Abstract
Coronary artery disease (CAD) is a disease which has become a leading cause of death worldwide. The polymorphisms in Interleukin-17 (IL-17A), including rs2275913, rs3819024, rs3819025, rs3748067, rs8193037, rs4711998, and rs8193036, have been found to be probably associated with the risk of CAD. However, the results were inconsistent and inconclusive. The present study performed a meta-analysis to get a more precise and comprehensive estimation of the association between the IL-17A polymorphisms and CAD risk. The Pubmed, Embase, Cochrane Central Register of Controlled Trials, Chinese National Knowledge Infrastructure, and Chinese Biomedical Literature Databases were searched for related studies. A total of six studies, including 3542 cases and 3212 controls, were identified for the meta-analysis. The main findings of the present meta-analysis show that the TT genotype of IL-17A rs3748067 is associated with a significant lower risk of CAD in the homozygous model odds ratio (OR) (OR = 0.37) in Asians. No significant association was found for rs2275913, rs3819024, rs3819025, rs8193037, rs4711998, and rs8193036 with CAD susceptibility in the overall analysis. However, subgroup analysis indicated a significant decreased risk of CAD for the GG genotype and G allele of rs2275913 in a small sample size group, and a higher risk of CAD for the GG genotype and G allele of rs8193037 in a heterozygous model (OR = 1.56), dominant model (OR = 1.54), and allelic model (OR = 1.47) in Asians. In conclusion, the current meta-analysis suggests a significant relationship between rs3748067, rs8193037, and CAD in Asians, while for rs2275913, rs3819024, rs3819025, rs4711998, rs8193036, no such relations were found. Thus, IL-17A rs3748067 and rs8193037 might be recommended as a predictor for susceptibility of CAD for Asians. However, the results of this meta-analysis are hypothesis-generating results which should be interpreted with caution because of the heterogeneity and publication bias among study designs.
Keywords:
IL-17A; rs3748067; rs2275913; rs8193037; coronary artery disease; polymorphism; meta-analysis 1. Introduction
It is well known that atherosclerosis is a chronic inflammatory disease, which has become the major cause of cardiovascular diseases (CVDs), including coronary artery disease (CAD), ischemic stroke, cerebral stroke, and peripheral vascular disease [1]. Among these CVDs, CAD is a disease which represents a leading cause of death worldwide. According to the Chinese Cardiovascular Disease Epidemiology Report 2015, the annual CAD mortality rate was 105.7 per 100,000 worldwide and 70.1 per 100,000 in China. As a principal clinical manifestation of atherosclerosis, the risk factors of CAD include age, smoking, diabetes mellitus, and hyperlipidemia, etc. The genetic mutant was also found to play important roles in CAD susceptibility.
Interleukin-17 (IL-17) is a novel family cytokine that consists of six protein members (from IL-17A to IL-17F), which plays an important role in many chronic inflammatory diseases [2,3]. It has been demonstrated that atherosclerotic plaque shows higher expression of IL-17A [4]. After myocardial ischemia/reperfusion injuries, the expression of IL-17A and IL-17F also increase [5]. Recent studies indicated the IL-17A polymorphisms be connected with the risk of developing atherosclerosis and CAD [6,7,8,9,10]. Several polymorphisms in IL-17A, such as rs2275913, rs3819024, rs3819025, rs3748067, rs8193037, rs4711998, and rs8193036 have been found to be related with the risk of CAD [6,7,8,9,10]. According to recent data, the AA and GA + AA genotype of rs2275913, CC, and TC + CC of rs3748067 are thought to be probably associated with the risk of CAD by some research [7,8]. However, the results were inconsistent and inconclusive [9,10,11]. Thus, in the present study, we included a total of six studies, with 3542 cases and 3212 controls, to get a more precise and comprehensive estimation of the association between these polymorphisms and CAD.
2. Experimental Section
2.1. Publication Search Strategy and Inclusion Criteria
We systematically searched the published studies in the following electronic databases: Pubmed, Embase, Cochrane Central Register of Controlled Trials (CENTRAL), Chinese National Knowledge Infrastructure (CNKI), and Chinese Biomedical Literature Database (CBM). The searched terms were (“Coronary artery disease”, or “CAD”), (“interleukin-17A” or “IL-17a”), and (“polymorphism” or “mutation” or “SNP” or “single nucleotide polymorphism”), without restrictions on language. The deadline for publication was 30 April 2016. All of the results from the databases were screened. Firstly, we screened the title. If the titles fulfilled our criteria, we then screened the abstract. We retrieved the full text if the abstract matched our interest. The references of all eligible studies were retrieved manually for other potentially relevant studies. We contacted the authors for related data that were unavailable in the original publications.
Inclusion criteria: (a) case-control design; (b) the association of interleukin-17A polymorphisms and CAD risks should be evaluated; (c) the data in the publication were sufficient for the present estimation. Studies were excluded if any of the following applies: (1) repeat publications, abstracts, letters or reviews; (2) studies not meeting all of the inclusion criteria.
2.2. Data Extraction
We extracted information from each eligible publication manually by two investigators independently. For each study, the extracted information included: first authors’ name, publishing year, country, ethnicity, genotyping method, the source of controls, sex and age match, and genotype numbers of cases and controls. If we met discrepancies during the data extraction, they would be resolved by a consensus achieved by the third author.
2.3. Quality Assessment
The quality of the included studies was evaluated according to the predefined scale for quality assessment [12]. The score scale includes the following aspects: source of cases, source of controls, specimens used for determining genotypes, total sample size, and evidence of Hardy-Weinberg equilibrium (HWE). The quality scores range from 0–15. Reports scoring <10 were classified as “low quality”, and those ≥10 as “high quality”. The quality evaluation was performed by two authors independently. Consensus was held to resolve any discrepancies in the assessment process.
2.4. Statistical Methods
χ2-test was used to evaluate the HWE of the control group polymorphism. If p < 0.05, it was considered to be deviated from HWE. To evaluate the association between IL-17A polymorphisms and CAD risk, the crude odds ratio (OR) with 95% confidence interval (CI) was used. The pooled ORs were calculated using homozygous, heterozygous, dominant, and allelic genetic model. The statistical significance was determined by the Z-test, and the p-value was adjusted using Bonferroni’s correction. p < 0.05 was considered to be statistically significant. Subgroup analysis was conducted by the sample size and ethnicity. Total samples minor than 1000 will be treated as small, and large, otherwise.
The statistical heterogeneity between studies was evaluated by an I-square statistical test, which was not dependent on the number of studies in the meta-analysis [13]. If there was an obvious heterogeneity among the studies (I2 > 50%), the random-effects model (the DerSimonian and Laird method) was used for the meta-analysis [14]. Otherwise, the fixed-effect model using the Mantel-Haenszel method was adopted [15]. Sensitivity analysis was performed to assess the effect of individual study on pooled results and the stability of results. The publication bias was detected with Begg’s funnel plot and Egger’s linear regression method, and p < 0.05 was considered to be statistically significant [16]. All statistical analyses were performed using the STATA 12.0 software (StataCorp, College Station, TX, USA) and Revman 5.3 (The Cochrane Collaboration, Oxford, UK).
3. Results and Discussion
3.1. Characteristics of Eligible Studies
A total of 68 studies were obtained from the literature search after duplicates were removed. Among them, 57 studies were excluded for irrelevance, three were master’s degree theses, two for predicting IL-17A polymorphisms and ischemic stroke. Finally, six studies met the criteria, including 3542 cases and 3212 controls [7,8,9,10,11,17]. Among them, five studies described the associations between IL-17A rs2275913 and CAD, and three studies described the associations between IL-17A rs8193037 and CAD, two studies for each described association between rs3819025, rs3748067, rs3819024 and CAD, and one study for each described association between IL-17A rs4711998, rs8193036, and CAD. The PRISMA flowchart is shown in Figure 1 and the information for included studies is presented in Table 1.
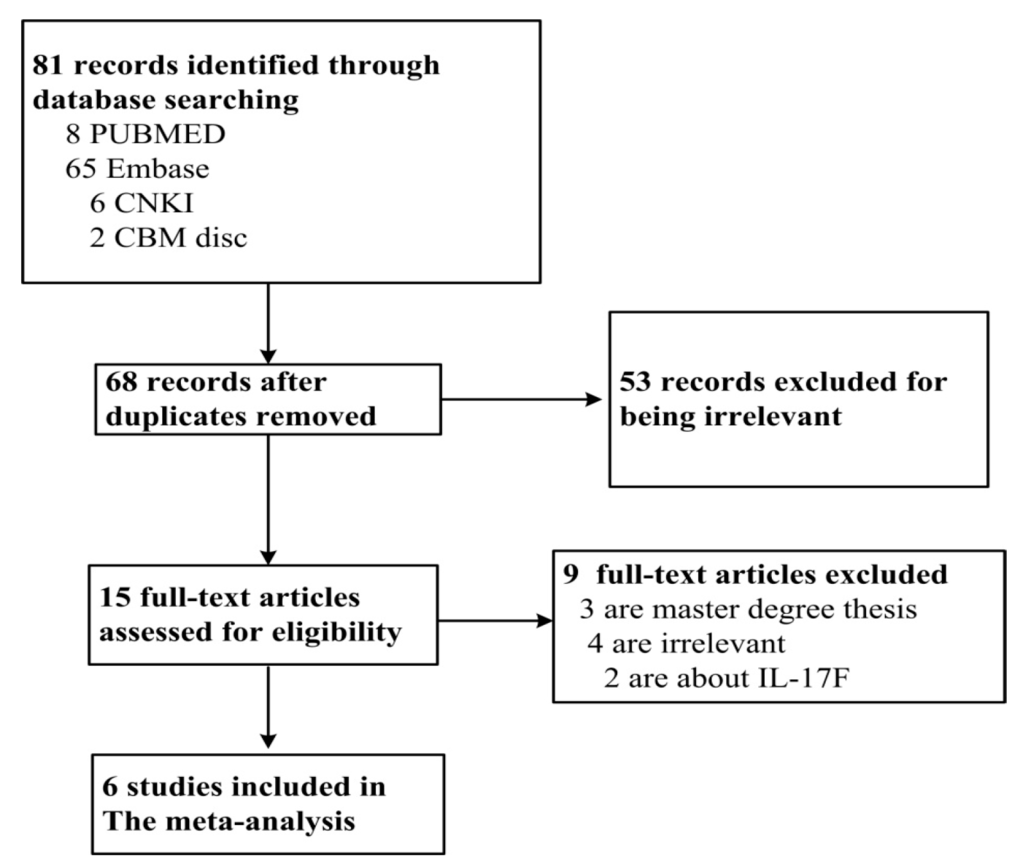
Figure 1.
PRISMA flowchart of study inclusion and exclusion.

Table 1.
Characteristics of eligible studies included in the meta-analysis.
3.2. Results of Meta-Analysis
The results of the meta-analysis for the associations between IL-17A rs2275913, rs3819024, rs3819025, rs8193037, rs3748067, rs4711998, rs8193036, and CAD risks were shown in Table 2, Figure 2, Figure 3 and Figure 4, and Supplementary Figures S1 and S2.

Table 2.
Pooled ORs and 95% CIs of the association between IL-17A rs2275913, rs3819024, rs3819025, rs8193037, rs3748067, rs4711998, rs8193036, and CAD risks.
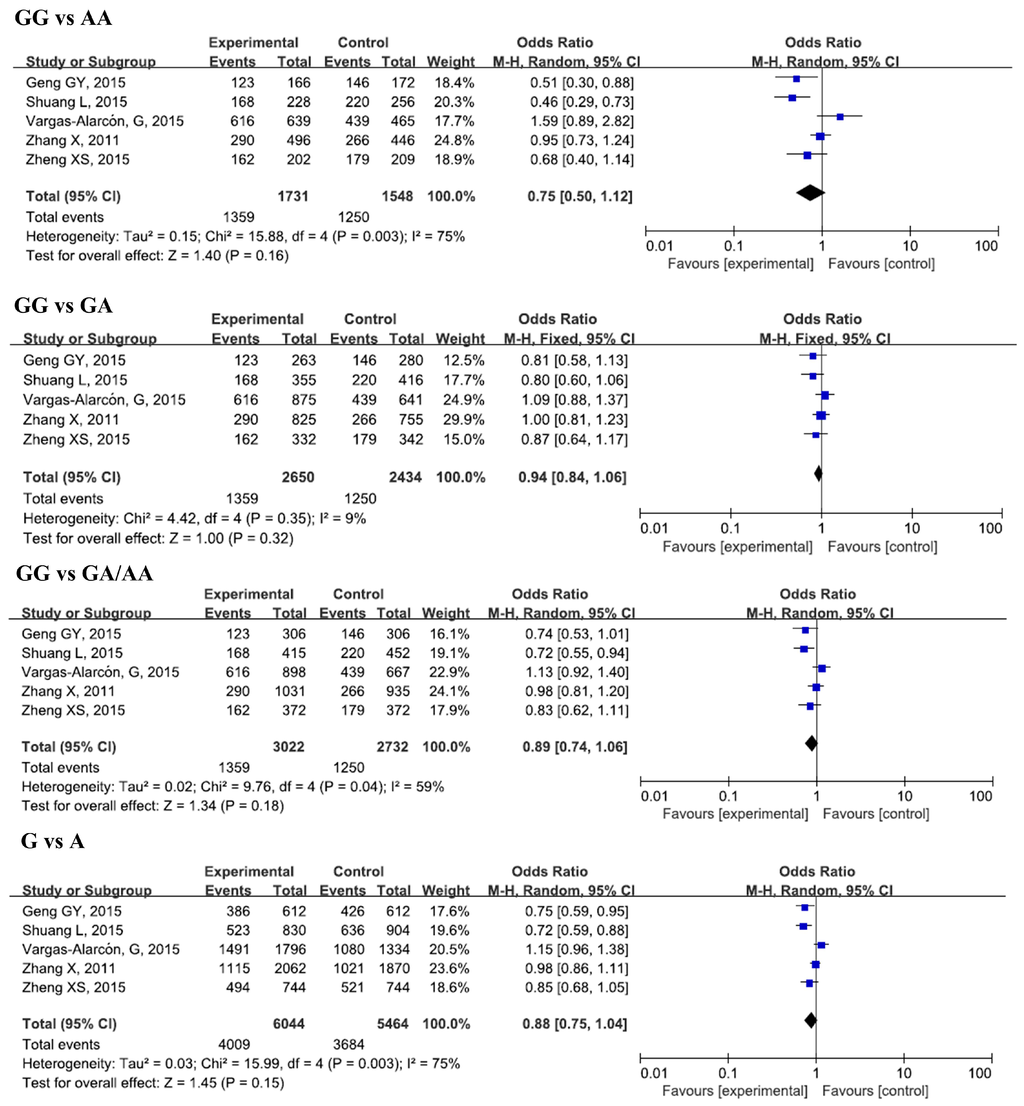
Figure 2.
Forest plots of odds ratios for the association of IL-17A rs2275913 with risk of CAD.
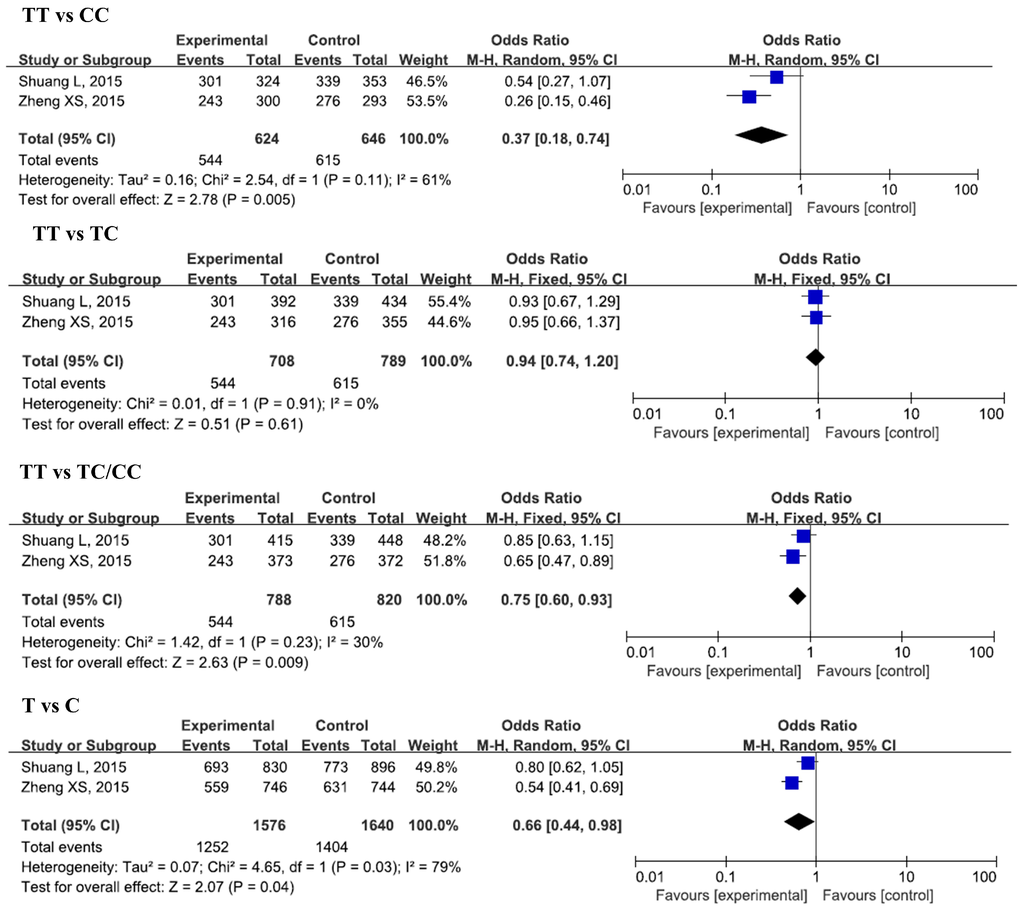
Figure 3.
Forest plots of odds ratios for the association of IL-17A rs3748067 with risk of CAD.
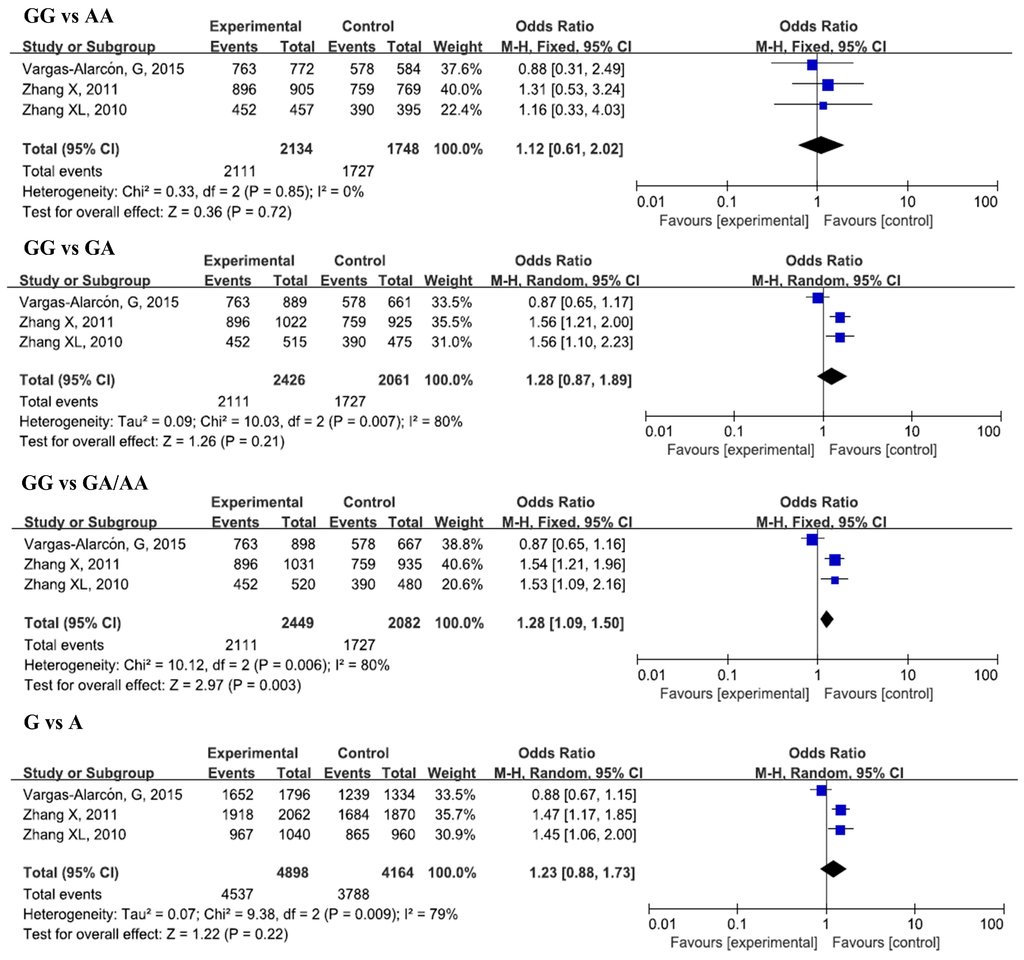
Figure 4.
Forest plots of odds ratios for the association of IL-17A rs8193037 with risk of CAD.
3.2.1. IL-17A rs2275913 and CAD
Five studies, with 3022 cases and 2732 controls, were included in this analysis. No significant associations were found between rs2275913 and CAD susceptibility in the overall analysis for all genetic models. However, when we conducted a subgroup analysis by sample size, significant decreases in risk of CAD for the GG genotype and G allele in a small sample size group in all genetic models was found (GG vs. AA, OR = 0.53, 95% CI = 0.40–0.71, padj < 0.0001; GG vs. GA/AA, OR = 0.76, 95% CI = 0.64–0.90, padj = 0.007; G vs. A, OR = 0.77, 95% CI = 0.68–0.87, padj < 0.0001). In the subgroup analysis by ethnicity, we did not find any remarkable differences between Asians and Caucasians after Bonferroni’s adjustment of p-value.
3.2.2. IL-17A rs8193037 and CAD
Three studies, with 2449 cases and 2082 controls, were included in this analysis. No significant associations were found in the overall analysis for all genetic models. In the subgroup analysis by ethnicity, a significant increase in CAD susceptibility was found for the GG genotype and G allele in a heterozygous model (GG vs. GA, OR = 1.56, 95% CI = 1.27–1.91, padj < 0.0001), dominant model (GG vs. GA/AA, OR = 1.54, 95% CI = 1.26–1.88, padj < 0.0001), and allelic model (G vs. A, OR = 1.47, 95% CI = 1.22–1.76, padj < 0.0001) in Asians.
3.2.3. IL-17A rs3748067 and CAD
Two studies, with 788 cases and 820 controls, were included in this analysis. A significant decrease in CAD risk was found in the homozygous model (TT vs. CC, OR = 0.37, 95% CI = 0.18–0.74, padj = 0.04). No significant associations were found between these SNPs and CAD risk in other genetic models.
3.2.4. IL-17A rs3819024, rs3819025 and CAD
For the association between rs3819024, rs3819025, and CAD, two studies were included for each of them (1929 cases and 1602 controls for rs3819024, and 1446 cases and 1383 controls for rs3819025). No significant relationship was found between these SNPs and CAD risk in all genetic models (Figures S1 and S2).
3.2.5. IL-17A rs4711998, rs8193036, and CAD
One study with 1031 cases and 935 controls was included in the analysis for rs4711998 and CAD; other study with 895 cases and 664 controls, was included in the analysis for rs8193036. No significant associations were found between these SNPs and CAD susceptibility in the overall analysis.
3.3. Sources of Heterogeneity
Since a significant heterogeneity was found for IL-17A rs2275913, we performed a subgroup analysis by sample size and ethnicity to explore the source of heterogeneity. The results indicate that the sample size was the source of heterogeneity for rs2275913 in all genetic models. Ethnicity also took the responsibility in heterogeneity and dominant models. For IL-17A rs8193037, ethnicity is the source of the heterogeneity in all genetic models.
3.4. Sensitivity Analysis
The influence of each study on the pooled ORs and 95% CIs was evaluated by excluding one single study at a time using STATA 12.0 software. No significant altered pooled ORs were found in all genetic models for IL-17A rs2275913 (data not shown).
3.5. Publication Bias
We performed the Begg’s funnel plot and Egger’s test to evaluate the publication bias. The p values for Begg’s and Egger’s tests are shown in Table 3. Obvious publication bias was observed for rs2275913 in heterozygous, dominant, and allelic models in Egger’s test. These results were also demonstrated by the shape of the funnel plot (only the heterozygous model results are shown, in Figure 5).

Table 3.
Egger’s and Begg’s test for the publication bias of rs2275913.
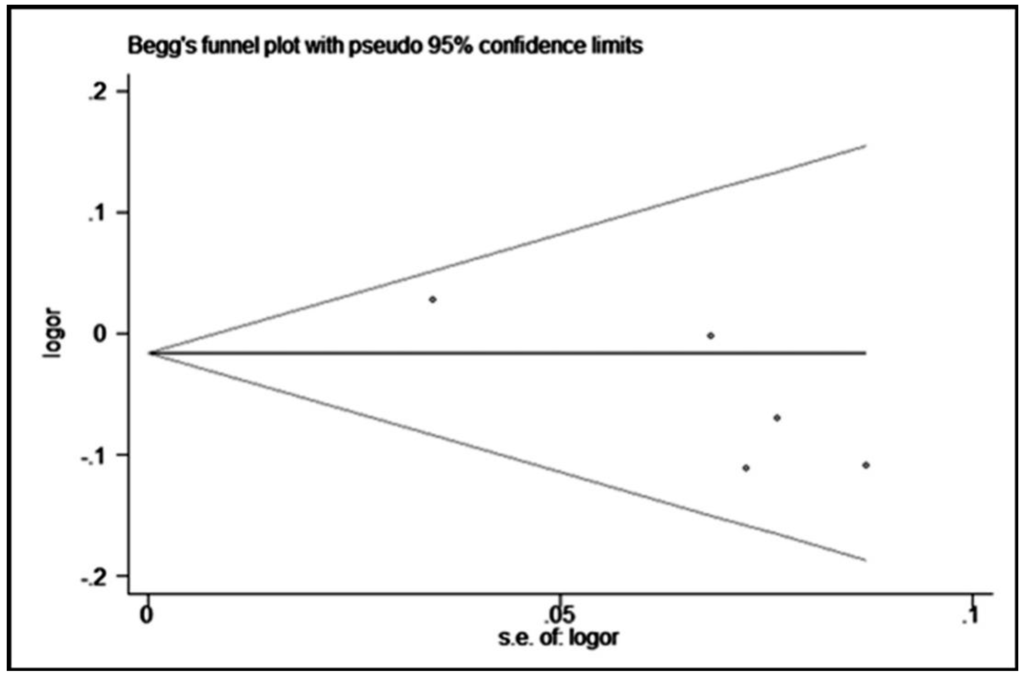
Figure 5.
Begg’s funnel plot with pseudo 95% confidence limits for a heterozygous model (GG vs. GA) of rs2275913.
4. Discussion
The main findings of the present meta-analysis show that the TT genotype of IL-17A rs3748067 was associated with a significant lower risk of CAD in a homozygous model (OR = 0.37) in Asians. No significant association was found for rs2275913, rs3819024, rs3819025, rs8193037, rs4711998, and rs8193036 with CAD susceptibility in overall analysis. However, a subgroup analysis indicated a significantly decreased risk of CAD for the GG genotype and G allele of rs2275913 in small sample size groups. A higher risk of CAD for the GG genotype and G allele of rs8193037 was observed in a heterozygous model (OR = 1.56), dominant model (OR = 1.54), and allelic model (OR = 1.47) in Asians.
Previous studies have reported the importance of inflammation in the risk of atherosclerosis [18]. IL-17 is produced by activated CD4+ T cells (Th17 cells) and other leukocytes. IL-17 family contains six members (IL-17A~F) [19]. IL-17A is one of the most important members, which induces the production of multiple cytokines, chemokines, and adhesion molecules in atherogenic cells. As a result, IL-17A was demonstrated to govern mononuclear cell accumulation and cell death, mobilization, recruitment, and activation of macrophages in atherosclerotic lesions, which eventually cause atherogenesis, atherosclerotic plaque disruption, and thrombosis [20,21]. Several studies indicated the correlation between IL-17A and CRP in different diseases, such as cardiovascular disease [22], inflammatory bowel disease [23], Cystic Echinococcosis [24], Behcet’s disease [25], etc. It was also reported that IL-17A triggered the inflammatory mediator Interleukin-1α (IL-1α) through NLR family pyrin domain containing 3 (NLRP3) inflammasome and NF-κB signal [26]. In ischemia/reperfusion injury in cardiac myocytes, enhanced IL-17A increased the pro-inflammatory targets, such as chemokine (C-X-C motif) ligand 1 and interleukin-6, through mitogen-activated protein kinases. Inhibition of the IL-17 pathway reduced necrosis and apoptosis in myocytes [5]. However, there are no studies investigating the relationship between these SNP and inflammation. Therefore, whether these SNPs in IL-17A have effects on these pro-inflammatory factors, as well as the pathways, still needs further investigation.
Recently, several SNPs in IL-17A have been studied to investigate their relations with CAD risk. rs2275913 has gained the greatest attention in these involved studies, some of which found a significant association between rs2275913 and CAD [7,8], while others denied it [9,10,11]. Our meta-analysis did not find any relations between rs2275913 and CAD in overall analysis, but lower risk of CAD was found in GG genotype in small sample size groups when subgroup analysis was conducted. Since the small sample studies sometimes have small-study effects [27], our results on the relationship between rs2275913 and CAD should be considered with caution.
In Asians, we also observed a higher risk of CAD for the GG genotype and G allele of rs8193037. However, only one study was included for the evaluation in Caucasians. Further studies are still needed to confirm the associations in other ethnicities. We also found a significant association between rs3748067 and CAD, but significant deviation from HWE (p < 0.05) was found for both of the two included studies. Furthermore, the two studies were all conducted among Asians. Therefore, further studies with larger sample sizes are required to confirm the results.
Significant heterogeneity was found in the meta-analysis for rs2275913, rs8193037, rs3748067, rs3819025, and rs3819024. Thus, we conducted subgroup analysis by sample size and ethnicity and found that both of them were the sources of the heterogeneity for rs2275913, and ethnicity was the source of the heterogeneity for rs8193037. However, only one study was included in the Caucasian group, and two studies were included in a large sample size group of rs2275913. And for rs8193037, a total of three studies were included, with two in Asians and one in Caucasians. Further studies are needed to confirm these results. In the sensitivity analysis, no significant changes were found after omitting each study at a time, indicating the relative stability and credibility of the results of our meta-analysis.
We found an obvious asymmetry in funnel plots, and a significant p-value for rs2275913 through Egger’s test in the present study. According to the Cochrane Handbook (version 5.1.0, Section 10), the cause for the asymmetry in funnel plots includes selection bias (publication bias, selective outcome reporting), small-sample effects, true heterogeneity, artifacts, and chance. In the present meta-analysis, most of the included studies were of small size. After a comprehensive literature search, all published studies were included in the present study. No large consortia or GWAS data are available yet. Therefore, the small-sample effects might be one reason for this bias. Furthermore, we have assessed the quality of all included studies, and they are all of high quality. However, only studies in English or Chinese have been searched and included. There might be studies in other languages which are not included in the present analysis. Based on the Cochrane Handbook, tests for funnel plot asymmetry should be used only when there are at least 10 studies included, because when there are fewer studies the power of the test is too low to distinguish chance from real asymmetry. In the present test, only five studies are included, which might be another reason for the asymmetry.
We included six studies in our present meta-analysis, while only one study was conducted in Caucasians, which estimated the associations between rs2275913, rs8193037, and rs3819024 with risk of CAD. However, no obvious relationships were found in all of these SNPs. The positive results in our meta-analysis are all related with Asians. Thus, the conclusion drawn in the present meta-analysis might be only of generalizability for Asians.
The results of the present meta-analysis should be interpreted carefully because of the following limitations. Firstly, the number of patients was relatively small, and may influence the outcomes. After a very comprehensive literature search from several different databases, only six studies including 3542 cases and 3212 controls were included in the present meta-analysis, and for rs8193037, rs3748067, rs3819025, and rs3819024, even fewer studies, each less than three were included. Secondly, the clinicopathological characteristics or disease subtypes are limited in most of these studies. Thirdly, the ethnicity of the subjects is predominantly Asiatic. Only one study, with 898 cases and 667 controls, was committed in Caucasians, and the last limitation is that CAD is a multi-factorial disease influenced by both genetic and environmental factors. The gene-gene and gene-environment interactions may play important roles in the functions of these polymorphisms, but most studies lack the information on environmental exposure and multiple SNPs in haplotypes.
5. Conclusions
In conclusion, the current meta-analysis suggests a lower risk of CAD for the TT genotype of IL-17A rs3748067 in Asians, and a higher risk of CAD for the GG genotype and G allele of rs8193037 in Asians. A decreased risk of CAD for the GG genotype and G allele of rs2275913 were observed in a small sample size group, while rs3819024, rs3819025, and rs4711998 are not associated with the risk of CAD. Thus, IL-17A rs3748067 and rs8193037 might be recommended as predictors for susceptibility of CAD in Asians. However, the results of this meta-analysis are hypothesis-generating results which should be interpreted with caution because of the heterogeneity and publication bias among study designs. Further study is needed to evaluate the association between these SNPs and CAD, especially in a large sample size, in Caucasians, and with clinicopathological characteristics.
Supplementary Materials
The following are available online at www.mdpi.com/1660-4601/13/7/660/s1 Figure S1: Forest plots of odds ratio for the association of IL-17A rs3819024 with risk of CAD, Figure S2: Forest plots of odds ratio for the association of IL-17A rs3819025 with risk of CAD.
Acknowledgments
The present study was supported by the Construct Program of the Key Discipline in Hunan Province, Natural Science Foundation of China (Grant No. 81300231), the Foundation of Hunan Educational Committee (Grant No. 5B031 and 15A023) and the Hunan Provincial Natural Science Foundation (Grant No. 2015JJ6010).
Author Contributions
Mei-Hua Bao and Jian-Ming Li planned and designed the experiments; Huai-Qing Luo, Liang Tang, Li-Ping Dong, Guang-Yi Li and Mei-Hua Bao performed experiments; Ju Xiang and Jie Zeng analyzed the data; Mei-Hua Bao and Yan Xiao wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Frostegård, J. Immunity, atherosclerosis and cardiovascular disease. BMC Med. 2013, 11, 117. [Google Scholar] [CrossRef] [PubMed]
- Cãtanã, C.; Neagoe, I.B.; Cozma, V.; Magdaş, C.; Tãbãran, F.; Dumitraşcu, D.L. Contribution of the IL-17/IL-23 axis to the pathogenesis of inflammatory bowel disease. World J. Gastroenterol. 2015, 21, 5823–5830. [Google Scholar] [PubMed]
- Isailovic, N.; Daigo, K.; Mantovani, A.; Selmi, C. Interleukin-17 and innate immunity in infections and chronic inflammation. J. Autoimmun. 2015, 60, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Jiang, Y.; Ma, T.; Zhu, F.; Gao, F.; Zhang, P.; Guo, C.; Wang, Q.; Wang, X.; Ma, C.; et al. A critical function of Th17 proinflammatory cells in the development of atherosclerotic plaque in mice. J. Immunol. 2010, 185, 5820–5827. [Google Scholar] [CrossRef] [PubMed]
- Barry, S.P.; Ounzain, S.; McCormick, J.; Scarabelli, T.M.; Chen-Scarabelli, C.; Saravolatz, L.I.; Faggian, G.; Mazzucco, A.; Suzuki, H.; Thiemermann, C.; et al. Enhanced IL-17 signalling following myocardial ischaemia/reperfusion injury. Int. J. Cardiol. 2013, 163, 326–334. [Google Scholar] [CrossRef] [PubMed]
- Bao, M.H.; Xiao, Y.; Zhang, Q.S.; Luo, H.Q.; Luo, J.; Zhao, J.; Li, G.Y.; Zeng, J.; Li, J.M. Meta-Analysis of miR-146a Polymorphisms Association with Coronary Artery Diseases and Ischemic Stroke. Int. J. Mol. Sci. 2015, 16, 14305–14317. [Google Scholar] [CrossRef] [PubMed]
- Geng, G.Y.; Liu, H.L.; Zhao, Y.J.; Wu, L.; Mao, L.; Ba, N. Correlation between polymorphisms in the IL-17A and IL-17F genes and development of coronary artery disease. Genet. Mol. Res. 2015, 14, 11488–11494. [Google Scholar] [CrossRef] [PubMed]
- Shuang, L.; Li, Z.; Chen, F.; Cui, X.; Ning, Y.; Su, Y.; Dong, M. Association between interleukin-17 gene polymorphisms and risk of coronary artery disease. Int. J. Clin. Exp. Pathol. 2015, 8, 11653–11658. [Google Scholar] [PubMed]
- Vargas-Alarcón, G.; Angeles-Martínez, J.; Villarreal-Molina, T.; Alvarez-León, E.; Posadas-Sánchez, R.; Cardoso-Saldaña, G.; Ramírez-Bello, J.; Pérez-Hernández, N.; Juárez-Rojas, J.G.; Rodríguez-Pérez, J.M.; et al. Interleukin-17A gene haplotypes are associated with risk of premature coronary artery disease in Mexican patients from the Genetics of Atherosclerotic Disease (GEA) study. PLoS ONE 2015, 10, e0114943. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Pei, F.; Zhang, M.; Yan, C.; Huang, M.; Wang, T.; Han, Y. Interleukin-17A gene variants and risk of coronary artery disease: A large angiography-based study. Clin. Chim. Acta 2011, 412, 327–331. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.S.; Wang, S.; Ni, M. Association between interleukin 17A gene polymorphisms and risk of coronary artery disease. Genet. Mol. Res. 2016, 15, 11653–11658. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Li, X.F.; Wang, Y.D.; Yuan, Y. Arg72Pro polymorphism of TP53 gene and the risk of skin cancer: A meta-analysis. PLoS ONE 2013, 8, e79983. [Google Scholar] [CrossRef] [PubMed]
- Higgins, J.P.; Thompson, S.G.; Deeks, J.J.; Altman, D.G. Measuring inconsistency in meta-analyses. BMJ 2003, 327, 557–560. [Google Scholar] [CrossRef] [PubMed]
- DerSimonian, R.; Laird, N. Meta-analysis in clinical trials revisited. Contemp. Clin. Trials 2015, 45, 139–145. [Google Scholar] [CrossRef] [PubMed]
- Mantel, N.; Haenszel, W. Statistical aspects of the analysis of data from retrospective studies of disease. J. Natl. Cancer Inst. 1959, 22, 719–748. [Google Scholar] [PubMed]
- Peters, J.L.; Sutton, A.J.; Jones, D.R.; Abrams, K.R.; Rushton, L. Comparison of two methods to detect publication bias in meta-analysis. JAMA 2006, 295, 676–680. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.L.; Han, Y.L.; Pei, F.; Yan, C.H.; Kang, J. Association of IL-17A -121G/A polymorphism and premature coronary artery disease in Han population of north China. Shandong Med. J. 2010, 50, 1–3. [Google Scholar]
- Weber, C.; Hristov, M. Atherogenesis and inflammation. From cellular mediators to regulatory mechanisms of inflammation in atherosclerosis. Hämostaseologie 2015, 35, 99, 101. [Google Scholar] [PubMed]
- Cua, D.J.; Tato, C.M. Innate IL-17-producing cells: The sentinels of the immune system. Nat. Rev. Immunol. 2010, 10, 479–489. [Google Scholar] [CrossRef] [PubMed]
- Eid, R.E.; Rao, D.A.; Zhou, J.; Lo, S.F.; Ranjbaran, H.; Gallo, A.; Sokol, S.I.; Pfau, S.; Pober, J.S.; Tellides, G. Interleukin-17 and interferon-gamma are produced concomitantly by human coronary artery-infiltrating T cells and act synergistically on vascular smooth muscle cells. Circulation 2009, 119, 1424–1432. [Google Scholar] [CrossRef] [PubMed]
- Erbel, C.; Chen, L.; Bea, F.; Wangler, S.; Celik, S.; Lasitschka, F.; Wang, Y.; Böckler, D.; Katus, H.A.; Dengler, T.J. Inhibition of IL-17A attenuates atherosclerotic lesion development in apoE-deficient mice. J. Immunol. 2009, 183, 8167–8175. [Google Scholar] [CrossRef] [PubMed]
- Todd, J.; Simpson, P.; Estis, J.; Torres, V.; Wub, A.H. Reference range and short- and long-term biological variation of interleukin (IL)-6, IL-17A and tissue necrosis factor-alpha using high sensitivity assays. Cytokine 2013, 64, 660–665. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, M.; Yuksel, M.; Ates, I.; Yaln, K.Z.M.; Kilic, H.; Ates, H.; Kayacetin, E. Are sTWEAK and IL-17A Levels in Inflammatory Bowel Disease Associated with Disease Activity and Etiopathogenesis. Inflamm. Bowel Dis. 2016, 22, 615–622. [Google Scholar] [CrossRef] [PubMed]
- Mezioug, D.; Touil-Boukoffa, C. Interleukin-17A correlates with interleukin-6 production in human cystic echinococcosis: A possible involvement of IL-17A in immunoprotection against Echinococcus granulosus infection. Eur. Cytokine Netw. 2012, 23, 112–119. [Google Scholar] [PubMed]
- Mesquida, M.; Molins, B.; Llorenç, V.; de la Maza, M.S.; Hernandez, M.V.; Espinosa, G.; Adán, A. Proinflammatory cytokines and C-reactive protein in uveitis associated with Behçet’s disease. Mediat. Inflamm. 2014, 2014, 396204. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Yu, N.; Zhang, R.; Zhang, S.; Wu, J. Interleukin-17A Induces IL-1â Secretion from RPE Cells via the NLRP3 Inflammasome. Investig. Ophthalmol. Vis. Sci. 2016, 57, 312–319. [Google Scholar] [CrossRef] [PubMed]
- Sterne, J.A.; Gavaghan, D.; Egger, M. Publication and related bias in meta-analysis: Power of statistical tests and prevalence in the literature. J. Clin. Epidemiol. 2000, 53, 1119–1129. [Google Scholar] [CrossRef]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).