Abstract
Background and Objectives: Staphylococcus aureus is a prevalent bacterium capable of inducing various infections, including skin and soft tissue infections, bloodstream infections, pneumonia, and surgical site infections. The emergence of antimicrobial resistance in S. aureus, particularly methicillin-resistant S. aureus, has raised substantial concerns within global healthcare settings. Prior to antibiotic prescription, the ideal approach is antimicrobial susceptibility testing (AST); however, this is frequently perceived as excessively complex and time-intensive. Lab-on-a-chip (LOC) technology holds promise in addressing these challenges and advancing fundamental microbiological research while also aiding in the development of therapeutic strategies. This systematic review aims to evaluate the potential utility of LOC for AST of S. aureus. Materials and Methods: This study adhered to the PRISMA guidelines. Various databases, including SCOPUS, PubMed/MEDLINE, SCIELO, and LILACS, in addition to gray literature sources, were employed in the review process. Results: Sixteen studies were included in this systematic review. All these studies detailed the effectiveness, rapidity, and predictability of LOC systems for assessing S. aureus susceptibility to various antibiotics. When comparing the LOC approach to traditional manual methods, it was evident that LOC requires a minimal quantity of reagents. Furthermore, most studies reported that the entire LOC procedure took 10 min to 7 h, with results being equally accurate as those obtained through traditional AST protocols. Conclusions: The potential application of LOC for AST of S. aureus is emphasized by its ability to provide rapid access to minimum inhibitory concentration data, which can substantially aid in selecting the most suitable antibiotics and dosages for treating challenging infections caused by this microorganism. Moreover, the rapid AST facilitated by LOC holds promise for enhancing the appropriateness and efficacy of therapy in clinical settings.
1. Introduction
The major threat posed by antimicrobial resistance to the world’s healthcare systems makes it difficult for modern medicine to control infectious diseases effectively. Infections with antimicrobial resistance cause 33,000 fatalities annually in the European Union and about 35,000 deaths annually in the USA [1,2,3]. Most bacterial-related deaths could be treated with antibiotics, but according to the World Health Organization, the number of deaths brought on by bacteria resistant to antimicrobials is expected to rise to almost 10 million per year by 2050 [4,5].
Staphylococcus aureus is a Gram-positive, human commensal bacterium that is regularly found on healthy people’s skin. S. aureus can cause a range of infections, including skin and soft tissue infections, bloodstream infections, pneumonia, and surgical site infections. It is a major pathogen that may originate bacteremia as well as more serious and difficult-to-treat osteoarticular infections and heart infections such as infective endocarditis [5,6,7]. Antimicrobial resistance in S. aureus, particularly methicillin-resistant S. aureus (MRSA), has been a significant concern in healthcare settings worldwide. MRSA infections can be challenging to treat as they are resistant to commonly used antibiotics like methicillin and other beta-lactam drugs [6,7].
The impact of deaths specifically attributed to antimicrobial resistance in S. aureus can vary depending on several factors, such as geographic location, healthcare settings, population demographics, and access to appropriate healthcare. It is difficult to provide precise global figures on deaths solely caused by antimicrobial resistance in S. aureus, as often these infections occur in conjunction with other underlying health conditions [5,6,7]. However, it is well-documented that MRSA infections can result in increased morbidity, mortality, and healthcare costs compared to infections caused by non-resistant strains of S. aureus. According to estimates from the Centers for Disease Control and Prevention (CDC) in the United States, MRSA infections were associated with approximately 11,000 deaths in 2017. These numbers include both healthcare-associated and community-associated MRSA infections [8]. To combat the impact of antimicrobial resistance in S. aureus and reduce associated deaths, healthcare facilities have implemented strategies such as improved infection control practices, screening and isolation of patients carrying MRSA, and appropriate antibiotic prescribing guidelines [1,2,4,6,8].
The vital first step in providing patients with appropriate care is the prompt and correct identification of the causal agent responsible for infection [9,10]. This initial stage directs patient treatment plans and the efficient use of antibiotics for bacterial infections. To reduce the spread of antibiotic resistance, proper medicine use is essential [11,12]. Studying certain bacterial phenotypes requires an understanding of how bacteria interact with cells in physiological settings. The minimum inhibitory concentration (MIC) of antibiotics, which defines the likelihood of a specific drug’s therapeutic efficacy against a given infection, is determined using antibiotic susceptibility testing. Conducting bacterial categorization studies is also helpful in many applications as complementary elements to support treatment decision-making. The collection of results from antibiotic susceptibility testing and bacterial classification can take up to 48 h or longer, which can cause delays in the prescription of appropriate antibiotics or the administration of inappropriate antibiotics before obtaining antibiotic susceptibility results, impacting morbidity, mortality, the gravity of infections, and the incidence of antibiotic resistance [13,14,15].
Microscopy, cell infection models, and more contemporary molecular, cellular, and immunological assays are examples of traditional microbiological in vitro techniques [13]. These techniques have given and will continue to give invaluable information that will help us understand the host-bacterial interaction’s molecular and cellular microbiology. Nevertheless, traditional methods can have certain drawbacks [14]. Microorganisms that cannot be cultivated in vitro still exist despite advancements in culture techniques. Ex vivo and in vitro models, on the other hand, are unable to accurately replicate the physiological environment [14,15]. As a result, outcomes obtained in vitro and in vivo are not necessarily comparable. Moreover, some phenotypes, like the formation of biofilms, are conventionally researched in simple settings that do not accurately reflect complicated physiological circumstances [14,16].
The recent development of lab on a chip, however, can help to address some of these issues and enhance fundamental microbiological research while also assisting in the creation of therapeutic strategies.
A lab on a chip refers to a miniaturized device that integrates multiple laboratory functions onto a single microchip or small platform. It aims to replicate and perform various laboratory processes and analyses in a portable and efficient manner. Lab-on-a-chip technology has gained significant attention in recent years due to its potential applications in healthcare diagnostics, environmental monitoring, and biological research [14,17]. The concept behind lab on a chip is to downscale and miniaturize laboratory processes, such as sample preparation, mixing, separation, detection, and analysis, onto a small chip or platform. This integration of multiple functions onto a single device offers several advantages, including reduced sample and reagent volumes, faster analysis times, improved precision, and increased automation [14,18].
Lab-on-a-chip devices typically consist of microfluidic channels, chambers, valves, sensors, and detection systems, all fabricated on a small chip using techniques such as lithography, etching, and bonding. These microfluidic channels allow precise manipulation and control of fluids, enabling the handling and processing of small volumes of samples and reagents [14,17,18].
Recently, the use of microfluidics for assessing antibiotic susceptibility has gained popularity. These devices provide quick, high-throughput, and inexpensive studies using fluidic channels with a diameter of a few micrometers [19]. A droplet-based microfluidic system that confines single bacteria and medications into plugs of small volume for single-drug antimicrobial susceptibility tests has been reported to shorten detection times [20]. An alternative method for quickly isolating microorganisms is to use an inertial microfluidic chip to perform an antibiotic susceptibility test followed by a hybridization-based RNA detection method [21]. Additionally, molecular diffusion has been used to detect microbial growth on the surface of numerous gradient zones created in hydrogels by drugs or drug combinations, and MIC data were achieved after 3 h of incubation [22]. It is important to note that prior to prescribing antibiotics, antimicrobial susceptibility testing is ideal but is typically viewed as being excessively complicated and time-consuming.
Considering what was previously described, this systematic review aims to assess the potential application of lab on a chip for antibiotic susceptibility testing of S. aureus.
2. Materials and Methods
2.1. Search Approach
This study’s analysis followed PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-analyses) guidelines [23]. Several databases, including SCOPUS, PubMed/MEDLINE, SCIELO, and LILACS, as well as the gray literature, were used in the review framework. Up to April 2023, searches using keywords and MeSH terms included the terms organ on a chip, lab on a chip, microphysiological systems, microfluidics, bioassays, minimal inhibitory concentration, antimicrobial resistance, antimicrobial susceptibility test, biofilms, Staphylococcus aureus, and studies published in all languages. The next exploration included searching databases using Boolean operators (AND, OR): “organ on a chip” OR “lab on a chip” OR “microphysiological systems” OR “microfluidics” OR “bioassays” OR “minimal inhibitory concentration” OR “antimicrobial resistance” OR “antimicrobial susceptibility test” OR “biofilms” AND “Staphylococcus aureus”.
2.2. Selection Criteria
The studies that were taken into consideration for this evaluation must use microfluidic platforms or lab-on-a-chip devices and 3D printing and/or bioprinting methods to organ-on-a-chip technology.
Abstracts, reviews, systematic reviews, meta-analyses, brief communications, conference articles, patents, case reports, and studies lacking critical information on the manufacturing process were also disregarded.
2.3. Question
This comprehensive review aims to answer the question, what potential clinical applicability of lab on a chip for antibiotic susceptibility testing exists in experiments with S. aureus?
P: experimentation with S. aureus
I: lab on a chip
C: control experiments
O: potential clinical applicability for antibiotic susceptibility testing
2.4. Review Course
To identify publications that might be suitable, two researchers examined the titles and abstracts. Given the probability of divergence in the choice of studies, a third author (MZ) could mediate. The statistical test Kappa was used to determine the importance of observer agreement (>90).
2.5. Data Compilation
The most significant information from the chosen studies was compiled into a table. Each researcher completed this method individually. After that, the data were compared. The data included the names of the authors, the publication date, the application of the lab-on-a-chip device, and information about its key characteristics, such as the materials used in building it, culture, bacterial strains and growth conditions, MICs, antibiotics utilized, and the main findings.
2.6. Risk of Bias
Two authors evaluated the quality and potential for bias in the included studies using a scale that had previously been published [24]. With the help of the tool, it is possible to assess the study using 15 different criteria, including the following: design (objective, sample, baseline characteristics, co-interventions), measures studied (measurement method, blinding (examiner, statistician), described reliability, level of agreement), statistical analysis (appropriate analysis, co-interventions, subgroup analysis, statistical significance, confidence intervals), and clinical significance.
3. Results
A total of 80 articles were identified after the initial search. Then, 53 publications were excluded (because they did not study S. aureus or, because they were aimed only at the identification of the microorganism, or they investigated genes or proteins and did not assess antimicrobial susceptibility). Four papers that appeared twice were not included. After reading the entire text, seven other studies were excluded since they did not meet the selection criteria. Finally, 16 studies [19,20,25,26,27,28,29,30,31,32,33,34,35,36,37,38] were included in this systematic review (Figure 1).
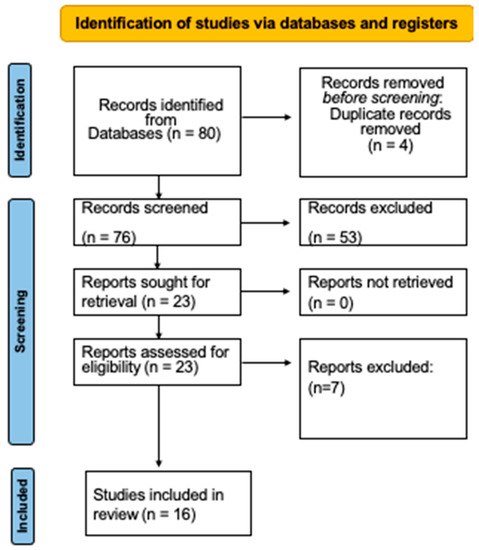
Figure 1.
Diagram of the selection process.
The features of the included studies are shown in Table 1. Between 2008 [20] and 2021 [25], the studies included were published.

Table 1.
Principal characteristics of the included studies.
As observed in Table 1, the studies evaluated different lab-on-a-chip technologies to mainly establish the susceptibility of S. aureus to different antibiotics. All studies described effective, rapid, and predictable systems to assess the susceptibility of S. aureus to different antibiotics. Comparing the lab-on-a-chip approach to the traditional manual methods, it was found that lab-on-a-chip needs a very small number of reagents. Besides, most studies reported that the entire antimicrobial susceptibility test procedure lasted from 10 min to 7 h, and the results are equally accurate as the results of the Clinical & Laboratory Standards Institute (CLSI) [39] and the European Committee on Antimicrobial Susceptibility Testing (EUCAST, Europe) [40]. It was also observed that lab on a chip has the potential to identify S. aureus with high sensitivity and speed in as little as 10 min by forming and measuring small volume plugs and is less dependent on the initial concentration and growth rate of bacteria in the sample [25]. This approach might make pre-incubation unnecessary because tests can be run on a single bacterium. It was also reported that this method avoids MIC measurements because it is not based on standard methodology.
Table 1 also shows that various materials and microfabrication processes can be used to create a lab on a chip during the study of S. aureus. As a result, silicon wafers can be produced with nanometer-scale chip features using photolithography. In microfluidic chips, common compartment types include reservoirs, chambers, and microchannels. Additionally, functional components such as valves, mixers, and pumps are made to transfer liquid in a particular manner. According to the findings of this review, polydimethylsiloxane silicone rubber is largely used in lab settings for the creation of the lab on a chip.
Different strains of S. aureus were explored in the same trial. Moreover, most of the experiments were performed considering the recommendations of CLSI [39] and EUCAST [40] with minor modifications.
The susceptibility of S. aureus to the following antibiotics was evaluated: gentamicin, ciprofloxacin, moxifloxacin, tobramycin, ceftazidime, meropenem, vancomycin, linezolid, chloramphenicol, flucloxacillin, ampicillin, spectinomycin, streptomycin, amikacin, norfloxacin, ofloxacin, tetracycline, oxacillin, ampicillin, cephalosporin, cephalotin, cefoxitin, levofloxacin, and erythromycin. Most studies also considered MIC ranges according to CLSI [39] and EUCAST [40] and evaluated the lab on a chip on several antibiotics at the same time. It is also important to note that microfluidics works with volumes in the microliter range, whereas standard approaches work with milliliter volumes.
As was described here, lab-on-a-chip devices for assessing antimicrobial susceptibility often utilize various types of sensors to detect and measure the response of microorganisms to antimicrobial agents. These sensors are integrated into the microfluidic platforms to provide rapid and accurate analysis of antimicrobial susceptibility. Some common types of sensors used in lab on a chip system for antimicrobial susceptibility testing include microfluidic optical sensors, microfluidic impedance sensors, microfluidic electrochemical sensors, microfluidic biosensors, microfluidic piezoelectric sensors, and microfluidic acoustic sensors.
The level of risk of bias was minimal across all experiments examined (Table 2). However, the aims, conception, and methods of each model varied. For these reasons, a quantitative assessment is challenging.

Table 2.
Risk of bias of the assessed investigations.
4. Discussion
An illustrative depiction of the lab-on-a-chip concept includes a small, rectangular chip or substrate that serves as a platform for miniaturized laboratory functions (Figure 2). The chip is typically made of materials such as glass, silicon, or polymer, and it contains a network of microfluidic channels, chambers, and functional components. On the chip, it can see a variety of tiny channels interconnected like a maze. These channels are designed to transport fluids, such as biological samples, reagents, or cell cultures. The microfluidic channels allow precise control over the flow of fluids, enabling various processes. Along the channels, it can visualize functional components, such as sensors, electrodes, and optical elements. These components are integrated directly into the chip and can measure parameters like pH, temperature, fluorescence, or electrical signals. In one part of the chip, there is a sample loading area, where a small volume of the biological sample is introduced into the system. The sample may contain bacteria, cells, or molecules that need to be analyzed or tested. By following the channels, multiple regions where the sample interacts with different reagents or undergoes various processes are noticed. For example, one region might be designated for mixing the sample with specific antibiotics or other test compounds. Further along, areas are seen where the sample is exposed to different conditions, such as varying temperatures or incubation times. These conditions mimic specific aspects of a laboratory environment but at a much smaller scale. In another section of the chip, there are sensing elements. These sensors can monitor changes in the sample’s properties, such as bacterial growth, fluorescence, or electrical conductivity. The sensors provide real-time data, allowing researchers to observe how the sample responds to different treatments. The final part of the chip contains output regions, where the results of the analysis or testing are displayed or recorded. This could be in the form of color changes, light signals, electrical readouts, or digital data.
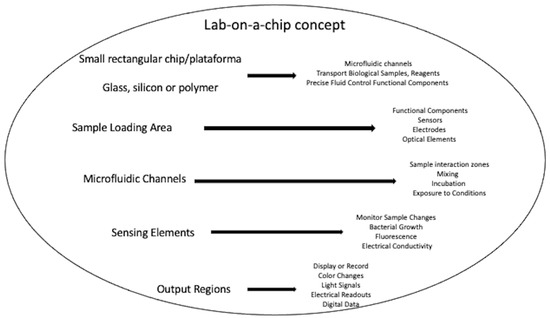
Figure 2.
Lab-on-a-chip concept.
Growth techniques for microorganisms, including S. aureus, for in vitro phenotypic testing have often lagged behind other technological advances. Even currently, with remarkable automation and robotics, the well plate is still referred to as the ultimate high-throughput platform [27]. Because of the complexities of the procedures involved in the assay, it is frequently limited to the use of 96-well plates when culturing microbial biofilms. Even though liquid-handling stations can help with the initial dispensing of cells into wells, eliminating the need for human interaction to complete the rest of the experiment is difficult [26,27]. The delicate nature of biofilms, the three-dimensional architecture, various morphologies, possible disturbance, and cell loss during the washing processes required in the test are some of the problems that impede the full automation of these assays [27,28,29]. To address all these challenges, a completely automated lab-on-a-chip platform for biofilm and planktonic culturing in nanoscale volumes has been developed. Antimicrobial susceptibility testing and ultra-high-throughput screening applications, such as those meant to prevent biofilm formation and those designed to assess established biofilms, are excellent for lab on a chip [25,26,27].
As noted in this systematic review, antimicrobial susceptibility testing of S. aureus is a clinically important application of lab-on-a-chi- in antibiotics research [17]. Slow-growing bacteria may take more than a day to be identified in phenotypic antimicrobial susceptibility testing because they must divide enough times to be seen under a microscope [17,19,25,26,27]. This review demonstrated that there are several methods to solve the detection time problem in antimicrobial susceptibility testing of S. aureus using a lab on a chip; however, this approach also has its challenges.
It was observed here that following the growth of S. aureus in the presence of antibiotics is a relevant technique [25,26,27,28,29,30,31,32,33,34,35,36,37,38]. Such assays have been found to produce MIC quickly [26,29,30,31,33]. Nevertheless, methodologies created on single-cell imaging necessitate high-resolution optics and time-lapse imaging at many locations [17]. This review also found that there are different ways to perform rapid microfluidic antimicrobial susceptibility testing of S. aureus. Although most of the studies included in this systematic review reported rapid assessments of up to two hours, shorter times of up to 10–30 min have also been described [25,33]. Solely microfluidic phenomolecular tests have been presented to produce antimicrobial susceptibility testing findings from clinical specimens in such a brief period; however, as was observed here, optical- or sensor-based microfluidic approaches have also shown effective results in a very short time [17].
Integrating various experimental processes into a single platform can also reduce the number of modifications performed by researchers, hence the danger of contamination and methodological unpredictability [14,41]. As was observed here, droplet-based devices are an excellent example of the integrative power of microfluidics [26]. This time savings and low technical manipulation are unquestionably essential considerations for the usage of microfluidic devices in a clinical setting, where correct management of infected patients with S. aureus demands predictable and rapid findings [14].
As was observed in this systematic review, it is also important to highlight that whereas conventional approaches are limited to performing analysis on flat 2D surfaces, including culture flasks, Petri dishes, or well plates, lab on a chip proposes a novel range of constituents with interesting features, the most notable of which being polydimethylsiloxane [14]. It is good for cellular and microbiological cultures due to its biocompatibility and gas permeability, but its optical transparency also enables microscope viewing and analysis. It is important to note that its main benefit is the ability to combine it with various materials, resulting in significant equipment composition diversity. Therefore, microfluidics allows for the application of diverse stimuli, such as fluid flow or tactic gradients, in heterogeneous or even three-dimensional settings [14,17,25,26,27,28,30,31,32,33].
As was described in this systematic review, the disparity in volume and S. aureus populations employed in microfluidics versus traditional approaches is also an important factor to recognize [14]. Microfluidics works with volumes in the microliter range, whereas standard approaches work with milliliter volumes. This minor volume transforms microfluidic strategies into portable implements, allowing for cost reductions in reagents, resources, and area, as well as more exact control of the study’s biological and physical parameters [14,26,29,30,31]. The quantity of bacteria in a lab on a chip is reduced to hundreds or even solitary bacterium, resulting in more exact results on a single-cell scale [14].
As was described here, various types of sensors were described in this systematic review. The choice of the sensor depends on the specific requirements of the antimicrobial susceptibility assay, such as sensitivity, detection limit, and compatibility with the microfluidic platform. As was observed here, the integration of these sensors into lab-on-a-chip systems enables rapid, automated, and miniaturized antimicrobial susceptibility testing, providing valuable tools for clinical diagnostics, research, and antimicrobial stewardship [41,42].
Lab-on-a-chip technology, Raman spectroscopy, and Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF MS) are all powerful tools with distinct capabilities for antibiotic susceptibility testing. Lab-on-a-chip technology excels in providing real-time, miniaturized, and integrated platforms for phenotypic and genotypic antibiotic susceptibility testing [41,42]. Raman spectroscopy offers label-free, non-destructive analysis and the ability to study subtle cellular changes related to antibiotic response, while MALDI-TOF MS provides high-throughput bacterial identification and potential insights into protein expression changes linked to antibiotic resistance. Each technology offers unique strengths for antibiotic susceptibility testing, and their potential varies depending on the specific application and research objectives [43]. Integrating these techniques and combining their capabilities may lead to comprehensive and accurate assessments of antimicrobial susceptibility in the future.
Lab-on-a-chip systems designed for assessing antimicrobial susceptibility typically require aseptic environments to ensure accurate and reliable results. The reason for this is that antimicrobial susceptibility testing involves evaluating the response of microorganisms to antibiotics, and any contamination from external sources could interfere with the test results and lead to misleading interpretations [41,42,43].
Although the great advantages of lab on a chip are evident, this alternative also presents some limitations. Despite the various benefits indicated for polydimethylsiloxane, several disadvantages have been reported. Its main restriction in the cellular sector is its ability to absorb tiny hydrophobic molecules or biomolecules, such as proteins, which can interfere with assay results [14,41]. Therefore, to avoid these events, methods that change the surface of polydimethylsiloxane are being established [42]. On the other hand, there is a clear lack of established protocols in a lab on a chip-based investigation [44]. Microorganisms such as S. aureus are mostly observed through microscopy techniques, which necessitate the use of reporter-labeled bacteria and/or specialized microscope equipment [14,45]. This limits lab-on-a-chip experiments to visual inspection and model microorganisms, and it is evident that molecular characterization is lacking [14]. However, as was described in this systematic review, the elaboration of droplet-based technologies has resulted in significant progress in this sector, allowing for the study of S. aureus at the single-cell level [26].
This systematic review has several shortcomings as well. The existing evidence is based on a few in vitro experiments that may have therapeutic applications. Besides, these studies had different objectives, designs, and methodologies, giving them great heterogeneity. The included investigations, on the other hand, exhibited a low degree of risk and have conceivable applicability in antibiotic susceptibility testing.
Antimicrobial susceptibility testing through lab-on-a-chip systems offers several advantages, such as rapid, miniaturized, and automated testing [41,42,43,44]. However, there are also challenges and perspectives that need to be considered for the successful implementation and widespread adoption of this technology. One of the significant challenges is the need for standardization of antimicrobial susceptibility testing protocols on lab-on-a-chip platforms [41,43]. Ensuring consistency in testing methods and interpretation of results is essential for reliable and comparable data across different systems and laboratories. Preparing samples for testing on a lab-on-a-chip system can be challenging, especially when dealing with complex clinical samples containing multiple pathogens or heterogeneous populations of microorganisms. Sample preparation techniques must be optimized to ensure accurate results [41,44]. Effective antimicrobial susceptibility testing requires the integration of multiple parameters, such as bacterial growth, cell viability, and specific markers of antibiotic resistance. Integrating different sensors and analytical techniques into a single chip while maintaining sensitivity and specificity can be demanding. To address the growing problem of multi-drug resistance, lab-on-a-chip systems need to incorporate multiplexing capabilities to assess the susceptibility of multiple antibiotics simultaneously [41,42]. Developing multiplexed assays on a small chip can be technically challenging. Handling the vast amount of data generated by lab-on-a-chip systems can be overwhelming. The development of automated data analysis algorithms and user-friendly interfaces is crucial to ensure accessible and actionable results [41,42,43,44]. In short, lab-on-a-chip technology offers numerous advantages, especially in terms of miniaturization, speed, and integration of lab processes. However, it also comes with challenges related to fabrication complexity, material compatibility, and scalability [46,47,48]. The choice to implement lab-on-a-chip technology should be based on the specific needs and constraints of the application or research project.
Future Perspective
The future perspective of lab-on-a-chip technology in the clinical field is promising, and it holds the potential to transform various aspects of healthcare and diagnostics:
- Point-of-Care Diagnostics: lab-on-a-chip devices are likely to become integral for point-of-care diagnostics. They can enable rapid and accurate testing at or near the patient’s location, reducing the need for centralized laboratories and providing quicker results for timely decision-making in emergency situations and routine healthcare [47].
- Personalized Medicine: lab-on-a-chip technology could play a crucial role in personalized medicine by allowing for the rapid and cost-effective analysis of an individual’s genetic makeup, enabling tailored treatment plans and medication choices based on a patient’s unique genetic profile [49].
- Early Disease Detection: The sensitivity and specificity of lab-on-a-chip devices make them ideal for early disease detection. They can detect biomarkers associated with diseases like cancer, diabetes, and infectious diseases at their earliest stages, increasing the chances of successful treatment and reducing healthcare costs [47].
- Remote Monitoring: Miniaturized sensors and microfluidic devices can be integrated into wearable or implantable devices, facilitating continuous monitoring of patients’ health parameters. This real-time data can be sent to healthcare providers, enabling proactive interventions and better management of chronic conditions [50].
- Drug Development: lab-on-a-chip technology can streamline drug development processes by allowing for high-throughput screening of potential drug candidates and the study of their effects on cells or tissues. This can accelerate the development of new drugs and therapies [51].
- Telemedicine and Telehealth: The portability of lab-on-a-chip devices makes them suitable for telemedicine and remote health consultations. Patients in remote or underserved areas can access high-quality diagnostics and medical advice through telehealth platforms [43,47].
- Reduced Healthcare Costs: By enabling early diagnosis, targeted treatments, and efficient monitoring, lab-on-a-chip technology can potentially reduce overall healthcare costs by preventing expensive late-stage interventions and hospitalizations [43,48].
- Global Health Impact: In resource-limited settings and developing countries, lab-on-a-chip devices can provide affordable and accessible diagnostic tools for diseases, helping to address global health challenges [52].
- Advanced Imaging and Analysis: Future lab-on-a-chip systems may incorporate advanced imaging and analysis techniques, such as artificial intelligence and machine learning, to enhance the accuracy and speed of diagnoses and data interpretation [53].
Research in the clinical assessment of S. aureus susceptibility using lab-on-a-chip technology has the potential to significantly impact the diagnosis and treatment of S. aureus infections. As observed in the current systematic review, lab-on-a-chip technology can be used to perform rapid AST for S. aureus isolates. Traditional AST methods can take 24–48 h [13,14,15], delaying the initiation of appropriate antibiotic therapy. Lab-on-a-chip can provide results within hours, allowing healthcare providers to select the most effective antibiotic sooner and reducing the risk of treatment failure and the spread of antibiotic-resistant strains. In addition to these promising results, lab-on-a-chip has other applications in clinical research on S. aureus. Lab-on-a-chip can be designed to identify the specific strain or type of S. aureus, including MRSA. Rapid and accurate identification is crucial for selecting the appropriate antibiotics, as MRSA is often resistant to common antibiotics [28]. Lab-on-a-chip technology can be used in surveillance efforts to monitor antibiotic resistance trends in S. aureus [54]. This data can help healthcare institutions and public health agencies track the prevalence of antibiotic-resistant strains, guiding antibiotic stewardship programs and infection control measures. Miniaturized lab-on-a-chip devices can also be used at the point of care, such as in clinics or emergency departments, to quickly assess S. aureus susceptibility [55]. This can lead to more targeted and effective treatment decisions, especially in cases of severe infections. Moreover, by rapidly determining the susceptibility of S. aureus to various antibiotics, lab-on-a-chip technology can support personalized treatment plans for patients [49]. This can optimize antibiotic selection and dosing, reducing the risk of adverse effects and improving patient outcomes. Lab-on-a-chip technology can aid in the development of novel antibiotics and therapies for S. aureus infections [27]. Researchers can use microfluidic devices to test the efficacy of new drug candidates and study the mechanisms of antibiotic resistance. In summary, research in the clinical assessment of S. aureus susceptibility using lab-on-a-chip technology has practical applications that can enhance the diagnosis and treatment of S. aureus infections, reduce antibiotic resistance, and improve patient care in various healthcare settings. It represents a promising approach to addressing the challenges posed by S. aureus and antibiotic resistance in clinical practice.
5. Conclusions
The potential use of lab-on-a-chip technology for assessing the antibiotic susceptibility of S. aureus is highlighted by its capability to quickly provide essential minimum inhibitory concentration data. This data can greatly assist in the selection of the most appropriate antibiotics and their dosages for addressing complex infections caused by this microorganism. Furthermore, the swift antibiotic susceptibility testing made possible by lab-on-a-chip holds great potential for improving the suitability and effectiveness of treatment in clinical environments. It increases efficiency and conserves both time and resources.
Author Contributions
Conceptualization, C.M.A.; methodology, C.M.A., M.Z.-G. and A.M.V.-B.; validation, C.M.A., M.Z.-G. and A.M.V.-B.; formal analysis, C.M.A. and A.M.V.-B.; investigation, C.M.A., M.Z.-G. and A.M.V.-B.; resources, C.M.A., M.Z.-G. and A.M.V.-B.; data curation, C.M.A. and A.M.V.-B.; writing—original draft preparation, C.M.A.; writing—review and editing, C.M.A., M.Z.-G. and A.M.V.-B.; supervision, C.M.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
This study did not involve humans.
Data Availability Statement
The data obtained in this review were pooled from the included investigations.
Acknowledgments
To the Universidad de Antioquia, Pontificia Universidad Bolivariana and Institución Universitaria Visión de Las Américas.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Aslam, B.; Khurshid, M.; Arshad, M.I.; Muzammil, S.; Rasool, M.; Yasmeen, N.; Shah, T.; Chaudhry, T.H.; Rasool, M.H.; Shahid, A.; et al. Antibiotic Resistance: One Health One World Outlook. Front. Cell. Infect. Microbiol. 2021, 11, 771510. [Google Scholar] [CrossRef] [PubMed]
- Ferri, M.; Ranucci, E.; Romagnoli, P.; Giaccone, V. Antimicrobial resistance: A global emerging threat to public health systems. Crit. Rev. Food Sci. Nutr. 2017, 57, 2857–2876. [Google Scholar] [CrossRef] [PubMed]
- Uddin, T.M.; Chakraborty, A.J.; Khusro, A.; Zidan, B.R.M.; Mitra, S.; Bin Emran, T.; Dhama, K.; Ripon, K.H.; Gajdács, M.; Sahibzada, M.U.K.; et al. Antibiotic resistance in microbes: History, mechanisms, therapeutic strategies and future prospects. J. Infect. Public Health 2021, 14, 1750–1766. [Google Scholar] [CrossRef] [PubMed]
- Hernando-Amado, S.; Coque, T.M.; Baquero, F.; Martínez, J.L. Defining and combating antibiotic resistance from One Health and Global Health perspectives. Nat. Microbiol. 2019, 4, 1432–1442. [Google Scholar] [CrossRef] [PubMed]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. WHO Pathogens Priority List Working Group. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Song, G.; Sun, M.; Wang, J.; Wang, Y. Prevalence and Therapies of Antibiotic-Resistance in Staphylococcus aureus. Front. Cell. Infect. Microbiol. 2020, 10, 107. [Google Scholar] [CrossRef] [PubMed]
- Silva-de-Jesus, A.C.; Ferrari, R.G.; Panzenhagen, P.; Conte-Junior, C.A. Staphylococcus aureus biofilm: The role in disseminating antimicrobial resistance over the meat chain. Microbiology 2022, 168, 001245. [Google Scholar]
- Lewis, N.; Leaptrot, D.; Witt, E.; Smith, H.; Hebden, J.N.; Wright, M.O. Health Care-Associated Infections Studies Project: An American Journal of Infection Control and National Healthcare Safety Network Data Quality Collaboration Case Study—Laboratory-Identified Event Reporting Validation. Am. J. Infect Control. 2023, 51, 1172–1174. [Google Scholar] [CrossRef]
- Sydnor, E.R.; Perl, T.M. Hospital epidemiology and infection control in acute-care settings. Clin. Microbiol. Rev. 2011, 24, 141–173. [Google Scholar] [CrossRef]
- Leekha, S.; Terrell, C.L.; Edson, R.S. General principles of antimicrobial therapy. Mayo Clin. Proc. 2011, 86, 156–167. [Google Scholar] [CrossRef]
- Braykov, N.P.; Morgan, D.J.; Schweizer, M.L.; Uslan, D.Z.; Kelesidis, T.; A Weisenberg, S.; Johannsson, B.; Young, H.; Cantey, J.; Srinivasan, A.; et al. Assessment of empirical antibiotic therapy optimisation in six hospitals: An observational cohort study. Lancet Infect. Dis. 2014, 14, 1220–1227. [Google Scholar] [CrossRef] [PubMed]
- Renner, L.D.; Zan, J.; Hu, L.I.; Martinez, M.; Resto, P.J.; Siegel, A.C.; Torres, C.; Hall, S.B.; Slezak, T.R.; Nguyen, T.H.; et al. Detection of ESKAPE Bacterial Pathogens at the Point of Care Using Isothermal DNA-Based Assays in a Portable Degas-Actuated Microfluidic Diagnostic Assay Platform. Appl. Environ. Microbiol. 2017, 83, e02449-16. [Google Scholar] [CrossRef] [PubMed]
- Balouiri, M.; Sadiki, M.; Ibnsouda, S.K. Methods for in vitro evaluating antimicrobial activity: A review. J. Pharm. Anal. 2016, 6, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Rodríguez, S.; García-Aznar, J.M.; Gonzalo-Asensio, J. Microfluidic devices for studying bacterial taxis, drug testing and biofilm formation. Microb. Biotechnol. 2022, 15, 395–414. [Google Scholar] [CrossRef] [PubMed]
- Bodor, A.; Bounedjoum, N.; Vincze, G.E.; Kis, E.; Laczi, K.; Bende, G.; Szilágyi, Á.; Kovács, T.; Perei, K.; Rákhely, G. Challenges of unculturable bacteria: Environmental perspectives. Rev. Environ. Sci. Biotechnol. 2020, 19, 1–22. [Google Scholar] [CrossRef]
- Lebeaux, D.; Chauhan, A.; Rendueles, O.; Beloin, C. From in vitro to in vivo Models of Bacterial Biofilm-Related Infections. Pathogens 2013, 2, 288–356. [Google Scholar] [CrossRef] [PubMed]
- Postek, W.; Pacocha, N.; Garstecki, P. Microfluidics for antibiotic susceptibility testing. Lab. Chip. 2022, 22, 3637–3662. [Google Scholar] [CrossRef]
- Kaprou, G.D.; Bergšpica, I.; Alexa, E.A.; Alvarez-Ordóñez, A.; Prieto, M. Rapid Methods for Antimicrobial Resistance Diagnostics. Antibiotics 2021, 10, 209. [Google Scholar] [CrossRef]
- Lee, W.B.; Chien, C.C.; You, H.L.; Kuo, F.C.; Lee, M.S.; Lee, G.B. An integrated microfluidic system for antimicrobial susceptibility testing with antibiotic combination. Lab Chip 2019, 19, 2699–2708. [Google Scholar] [CrossRef]
- Boedicker, J.Q.; Li, L.; Kline, T.R.; Ismagilov, R.F. Detecting bacteria and determining their susceptibility to antibiotics by stochastic confinement in nanoliter droplets using plug-based microfluidics. Lab Chip 2008, 8, 1265–1272. [Google Scholar] [CrossRef]
- Hou, H.W.; Bhattacharyya, R.P.; Hung, D.T.; Han, J. Direct detection and drug-resistance profiling of bacteremias using inertial microfluidics. Lab Chip 2015, 15, 2297–2307. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Liu, Z.; Hu, C.; Ren, K. Cell-on-hydrogel platform made of agar and alginate for rapid, low-cost, multidimensional test of antimicrobial susceptibility. Lab Chip 2016, 16, 3130–3138. [Google Scholar] [CrossRef] [PubMed]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G.; PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA Statement. Open Med. 2009, 3, e123–e130. [Google Scholar] [PubMed]
- Ehsani, S.; Mandich, M.A.; El-Bialy, T.H.; Flores-Mir, C. Frictional resistance in self-ligating orthodontic brackets and conventionally ligated brackets: A systematic review. Angle Orthod. 2009, 79, 592–601. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.B.; Chien, C.C.; You, H.L.; Kuo, F.C.; Lee, M.S.; Lee, G.B. Rapid antimicrobial susceptibility tests on an integrated microfluidic device for precision medicine of antibiotics. Biosens. Bioelectron. 2021, 176, 112890. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.-K.; Cheng, H.-W.; Liao, C.-C.; Lin, S.-J.; Chen, Y.-Z.; Wang, J.-K.; Wang, Y.-L.; Huang, N.-T. Bacteria encapsulation and rapid antibiotic susceptibility test using a microfluidic microwell device integrating surface-enhanced Raman scattering. Lab Chip 2020, 20, 2520–2528. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, A.; Torres, N.S.; Leung, K.P.; Lopez-Ribot, J.L.; Ramasubramanian, A.K. nBioChip, a Lab-on-a-Chip Platform of Mono- and Polymicrobial Biofilms for High-Throughput Downstream Applications. mSphere 2017, 2, e00247-17. [Google Scholar] [CrossRef]
- Abeyrathne, C.D.; Huynh, D.H.; Mcintire, T.W.; Nguyen, T.C.; Nasr, B.; Zantomio, D.; Chana, G.; Abbott, I.; Choong, P.; Catton, M.; et al. Lab on a chip sensor for rapid detection and antibiotic resistance determination of Staphylococcus aureus. Analyst 2016, 141, 1922–1929. [Google Scholar] [CrossRef]
- Hou, Z.; An, Y.; Hjort, K.; Hjort, K.; Sandegren, L.; Wu, Z. Time lapse investigation of antibiotic susceptibility using a microfluidic linear gradient 3D culture device. Lab Chip 2014, 14, 3409–3418. [Google Scholar] [CrossRef]
- Choi, J.; Jung, Y.-G.; Kim, J.; Kim, S.; Jung, Y.; Na, H.; Kwon, S. Rapid antibiotic susceptibility testing by tracking single cell growth in a microfluidic agarose channel system. Lab Chip 2013, 13, 280–287. [Google Scholar] [CrossRef]
- Kalashnikov, M.; Lee, J.C.; Campbell, J.; Sharon, A.; Sauer-Budge, A.F. A microfluidic platform for rapid, stress-induced antibiotic susceptibility testing of Staphylococcus aureus. Lab Chip 2012, 12, 4523–4532, Erratum in Lab Chip 2013, 13, 4890. [Google Scholar] [CrossRef] [PubMed]
- Wistrand-Yuen, P.; Malmberg, C.; Fatsis-Kavalopoulos, N.; Lübke, M.; Tängdén, T.; Kreuger, J. A Multiplex Fluidic Chip for Rapid Phenotypic Antibiotic Susceptibility Testing. mBio 2020, 11, e03109-19. [Google Scholar] [CrossRef] [PubMed]
- Spencer, D.C.; Paton, T.F.; Mulroney, K.T.; Inglis, T.J.J.; Sutton, J.M.; Morgan, H. A fast impedance-based antimicrobial susceptibility test. Nat. Commun. 2020, 11, 5328. [Google Scholar] [CrossRef] [PubMed]
- Yi, Q.; Cai, D.; Xiao, M.; Nie, M.; Cui, Q.; Cheng, J.; Li, C.; Feng, J.; Urban, G.; Xu, Y.C.; et al. Direct antimicrobial susceptibility testing of bloodstream infection on SlipChip. Biosens. Bioelectron. 2019, 135, 200–207. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Chan, C.W.; Wang, Y.; Yao, X.; Mu, X.; Lu, X.; Zhou, J.; Cai, Z.; Ren, K. Reliable and reusable whole polypropylene plastic microfluidic devices for a rapid, low-cost antimicrobial susceptibility test. Lab Chip 2019, 19, 2915–2924. [Google Scholar] [CrossRef] [PubMed]
- Kang, W.; Sarkar, S.; Lin, Z.S.; McKenney, S.; Konry, T. Ultrafast Parallelized Microfluidic Platform for Antimicrobial Susceptibility Testing of Gram Positive and Negative Bacteria. Anal. Chem. 2019, 91, 6242–6249. [Google Scholar] [CrossRef] [PubMed]
- Hou, Z.; An, Y.; Wu, Z. Dynamic Antibiotic Susceptibility Test via a 3D Microfluidic Culture Device. Methods Mol. Biol. 2017, 1572, 365–377. [Google Scholar] [PubMed]
- Malmberg, C.; Yuen, P.; Spaak, J.; Cars, O.; Tängdén, T.; Lagerbäck, P. A Novel Microfluidic Assay for Rapid Phenotypic Antibiotic Susceptibility Testing of Bacteria Detected in Clinical Blood Cultures. PLoS ONE 2016, 11, e0167356. [Google Scholar] [CrossRef]
- CLSI. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically; CLSI Document M07-A11; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
- Desmet, S.; Verhaegen, J.; Glupzcynski, Y.; Van Eldere, J.; Melin, P.; Goossens, H.; Piérard, D.; Declercq, P.; Lagrou, K.; Boel, A.; et al. Development of a national EUCAST challenge panel for antimicrobial susceptibility testing. Clin. Microbiol. Infect. 2016, 22, 704–710. [Google Scholar] [CrossRef]
- Halldorsson, S.; Lucumi, E.; Gómez-Sjöberg, R.; Fleming, R.M.T. Advantages and challenges of microfluidic cell culture in polydimethylsiloxane devices. Biosens. Bioelectron. 2015, 63, 218–231. [Google Scholar] [CrossRef]
- You, J.B.; Lee, B.; Choi, Y.; Lee, C.-S.; Peter, M.; Im, S.G.; Lee, S.S. Nanoadhesive layer to prevent protein absorption in a poly(dimethylsiloxane) microfluidic device. Biotechniques 2020, 69, 404–409. [Google Scholar] [CrossRef] [PubMed]
- Daniel, F.; Kesterson, D.; Lei, K.; Hord, C.; Patel, A.; Kaffenes, A.; Congivaram, H.; Prakash, S. Application of Microfluidics for Bacterial Identification. Pharmaceuticals 2022, 15, 1531. [Google Scholar] [CrossRef] [PubMed]
- Ardila, C.M.; Jiménez-Arbeláez, G.A.; Vivares-Builes, A.M. Potential Clinical Application of Organs-on-a-Chip in Periodontal Diseases: A Systematic Review of In Vitro Studies. Dent. J. 2023, 11, 158. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, S.; Huiszoon, R.C.; Chu, S.; Bentley, W.E.; Ghodssi, R. Microsystems for biofilm characterization and sensing—A review. Biofilm 2019, 2, 100015. [Google Scholar] [CrossRef] [PubMed]
- Ezrre, S.; Reyna, M.A.; Anguiano, C.; Avitia, R.L.; Márquez, H. Lab-on-a-Chip Platforms for Airborne Particulate Matter Applications: A Review of Current Perspectives. Biosensors 2022, 24, 191. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, J.; Yin, J.; Lv, S.; Wang, B.; Mu, Y. Advanced “lab-on-a-chip” to detect viruses—Current challenges and future perspectives. Biosens. Bioelectron. 2020, 163, 112291. [Google Scholar] [CrossRef] [PubMed]
- Cunha, M.L.; da Silva, S.S.; Stracke, M.C.; Zanette, D.L.; Aoki, M.N.; Blanes, L. Sample Preparation for Lab-on-a-Chip Systems in Molecular Diagnosis: A Review. Anal. Chem. 2022, 94, 41–58. [Google Scholar] [CrossRef]
- Ingber, D.E. Human organs-on-chips for disease modelling, drug development and personalized medicine. Nat. Rev. Genet. 2022, 23, 467–491. [Google Scholar] [CrossRef]
- Zhang, C.; Su, Y.; Hu, S.; Jin, K.; Jie, Y.; Li, W.; Nathan, A.; Ma, H. An Impedance Sensing Platform for Monitoring Heterogeneous Connectivity and Diagnostics in Lab-on-a-Chip Systems. ACS Omega. 2020, 5, 5098–5104. [Google Scholar] [CrossRef]
- Weigl, B.H.; Bardell, R.L.; Cabrera, C.R. Lab-on-a-chip for drug development. Adv. Drug. Deliv. Rev. 2003, 55, 349–377. [Google Scholar] [CrossRef]
- Gebauer, A.; Schmidt, S.; Hoffmann, W. Status and perspective of lab-on-a-chip systems for common diseases: A systematic review from 2003 to 2013. Pers. Med. 2016, 13, 71–91. [Google Scholar] [CrossRef]
- Vargas-Ordaz, E.J.; Gorelick, S.; York, H.M.; Liu, B.; Halls, M.L.; Arumugam, S.; Neild, A.; de Marco, A.; Cadarso, V.J. Three-dimensional imaging on a chip using optofluidics light-sheet fluorescence microscopy. Lab Chip 2021, 21, 2945–2954. [Google Scholar] [CrossRef]
- Peter, H.; Wienke, J.; Bier, F.F. Lab-on-a-Chip Multiplex Assays. Methods Mol. Biol. 2017, 1546, 283–294. [Google Scholar]
- Schulz, M.; Calabrese, S.; Hausladen, F.; Wurm, H.; Drossart, D.; Stock, K.; Sobieraj, A.M.; Eichenseher, F.; Loessner, M.J.; Schmelcher, M.; et al. Point-of-care testing system for digital single cell detection of MRSA directly from nasal swabs. Lab Chip 2020, 20, 2549–2561. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).