen cancer tissues. PLoS ONE 2013, 8, e64393. [Google Scholar]
- Hedegaard, J.; Thorsen, K.; Lund, M.K.; Hein, A.-M.K.; Hamilton-Dutoit, S.J.; Vang, S.; Nordentoft, I.; Birkenkamp-Demtröder, K.; Kruhøffer, M.; Hager, H. Next-generation sequencing of RNA and DNA isolated from paired fresh-frozen and formalin-fixed paraffin-embedded samples of human cancer and normal tissue. PLoS ONE 2014, 9, e98187. [Google Scholar]
- Ojha, R.; Nandani, R.; Pandey, R.K.; Mishra, A.; Prajapati, V.K. Emerging role of circulating microRNA in the diagnosis of human infectious diseases. J. Cell. Physiol. 2019, 234, 1030–1043. [Google Scholar] [CrossRef]
- Vescovo, V.D.; Denti, M.A. microRNA and lung cancer. MicroRNA Cancer 2015, 153–177. [Google Scholar]
- Precazzini, F.; Detassis, S.; Imperatori, A.S.; Denti, M.A.; Campomenosi, P. Measurements methods for the development of MicroRNA-based tests for cancer diagnosis. Int. J. Mol. Sci. 2021, 22, 1176. [Google Scholar] [CrossRef]
- Zhao, P.; Zhao, L.; Zhang, T.; Wang, H.; Qin, C.; Yang, S.; Xia, X. Changes in microRNA expression induced by rabies virus infection in mouse brains. Microb. Pathog. 2012, 52, 47–54. [Google Scholar] [CrossRef]
- Qi, Y.; Cui, L.; Ge, Y.; Shi, Z.; Zhao, K.; Guo, X.; Yang, D.; Yu, H.; Cui, L.; Shan, Y. Altered serum microRNAs as biomarkers for the early diagnosis of pulmonary tuberculosis infection. BMC Infect. Dis. 2012, 12, 384. [Google Scholar] [CrossRef] [Green Version]
- Biswas, S.; Haleyurgirisetty, M.; Lee, S.; Hewlett, I.; Devadas, K. Development and validation of plasma miRNA biomarker signature panel for the detection of early HIV-1 infection. EBioMedicine 2019, 43, 307–316. [Google Scholar] [CrossRef] [Green Version]
- Chen, T.; Wang, Z.-Y.; Li, C.-C. miRNA-22 as a Candidate Diagnostic Biomarker for Coronary Slow Flow. Cardiol. Res. Pract. 2020, 2020, 7490942. [Google Scholar] [CrossRef]
- Grasso, M.; Piscopo, P.; Confaloni, A.; Denti, M.A. Circulating miRNAs as biomarkers for neurodegenerative disorders. Molecules 2014, 19, 6891–6910. [Google Scholar] [CrossRef]
- Schulte, C.; Zeller, T. microRNA-based diagnostics and therapy in cardiovascular disease—Summing up the facts. Cardiovasc. Diagn. Ther. 2015, 5, 17. [Google Scholar]
- Agrawal, S.; Tapmeier, T.T.; Rahmioglu, N.; Kirtley, S.; Zondervan, K.T.; Becker, C.M. The miRNA mirage: How close are we to finding a non-invasive diagnostic biomarker in endometriosis? A systematic review. Int. J. Mol. Sci. 2018, 19, 599. [Google Scholar] [CrossRef] [Green Version]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529. [Google Scholar] [CrossRef] [Green Version]
- Michael, M.Z.; O’Connor, S.M.; van Holst Pellekaan, N.G.; Young, G.P.; James, R.J. Reduced Accumulation of Specific MicroRNAs in Colorectal Neoplasia11Note: Susan M. O’Connor and Nicholas G. van Holst Pellekaan contributed equally to this work. Mol. Cancer Res. 2003, 1, 882–891. [Google Scholar]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A. MicroRNA expression profiles classify human cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef]
- Houzet, L.; Yeung, M.L.; de Lame, V.; Desai, D.; Smith, S.M.; Jeang, K.-T. MicroRNA profile changes in human immunodeficiency virus type 1 (HIV-1) seropositive individuals. Retrovirology 2008, 5, 118. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Wu, C.; Che, X.; Wang, L.; Yu, D.; Zhang, T.; Huang, L.; Li, H.; Tan, W.; Wang, C. Circulating microRNAs, miR-21, miR-122, and miR-223, in patients with hepatocellular carcinoma or chronic hepatitis. Mol. Carcinog. 2011, 50, 136–142. [Google Scholar] [CrossRef]
- Wu, W.; Lee, C.; Cho, C.; Fan, D.; Wu, K.; Yu, J.; Sung, J. MicroRNA dysregulation in gastric cancer: A new player enters the game. Oncogene 2010, 29, 5761–5771. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; He, Q.; Han, C.; Gu, H.; Jin, L.; Li, Q.; Mei, Y.; Wu, M. p53-facilitated miR-199a-3p regulates somatic cell reprogramming. Stem Cells 2012, 30, 1405–1413. [Google Scholar] [CrossRef]
| S. No | Study/Author | Year | MiRNA | Regulation | Functions |
|---|---|---|---|---|---|
| 1. | Ketprasit et al. [27] | 2020 | MiR-150-5p andMiR-15b-5p | Upregulated level of extracellular derived miRNA in P. vivax infected patient | Adherens junction and TGF-beta signalling pathway. |
| let-7a-5p | Upregulated in both P. vivax- and P. falciparum infected patients | Adherens junction | |||
| 2. | Aarón Martin-Alonso et al. [34] | 2018 | miR-19a-3p, miR-27a-5p, and miR-142-3p | It is upregulated in CM infected mice’s brains compared to NI and NCM. | Play a significant role in several pathways relevant to CM, including the TGF-β and endocytosis pathways. |
| 3. | Cohen et al. [33] | 2018 | miR-146a and miR-193b | Upregulated in microvesicles from cerebral malaria-infected mice | Cerebral pathology |
| 4. | Chamnanchanunt et al. [3] | 2015 | MiR-145 and miR-16 | Down-regulated in serum of P. vivax infected patients | Not defined |
| miR-223, miR-226-3p | No change | Not defined | |||
| 5. | LaMonte et al. [19] | 2012 | MiRNA-145, MiRNA-223 and let-7i | It is upregulated in HbAS and HbSS erythrocytes of P. falciparum infected patients. | Integrated into parasite mRNAs and resulted in translational inhibition. |
| 6. | El-Assaad et al. [32] | 2011 | MiR-27a, miR-150 | Upregulated in the brain tissue of PbA infected mice | Cell proliferation, development, and differentiation. |
| let-7i | Upregulated in the brain tissue of PbA infected mice | Cellular proliferation and the innate immune response | |||
| 7. | Rathjen et al. [20] | 2006 | MiR-145 | Upregulated in both infected and healthy red blood cells | Differentiation of erythroid cells. |
| S. No. | Disease | Type of Disease | Effect on MiRNAs | References |
|---|---|---|---|---|
| 1. | Chronic Lymphocytic Leukemia | Non-infectious | Altered miRNA expression pattern in patients who have chronic lymphocytic leukaemia. | Calin et al., 2002 [54] |
| 2. | Colorectal Neoplasia | Non-infectious | Murine miRNAs (miR-143 and miR-145) showed reduced steady-state concentrations at different cancer stages of colorectal neoplasia. | Michael et al., 2003 [55] |
| 3. | Human cancers | Non-infectious | Downregulation of miRNAs in tumours compared with normal tissues was observed. The potential of miRNA profiling in cancer diagnosis was highlighted. | Lu et al., 2005 [56] |
| 4. | Sepsis | Viral infection | miR-15a, miR-122, miR-4661, miR-483-5p, miR-342-5p miR-297, miR-181b, and miR-193 were, while miR-486, miR182, miR-4772, miR-574-5p, and miR-133a were found to be upregulated. | Ojha R et al., 2019 [44] |
| 5. | Human Immunodeficiency Virus | Viral infection | miR-33a-5p, hsa-miR-223, hsa-miR-146a-5p, and miR-29b-3p were downregulated in the infected individuals. | Houzet et al., 2008 [57] |
| 6. | Hepatitis | Viral infection | MiR-34, miR-4485, miR-92b-5p, miR-200b-5p, miR-29b and miR-192b-5p were upregulated, whereas miR-125, miR-330-3p, miR-1468, and miR-3180 were downregulated in infected individuals. | Xu et al., 2011 [58] |
| 7. | Tuberculosis | Bacterial infection | miR-889, miR-576-3p and miR-361-5p were elevated in tuberculosis-infected serum | Qi et al., 2012 [48] |
| 8. | Helicobacter pylori | Bacterial infection | The level of miR-17-p, miR-106a, miR-21, andmiR-106b were upregulated, and the expression of let-7a was downregulated. | W. K. K. Wu et al., 2010 [59] |
| 9. | Malaria | Protozoan infection | miRNA-16, miRNA-106, miRNA-91, MiRNA-451, miRNA-144, miRNA-7b, miRNA-142, miRNA-92 let-7a, and let-7f, were downregulated, whereas miRNA-19b and miRNA-223 were upregulated in the RBCs. | LaMonte et al., 2012 [17] |
| 10. | Trypanosomiasis | Protozoan infection | the expression level of miR-338 and miR-193b remarkably increased in patients affected with trypanosomiasis, whereas the expression of miR144 decreases at the time of infection | J. Wang, He, et al., 2012; Liu et al., 2011 [60] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kataria, P.; Surela, N.; Chaudhary, A.; Das, J. MiRNA: Biological Regulator in Host-Parasite Interaction during Malaria Infection. Int. J. Environ. Res. Public Health 2022, 19, 2395. https://doi.org/10.3390/ijerph19042395
Kataria P, Surela N, Chaudhary A, Das J. MiRNA: Biological Regulator in Host-Parasite Interaction during Malaria Infection. International Journal of Environmental Research and Public Health. 2022; 19(4):2395. https://doi.org/10.3390/ijerph19042395
Chicago/Turabian StyleKataria, Poonam, Neha Surela, Amrendra Chaudhary, and Jyoti Das. 2022. "MiRNA: Biological Regulator in Host-Parasite Interaction during Malaria Infection" International Journal of Environmental Research and Public Health 19, no. 4: 2395. https://doi.org/10.3390/ijerph19042395
APA StyleKataria, P., Surela, N., Chaudhary, A., & Das, J. (2022). MiRNA: Biological Regulator in Host-Parasite Interaction during Malaria Infection. International Journal of Environmental Research and Public Health, 19(4), 2395. https://doi.org/10.3390/ijerph19042395





