Abstract
Fibroblast growth factor receptor 4 (FGFR4) is involved in multiple physiological and pathological processes. Several genetic variants of FGFR4 have been shown to be associated with tumor progression in many cancers. However, its association, such as genetic variants and expression levels, with lung cancer is controversial. The present study examined the relationship between four single-nucleotide polymorphisms (SNPs; rs2011077 T/C, rs351855 G/A, rs7708357 G/A, and rs1966265 A/G) of FGFR4 and the risk of lung adenocarcinoma with the epidermal growth factor receptor (EGFR) mutation status in a Taiwanese cohort. The results demonstrated that FGFR4 rs2011077 (odds ratio (OR) = 0.348, 95% confidence interval (CI) = 0.136–0.891, p = 0.024), and rs351855 (OR = 0.296, 95% CI = 0.116–0.751, p = 0.008) showed an inverse association with distant metastasis in wild-type EGFR lung adenocarcinoma. Furthermore, a database analysis using The Cancer Genome Atlas revealed that the higher FGFR4 expression level was correlated with poor survival rates in wild-type EGFR lung adenocarcinoma. In conclusion, the data suggest that FGFR4 SNPs may help in identifying patient subgroups at low-risk for tumor metastasis, among carriers of lung adenocarcinoma bearing wild-type EGFR.
1. Introduction
Lung cancer has the highest global incidence and mortality rate of all cancers [1]. According to statistical data from Taiwan’s Ministry of Health and Welfare, the leading causes of cancer-related death in 2018 were tracheal, bronchial, and lung cancer. Lung adenocarcinoma, accounting for approximately 50% of lung cancer cases, occurs frequently in the female and nonsmoking populations [2,3]. In addition to environmental risk factors such as smoking and exposure to certain chemicals, genetic variation also increases the risk of lung cancer. Up to 30% of patients with lung adenocarcinoma in the Asian population have been shown to harbor epidermal growth factor receptor (EGFR) mutations [4,5]. Although EGFR inhibitors are highly efficacious in treating lung cancer, not all lung adenocarcinomas respond to this therapy. Therefore, the identification of other biomarkers and novel treatment strategies for lung adenocarcinoma is an unmet need.
Fibroblast growth factor receptors (FGFRs), which comprise five members (FGFR1-5), belong to the receptor tyrosine kinase family [6]. FGFRs participate in multiple physiological and pathological processes, such as proliferation, differentiation, metabolism, and migration [6]. Dysregulation of FGFR signaling cascades has been shown to be highly associated with tumor development in numerous human cancers [7]. Single-nucleotide polymorphisms (SNPs), the most common type of genetic variation, are reported to be significantly associated with cancer morbidity and mortality [8]. Numerous SNPs in FGFR genes, including FGFR4, have been identified as being significantly associated with cancer susceptibility and progression [7,9]. The human FGFR4 gene is located on chromosomal locus 5q35 and contains 18 exons [10]. Recent studies have focused on the clinical significance of FGFR4 genetic variations [10]. FGFR4 SNPs have been shown to be significantly associated with poor survival rates and worse prognosis in prostate cancer [11,12], head and neck cancer [13,14], breast cancer [15,16,17,18,19], ovarian cancer [20], hepatocellular carcinoma [21], uterine cervical cancer [22], urothelial cell carcinoma [23], pituitary tumors [24,25], soft tissue sarcoma [26], and oral squamous cell carcinoma [27,28,29]. However, the effects of FGFR4 SNPs on the risk of lung cancer seem to be controversial. The most common FGFR4 genetic variant in lung cancer is the SNP rs351855 G/A, which is in the exon 9 coding region of the FGFR4 gene. The FGFR4 SNP rs351855 induces the substitution of glycine for arginine at codon 388 (G388R) in the transmembrane domain of the FGFR4 protein receptor [30,31]. The FGFR4-G388R SNP has been shown to be significantly correlated with poor outcomes, including lymph node involvement and reduced overall survival [32,33], and is associated with worse survival in node-positive patients [34]. Conversely, this SNP was also reportedly correlated with better outcomes, including longer overall survival and reduced cancer risk in patients with non-small-cell lung carcinoma [35]. In addition, no association between FGFR4 variants and clinical outcomes has been found [36]. These conflicting results require further examination using a greater variety of research designs.
The role of FGFR4 gene polymorphisms in the clinicopathological characteristics of lung adenocarcinoma in the Taiwanese cohort remains unclear. The prevalence of EGFR mutations is relatively high in the Taiwanese population with lung adenocarcinoma [2,4]. Therefore, we examined the association between the FGFR4 SNPs and EGFR mutation in Taiwanese patients with lung adenocarcinoma. We explored four SNPs of FGFR4, namely rs2011077, rs351855, rs7708357, and rs1966265, to examine the pathophysiological effects on tumorigenesis of several cancers. Furthermore, we studied the effect of FGFR4 SNPs on the clinicopathological characteristics of lung adenocarcinoma with and without EGFR mutation.
2. Materials and Methods
2.1. Participant Selection and Specimen Collection
This study included 277 patients with lung adenocarcinoma at Cheng-Ching General Hospital in Taichung, Taiwan between 2012 and 2015. Each patient provided signed informed consent before initiation of the study. From all participants, tumor tissue specimens and whole-blood specimens were collected for EGFR gene sequencing and FGFR4 gene polymorphism analysis, respectively. The clinical information of the enrolled patients and the examined lifestyle variables (e.g., cigarette smoking) were obtained from medical records and questionnaires, respectively. The adenocarcinoma was staged in accordance with the TNM Classification of Malignant Tumors (where TNM refers to tumor/node/metastasis) of the American Joint Committee on Cancer, where T1–T4 describes primary tumor sizes and the invasion of adjacent tissues, N describes the regional lymph nodes involved, and M describes distant metastasis [37]. The study protocol was approved by the Institutional Review Board of Cheng-Ching General Hospital (No. HP120009; 22 September 2012).
2.2. Genomic DNA Extraction and EGFR Gene Sequencing
Genomic DNA from tumor tissues and whole-blood specimens of patients with lung adenocarcinoma were prepared as described previously [38]. DNA was extracted using the QIAamp Fast DNA Tissue Kit and the QIAamp DNA Blood Mini Kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions. DNA from the tumor tissues was used as a template. Exons 18–21 of the EGFR gene were amplified using the polymerase chain reaction (PCR) and then subjected to the DNA sequencing reaction using the ABI PRISM 3130XL System (Applied Biosystems, Foster City, CA, USA) as described previously [3].
2.3. Genotyping of FGFR4 Polymorphisms
The four SNPs investigated, namely rs2011077, rs351855, rs7708357, and rs1966265, in the FGFR4 genome region have been well defined and suggested to be associated with the risk of various cancers susceptibility [22,23,29,39]. The peripheral blood samples from lung cancer patients were collected for genomic DNA extraction. The whole blood samples were placed in EDTA containing tubes and were centrifuged at 3000 rpm, for 10 min as described previously [40]. Real-time PCR genotyping with the TaqMan SNP Genotyping Assay and the ABI StepOne Real-Time PCR System were used to examine the SNPs (Applied Biosystems, Foster City, CA, USA).
2.4. Statistical Analysis
Differences in the distributions of clinical characteristics and genotype frequencies between patients with lung adenocarcinoma harboring wild-type EGFR and mutant-type EGFR were analyzed using the Mann–Whitney U test and Fisher’s exact test. The association between the genotype frequencies and risk of EGFR types was estimated using multiple logistic regression models after controlling for other variables and is presented as odds ratios (ORs) with corresponding 95% confidence intervals (CIs). A p-value of <0.05 was considered to be statistically significant. All analyses were generated using SAS software, version 9.1 of the SAS System (SAS Institute Inc., Cary, NC, USA).
3. Results
3.1. Characteristics of the Study Population
A total of 277 patients with lung adenocarcinoma, divided into those harboring wild-type EGFR (n = 108, 40%) and those harboring mutant-type EGFR (n = 169, 60%) were included in this study. Table 1 shows the clinical characteristics of the enrolled patients. As Table 1 shows, the two groups, the wild-type EGFR group and the mutant-type EGFR group, differed significantly with respect to gender (p < 0.001), cigarette smoking (p < 0.001), and cell differentiation (p = 0.002). The EGFR mutant-type group had a higher proportion of women (n = 109, 64.5%), nonsmokers (n = 131, 77.5%), and cases of well differentiated (n = 21, 12.4%) and moderately differentiated tumors (n = 138, 81.7%) compared with the EGFR wild-type group (Table 1). The other clinicopathological characteristics, such as tumor stage, tumor T status, lymph node status, and distant metastasis, were similar between groups (Table 1).

Table 1.
Clinical characteristics in lung adenocarcinoma patients with the epidermal growth factor receptor (EGFR) wild-type or mutation-type.
3.2. Genotype Distributions of FGFR4 Polymorphisms in Patients with Lung Adenocarcinoma
To explore the effects of FGFR4 polymorphisms on lung adenocarcinoma risk, a whole-blood specimen from each of the 277 patients was collected and examined for genotype frequencies of the four FGFR4 SNPs (rs2011077, rs351855, rs7708357, and rs1966265). The genotype distributions and associations between the four FGFR4 polymorphisms and lung adenocarcinoma are shown in Table 2. Among the patients, the alleles with the highest distribution frequency for rs2011077, rs351855, rs7708357, and rs1966265 were heterozygous for T/C, heterozygous for G/A, homozygous for G/G, and heterozygous for A/G, respectively. To reduce the influence of possible confounding variables, we used adjusted ORs with 95% CIs—estimated using the multiple logistic regression model after normalizing for age, gender, and smoking habits—to evaluate the distribution of SNPs. As Table 2 shows, no significant differences in the genotype distributions of the four FGFR4 SNPs between patients with wild-type EGFR versus those with mutant-type EGFR lung adenocarcinoma were observed.

Table 2.
Distribution frequency of fibroblast growth factor receptor 4 (FGFR4) genotypes of patients with lung adenocarcinoma and multiple logistic regression analysis of EGFR mutation association.
3.3. Associations between FGFR4 SNPs rs2011077 and rs351855 and Distant Metastasis among Lung Adenocarcinoma Patients with the Wild-Type EGFR Gene
Next, we examined the genotype frequencies of each of the four FGFR4 SNPs and the clinicopathological characteristics of the patients: Whole tumor stage, tumor T status (primary tumor size), lymph node status, existence of distant metastasis, and cell differentiation (the histopathological grading status). As Table 3 shows, we observed an inverse association between FGFR4 rs2011077 and distant metastasis, although it was not significant (n = 277, p = 0.061). We further performed a subgroup analysis based on the EGFR status: Wild-type (n = 108) and mutant-type (n = 169). Compared with the homozygous TT genotype, the existence of at least one allele of SNP rs2011077 C genotype (TC and CC) was associated with a significantly lower risk of distant metastasis (OR: 0.348, 95% CI: 0.136–0.891, p = 0.024) in the patients harboring wild-type EGFR (n = 108; Table 3). Patients with the GG genotype were taken as a reference, and the existence of at least one carrier allele in FGFR4 rs351855 had a significantly reduced risk for distant metastasis in all patients (n = 277, OR = 0.523, 95% CI = 0.304–0.899, p = 0.018; Table 4). In the EGFR wild-type group, a significantly reduced presence of distant metastasis was also observed in patients with the FGFR4 rs351855 A genotype (GA and AA) compared with those with GG homozygotes (n = 108, OR = 0.296, 95% CI = 0.116–0.751, p = 0.008; Table 4). In addition, no significant association was found between the other two FGFR4 polymorphisms and the clinicopathological characteristics (data not shown).

Table 3.
Clinicopathologic characteristics of lung adenocarcinoma patients with EGFR mutation, stratified by polymorphic genotypes of FGFR4 rs2011077.

Table 4.
Clinicopathologic characteristics of lung adenocarcinoma patients with EGFR mutation, stratified by polymorphic genotypes of FGFR4 rs351855.
3.4. Clinical Relevance of FGFR4 Levels in Lung Adenocarcinoma Patients with the Wild-Type EGFR Gene Obtained from the Cancer Genome Atlas (TCGA) Databases
Considering the potential effects of FGFR4 expression levels in the clinicopathological characteristics in lung adenocarcinoma, we further analyzed the correlations between the FGFR4 expression level and the survival rate in 492 lung adenocarcinoma patients from The Cancer Genome Atlas (TCGA) database. In the TCGA database, 380 patients were early stage (stage I + II). A total of 163 and 24 patients were lymph node metastasis (33.1%) and distant metastasis (4.88%), respectively. As shown in Figure 1, we found that patients in wild-type EGFR lung adenocarcinoma with higher FGFR4 expression had shorter overall survival times and five-years survival times compared with those with lower FGFR4 expression (p = 0.008; Figure 1A; p = 0.006; Figure 1B). No significant differences in the FGFR4 expression and survival rates were observed in all patients with lung adenocarcinoma and in lung adenocarcinoma patients with mutant-type EGFR. Therefore, the expression level of FGFR4 may provide clinical significance for the survival rate of lung adenocarcinoma patients with wild-type EGFR.
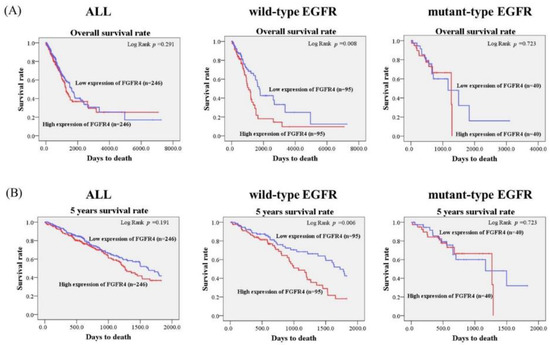
Figure 1.
Association of FGFR4 mRNA level and survival rates in lung adenocarcinoma patients from The Cancer Genome Atlas (TCGA) database. (A) The effects of FGFR4 mRNA expression on the overall survival of patients with lung adenocarcinoma were evaluated with the Kaplan-Meier plot using the Log-rank test. (B) The effects of FGFR4 mRNA expression on the five-year survival of patients with lung adenocarcinoma were evaluated with the Kaplan-Meier plot using the Log-rank test.
4. Discussion
In the present study, we found no significantly different frequencies of the FGFR4 SNPs rs2011077, rs351855, rs7708357, and rs1966265 in patients with wild-type EGFR and mutant-type EGFR lung adenocarcinoma. The existence of at least one allele of the rs2011077 C genotype or the rs351855 A genotype was associated with a significantly reduced presence of distant metastasis in the EGFR wild-type group. The data indicate that both FGFR4 SNPs rs2011077 and rs351855 may be associated with reduced presence of distant metastasis in Taiwanese patients with lung adenocarcinoma, especially those with the wild-type EGFR gene. FGFR4 SNPs may help in identifying patient subgroups at low-risk for tumor metastasis, among carriers of lung adenocarcinoma bearing wild-type EGFR.
Previous studies have shown that the Asia-Pacific patients with lung adenocarcinoma had the higher EGFR mutation frequency (47% (5958/12819)) and the Oceania patients with lung adenocarcinoma showed the lower EGFR mutation frequency (12% (69/570)) [41]. EGFR mutations have been found to occur more frequently in the female and nonsmoking populations and mainly occur in patients with lung adenocarcinoma in the Asian population [4,41,42]. Hsu et al., reported that the EGFR mutation rate in Taiwan is higher than 50%, based on data from the National Taiwan Lung Cancer Registry [2]. Consistent with these findings, we noted a higher proportion of women, never-smokers, and cases of well or moderately differentiated tumors in the collected samples. Moreover, most of the specimens were positive for EGFR mutations. Two EGFR mutations, an in-frame deletion within exon 19 and the L858R point mutation in exon 22, account for approximately 90% of overall EGFR mutations in lung adenocarcinoma [43,44]. We examined the exons 18–21 of the EGFR gene using direct sequencing to examine EGFR mutations in this study. Indeed, a high frequency of EGFR mutations (60%) was found in our cohort. No significant association between the four FGFR4 SNPs and EGFR wild-type and EGFR mutant-type lung adenocarcinoma was found after adjustment for gender, cigarette smoking, and cell differentiation.
Increasing evidence indicates that upregulation of FGFR4 expression levels participate in tumorigenesis and cancer progression; thus, FGFR4 has been proven as a therapeutic target for several cancers [10]. Using in silico analysis of the TCGA database, we found that lung adenocarcinoma patients bearing wild-type EGFR with the higher FGFR4 expression level is correlated with poor overall survival rates and five-years survival rates compared with those with lower FGFR4 expression patients. The increased FGFR4 expression is correlated with worse overall survival in head and neck squamous cell carcinoma [13]. Moreover, FGFR4 rs351855 SNP is associated with the increased FGFR4 protein expression and a worse prognosis [15]. However, the previous study has found that FGFR4 mRNA expression levels are not associated with FGFR4 genotypes in healthy lungs of lung cancer patients [32]. A further study is the association between various FGFR4 SNPs and FGFR4 expression levels in lung adenocarcinoma.
We observed that the FGFR4 SNP rs2011077 T/C was significantly inversely associated with distant metastasis. In addition, after the patients were stratified by the EGFR status, both FGFR4 SNPs rs2011077 T/C and rs351855 G/A were found to be inversely associated with distant metastasis. The FGFR4 SNP rs351855 G/A, which causes a substitution of glycine for arginine at codon 388 (G388R), has been proven to be related to worse prognosis in many cancers. Fang et al. revealed that the FGFR4 rs351855 A genotype is correlated with longer overall survival and reduced cancer risk in patients with Stage III (A + B) or IV non-small-cell lung carcinoma [35]. The meta-analytic evidence has demonstrated a modest reduction in the risk of lung cancer [30]. Moreover, no significant association between this FGFR4 genetic variant and clinical outcomes was found [36]. By contrast, several studies have reported that patients carrying the FGFR4 rs351855 A genotype had poorer outcomes, including lymph node involvement and reduced overall survival [32,33,34]. Using the molecular approach, the G388R substitution in FGFR4 was found to cause a conformational change in the FGFR4 receptor, which then recruits the signal transducer and activator of transcription 3 (STAT3) to the inner cell membrane. The evidence demonstrated that the rs351855 SNP is a significant risk for cancer prognosis and tumor progression [45]. Moreover, a study using FGFR4 rs351855-A/A-knockin transgenic mice to investigate lung cancer observed a marked decrease in the number of tumor-infiltrating CD8-positive T cells and a significant increase in regulatory T cell proportions compared with those from the G/G-knockin mice [46]. These results suggest that FGFR4 rs351855 alone could exhibit an immune evasive phenotype of the tumor microenvironment in lung cancer progression [46]. The fact that other studies have reached the opposite conclusion may be due to unknown causes other than FGFR4 variants. The possibilities of other genetic variations and a greater variety of abnormal signaling molecules are worthy of further investigation.
5. Conclusions
Our findings concerning FGFR4 SNPs rs2011077 and rs351855 may suggest a lower risk of tumor distant metastasis and FGFR4 gene expression levels may help the prediction of survival rates in Taiwanese lung adenocarcinoma patients, especially those bearing the wild-type EGFR gene. The potential mechanisms involved in dysregulated FGFR4 signaling cascades in Taiwanese patients with lung adenocarcinoma, particularly those with the wild-type EGFR gene, warrant further exploration. Further large-scale prospective studies are required to verify the significance of FGFR4 genetic polymorphisms in lung adenocarcinoma.
Author Contributions
Conceptualization, J.-P.L., H.-C.H., M.-J.H., and S.-F.Y.; methodology, H.H., Y.-C.H., and P.-J.Y.; formal analysis, C.-Y.C., Y.-H.C., and S.-F.Y.; resources, T.C.-Y.T. and S.-F.Y.; writing—original draft preparation, J.-P.L., H.-C.H., and S.-F.Y.; writing—review and editing, J.-P.L., M.-J.H., and S.-F.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. CA A Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.H.; Tseng, C.H.; Chiang, C.J.; Hsu, K.H.; Tseng, J.S.; Chen, K.C.; Wang, C.L.; Chen, C.Y.; Yen, S.H.; Chiu, C.H.; et al. Characteristics of young lung cancer: Analysis of Taiwan's nationwide lung cancer registry focusing on epidermal growth factor receptor mutation and smoking status. Oncotarget 2016, 7, 46628–46635. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.Y.; Yang, T.Y.; Chen, K.C.; Li, Y.J.; Hsu, K.H.; Tsai, C.R.; Chen, C.Y.; Hsu, C.P.; Hsia, J.Y.; Chuang, C.Y.; et al. EGFR l858r mutation and polymorphisms of genes related to estrogen biosynthesis and metabolism in never-smoking female lung adenocarcinoma patients. Clin. Cancer Res. 2011, 17, 2149–2158. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Au, J.S.; Thongprasert, S.; Srinivasan, S.; Tsai, C.M.; Khoa, M.T.; Heeroma, K.; Itoh, Y.; Cornelio, G.; Yang, P.C. A prospective, molecular epidemiology study of EGFR mutations in asian patients with advanced non-small-cell lung cancer of adenocarcinoma histology (pioneer). J. Thorac. Oncol. 2014, 9, 154–162. [Google Scholar] [CrossRef]
- Li, S.; Choi, Y.L.; Gong, Z.; Liu, X.; Lira, M.; Kan, Z.; Oh, E.; Wang, J.; Ting, J.C.; Ye, X.; et al. Comprehensive characterization of oncogenic drivers in asian lung adenocarcinoma. J. Thorac. Oncol. 2016, 11, 2129–2140. [Google Scholar] [CrossRef]
- Babina, I.S.; Turner, N.C. Advances and challenges in targeting fgfr signalling in cancer. Nat. Rev. Cancer 2017, 17, 318–332. [Google Scholar] [CrossRef]
- Tiong, K.H.; Mah, L.Y.; Leong, C.O. Functional roles of fibroblast growth factor receptors (FGFRS) signaling in human cancers. Apoptosis 2013, 18, 1447–1468. [Google Scholar] [CrossRef]
- Deng, N.; Zhou, H.; Fan, H.; Yuan, Y. Single nucleotide polymorphisms and cancer susceptibility. Oncotarget 2017, 8, 110635–110649. [Google Scholar] [CrossRef]
- Quintanal-Villalonga, A.; Ferrer, I.; Molina-Pinelo, S.; Paz-Ares, L. A patent review of FGFR4 selective inhibition in cancer (2007–2018). Expert Opin. Ther. Pat. 2019, 29, 429–438. [Google Scholar] [CrossRef]
- Lang, L.; Teng, Y. Fibroblast growth factor receptor 4 targeting in cancer: New insights into mechanisms and therapeutic strategies. Cells 2019, 8, 31. [Google Scholar] [CrossRef]
- Ma, Z.; Tsuchiya, N.; Yuasa, T.; Inoue, T.; Kumazawa, T.; Narita, S.; Horikawa, Y.; Tsuruta, H.; Obara, T.; Saito, M.; et al. Polymorphisms of fibroblast growth factor receptor 4 have association with the development of prostate cancer and benign prostatic hyperplasia and the progression of prostate cancer in a Japanese population. Int. J. Cancer 2008, 123, 2574–2579. [Google Scholar] [CrossRef] [PubMed]
- FitzGerald, L.M.; Karlins, E.; Karyadi, D.M.; Kwon, E.M.; Koopmeiners, J.S.; Stanford, J.L.; Ostrander, E.A. Association of FGFR4 genetic polymorphisms with prostate cancer risk and prognosis. Prostate Cancer Prostatic Dis. 2009, 12, 192–197. [Google Scholar] [CrossRef] [PubMed]
- Wimmer, E.; Ihrler, S.; Gires, O.; Streit, S.; Issing, W.; Bergmann, C. Fibroblast growth factor receptor 4 single nucleotide polymorphism gly388arg in head and neck carcinomas. World J. Clin. Oncol. 2019, 10, 136–148. [Google Scholar] [CrossRef] [PubMed]
- Farnebo, L.; Tiefenbock, K.; Ansell, A.; Thunell, L.K.; Garvin, S.; Roberg, K. Strong expression of survivin is associated with positive response to radiotherapy and improved overall survival in head and neck squamous cell carcinoma patients. Int. J. Cancer 2013, 133, 1994–2003. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; You, Z.; Sun, S.; Wang, Y.; Zhang, X.; Pang, D.; Jiang, Y. Prognostic implications of fibroblast growth factor receptor 4 polymorphisms in primary breast cancer. Mol. Carcinog. 2018, 57, 988–996. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Qi, H.; Zhang, L.; Li, H.; Shao, J.; Chen, H.; Zhong, M.; Shi, X.; Ye, T.; Li, Q. Effects of FGFR gene polymorphisms on response and toxicity of cyclophosphamide-epirubicin-docetaxel-based chemotherapy in breast cancer patients. BMC Cancer 2018, 18, 1038. [Google Scholar] [CrossRef]
- Jiang, Y.; Sun, S.; Wei, W.; Ren, Y.; Liu, J.; Pang, D. Association of FGFR3 and FGFR4 gene polymorphisms with breast cancer in chinese women of heilongjiang province. Oncotarget 2015, 6, 34023–34029. [Google Scholar] [CrossRef][Green Version]
- Marme, F.; Werft, W.; Benner, A.; Burwinkel, B.; Sinn, P.; Sohn, C.; Lichter, P.; Hahn, M.; Schneeweiss, A. FGFR4 arg388 genotype is associated with pathological complete response to neoadjuvant chemotherapy for primary breast cancer. Ann. Oncol. 2010, 21, 1636–1642. [Google Scholar] [CrossRef]
- Thussbas, C.; Nahrig, J.; Streit, S.; Bange, J.; Kriner, M.; Kates, R.; Ulm, K.; Kiechle, M.; Hoefler, H.; Ullrich, A.; et al. FGFR4 arg388 allele is associated with resistance to adjuvant therapy in primary breast cancer. J. Clin. Oncol. 2006, 24, 3747–3755. [Google Scholar] [CrossRef]
- Marme, F.; Hielscher, T.; Hug, S.; Bondong, S.; Zeillinger, R.; Castillo-Tong, D.C.; Sehouli, J.; Braicu, I.; Vergote, I.; Isabella, C.; et al. Fibroblast growth factor receptor 4 gene (FGFR4) 388arg allele predicts prolonged survival and platinum sensitivity in advanced ovarian cancer. Int. J. Cancer 2012, 131, E586–E591. [Google Scholar] [CrossRef]
- Yang, Y.; Zhou, Y.; Lu, M.; An, Y.; Li, R.; Chen, Y.; Lu, D.-R.; Jin, L.; Zhou, W.-P.; Qian, J.; et al. Association between fibroblast growth factor receptor 4 polymorphisms and risk of hepatocellular carcinoma. Mol. Carcinog. 2012, 51, 515–521. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.H.; Yang, S.F.; Liu, Y.F.; Lin, W.L.; Han, C.P.; Wang, P.H. Association of fibroblast growth factor receptor 4 genetic polymorphisms with the development of uterine cervical cancer and patient prognosis. Reprod. Sci. 2018, 25, 86–93. [Google Scholar] [CrossRef] [PubMed]
- Tsay, M.D.; Hsieh, M.J.; Lee, C.Y.; Wang, S.S.; Chen, C.S.; Hung, S.C.; Lin, C.Y.; Yang, S.F. Involvement of FGFR4 gene variants on the clinicopathological severity in urothelial cell carcinoma. Int. J. Environ. Res. Public Health 2019, 17. [Google Scholar] [CrossRef]
- Tateno, T.; Asa, S.L.; Zheng, L.; Mayr, T.; Ullrich, A.; Ezzat, S. The FGFR4-g388r polymorphism promotes mitochondrial stat3 serine phosphorylation to facilitate pituitary growth hormone cell tumorigenesis. PLoS Genet. 2011, 7, e1002400. [Google Scholar] [CrossRef]
- Ezzat, S.; Wang, R.; Pintilie, M.; Asa, S.L. FGFR4 polymorphic alleles modulate mitochondrial respiration: A novel target for somatostatin analog action in pituitary tumors. Oncotarget 2017, 8, 3481–3494. [Google Scholar] [CrossRef] [PubMed]
- Morimoto, Y.; Ozaki, T.; Ouchida, M.; Umehara, N.; Ohata, N.; Yoshida, A.; Shimizu, K.; Inoue, H. Single nucleotide polymorphism in fibroblast growth factor receptor 4 at codon 388 is associated with prognosis in high-grade soft tissue sarcoma. Cancer 2003, 98, 2245–2250. [Google Scholar] [CrossRef] [PubMed]
- Dutra, R.L.; de Carvalho, M.B.; Dos Santos, M.; Mercante, A.M.; Gazito, D.; de Cicco, R.; Group, G.; Tajara, E.H.; Louro, I.D.; da Silva, A.M. FGFR4 profile as a prognostic marker in squamous cell carcinoma of the mouth and oropharynx. PLoS ONE 2012, 7, e50747. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.Y.; Rho, Y.S.; Kwon, K.H.; Chung, E.J.; Kim, J.H.; Park, I.S.; Lee, D.J. ECRG1 and FGFR4 single nucleotide polymorphism as predictive factors for nodal metastasis in oral squamous cell carcinoma. Cancer Biomark 2012, 12, 115–124. [Google Scholar] [CrossRef]
- Chou, C.H.; Hsieh, M.J.; Chuang, C.Y.; Lin, J.T.; Yeh, C.M.; Tseng, P.Y.; Yang, S.F.; Chen, M.K.; Lin, C.W. Functional FGFR4 gly388arg polymorphism contributes to oral squamous cell carcinoma susceptibility. Oncotarget 2017, 8, 96225–96238. [Google Scholar] [CrossRef]
- Xiong, S.W.; Ma, J.; Feng, F.; Fu, W.; Shu, S.R.; Ma, T.; Wu, C.; Liu, G.C.; Zhu, J. Functional FGFR4 gly388arg polymorphism contributes to cancer susceptibility: Evidence from meta-analysis. Oncotarget 2017, 8, 25300–25309. [Google Scholar] [CrossRef]
- Xu, W.; Li, Y.; Wang, X.; Chen, B.; Wang, Y.; Liu, S.; Xu, J.; Zhao, W.; Wu, J. FGFR4 transmembrane domain polymorphism and cancer risk: A meta-analysis including 8555 subjects. Eur. J. Cancer 2010, 46, 3332–3338. [Google Scholar] [CrossRef] [PubMed]
- Spinola, M.; Leoni, V.; Pignatiello, C.; Conti, B.; Ravagnani, F.; Pastorino, U.; Dragani, T.A. Functional FGFR4 gly388arg polymorphism predicts prognosis in lung adenocarcinoma patients. J. Clin. Oncol. 2005, 23, 7307–7311. [Google Scholar] [CrossRef] [PubMed]
- Falvella, F.S.; Frullanti, E.; Galvan, A.; Spinola, M.; Noci, S.; De Cecco, L.; Nosotti, M.; Santambrogio, L.; Incarbone, M.; Alloisio, M.; et al. FGFR4 gly388arg polymorphism may affect the clinical stage of patients with lung cancer by modulating the transcriptional profile of normal lung. Int. J. Cancer 2009, 124, 2880–2885. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, H.; Okuda, K.; Kawano, O.; Yukiue, H.; Yano, M.; Fujii, Y. Fibroblast growth factor receptor 4 mutation and polymorphism in japanese lung cancer. Oncol. Rep. 2008, 20, 1125–1130. [Google Scholar] [CrossRef]
- Fang, H.M.; Tian, G.; Zhou, L.J.; Zhou, H.Y.; Fang, Y.Z. FGFR4 genetic polymorphisms determine the chemotherapy response of chinese patients with non-small cell lung cancer. Acta Pharmacol. Sin. 2013, 34, 549–554. [Google Scholar] [CrossRef][Green Version]
- Matakidou, A.; El Galta, R.; Rudd, M.F.; Webb, E.L.; Bridle, H.; Eisen, T.; Houlston, R.S. Further observations on the relationship between the FGFR4 gly388arg polymorphism and lung cancer prognosis. Br. J. Cancer 2007, 96, 1904–1907. [Google Scholar] [CrossRef]
- Goldstraw, P.; Chansky, K.; Crowley, J.; Rami-Porta, R.; Asamura, H.; Eberhardt, W.E.; Nicholson, A.G.; Groome, P.; Mitchell, A.; Bolejack, V.; et al. The iaslc lung cancer staging project: Proposals for revision of the tnm stage groupings in the forthcoming (eighth) edition of the tnm classification for lung cancer. J. Thorac. Oncol. 2016, 11, 39–51. [Google Scholar] [CrossRef]
- Chou, Y.-E.; Hsieh, M.-J.; Hsin, C.-H.; Chiang, W.-L.; Lai, Y.-C.; Lee, Y.-H.; Huang, S.-C.; Yang, S.-F.; Lin, C.-W. CD44 gene polymorphisms and environmental factors on oral cancer susceptibility in Taiwan. PLoS ONE 2014, 9, e93692. [Google Scholar] [CrossRef]
- Sheu, M.J.; Hsieh, M.J.; Chiang, W.L.; Yang, S.F.; Lee, H.L.; Lee, L.M.; Yeh, C.B. Fibroblast growth factor receptor 4 polymorphism is associated with liver cirrhosis in hepatocarcinoma. PLoS ONE 2015, 10, e0122961. [Google Scholar] [CrossRef]
- Hua, K.T.; Liu, Y.F.; Hsu, C.L.; Cheng, T.Y.; Yang, C.Y.; Chang, J.S.; Lee, W.J.; Hsiao, M.; Juan, H.F.; Chien, M.H.; et al. 3'utr polymorphisms of carbonic anhydrase ix determine the mir-34a targeting efficiency and prognosis of hepatocellular carcinoma. Sci. Rep. 2017, 7, 4466. [Google Scholar] [CrossRef]
- Midha, A.; Dearden, S.; McCormack, R. Egfr mutation incidence in non-small-cell lung cancer of adenocarcinoma histology: A systematic review and global map by ethnicity (mutmapii). Am. J. Cancer Res. 2015, 5, 2892–2911. [Google Scholar] [PubMed]
- de Groot, P.M.; Wu, C.C.; Carter, B.W.; Munden, R.F. The epidemiology of lung cancer. Transl. Lung Cancer Res. 2018, 7, 220–233. [Google Scholar] [CrossRef] [PubMed]
- Rosell, R.; Moran, T.; Queralt, C.; Porta, R.; Cardenal, F.; Camps, C.; Majem, M.; Lopez-Vivanco, G.; Isla, D.; Provencio, M.; et al. Screening for epidermal growth factor receptor mutations in lung cancer. N. Engl. J. Med. 2009, 361, 958–967. [Google Scholar] [CrossRef] [PubMed]
- Hong, W.; Wu, Q.; Zhang, J.; Zhou, Y. Prognostic value of EGFR 19-del and 21-l858r mutations in patients with non-small cell lung cancer. Oncol. Lett. 2019, 18, 3887–3895. [Google Scholar] [CrossRef] [PubMed]
- Ulaganathan, V.K.; Sperl, B.; Rapp, U.R.; Ullrich, A. Germline variant FGFR4 p.G388r exposes a membrane-proximal stat3 binding site. Nature 2015, 528, 570–574. [Google Scholar] [CrossRef]
- Kogan, D.; Grabner, A.; Yanucil, C.; Faul, C.; Ulaganathan, V.K. Stat3-enhancing germline mutations contribute to tumor-extrinsic immune evasion. J. Clin. Investig. 2018, 128, 1867–1872. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).