Simple Summary
Phenylpropanoids are specialized metabolites playing crucial roles in plant developmental processes and in plant defense towards pathogens. The attachment of sugar moieties to these small hydrophobic molecules renders them more hydrophilic and increases their solubility. The UDP-glycosyltransferase 72 family (UGT72) of plants has been shown to glycosylate mainly two classes of phenylpropanoids, (i) the monolignols that are the building blocks of lignin, the second most abundant polymer after cellulose, and (ii) the flavonoids, which play determinant roles in plant interactions with other organisms and in response to stress. The purpose of this review is to bring an overview of the current knowledge of the UGT72 family and to highlight its role in the homeostasis of these molecules. Potential applications in pharmacology and in wood, paper pulp, and bioethanol production are given within the perspectives.
Abstract
Plants have developed the capacity to produce a diversified range of specialized metabolites. The glycosylation of those metabolites potentially decreases their toxicity while increasing their stability and their solubility, modifying their transport and their storage. The UGT, forming the largest glycosyltransferase superfamily in plants, combine enzymes that glycosylate mainly hormones and phenylpropanoids by using UDP-sugar as a sugar donor. Particularly, members of the UGT72 family have been shown to glycosylate the monolignols and the flavonoids, thereby being involved in their homeostasis. First, we explore primitive UGTs in algae and liverworts that are related to the angiosperm UGT72 family and their role in flavonoid homeostasis. Second, we describe the role of several UGT72s glycosylating monolignols, some of which have been associated with lignification. In addition, the role of other UGT72 members that glycosylate flavonoids and are involved in the development and/or stress response is depicted. Finally, the importance to explore the subcellular localization of UGTs to study their roles in planta is discussed.
1. Introduction
In order to adapt to terrestrial environments and biotic interactions, plants have developed the capacity to produce a wide range of specialized compounds. These compounds can undergo various chemical modifications, such as hydroxylation, methylation, or glycosylation. This capacity has been allowed by the diversification of enzymes able to add new functional groups to molecules. Among these modifications, the glycosylation reaction can occur on –OH, –COOH, –NH2, –SH, and C–C groups in diverse molecules such as proteins, carbohydrates, primary and specialized metabolites, as well as xenobiotics [1]. Glycosylation reaction can be carried out with one or several sugars (oligosaccharides) moieties from activated donors, which generate a great diversity of glycosylated compounds. This conjugation reduces the toxicity of the substrate and increases its stability and its solubility owing to the high polarity of the sugar moiety [1]. Glycosylated compounds are stored mainly in the vacuole and their activity may be restored by a single deglycosylation step when required [2,3]. In addition, glycosylation of molecules can modify their interaction with signal receptors, transport proteins, or degradation systems [4,5].
The ubiquitous glycosyltransferases (GTs) are classified in 114 superfamilies on the basis of the similarity of their amino acid sequences [6,7]. The proportion of GT genes in each GT superfamily differs among organisms in function of the peculiarities of their metabolism [8]. GT1, comprising UGTs, is the largest GT superfamily in plants and clusters enzymes glycosylating low molecular weight molecules and making a β-bond using UDP-sugar as sugar donor. UGTs represent about 25% and 35% of GT genes in Arabidopsis thaliana and in Oryza sativa, respectively [8]. The diversity of UGTs allows plants to produce and regulate a myriad of specialized metabolites [9,10].
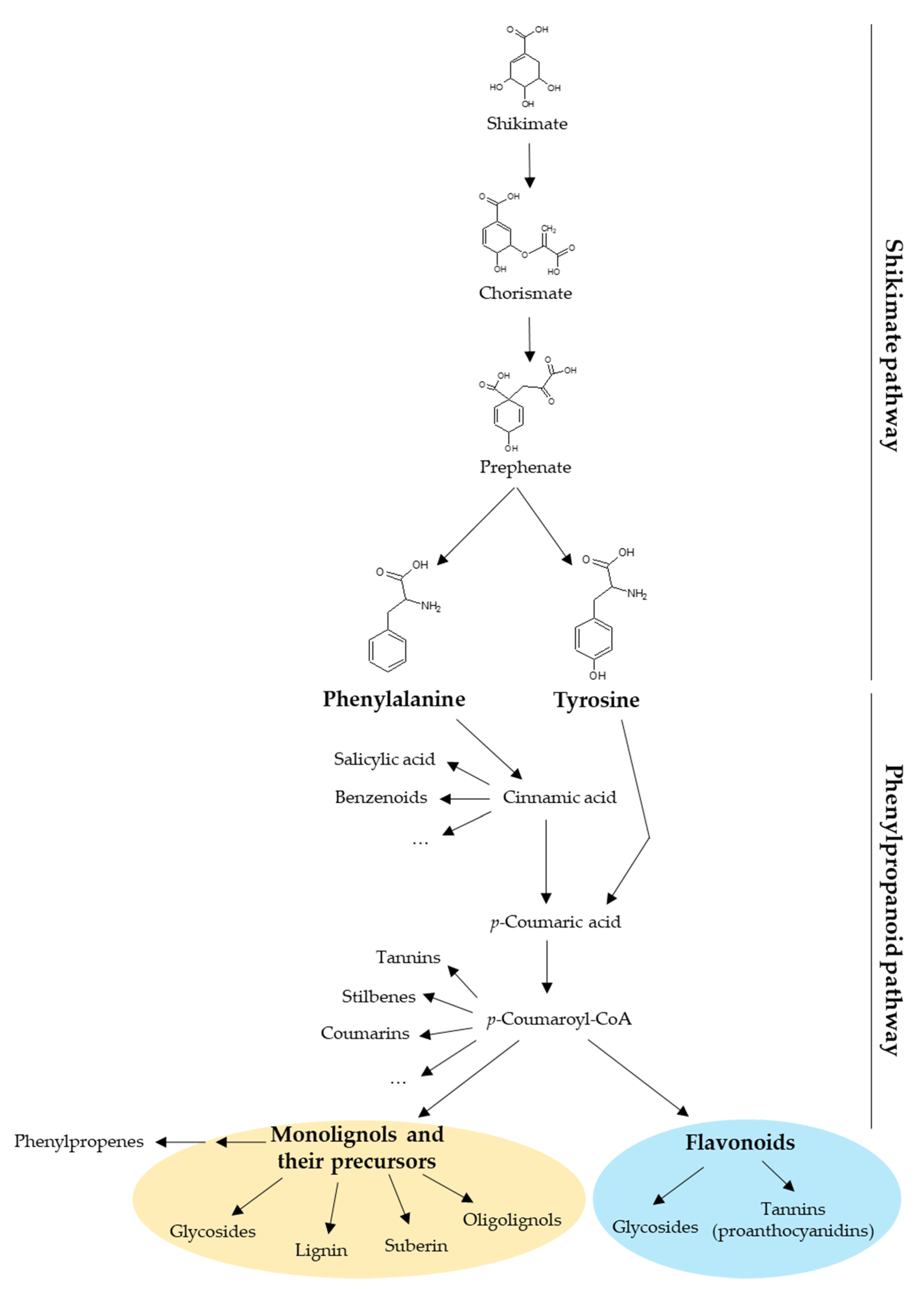
Plant UGTs can glycosylate phytohormones such as abscisic acid, auxins, brassinosteroids, and salicylic acid, as well as terpenoids. However, most of their substrates derive from the phenylpropanoid pathway whose precursors are the aromatic amino acids phenylalanine and tyrosine (Figure 1) [11]. Besides being building blocks for proteins, these two amino acids are also precursors of a large number of plant products with specific functions in plant growth, development, and stress response [12,13]. Phenylalanine and tyrosine are generated by the shikimate pathway in bacteria, fungi, and plants [12,14]. In plants, the shikimate pathway takes place mainly in the chloroplastic compartment and the produced amino acids are exported to the cytosol [12,13]. The first step of the phenylpropanoid pathway is the deamination of phenylalanine by the phenylalanine ammonia-lyase (PAL) producing cinnamic acid. The second step is the hydroxylation of cinnamic acid at the C4 position (aromatic nucleus) by cinnamate 4-hydroxylase (C4H) leading to p-coumaric acid [15]. Alternatively, tyrosine can be deaminated by bifunctional phenylalanine/tyrosine ammonia-lyase (PTAL) in some plant species, which produces both cinnamic acid from phenylalanine and p-coumaric acid from tyrosine [16]. During the third step, a coenzyme A (CoA) is added to p-coumaric acid by a 4-coumarate: CoA ligase (4CL) to form p-coumaroyl-CoA [15,17,18], which is the last common precursor of monolignols, stilbenes, coumarins, and flavonoids (Figure 1) [19]. In addition, phenylpropenes are synthetized by acetylation and subsequent reduction of monolignols [20]. All these compounds may be glycosylated by UGTs [11]. Monolignol biosynthesis and polymerization have been extensively studied because they are the main components of lignin [2]. In that context, monolignol glycosylation by UGTs may regulate their flux to the cell wall and therefore lignification.
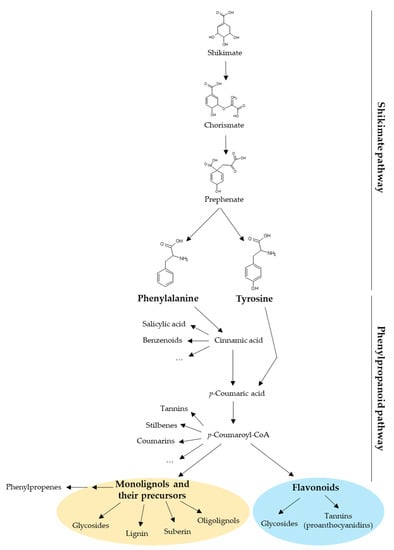
Figure 1.
Diversity of compounds produced by the shikimate and the phenylpropanoid pathways in plants. The shikimate pathway yields phenylalanine and tyrosine, which are the precursors of phenylpropanoids. This review focuses on two phenylpropanoid families: the monolignols (yellow circle) and the flavonoids (blue circle). CoA, coenzyme A.
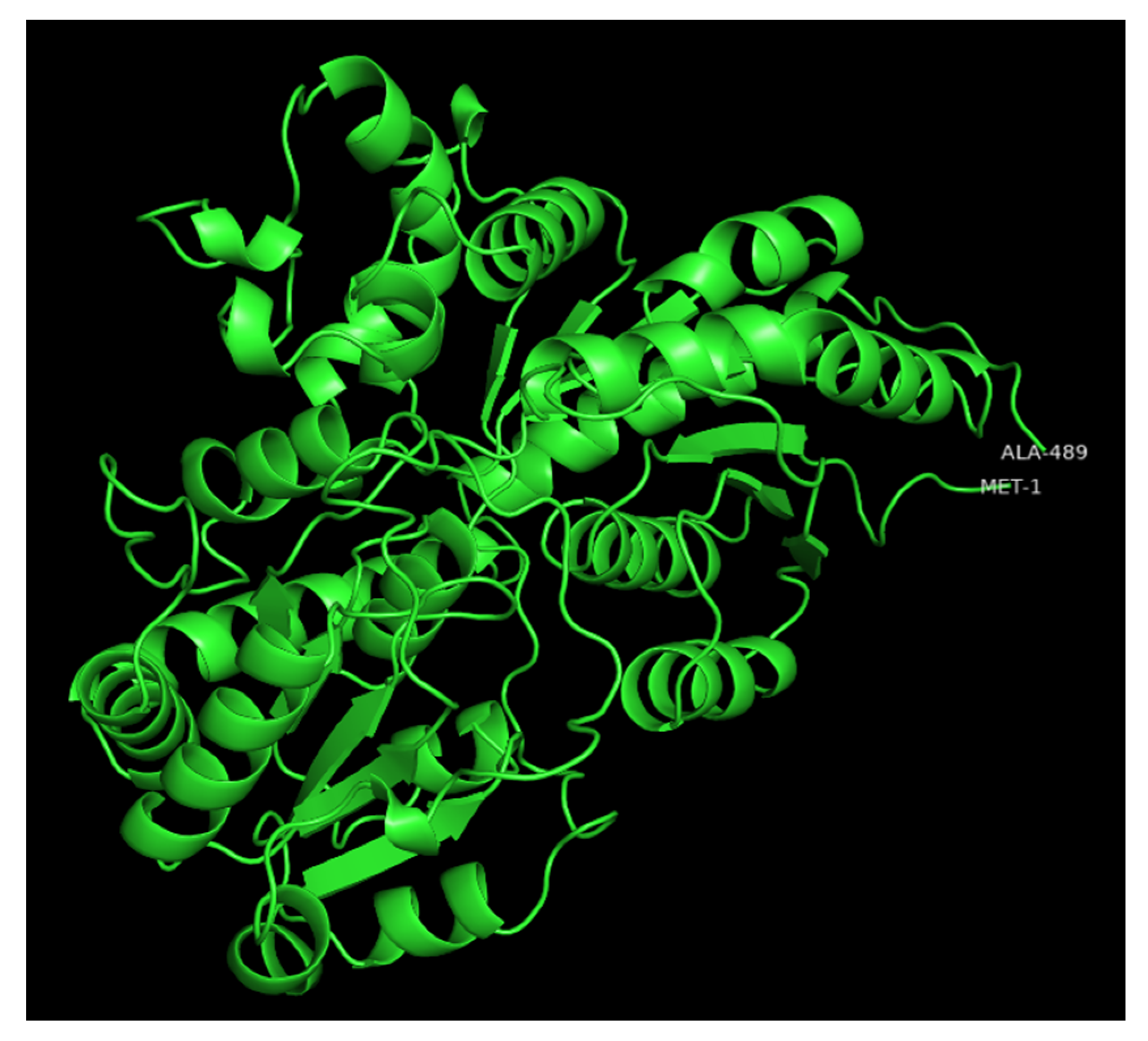
UGTs are classified into families in which members share more than 45% of amino acid sequence identity [21]. However, the prediction of substrate specificity based on primary sequence is complex as divergent families can recognize common substrates and closely related UGTs can have different substrate affinities [5,22]. X-ray crystal three-dimensional (3D) structures of a number of plant UGTs have been reported [23,24] and show that their secondary and tertiary structures are conserved [5,25,26]. As an example of the various regions of UGTs, a modeled 3D structure of the poplar UGT72A2 [27] is depicted in Figure 2.
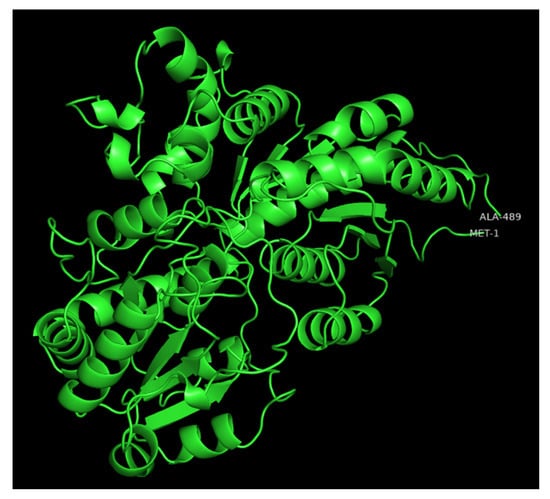
Figure 2.
Modeled 3D structure of poplar UGT72A2 obtained with the intensive mode of the Phyre2 web portal [28] and visualized with PyMOL2. The first and last residues are indicated on the polypeptide chain.
Plant UGTs display a typical GT-B fold which consists of two Rossman fold domains connected by a linker and forming a cleft [7]. The C-terminal domain contains a conserved motif involved in most of the interactions with the UDP-sugar donor. In plants, this consensus sequence is named plant secondary product glycosyltransferase (PSPG) domain and is composed of 44 amino acids [29] whose 10 have a well-known role in the fixation of the UDP-sugar, mainly UDP-glucose [5,30,31,32,33]. In contrast, the acceptor molecule binding site, mainly composed of N-terminal residues, is more variable than the C-terminal part [21]. Different amino acids maintain a hydrophobic environment in the pocket and form a deep narrow cleft. For instance, in the UGT72B1 structure, the substrate is enclosed by six hydrophobic residues (I-86, L-118, F-119, F-148, L-183, and L-197), in addition to E-83 which closes the catalytic site [33]. The correct positioning of the substrate in the pocket is crucial for its glycosylation. The functional group of the sugar acceptor must indeed be close to the first carbon of the sugar, where the link has to be made [5,34]. According to the UGT72B1 model, the H-19 residue is positioned near the sugar acceptor to act as a Brønsted base and deprotonates it before glycosylation, while Q-389 interacts with the glucose moiety of the UDP-sugar [33]. We have recently published a molecular docking summarizing the main interactions between these residues and selected flavonoids using poplar UGT72A2 as a model [27]. According to the opening mechanism of the cleft and the importance of the positioning of the substrates, the physicochemical information about substrates (size, functional group, hydrophobicity, etc.) allows predicting their recognition by UGTs, as well as the regiospecificity of the enzyme [22]. This review focuses on the UGT72 family, whose members are known for their role in the homeostasis of two groups of compounds, the monolignols and the flavonoids (and their derivatives), and thereby on plant development [35,36,37,38,39,40].
2. Diversification of UGTs from Algae to Vascular Plants
About 450 million years ago, embryophytes, deriving from green algae emerged from the aquatic habitat and colonized the terrestrial environment thanks to the acquisition of novel biochemical pathways, phytohormones, and signaling pathways [41,42]. Especially, the number of genes coding for enzymes involved in secondary metabolism through the evolution of embryophytes significantly expanded, notably within the families of cytochromes P450, glycosyl hydrolases, and glycosyltransferases [43].
As shown in Table 1, the number of UGTs increased during evolution, consisting of five in Chlamydomonas reinhardtii to more than 200 in trees such as Pinus taeda (gymnosperm) and Populus trichocarpa (angiosperm). This divergence suggests an early and continuous expansion of the UGT family in plants which may be related to their constant adaptation to the terrestrial environment and biotic interactions by producing and regulating specialized metabolites [9,10]. Across the embryophytes evolution and especially in gymnosperms and angiosperms, several whole-genome and tandem duplication events happened, diversifying some gene families such as UGTs and leading to sub- and neo-functionalization [8,44].

Table 1.
UGTs occurrence in different species from green algae to higher plants.
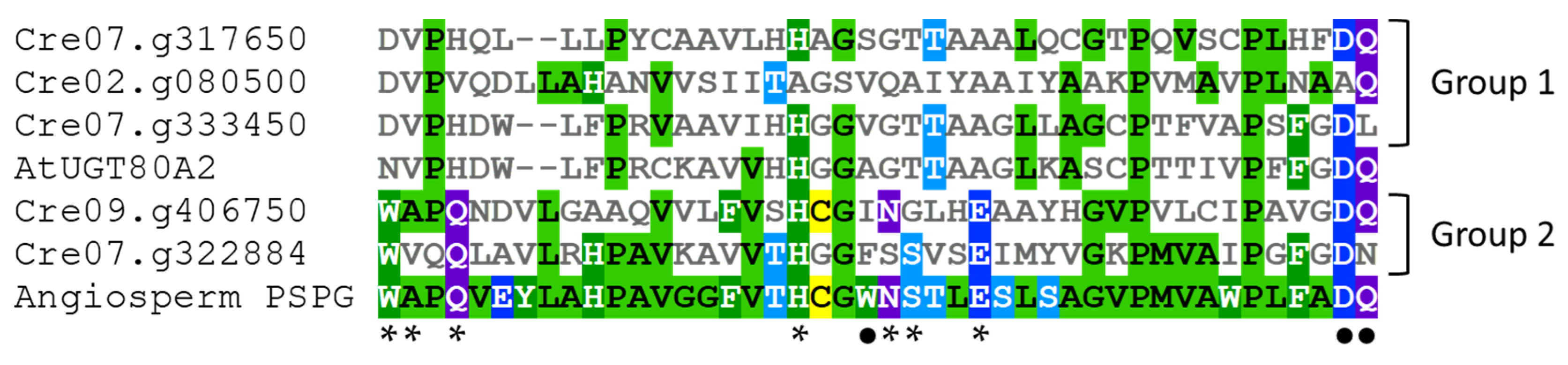
An alignment of the PSPG domain of the five UGTs from C. reinhardtii with the PSPG consensus sequence for angiosperm UGTs (from 246 protein sequences according to [47]) allows distinguishing two UGT groups in green algae (Figure 3). The PSPG sequences of the group 1 (Cre02.g080500, Cre07.g317650 and Cre07.g333450) show few similarities compared to the angiosperm PSPG consensus and, among the 10 residues with a well-known role in UDP-sugar fixation, only H-19, D-43 (for Cre07.g317650 and Cre07.g333450) and Q-44 (for Cre02.g080500 and Cre07.g317650) are conserved. The H-19 is known to bind to UDP phosphates [32], whereas the last D and Q residues make a hydrogen bond with glucose and are involved in UDP-glucose recognition [30,31,33]. Cre07.g333450, named UGT711A1, is close to the angiosperm UGT80 family (Figure 3) and to fungi UGT families (UGT51-UGT57 and UGT60) presenting a sterol β-glucosyltransferase or a putative sterol β-glucosyltransferase activity [48,49], suggesting that this C. reinhardtii UGT recognizes sterols. Glycosylation of sterols is involved in the regulation of the structure and the permeability of the plasma membrane [50,51].
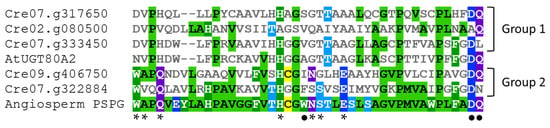
Figure 3.
PSPG sequences alignment of the five C. reinhardtii UGTs with the PSPG sequence of the Arabidopsis UGT80A2 and the PSPG consensus sequence of angiosperm UGTs (according to [44]). The sequences were aligned using MView [53]. Amino acids are colored depending on their similarity. *: well-conserved amino acids with known function on UDP-sugar binding and enzyme conformation [5,31,32,33]. •: well-conserved amino acids with known function on UDP-glucose recognition [30,31].
In contrast, the PSPG sequences of the UGTs of C. reinhardtii from group 2 (Cre07.g322884 and Cre09.g406750) present most of the amino acids required for UDP-sugar fixation. However, together with the members of group 1, these two UGTs lack W-22 (involved in UDP-glucose recognition), suggesting different sugar donor specificities for these enzymes [31,52]. Other variations such as the absence of S-24 in Cre09.g406750 and of A-2, N-23, and Q-44 in Cre07.g322884 reveal some structural and sugar specificity differences in these green algae UGTs as compared to the majority of angiosperm UGTs [31,32]. For instance, the presence of an N as the last residue of the PSPG domain may allow the binding of UDP-rhamnose as demonstrated for the Arabidopsis UGT78D1 [31].
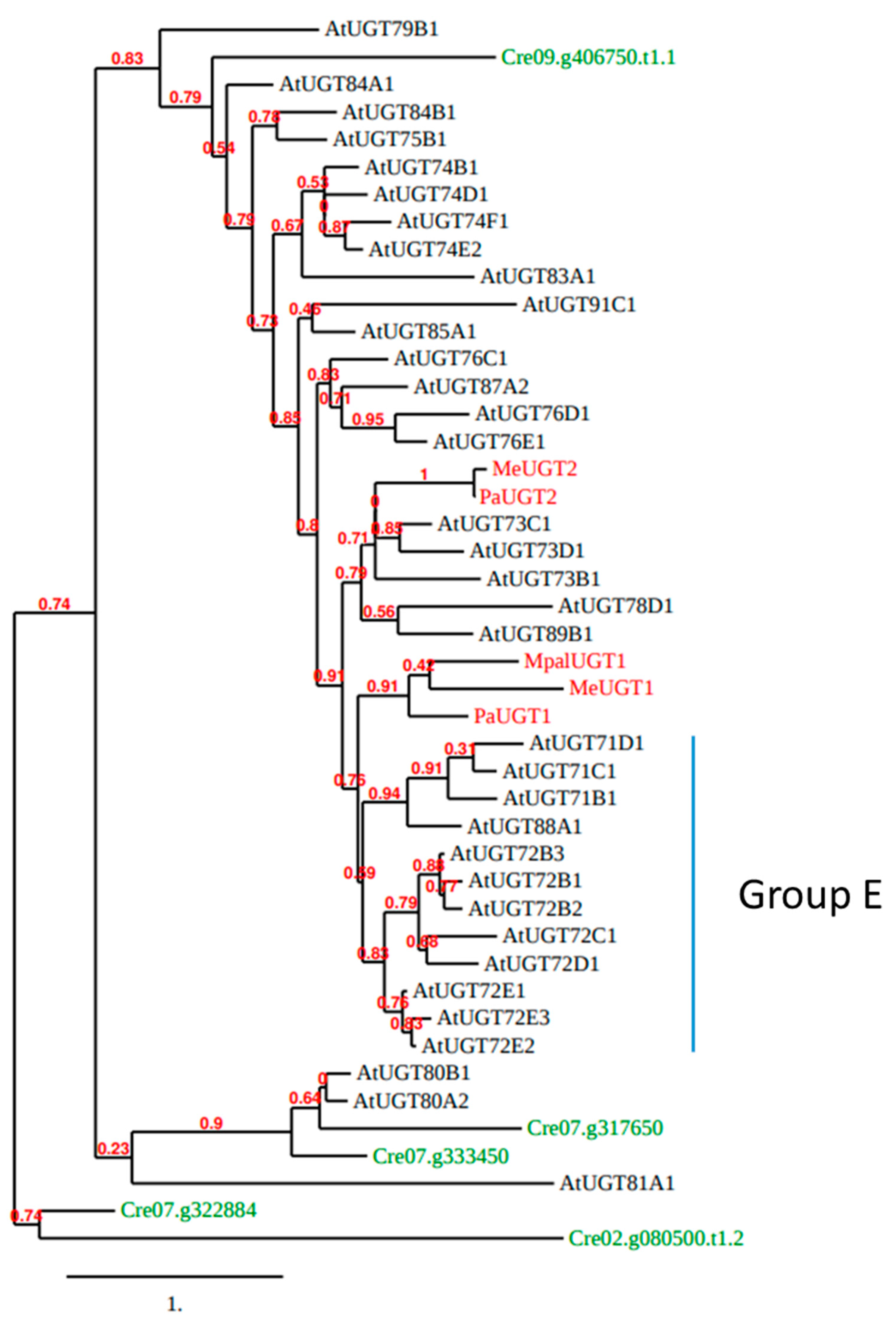
To investigate further the evolution of UGT72s, a phylogenetic analysis of Arabidopsis UGTs was performed (Figure 4). We included one representative of each UGT subfamily, but all UGT72s, as well as the five UGTs of C. reinhardtii and five functionally described UGTs from liverworts (MeUGT1 and MeUGT2 from Marchantia emarginata, MpalUGT1 from M. paleacea, as well as PaUGT1 and PaUGT2 from Plagiochasma appendiculatum) [54,55]. According to Figure 4, Cre07.g333450 and Cre07.g317650 (Group 1 from Figure 3) are clustered with Arabidopsis UGT80s and Cre09.g406750 (Group 2 from Figure 3) is the only green algae UGT that clusters with all other Arabidopsis UGTs. The liverwort MeUGT1, MpalUGT1, and PaUGT1 classify with the group E of Arabidopsis UGTs (gathering UGT71, UGT72, and UGT88 families according to [21]), while MeUGT2 and PaUGT2 are closer to UGT73s.
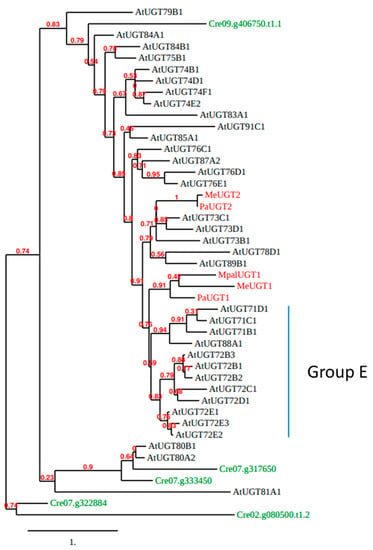
Figure 4.
Phylogenetic tree of whole polypeptide sequences of Arabidopsis UGTs (black), five functionally described UGTs (MeUGT1, MeUGT2, MpalUGT1, PaUGT1, and PaUGT2) from 3 species of liverworts (M. emarginata, M. paleacea, and P. appendiculatum; red) and the five C. reinhardtii UGTs (green). This tree was generated by the approximate likelihood-ratio test method [56,57]. Scale bar: expected number of amino acid substitutions per site.
The five UGTs from liverwort glycosylate flavonoids. MeUGT1 and MpalUGT1 glycosylate four flavonols (quercetin, kaempferol, isorhamnetin, and myricetin) to form 3-O-glucosides [55]. PaUGT1 glycosylates quercetin, kaempferol, and isorhamnetin, the flavones apigenin, luteolin, chrysoeriol, and diosmetin, the flavanones naringenin and eriodictyol, and the dihydrochalcone phloretin to form mainly 7-O-glucosides, but also 3-O-glucosides [54]. MeUGT2 and PaUGT2 also glycosylate flavonoids [54,55]. Among flavonoids, UGT72s (see the next chapter) glycosylate quercetin, kaempferol, myricetin, apigenin, luteolin, and naringenin indicate that flavonoid glycosylation may be conserved as an ancestral function in plant UGTs.
3. Functional Characteristics of the UGT72 Family
3.1. Substrate Identification of UGT72s
Table 2 summarizes the different known in vitro substrates reported for 30 recombinant UGT72 proteins distributed into 12 different angiosperm species. UGT72 members recognize flavonoids, monolignols, and their precursors/derivatives as substrates. In addition, several of them O/N-glycosylate xenobiotics such as chlorophenols and chloroanilines. UGT phylogeny, based on sequence identities and substrate specificities of the enzymes, is not correlated as diverging UGTs can recognize identical substrates. However, the analysis of the 3D structure of the enzyme may predict the substrate specificity [5].
Concerning monolignols and/or their precursors or derivatives, the A. thaliana UGT72E1 and UGT72E2 use coniferaldehyde and sinapaldehyde, and UGT72E2 and UGT72E3 glycosylate ferulic acid, sinapic acid, caffeic acid, coniferyl alcohol, and sinapyl alcohol [58]. In addition, UGT72E2 can also glycosylate p-, m-, and o-coumaric acid, as well as vanillin [58,59]. UGT72B1 glycosylates coniferaldehyde, p-coumaraldehyde, coniferyl alcohol, p-coumaryl alcohol, and vanillin, whereas UGT72B3 uses coniferaldehyde and sinapaldehyde [39,59]. The Camellia sinensis UGT72AM1 glycosylates coniferaldehyde (Table 2) [60]. In Populus tremula x P. alba, UGT72AZ2 glycosylates ferulic acid and sinapic acid, UGT72B37 uses p-coumaraldehyde, coniferaldehyde, sinapaldehyde, coniferyl alcohol, and sinapyl alcohol, while UGT72B39 glycosylates coniferyl alcohol [61]. In Solanum lycopersicum, SlUGT5 glycosylates cinnamyl alcohol [62]. Finally, the Vanilla planifolia UGT72U1 and the Vitis vinifera UGT72B27 glycosylate vanillin [63,64]. Although not always tested (Table 2), UGT72s glycosylate monolignols and their precursors/derivatives at the C4 position on the contrary to UGT84s which glycosylate hydroxycinnamic acids on the alcohol group to form esters as demonstrated in [35].
Concerning flavonoids, the most frequently reported in vitro substrates for UGT72 members are the flavonols quercetin, kaempferol, and myricetin (Table 2). However, the glycosylation pattern can be different among UGT72s, as LjUGT72AH1, LjUGT72Z2, LjUGT72V3, GmUGT72X4, and GmUGT72Z3 form 3-O-glycosides [65,66], while MtUGT72L1 forms 3′-O-glycoside [67]. In addition, LjUGT72AD1 forms both 3-O-glycosides and 7-O-glycosides, while CsUGT72AM1 and HpUGT72B11 perform multi-site glycosylation on 3, 7, and 4′ position [60,66,68].

Table 2.
Known in vitro substrates of 30 recombinant UGT72 proteins distributed into 12 angiosperm species. The regiospecificity of the glycosylation is noted between brackets when determined. All UGT72s use UDP-glucose as sugar donor. In addition, UGT72B1 uses UDP-5-thioglucose [22], UGT72B3, GmUGT72X4, and GmUGT72Z3 use UDP-galactose [22,65] and UGT72B3 uses UDP-xylose [22]. CTP, chlorothiophenol; DCA, dichloroaniline; DCP: dichlorophenol; DHCA: dihydroconiferyl alcohol; HPPA: hydroxyphenylpyruvic acid; TCA: trichloroaniline; TCP: trichlorophenol; TFMP, trifluoromethylphenol. -: no substrate known.
Table 2.
Known in vitro substrates of 30 recombinant UGT72 proteins distributed into 12 angiosperm species. The regiospecificity of the glycosylation is noted between brackets when determined. All UGT72s use UDP-glucose as sugar donor. In addition, UGT72B1 uses UDP-5-thioglucose [22], UGT72B3, GmUGT72X4, and GmUGT72Z3 use UDP-galactose [22,65] and UGT72B3 uses UDP-xylose [22]. CTP, chlorothiophenol; DCA, dichloroaniline; DCP: dichlorophenol; DHCA: dihydroconiferyl alcohol; HPPA: hydroxyphenylpyruvic acid; TCA: trichloroaniline; TCP: trichlorophenol; TFMP, trifluoromethylphenol. -: no substrate known.
| UGT72 | Accession | Species | Monolignol Pathway | Flavonoids | Other Phenolics | Other Compounds | References |
|---|---|---|---|---|---|---|---|
| UGT72B1 | At4G01070 | A. thaliana | Coniferaldehyde p-coumaraldehyde Coniferyl alcohol DHCA p-coumaryl alcohol Vanillin | - | Umbelliferone 4-methyl-umbelliferone Scopoletin (7-O) Esculetin (6-O) Hydroxybenzoic acids (3-O, 4-O, 5-O) 4-HPPA 4-nitrophenol Phenol 2,4-DCP 3,4-DCP 2,4,5-TCP 2,3,4-TCP 2,3,6-TCP 2-chloro-4-TFMP 1-naphthol Triclosan Alternariol | 3,4-DCA 2,4-DCA 2,4,5-TCA 3,4,5-TCA Picloram | [22,33,39,59,69,70,71,72,73] |
| UGT72B3 | At1G01420 | A. thaliana | Coniferaldehyde Sinapaldehyde | Quercetin Fisetin Kaempferol | 4-methyl-umbelliferone Scopoletin (7-O) Esculetin (7-O) Umbelliferone 4-acetic acid 2,4,5-TCP 2-chloro-4-TFMP | - | [22,39,70,72] |
| UGT72C1 | At4G36770 | A. thaliana | - | - | Scopoletin (7-O) Esculetin (6-O) 2,4,5-TCP | - | [22,70] |
| UGT72D1 | At2G18570 | A. thaliana | - | Luteolin Quercetin Fisetin Kaempferol Taxifolin Catechin Genistein | 4-methyl-umbelliferone Scopoletin (7-O) Esculetin (6-O) Dihydroxylbenzoic acids 2,4,5-TCP | - | [22,70,74] |
| UGT72E1 | At3G50740 | A. thaliana | Coniferaldehyde (4-O) Sinapaldehyde (4-O) | Quercetin Fisetin Kaempferol | - | - | [22,36,58] |
| UGT72E2 | At5G66690 | A. thaliana | Ferulic acid (4-O) Sinapyl alcohol (4-O) Sinapic acid (4-O) Caffeic acid (4-O) p-coumaric acid (4-O) o-coumaric acid m-coumaric acid Coniferaldehyde (4-O) Sinapaldehyde (4-O) Coniferyl alcohol (4-O) Vanillin | - | Scopoletin (7-O) Esculetin (6-O) 2,4,5-TCP | 3,4-DCA | [22,35,36,58,59,70,75] |
| UGT72E3 | At5G26310 | A. thaliana | Sinapic acid (4-O) Caffeic acid (4-O) Ferulic acid (4-O) Coniferyl alcohol Sinapyl alcohol | - | Scopoletin (7-O) Esculetin (6-O) | - | [22,35,36,58,70] |
| UGT72AM1 | KY399734 | C. sinensis | Coniferaldehyde (4-O) | Kaempferol (3-O) Quercetin (3-O) Myricetin (3-O) Naringenin (7-O, 4′-O) Eriodictyol Dihydromyricetin Cyanidin (3-O) | - | - | [60,76] |
| UGT72X4 | GLYMA8G338100 | G. max | - | Quercetin (3-O) Kaempferol (3-O) Myricetin (3-O) | - | - | [65] |
| UGT73 | GLYMA8G338200 | G. max | - | Quercetin (3-O) Kaempferol (3-O) Myricetin (3-O) | - | - | [65] |
| UGT72B11 | EU561016 | H. pilosella | - | Baicalein (7-O) Quercetin (3-O, 4′-O) Kaempferol (3-O, 7-O) Apigenin (7-O) Luteolin (7-O, 4′-O) Naringenin (7-O) Eriodictyol Scutellarein Chrysin Myricetin Morin | Umbelliferone Esculetin Catechol Resorcinol Hydroquinone | - | [68] |
| UGT72AD1 | AP009657 | L. japonicus | - | Kaempferol (3-O, 7-O) Quercetin (3-O) Myricetin (3-O) | - | - | [66] |
| UGT72AF1 | KT895083 | L. japonicus | - | Apigenin Daidzein Genistein | - | - | [66] |
| UGT72AH1 | AOG18241 | L. japonicus | - | Kaempferol Quercetin Myricetin | - | - | [66] |
| UGT72V3 | KT895088 | L. japonicus | - | Kaempferol Quercetin Myricetin Luteolin Daidzein Genistein | - | - | [66] |
| UGT72Z2 | KP410264 | L. japonicus | - | Kaempferol (3-O) Quercetin (3-O) Myricetin (3-O) | - | - | [66] |
| UGT72L1 | ACC38470 | M. truncatula | - | Epicatechin (3′-O) Epigallocatechin | - | - | [67] |
| UGT72AX1 | Nbv6. 1trP17460 | N. benthamiana | - | Kaempferol | Carvacrol Hydroquinone Scopoletin Carveol Alternariol | 3-cis-hexenol 1-octen-3-ol Benzyl alcohol Lavandulol 2-phenylethanol Farnesol Perillyl alcohol β-ionol Geraniol | [73,77] |
| UGT72AY1 | Nbv6. 1trP2283 | N. benthamiana | - | Kaempferol Malvidin | Carvacrol Hydroquinone Scopoletin Carveol Alternariol | Farnesol α-ionol β-ionol 2-phenylethanol Geraniol 3-cis-hexenol 1-octen-3-ol Perillyl alcohol Benzyl alcohol Lavandulol Tyrosol Myrtenol 3-oxo-α-ionol Mandelic acid Mandelonitrile Furanmethanethiol Sotolone Maple furanone Furaneol Homofuraneol | [73,77,78] |
| UGT72B34 | Nbv6. 1trP17549 | N. benthamiana | - | Kaempferol | Carvacrol Hydroquinone Scopoletin Carveol | Geraniol 3-cis-hexenol Perillyl alcohol 1-octen-3-ol Benzyl alcohol Lavandulol Tyrosol Myrtenol Farnesol 2-phenylethanol | [77] |
| UGT72B35 | Nbv6. 1trP72850 | N. benthamiana | - | Kaempferol | Carvacrol Hydroquinone Scopoletin | Benzyl alcohol Tyrosol Furanmethanethiol α-bisabolol 1-octen-3-ol 2-phenylethanol | [77] |
| PtGT1 | HM776516 | P. tomentosa | - | - | - | - | [38] |
| UGT72AZ1 | Potri- 7G030300 | P. tremula x P. alba | - | - | - | - | [61] |
| UGT72AZ2 | Potri- 7G030400 | P. tremula x P. alba | Ferulic acid Sinapic acid | - | - | - | [61] |
| UGT72A2 | Potri- 7G030500 | P. tremula x P. alba | - | - | - | - | [27,61] |
| UGT72B37 | Potri- 14G096100 | P. tremula x P. alba | p-coumaraldehyde Coniferaldehyde Sinapaldehyde Coniferyl alcohol Sinapyl alcohol | - | - | - | [61] |
| UGT72B39 | Potri- 2G168600 | P. tremula x P. alba | Coniferyl alcohol | - | - | - | [61] |
| SlUGT5 | HM209439 | S. lycopersicum | Cinnamyl alcohol | Kaempferol | Methyl salicylate Guaiacol Eugenol Hydroquinone Salicyl alcohol (7-O) | Benzyl alcohol | [62] |
| UGT72U1 | Not available | V. planifolia | Vanillin | - | - | - | [63] |
| UGT72B27 | AM483418 | V. vinifera | Vanillin | - | Trans-resveratrol (3-O, 4′-O) Thymol Carvacrol Eugenol Guaiacol 4-methylguaiacol Syringol 4-methylsyringol m-cresol o-cresol Alternariol | Menthol Sotolone Furaneol Homofuraneol | [64,73,78,79] |
3.2. Possible Roles of UGT72s in Monolignol Homeostasis and in the Regulation of Lignification
3.2.1. Monolignol Homeostasis
The role of UGT72s in monolignol homeostasis has been investigated in A. thaliana and in poplar. In leaves of A. thaliana, the overexpression of UGT72E2 and UGT72E3 induces the accumulation of both coniferin and syringin as compared to the wild type where they are not or poorly detected, while the overexpression of UGT72E1 induces a small accumulation of coniferin. Similarly, in light-grown roots, the overexpression of UGT72E2 and UGT72E3 induces an increase in both coniferin and syringin content as compared to the wild type, and the overexpression of UGT72E1 doubles the amount of coniferin [36,37]. The downregulation of UGT72E2 induces a decrease in monolignol glucosides content in the light-grown roots as compared to the wild type (Table 3) [36]. However, the simultaneous downregulation of the three Arabidopsis UGT72E results in a more pronounced reduction of both coniferin and syringin content than the single UGT72E2 downregulation, suggesting possible redundancy of their function [36]. Furthermore, the overexpression of UGT72E2 and UGT72E3 in Arabidopsis induces a decrease in the amount of sinapoyl malate, as well as the accumulation of ferulic acid glucoside in leaves. In addition, the overexpression of UGT72E3 induces an accumulation of sinapic acid glucoside. These results show modifications in the allocation of phenylpropanoids following alteration of the expression of UGT72s (Table 3). However, no significant difference was evidenced in the total soluble phenolics content in leaves [36,37].

Table 3.
Gene expression profile and functional characterization of UGT72s associated with monolignol glycosylation. ns: not studied./: no phenotype observed.
Because of their roles in monolignol homeostasis, UGT72Es may be involved in the resistance to pathogens. In fact, A. thaliana plants overexpressing UGT72E2 infected with the fungus V. longisporum showed a 5-fold increase in the coniferin amount, and their susceptibility to the pathogen was reduced (as the transgenic plants were 2-fold less stunted and as 10-fold less fungal DNA content was detected by real-time PCR, when compared to the wild type; Table 3) [3]. In vitro assays showed that treatment with 100 µM coniferyl alcohol reduces fungal growth which was not observed after a treatment with 100 µM coniferin. However, in the coniferin treatment, the melanization (the fungal last developmental stage) is delayed with only minor black spots compared to the control. As proposed by these authors, coniferin may be hydrolyzed by a β-glucosidase into coniferyl alcohol during the cell lysis. Afterwards, coniferyl alcohol may inhibit V. longisporum growth and be further oxidized into ferulic acid, which is toxic for the pathogen [3].
The overexpression of UGT72B1 induces an increase in coniferin content. However, the knock-out mutant of UGT72B1 also accumulates more coniferin than the wild type (Table 3) [39]. This unexpected result was linked to an increase in the expression of genes of the phenylpropanoid pathway in this mutant (i.e., CCR2, COMT1, COMT2, HCT2, CAD1, CAD5, CAD8, 4CL, C4H, CCoAOMT1, and PAL1). Moreover, UGT72B3 and UGT72E2 are also upregulated in the ugt72b1 mutant and could compensate for the UGT72B1 defection [39]. In that case, UGT72B1 may partake in the gene expression regulation of the phenylpropanoid pathway by a mechanism that remains to be determined. In addition, the mutant exhibits an accumulation of anthocyanins in the shoot tips which is explained by the upregulation of two important genes of the flavonoid biosynthesis, CHS and DFR [39]. Hence, UGT72B1 appears to have a role in both monolignol and flavonoid homeostasis.
In poplar (P. tremula × P. alba), the overexpression of UGT72AZ1 and UGT72AZ2 triggers the accumulation of coniferin, and the overexpression of UGT72AZ1 causes also the accumulation of syringin (Table 3). However, the corresponding recombinant proteins do not use monolignols. In contrast, UGT72B37 and UGT72B39 glycosylate monolignols in vitro, but the overexpression of the corresponding genes in poplar does not result in a higher accumulation of monolignol glucosides [61]. These differences in substrate specificity in vitro and in vivo may be due to the substrate availability in planta.
3.2.2. Regulation of Lignification
The involvement of monolignol glucosylation in lignification has been frequently discussed [80,81,82,83]. On the one hand, this process may have a role in monolignol transport in the cell. The mechanisms of subcellular monolignol transport are still under debate, and indirect experiments have evidenced both passive and ATP-dependent transport [80,81,84,85]. Molecular dynamic simulations support that most of the phenylpropanoids involved in lignification can passively cross the membranes, but not their glycosylated derivatives [81]. An ATP-dependent transport of both monolignols and monolignol glucosides has also been evidenced in isolated membrane vesicles from Arabidopsis [80]. As shown by these authors, only aglycon monolignols can cross the plasma membrane, while only glycosylated monolignols can cross the tonoplast, suggesting that monolignol glucosylation may determine the allocation of monolignols in cells [80]. On the other hand, monolignol glycosides may be directly incorporated into lignin as shown by biomimetic in vitro assays (dehydrogenative polymerization catalyzed by horseradish peroxidase in the presence of coniferin and syringin with or without commercial almond β-glucosidase) and nuclear magnetic resonance (NMR) spectroscopy analysis of cell wall and lignin fractions of wood from gymnosperms and angiosperms [83]. This incorporation could possibly intervene into the lignin–carbohydrate complex (linkages between lignin and hemicelluloses/cellulose), which adds complexity for the understanding of the role of glycosylation in the lignification process [83].
Candidates for lignin polymerization regulation have been searched among Arabidopsis and poplar UGT72s that glycosylate monolignols and/or their precursors. In Arabidopsis, GUS assays showed that pUGT72E2- and pUGT72E3-driven expressions are associated with vascular tissues in seedlings (roots, cotyledons, and apical meristem) and in flowers. In addition, pUGT72E2-driven expression is associated with vascular tissues in leaves [37]. pUGT72B1-driven expression was mainly found in the developing xylem, the pith, and the cortex of young floral stems while it is only expressed in the xylem of old floral stems (Table 3) [39]. These results suggest that UGT72E2, UGT72E3, and UGT72B1 may be associated with vascular tissue development, and possibly with lignification. In addition, the transcriptomic analysis of the triple lac4 lac11 lac17 Arabidopsis mutant characterized by a highly disrupted lignin deposition revealed a relation between the expression of UGT72Es and that of laccases (LAC) and peroxidases (PRX) associated with lignin polymerization. In the leaves of this mutant, coniferin and syringin contents are 4-fold higher than the wild type, and the expression of both UGT72E2 and UGT72E3 is 5.2-fold and 2-fold increased, respectively [86]. Another transcriptomic study showed that Arabidopsis transgenic lines overexpressing MYB58 or MYB63 upregulate monolignol biosynthesis genes as well as UGT72E2. These transgenic lines accumulate monolignol glucosides and display ectopic lignification in the epidermis, cortex, and pith [87]. Finally, PRX49 and PRX72, two genes encoding peroxidases likely involved in lignification [88,89], are co-expressed with UGT72E1/UGT72E2 and with the three UGT72Es, respectively [90].
Functional characterization of mutants has evidenced phenotypes to clarify this question. Although single ugt72e1, ugt72e2 or ugt72e3 Arabidopsis mutants do not show any difference in lignin quantity in the floral stem when using the acetyl bromide method, a higher proportion of lignin in the xylem and interfascicular fiber cell walls of the ugt72e3 mutant was evidenced by Raman microspectroscopy and safranin O ratiometric imaging technique, as compared to the wild type [40]. This difference was only observed in the young part and not in the old part of the floral stem and indicates a role of UGT72E3 in cell wall lignification during vascular cells development. Moreover, a higher capacity of incorporation of the three fluorescently labeled monolignols into lignin was found in the ugt72e3 young stem as compared to the wild type. This phenotype was related to an increased expression of lignin-specific PRX71 and LAC17 [40]. The ugt72b1 mutant exhibits a higher lignin content in the whole floral stem, as well as an ectopic lignification in the interfascicular fibers and the pith as compared to the wild type and the rescued line (insertion of the UGT72B1 cDNA under the control of the native gene promoter) [39]. In addition, the mutation leads to a 4-fold increase in the thickness of the pith cell wall. Like monolignol biosynthesis genes, genes potentially involved in monolignol transport across the plasma membrane and in lignin polymerization were upregulated (i.e., PRX34, PRX37, PRX71, PER64, LAC5, LAC12, LAC15, ABCG29, ABCG40, RBOHA and RBOHD) [39]. The authors suggest that the UGT72B1 mutation, which depletes the content of monolignol glucosides, may trigger a signal upregulating the genes involved in monolignol biosynthesis, transport, and polymerization, generating an overproduction of monolignols, hence an increased and ectopic lignification. Consequently, the increase in coniferin in the mutant may be a secondary effect of this monolignol overproduction [39]. The alteration of the monolignol biosynthesis and of lignification may interfere with other developmental processes, explaining the repression of the shoot growth of the ugt72b1 mutant [39].
In poplar, the UGT72s encoding proteins glycosylating monolignols and/or their precursors were expressed in vascular tissues of stem or roots (Table 3). In 4-month-old poplar, UGT72AZ1 is mainly expressed in phloem of stem and leaf, UGT72B37 and UGT72B39 are expressed in the primary xylem and in the secondary xylem of the stem, respectively, and UGT72AZ2 is expressed in the cortical region, in the phloem, and in the differentiating xylem of the root [61]. However, although the overexpression of UGT72AZ1 and UGT72AZ2 in P. tremula × P. alba increases the monolignol glucosides content in the leaf, there was no modification of total lignin content in the stem nor in the root [61]. It is possible that the effect of the increase in monolignol glucosides on lignification is compensated by other processes, such as hydrolysis catalyzed by β-glucosidases or incorporation of the glucosides in the growing lignin polymer, or that UGT72AZ1 and UGT72AZ2 are involved in other processes such as for instance the regulation of oligolignol biosynthesis or the defense towards pathogens. The P. tomentosa PtGT1 (orthologous to UGT72AZ2), when ectopically expressed in tobacco, induces a 60% increase in Klason lignin content in the stem of a 2-month-old plant (Table 3). However, as no enzymatic activity against monolignols was detected for the recombinant PtGT1 (Table 2), the effect of the PtGT1 ectopic expression on lignin content was suggested to be indirect [38].
3.3. UGT72s Involved in Flavonoid Homeostasis
Flavonoids have many roles in plants, especially in stress responses and development [91]. As several UGT72s glycosylate flavonoids, they can be critical in the regulation of these processes. Indeed, glycosylation may modify the activity, transport, accumulation, and biosynthesis of flavonoids [92]. For example, Pang et al. (2008) have demonstrated in vitro that the M. truncatula UGT72L1 can 3′-O-glycosylate epicatechin and epigallocatechin [67]. Epicatechin and epigallocatechin (as well as catechin) are the main components of proanthocyanidins, or condensed tannins which are found in fruits, flowers, bark, and seeds of many plants and whose astringency is known to protect plants against pathogens and herbivores [93]. Moreover, they provide UV protection and antioxidant activity to the plant and are involved in defense against biotic and abiotic stresses [94]. In the ugt72l1 mutant, the reduction of both epicatechin and epicatechin 3′-O-glucoside content triggers a decrease in extractable proanthocyanidins in seeds. In contrast, the overexpression of UGT72L1 in M. truncatula hairy roots increases both the extractable and non-extractable proanthocyanidins content and decreases the anthocyanins content (Table 4) [95]. GUS assays evidenced that UGT72L1 is expressed in the mid-rib of the rosette leaves, the peduncles of the siliques and the inflorescence, and developing seeds of A. thaliana, in a similar way to genes involved in proanthocyanidins biosynthesis (e.g., ANR, CHS, and TTG1; Table 4) [67]. UGT72L1 could regulate proanthocyanidin biosynthesis by directing the flux of epicatechin into the vacuole [95]. Indeed, the MATE1 transporter, which is involved in the epicatechin transport across the tonoplast like its Arabidopsis homologous TT12, has been demonstrated to be specific to epicatechin 3′-O-glucoside [96]. Moreover, as suggested by Pang and colleagues [67], this glycosylation may protect the plant against free epicatechins and help to direct monomers polymerization into the accurate 4–8 linkage.

Table 4.
Gene expression profile and functional characterization of UGT72s associated with flavonoid homeostasis. ns: not studied./: no phenotype observed.
The majority of UGT72s glycosylating flavonoids in vitro have not been studied for their role in stress tolerance. However, a transcriptomic analysis reveals, for instance, that UGT72B3 is upregulated in Arabidopsis aerial parts when temperature decreases from 20 °C to 4 °C, while UGT72E1 is downregulated in the same conditions. UGT72E1 and UGT72D1 are upregulated in roots after infection by Plasmodiophora brassicae. UGT72D1 is downregulated during drought [97].
In Populus tremula x P. alba, UGT72A2 is mainly expressed in young leaf and stem (Table 4). The downregulation of UGT72A2 triggers leaf yellowing and necrosis under standard growing conditions in comparison to the wild type [27]. This phenotype was associated with oxidative stress in leaves characterized by higher lipid peroxidation and the decrease in compounds involved in oxidative stress response when compared to the wild type. Especially the total flavonoid content, the anthocyanin content and the total phenolic content in leaves are lower in the UGT72A2-depleted lines than in the wild type. Consistently, pUGT72A2::GUS was significantly transactivated by the poplar transcription factor MYB119 regulating the biosynthesis of flavonoids. Moreover, the leaves of the UGT72A2-depleted lines show a decreased soluble peroxidase activity and a decrease in the NADPH to NADP+ ratio, both indicating an alteration of the redox scavenging system [27]. Curiously, the downregulation of UGT72A2 improves the tolerance to methyl viologen in leaves, an herbicide enhancing the production of superoxide ions in the chloroplasts [27]. This increased tolerance may be linked to the observed accumulation of proanthocyanidins which was previously demonstrated to protect poplar leaves against oxidative stress induced by methyl viologen (Table 4) [27,98]. This functional characterization of UGT72A2 reveals the function that UGT72s may have on flavonoid homeostasis, as well as on ROS scavenging and stress tolerance.
Flavonoids are also involved in developmental processes, especially by interacting with hormone signalings [91,99] and UGT72s may also be involved in those processes. In L. japonicus, UGT72AD1 and UGT72Z2 glycosylate in vitro kaempferol, quercetin, and myricetin, both forming 7-O-glycosides, while 3-O-glycosides were detected for UGT72AD1 only [66]. They are mainly expressed during the later stages of seed development in a similar manner to FLS, MYB11, and MYB14, which are involved in flavonoid biosynthesis [66]. The overexpression of UGT72AD1 and UGT72Z2 in L. japonicus hairy roots does not significantly modify the total flavonoid content. However, the flavonol content is increased in all of the transgenic lines compared to the wild type. Especially, kaempferol 3-O-rhamnoside-7-O-rhamnoside, kaempferol 3-O-glucoside-7-O-rhamnoside, and two additional flavonol hexosides are more accumulated. These results suggest that UGT72AD1 and UGT72Z2 may be involved in the kaempferol glycosides homeostasis and especially in kaempferol rhamnosides homeostasis (Table 4) [99]. A previous study had shown that increased kaempferol 3-O-rhamnoside-7-O-rhamnoside content inhibits polar auxin transport, affecting the growth and the gravitropism in Arabidopsis [99]. In accordance, transgenic Arabidopsis lines overexpressing UGT72AD1 and UGT72Z2 showed a significant inhibition of root growth (Table 4), suggesting a role of these genes in auxin homeostasis and developmental regulation in L. japonicus [66].
3.4. The Subcellular Localization of UGT72s Provides Information on Their Functions
UGT72s have been localized in different subcellular compartments. For instance, UGT72L1 fused to the green fluorescent protein (GFP) was detected in the cytosol in M. truncatula [95]. The first plant UGT localized into the nucleus was UGT72E1-GFP which interacts with the Arabidopsis MAP kinase kinase kinase SIS8, involved in sugar signaling [100]. In poplar, five UGT72s fused to GFP (UGT72AZ1, UGT72AZ2, UGT72B36, UGT72B37, and UGT72B39) are localized both in the nucleus and associated with the endoplasmic reticulum (ER). The localization of UGT72s in the nucleus may reveal a specialization of these UGTs in the phenylpropanoid homeostasis in this specific compartment. There is no report of monolignol glycosides in the nucleus and their potential function. However, some flavonoids are localized in the nucleus in several plant species, suggesting that this localization is also possible for monolignols [101,102,103]. It has been proposed that flavonoids may interfere with some nuclear proteins involved in DNA organization, signaling pathway, and gene expression [103,104,105], and these processes may be regulated by UGT72s.
The Polygonum tinctorium indoxyl-β-D-glucoside synthase (IGS), a member of the UGT72B sub-family, was localized in both cytosolic and microsomal fractions, suggesting a reversible binding of the protein to the membranes. The ER localization was confirmed using ultracentrifugation with a sucrose density gradient [106]. The Pyrus bretschneideri PbUGT72AJ2 fused to GFP was also located mainly in the cytosol and cytomembrane compartment [107]. The enzymatic steps of the phenylpropanoid pathway are known to occur in the cytosol and several enzymes of the monolignol biosynthesis pathway, such as HCT and the P450 proteins C4H, C3′H, and F5H are, at least partly, associated with the ER [108,109]. Assembly and disassembly of a given metabolon, for instance, a UGT coupled to a P450 protein would provide additional flexibility in specialized metabolite biosynthesis. Finally, UGT72A2-GFP is localized in the chloroplast [61]. The chloroplast localization of UGT72A2 fits with its function in flavonoid and ROS homeostasis within the chloroplast. Chloroplasts are indeed storage and even biosynthesis organelles for some phenylpropanoids such as kaempferol, quercetin, and catechin in different plant species [110,111,112]. These compounds may be involved in ROS scavenging in this compartment [113]. The subcellular localization of the majority of UGT72s has not been investigated yet, which could improve our knowledge of the biological functions in this family.
4. Challenges and Perspectives in UGT Research
Our knowledge of the glycosyltransferase activity of UGT72s remains limited because the enzymatic assays are performed in vitro and do not reflect the availability of the appropriate substrates in planta. In addition, since most of the recombinant proteins are produced using E. coli as an expression host, the significance of several post-translational modifications occurring in plants is not adequately addressed. For instance, various patterns of protein N-glycosylation modified the affinity of human UGT2B7 towards zidovudine and morphine [114]. Such an experimental limitation may be overcome by studying recombinant proteins in yeast, as shown by the expression of UGT72E2 and UGT72B1 in Saccharomyces cerevisiae or Schizosaccharomyces pombe engineered with the vanillin pathway, thereby allowing to produce vanillin β-D-glucoside which is less toxic than vanillin [59]. Otherwise, the investigated UGT may be extracted from a transgenic plant overexpressing its coding gene and to compare its glycosylation activity to proteins extracted from a control plant. This experimental setup was successfully applied to confirm in planta the activity of UGT92G6 detected in vitro with caffeic acid [115]. Chen and colleagues evidenced the activity of UGT78H2 extracted from transiently modified tobacco leaves towards quercetin [116].
The glycosyltransferase activity of UGT72s against monolignols and flavonoids opens avenues across numerous applications. The glycosylation of these compounds is extensively studied in the food and pharmacology sectors, because the biological functionalities of small molecules may be enhanced by increasing their hydrophilicity and stability through glycosylation [117]. For instance, syringin is known for its anti-inflammatory, anti-nociceptive, immune-modulatory, and anti-diabetic effects [118,119,120]. Despite the biosynthesis pathway of syringin being well conserved in the plant kingdom, few plant species naturally accumulate large quantities of syringin. In order to improve the biosynthesis of syringin in plants, a study has investigated the creation of a chimeric protein between UGT72E2 and UGT72E3 where the high glycosyltransferase activity of UGT72E2 is coupled with the higher affinity for sinapyl alcohol of UGT72E3. When this chimeric UGT72E3/2 gene is overexpressed in Arabidopsis plants, there is 145% more syringin in leaves than in the transgenic Arabidopsis overexpressing UGT72E3 [121]. Flavonoids are also important in promoting health molecules because of their antioxidant properties and their beneficial effect against, for instance, hypertension, inflammation, bacterial infections, cerebral disorder, atherosclerosis, and cancer [122]. The purple-leaf tea variety ‘Zijuan’, accumulating high levels of anthocyanidins and anthocyanins, has been selected for its antiproliferative effects on colorectal carcinoma cells [123]. Molecular analyses have shown that UGT72AM1 is 4.2-fold more expressed in this variety than in the wild type (the ‘Longjing43′ tea variety). As the protein coded by this gene can glycosylate in vitro kaempferol, quercetin, myricetin, naringenin, eriodictyol, and cyanidin, its higher expression may be linked to the higher content of glycosylated flavonoids in the ‘Zijuan’ variety [60,76].
The monolignol biosynthesis pathway and the impact of its modification on biomass recalcitrance is extensively studied to improve cellulose extraction for the production of paper pulp and bioethanol. Actually, lignin is the main limiting factor in these sectors because it restricts polysaccharides’ accessibility and inhibits enzymatic activity. Reducing the lignin content in the cell wall and/or altering its composition are considered as two strategies to improve pulping and saccharification [124]. The investigation of the role of UGT72s in monolignol homeostasis has revealed an additional novel regulation process for lignification in Arabidopsis [39,40]. In poplar, the role of UGT72s in lignin regulation is less clear and is worth further investigation according to the importance of trees in wood, paper pulp, and bioethanol production.
5. Conclusions
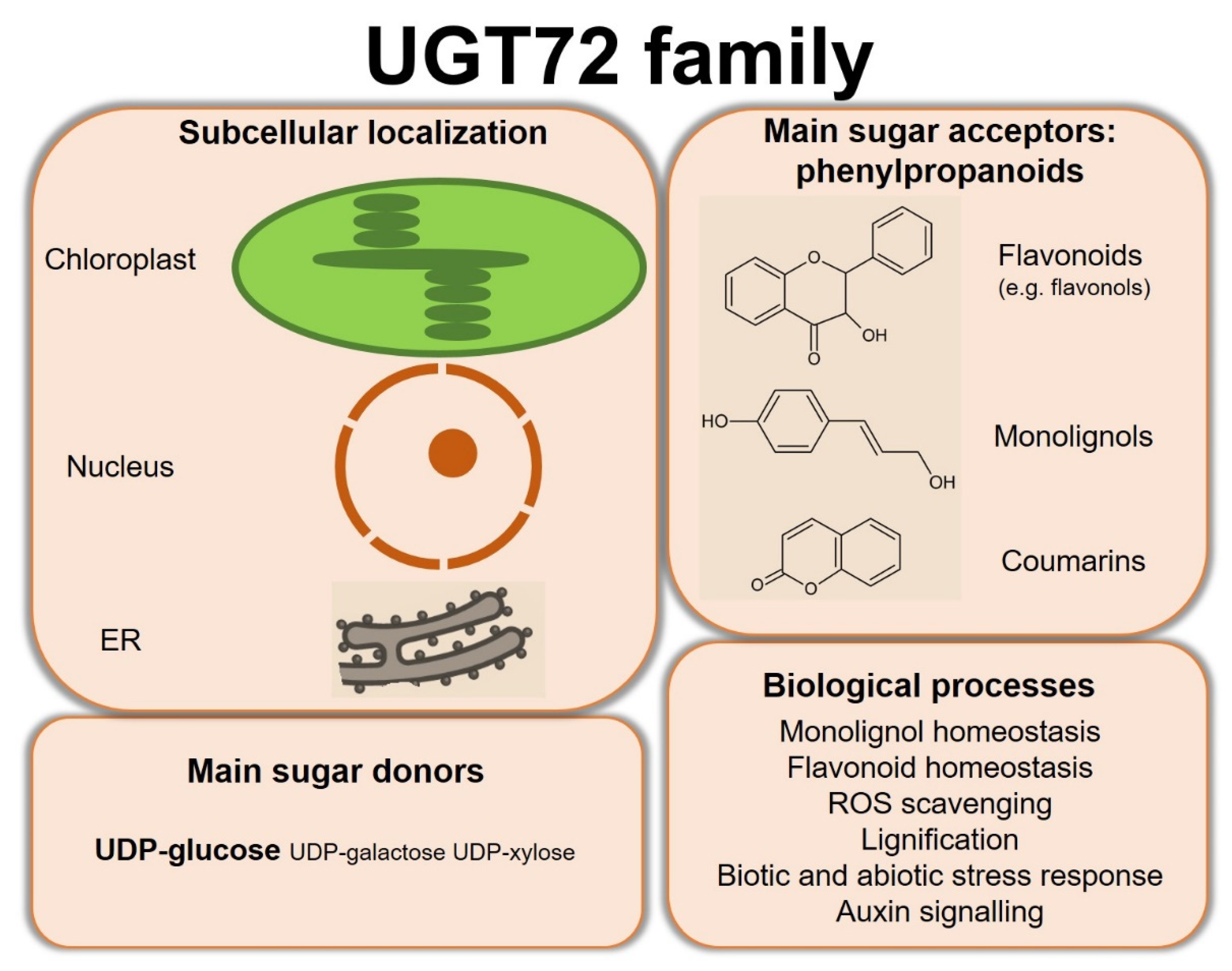
Substrates of UGT72 enzymes, using mainly UDP-glucose as sugar donor, cover a wide range of specialized metabolites and xenobiotics. Figure 5 summarizes the main properties of the UGT72 family. In particular, several members of this family have been shown to glycosylate phenylpropanoids including flavonoids, monolignols, and coumarins. Expression and functional studies highlighted a role for these UGT72s in various biological processes such as monolignol homeostasis, flavonoid homeostasis, ROS homeostasis, lignification, response to biotic and abiotic stresses, and auxin signaling. While the subcellular localization of some UGT72s was determined, such as in chloroplast or related to the endoplasmic reticulum, this biological feature is too often not investigated despite its importance regarding the function of the enzyme.
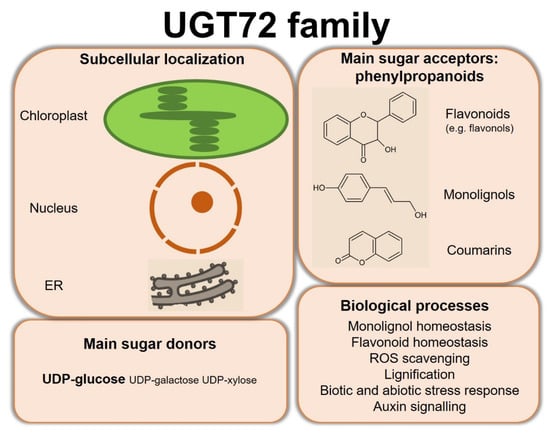
Figure 5.
Summary of the subcellular localization and main substrates of UGT72s, as well as the biological processes in which investigated enzymes are involved. ER, endoplasmic reticulum.
Author Contributions
Conceptualization, N.S., M.E.J., M.B. (Marie Baucher) and M.B. (Marc Behr); investigation, N.S., M.E.J., M.B. (Marie Baucher) and M.B. (Marc Behr); writing—original draft preparation, N.S.; writing—review and editing, all authors; supervision, M.B. (Marie Baucher); project administration, M.B. (Marie Baucher); funding acquisition, M.B. (Marie Baucher). All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Belgian Fonds de la Recherche Scientifique (FRS-FNRS), grant number T.0068.18. N. Speeckaert was the recipient of pre-doctoral grants from the Belgian Fonds pour la Formation à la Recherche dans l’Industrie et l’Agriculture (FRIA). M. Baucher is a Senior Research Associate of the FRS-FNRS; M. Behr is a Postdoctoral Researcher of the FRS-FNRS.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Bowles, D.; Lim, E.-K.; Poppenberger, B.; Vaistij, F.E. Glycosyltransferases of lipophilic small molecules. Annu. Rev. Plant Biol. 2006, 57, 567–597. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chantreau, M.; Sibout, R.; Hawkins, S. Plant cell wall lignification and monolignol metabolism. Front. Plant Sci. 2013, 4, 220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- König, S.; Feussner, K.; Kaever, A.; Landesfeind, M.; Thurow, C.; Karlovsky, P.; Gatz, C.; Polle, A.; Feussner, I. Soluble phenylpropanoids are involved in the defense response of Arabidopsis against Verticillium longisporum. New Phytol. 2014, 202, 823–837. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.; Vogt, T. Glycosyltransferases in secondary plant metabolites tranquilizers and stimulant controllers. Planta 2001, 213, 164–174. [Google Scholar] [CrossRef]
- Osmani, S.A.; Bak, S.; Møller, B.L. Substrate specificity of plant UDP-dependent glycosyltransferases predicted from crystal structures and homology modeling. Phytochemistry 2009, 70, 325–347. [Google Scholar] [CrossRef]
- CAZY-Carbohydrate-Active Enzymes. Available online: http://www.cazy.org/ (accessed on 31 January 2022).
- Lairson, L.L.; Henrissat, B.; Davies, G.J.; Withers, S.G. Glycosyltransferases: Structures, functions, and mechanisms. Annu. Rev. Biochem. 2008, 77, 521–555. [Google Scholar] [CrossRef] [Green Version]
- Yonekura-Sakakibara, K.; Hanada, K. An evolutionary view of functional diversity in family 1 glycosyltransferases. Plant J. 2011, 66, 182–193. [Google Scholar] [CrossRef]
- Caputi, L.; Malnoy, M.; Goremykin, V.; Nikiforova, S.; Martens, S. A genome-wide phylogenetic reconstruction of family 1 UDP-glycosyltransferases revealed the expansion of the family during the adaptation of plants to life on land. Plant J. 2012, 69, 1030–1042. [Google Scholar] [CrossRef]
- Bock, K.W. The UDP-glycosyltransferase (UGT) superfamily expressed in humans, insects and plants: Animal-plant arms-race and co-evolution. Biochem. Pharmacol. 2016, 99, 11–17. [Google Scholar] [CrossRef]
- Wilson, A.E.; Tian, L. Phylogenomic analysis of UDP-dependent glycosyltransferases provides insights into the evolutionary landscape of glycosylation in plant metabolism. Plant J. 2019, 100, 1273–1288. [Google Scholar] [CrossRef]
- Maeda, H.; Dudareva, N. The shikimate pathway and aromatic amino acid biosynthesis in plants. Annu. Rev. Plant Biol. 2012, 63, 73–105. [Google Scholar] [CrossRef] [PubMed]
- Qian, Y.; Lynch, J.H.; Guo, L.; Rhodes, D.; Morgan, J.A.; Dudareva, N. Completion of the cytosolic post-chorismate phenylalanine biosynthetic pathway in plants. Nat. Commun. 2019, 10, 15. [Google Scholar] [CrossRef] [PubMed]
- Yoo, H.; Widhalm, J.R.; Qian, Y.; Maeda, H.; Cooper, B.R.; Jannasch, A.S.; Gonda, I.; Lewinsohn, E.; Rhodes, D.; Dudareva, N. An alternative pathway contributes to phenylalanine biosynthesis in plants via a cytosolic tyrosine:phenylpyruvate aminotransferase. Nat. Commun. 2013, 4, 2833. [Google Scholar] [CrossRef] [Green Version]
- Vanholme, R.; De Meester, B.; Ralph, J.; Boerjan, W. Lignin biosynthesis and its integration into metabolism. Curr. Opin. Biotechnol. 2019, 56, 230–239. [Google Scholar] [CrossRef] [PubMed]
- Barros, J.; Dixon, R.A. Plant phenylalanine/tyrosine ammonia-lyases. Trends Plant Sci. 2020, 25, 66–79. [Google Scholar] [CrossRef] [PubMed]
- Boerjan, W.; Ralph, J.; Baucher, M. Lignin biosynthesis. Annu. Rev. Plant Biol. 2003, 54, 519–546. [Google Scholar] [CrossRef]
- Xie, M.; Zhang, J.; Tschaplinski, T.J.; Tuskan, G.A.; Chen, J.-G.; Muchero, W. Regulation of lignin biosynthesis and its role in growth-defense tradeoffs. Front. Plant Sci. 2018, 9, 1427. [Google Scholar] [CrossRef] [Green Version]
- Vogt, T. Phenylpropanoid biosynthesis. Mol. Plant 2010, 3, 2–20. [Google Scholar] [CrossRef] [Green Version]
- Atkinson, R.G. Phenylpropenes: Occurrence, distribution, and biosynthesis in fruit. J. Agric. Food Chem. 2018, 66, 2259–2272. [Google Scholar] [CrossRef]
- Ross, J.; Li, Y.; Lim, E.; Bowles, D.J. Higher plant glycosyltransferases. Genome Biol. 2001, 2, REVIEWS3004. [Google Scholar] [CrossRef]
- Yang, M.; Fehl, C.; Lees, K.V.; Lim, E.K.; Offen, W.A.; Davies, G.J.; Bowles, D.J.; Davidson, M.G.; Roberts, S.J.; Davis, B.G. Functional and informatics analysis enables glycosyltransferase activity prediction. Nat. Chem. Biol. 2018, 14, 1109–1117. [Google Scholar] [CrossRef] [PubMed]
- RCSB-Protein Data Bank. Available online: https://www.rcsb.org/ (accessed on 31 January 2022).
- Nair, P.C.; Meech, R.; Mackenzie, P.I.; McKinnon, R.A.; Miners, J.O. Insights into the UDP-sugar selectivities of human UDP-glycosyltransferases (UGT): A molecular modeling perspective. Drug Metab. Rev. 2015, 47, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Walker, S. Remarkable structural similarities between diverse glycosyltransferases. Chem. Biol. 2002, 9, 1287–1296. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.-Y.; Li, X. Identification of a residue responsible for UDP-sugar donor selectivity of a dihydroxybenzoic acid glycosyltransferase from Arabidopsis natural accessions. Plant J. 2016, 89, 195–203. [Google Scholar] [CrossRef]
- Behr, M.; Speeckaert, N.; Kurze, E.; Morel, O.; Prévost, M.; Mol, A.; Adamou, N.M.; Baragé, M.; Renaut, J.; Schwab, W.; et al. Leaf necrosis resulting from down-regulation of poplar glycosyltransferase UGT72A2. Tree Physiol. 2021, in press. [Google Scholar] [CrossRef]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [Green Version]
- Paquette, S.; Møller, B.L.; Bak, S. On the origin of family 1 plant glycosyltransferases. Phytochemistry 2003, 62, 399–413. [Google Scholar] [CrossRef]
- Kubo, A.; Arai, Y.; Nagashima, S.; Yoshikawa, T. Alteration of sugar donor specificities of plant glycosyltransferases by a single point mutation. Arch. Biochem. Biophys. 2004, 429, 198–203. [Google Scholar] [CrossRef]
- Shao, H.; He, X.; Achnine, L.; Blount, J.W.; Dixon, R.A.; Wang, X. Crystal structures of a multifunctional triterpene/flavonoid glycosyltransferase from Medicago truncatula. Plant Cell 2005, 17, 3141–3154. [Google Scholar] [CrossRef] [Green Version]
- Offen, W.; Martinez-Fleites, C.; Yang, M.; Kiat-Lim, E.; Davis, B.G.; Tarling, C.A.; Ford, C.M.; Bowles, D.J.; Davies, G.J. Structure of a flavonoid glucosyltransferase reveals the basis for plant natural product modification. EMBO J. 2006, 25, 1396–1405. [Google Scholar] [CrossRef] [Green Version]
- Brazier-Hicks, M.; Offen, W.A.; Gershater, M.C.; Revett, T.J.; Lim, E.-K.; Bowles, D.J.; Davies, G.J.; Edwards, R. Characterization and engineering of the bifunctional N- and O-glucosyltransferase involved in xenobiotic metabolism in plants. Proc. Natl. Acad. Sci. USA 2007, 104, 20238–20243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, R.; Rawat, V.; Suresh, C.G. Genome-wide identification and tissue-specific expression analysis of UDP-glycosyltransferases genes confirm their abundance in Cicer arietinum (Chickpea) genome. PLoS ONE 2014, 9, e109715. [Google Scholar] [CrossRef] [PubMed]
- Lim, E.K.; Li, Y.; Parr, A.; Jackson, R.; Ashford, D.A.; Bowles, D.J. Identification of glucosyltransferase genes involved in sinapate metabolism and lignin synthesis in Arabidopsis. J. Biol. Chem. 2001, 276, 4344–4349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lanot, A.; Hodge, D.; Jackson, R.G.; George, G.L.; Elias, L.; Lim, E.K.; Vaistij, F.E.; Bowles, D.J. The glucosyltransferase UGT72E2 is responsible for monolignol 4-O-glucoside production in Arabidopsis thaliana. Plant J. 2006, 48, 286–295. [Google Scholar] [CrossRef] [PubMed]
- Lanot, A.; Hodge, D.; Lim, E.K.; Vaistij, F.E.; Bowles, D.J. Redirection of flux through the phenylpropanoid pathway by increased glucosylation of soluble intermediates. Planta 2008, 228, 609–616. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.W.; Wang, W.C.; Jin, S.H.; Wang, J.; Wang, B.; Hou, B.K. Over-expression of a putative poplar glycosyltransferase gene, PtGT1, in tobacco increases lignin content and causes early flowering. J. Exp. Bot. 2012, 63, 2799–2808. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.S.; Huang, X.X.; Li, Q.; Cao, Y.; Bao, Y.; Meng, X.F.; Li, Y.J.; Fu, C.; Hou, B.K. UDP-glycosyltransferase 72B1 catalyzes the glucose conjugation of monolignols and is essential for the normal cell wall lignification in Arabidopsis thaliana. Plant J. 2016, 88, 26–42. [Google Scholar] [CrossRef] [Green Version]
- Baldacci-Cresp, F.; Le Roy, J.; Huss, B.; Lion, C.; Créach, A.; Spriet, C.; Duponchel, L.; Biot, C.; Baucher, M.; Hawkins, S.; et al. UDP-glycosyltransferase 72e3 plays a role in lignification of secondary cell walls in Arabidopsis. Int. J. Mol. Sci. 2020, 21, 6094. [Google Scholar] [CrossRef]
- Bowman, J.L.; Kohchi, T.; Yamato, K.T.; Jenkins, J.; Shu, S.; Ishizaki, K.; Yamaoka, S.; Nishihama, R.; Nakamura, Y.; Berger, F.; et al. Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell 2017, 171, 287–304.e15. [Google Scholar] [CrossRef] [Green Version]
- de Vries, J.; Archibald, J.M. Plant evolution: Landmarks on the path to terrestrial life. New Phytol. 2018, 217, 1428–1434. [Google Scholar] [CrossRef]
- Chae, L.; Kim, T.; Nilo-Poyanco, R.; Rhee, S.Y. Genomic signatures of specialized metabolism in plants. Science 2014, 344, 510–513. [Google Scholar] [CrossRef] [PubMed]
- Clark, J.W.; Donoghue, P.C.J. Whole-genome duplication and plant macroevolution. Trends Plant Sci. 2018, 23, 933–945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- JGI Phytozome 13-The Plant Genomics Resource. Available online: https://phytozome-next.jgi.doe.gov/ (accessed on 31 January 2022).
- Kumar, V.; Hainaut, M.; Delhomme, N.; Mannapperuma, C.; Immerzeel, P.; Street, N.R.; Henrissat, B.; Mellerowicz, E.J. Poplar carbohydrate-active enzymes: Whole-genome annotation and functional analyses based on RNA expression data. Plant J. 2019, 99, 589–609. [Google Scholar] [CrossRef] [PubMed]
- Louveau, T.; Osbourn, A. The sweet side of plant-specialized metabolism. Cold Spring Harb. Perspect. Biol. 2019, 11, a034744. [Google Scholar] [CrossRef] [PubMed]
- Grille, S.; Zaslawski, A.; Thiele, S.; Plat, J.; Warnecke, D. The functions of steryl glycosides come to those who wait: Recent advances in plants, fungi, bacteria and animals. Prog. Lipid Res. 2010, 49, 262–288. [Google Scholar] [CrossRef] [PubMed]
- Stucky, D.F.; Arpin, J.C.; Schrick, K. Functional diversification of two UGT80 enzymes required for steryl glucoside synthesis in Arabidopsis. J. Exp. Bot. 2015, 66, 189–201. [Google Scholar] [CrossRef] [Green Version]
- Mishra, M.K.; Singh, G.; Tiwari, S.; Singh, R.; Kumari, N.; Misra, P. Characterization of Arabidopsis sterol glycosyltransferase TTG15/UGT80B1 role during freeze and heat stress. Plant Signal. Behav. 2015, 10, e1075682. [Google Scholar] [CrossRef] [Green Version]
- Stonik, V.A.; Stonik, I.V. Sterol and sphingoid glycoconjugates from microalgae. Mar. Drugs 2018, 16, 514. [Google Scholar] [CrossRef] [Green Version]
- Noguchi, A.; Horikawa, M.; Fukui, Y.; Fukuchi-Mizutani, M.; Iuchi-Okada, A.; Ishiguro, M.; Kiso, Y.; Nakayama, T.; Onoa, E. Local differentiation of sugar donor specificity of flavonoid glycosyltransferase in Lamiales. Plant Cell 2009, 21, 1556–1572. [Google Scholar] [CrossRef] [Green Version]
- MView. Available online: https://www.ebi.ac.uk/Tools/msa/mview/ (accessed on 31 January 2022).
- Zhu, T.; Liu, H.; Wang, P.; Ni, R.; Sun, C.; Yuan, J.; Niu, M.; Lou, H.; Cheng, A. Functional characterization of UDP-glycosyltransferases from the liverwort Plagiochasma appendiculatum and their potential for biosynthesizing flavonoid 7-O-glucosides. Plant Sci. 2020, 299, 110577. [Google Scholar] [CrossRef]
- Yuan, J.; Xiong, R.; Zhu, T.; Ni, R.; Fu, J.; Lou, H.; Cheng, A. Cloning and functional characterization of three flavonoid O-glucosyltransferase genes from the liverworts Marchantia emarginata and Marchantia paleacea. Plant Physiol. Biochem. 2021, 166, 495–504. [Google Scholar] [CrossRef] [PubMed]
- Phylogeny.fr-Robust Phylogenetic Analysis for the Non-Specialist. Available online: http://www.phylogeny.fr/ (accessed on 31 January 2022).
- Dereeper, A.; Guignon, V.; Blanc, G.; Audic, S.; Buffet, S.; Chevenet, F.; Dufayard, J.F.; Guindon, S.; Lefort, V.; Lescot, M.; et al. Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008, 36, 465–469. [Google Scholar] [CrossRef] [PubMed]
- Lim, E.-K.; Jackson, R.G.; Bowles, D.J. Identification and characterisation of Arabidopsis glycosyltransferases capable of glucosylating coniferyl aldehyde and sinapyl aldehyde. FEBS Lett. 2005, 579, 2802–2806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hansen, E.H.; Møller, B.L.; Kock, G.R.; Bünner, C.M.; Kristensen, C.; Jensen, O.R.; Okkels, F.T.; Olsen, C.E.; Motawia, M.S.; Hansen, J. De novo biosynthesis of vanillin in fission yeast (Schizosaccharomyces pombe) and baker’s yeast (Saccharomyces cerevisiae). Appl. Environ. Microbiol. 2009, 75, 2765–2774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, X.; Dai, X.; Gao, L.; Guo, L.; Zhuang, J.; Liu, Y.; Ma, X.; Wang, R.; Xia, T.; Wang, Y. Functional analysis of an uridine diphosphate glycosyltransferase involved in the biosynthesis of polyphenolic glucoside in tea plants (Camellia sinensis). J. Agric. Food Chem. 2017, 65, 10993–11001. [Google Scholar] [CrossRef]
- Speeckaert, N.; Adamou, N.M.; Hassane, H.A.; Baldacci-Cresp, F.; Mol, A.; Goeminne, G.; Boerjan, W.; Duez, P.; Hawkins, S.; Neutelings, G.; et al. Characterization of the UDP-glycosyltransferase UGT72 family in poplar and identification of genes involved in the glycosylation of monolignols. Int. J. Mol. Sci. 2020, 21, 5018. [Google Scholar] [CrossRef]
- Louveau, T.; Leitao, C.; Green, S.; Hamiaux, C.; Van Der Rest, B.; Dechy-Cabaret, O.; Atkinson, R.G.; Chervin, C. Predicting the substrate specificity of a glycosyltransferase implicated in the production of phenolic volatiles in tomato fruit. FEBS J. 2011, 278, 390–400. [Google Scholar] [CrossRef] [Green Version]
- Gallage, N.J.; Hansen, E.H.; Kannangara, R.; Olsen, C.E.; Motawia, M.S.; Jørgensen, K.; Holme, I.; Hebelstrup, K.; Grisoni, M.; Møller, B.L. Vanillin formation from ferulic acid in Vanilla planifolia is catalysed by a single enzyme. Nat. Commun. 2014, 5, 4037. [Google Scholar] [CrossRef] [Green Version]
- Härtl, K.; Huang, F.C.; Giri, A.P.; Franz-Oberdorf, K.; Frotscher, J.; Shao, Y.; Hoffmann, T.; Schwab, W. Glucosylation of smoke-derived volatiles in grapevine (Vitis vinifera) is catalyzed by a promiscuous resveratrol/guaiacol glucosyltransferase. J. Agric. Food Chem. 2017, 65, 5681–5689. [Google Scholar] [CrossRef]
- Yin, Q.; Shen, G.; Di, S.; Fan, C.; Chang, Z.; Pang, Y. Genome-wide identification and functional characterization of UDP-glucosyltransferase genes involved in flavonoid biosynthesis in Glycine max. Plant Cell Physiol. 2017, 58, 1558–1572. [Google Scholar] [CrossRef]
- Yin, Q.; Shen, G.; Chang, Z.; Tang, Y.; Gao, H.; Pang, Y. Involvement of three putative glucosyltransferases from the UGT72 family in flavonol glucoside/rhamnoside biosynthesis in Lotus japonicus seeds. J. Exp. Bot. 2017, 68, 597–612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pang, Y.; Peel, G.J.; Sharma, S.B.; Tang, Y.; Dixon, R.A. A transcript profiling approach reveals an epicatechin-specific glucosyltransferase expressed in the seed coat of Medicago truncatula. Proc. Natl. Acad. Sci. USA 2008, 105, 14210–14215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Witte, S.; Moco, S.; Vervoort, J.; Matern, U.; Martens, S. Recombinant expression and functional characterisation of regiospecific flavonoid glucosyltransferases from Hieracium pilosella L. Planta 2009, 229, 1135–1146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, E.K.; Doucet, C.J.; Li, Y.; Elias, L.; Worrall, D.; Spencer, S.P.; Ross, J.; Bowles, D.J. The activity of Arabidopsis glycosyltransferases toward salicylic acid, 4-hydroxybenzoic acid, and other benzoates. J. Biol. Chem. 2002, 277, 586–592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, E.K.; Baldauf, S.; Li, Y.; Elias, L.; Worrall, D.; Spencer, S.P.; Jackson, R.G.; Taguchi, G.; Ross, J.; Bowles, D.J. Evolution of substrate recognition across a multigene family of glycosyltransferases in Arabidopsis. Glycobiology 2003, 13, 139–145. [Google Scholar] [CrossRef]
- Loutre, C.; Dixon, D.P.; Brazier, M.; Slater, M.; Cole, D.J.; Edwards, R.; Group, C.P.; Sciences, B.; Road, F. Isolation of a glucosyltransferase from Arabidopsis thaliana active in the metabolism of the persistent pollutant. Plant J. 2003, 34, 485–493. [Google Scholar] [CrossRef]
- Brazier-Hicks, M.; Gershater, M.; Dixon, D.; Edwards, R. Substrate specificity and safener inducibility of the plant UDP-glucose-dependent family 1 glycosyltransferase super-family. Plant Biotechnol. J. 2018, 16, 337–348. [Google Scholar] [CrossRef]
- Scheibenzuber, S.; Hoffmann, T.; Effenberger, I.; Schwab, W.; Asam, S.; Rychlik, M. Enzymatic synthesis of modified Alternaria mycotoxins using a whole-cell biotransformation system. Toxins 2020, 12, 264. [Google Scholar] [CrossRef] [Green Version]
- Chapelle, A. Caractérisation de Gènes de β-Glucosidase et d’UDP-Glycosyltransférase Potentiellement Impliqués dans la Lignification Chez Arabidopsis thaliana. Ph.D. Thesis, l’Université Paris 11, Orsay, Paris, France, 2009. Thèse Pour L’obtention du Grade Docteur en Sci. [Google Scholar]
- Han, D.Y.; Lee, H.R.; Kim, B.G.; Ahn, J.H. Biosynthesis of ferulic acid 4-O-glucoside and feruloyl glucoside using Escherichia coli harboring regioselective glucosyltransferases. Appl. Biol. Chem. 2016, 59, 481–484. [Google Scholar] [CrossRef]
- He, X.; Zhao, X.; Gao, L.; Shi, X.; Dai, X.; Liu, Y.; Xia, T.; Wang, Y. Isolation and characterization of key genes that promote flavonoid accumulation in purple-leaf tea (Camellia sinensis L.). Sci. Rep. 2018, 8, 130. [Google Scholar] [CrossRef] [Green Version]
- Sun, G.; Strebl, M.; Merz, M.; Blamberg, R.; Huang, F.C.; McGraphery, K.; Hoffmann, T.; Schwab, W. Glucosylation of the phytoalexin N-feruloyl tyramine modulates the levels of pathogen-responsive metabolites in Nicotiana benthamiana. Plant J. 2019, 100, 20–37. [Google Scholar] [CrossRef] [PubMed]
- Effenberger, I.; Hoffmann, T.; Jonczyk, R.; Schwab, W. Novel biotechnological glucosylation of high-impact aroma chemicals, 3(2H)- and 2(5H)-furanones. Sci. Rep. 2019, 9, 10943. [Google Scholar] [CrossRef] [Green Version]
- Kurze, E.; Ruß, V.; Syam, N.; Effenberger, I.; Jonczyk, R.; Liao, J.; Song, C.; Hoffmann, T.; Schwab, W. Glucosylation of (±)-menthol by uridine-diphosphate-sugar dependent glucosyltransferases from plants. Molecules 2021, 26, 5511. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.; Liu, C. ATP-binding cassette-like transporters are involved in the transport of lignin precursors across plasma and vacuolar membranes. Proc. Natl. Acad. Sci. USA 2010, 107, 22728–22733. [Google Scholar] [CrossRef] [Green Version]
- Vermaas, J.V.; Dixon, R.A.; Chen, F.; Mansfield, S.D.; Boerjan, W.; Ralph, J.; Crowley, M.F.; Beckham, G.T. Passive membrane transport of lignin-related compounds. Proc. Natl. Acad. Sci. USA 2019, 116, 23117–23123. [Google Scholar] [CrossRef] [PubMed]
- Perkins, M.; Smith, R.A.; Samuels, L. The transport of monomers during lignification in plants: Anything goes but how? Curr. Opin. Biotechnol. 2019, 56, 69–74. [Google Scholar] [CrossRef]
- Miyagawa, Y.; Tobimatsu, Y.; Ying Lam, P.; Mizukami, T.; Sakurai, S.; Kamitakahara, H.; Takano, T. Possible mechanisms for generation of phenyl-glycoside-type lignin-carbohydrate linkages in lignification with monolignol glucosides. Plant J. 2020, 104, 156–170. [Google Scholar] [CrossRef] [PubMed]
- Alejandro, S.; Lee, Y.; Tohge, T.; Sudre, D.; Osorio, S.; Park, J.; Bovet, L.; Lee, Y.; Geldner, N.; Fernie, A.R.; et al. AtABCG29 is a monolignol transporter involved in lignin biosynthesis. Curr. Biol. 2012, 22, 1207–1212. [Google Scholar] [CrossRef] [Green Version]
- Väisänen, E.; Takahashi, J.; Obudulu, O.; Bygdell, J.; Karhunen, P.; Blokhina, O.; Laitinen, T.; Teeri, T.H.; Wingsle, G.; Fagerstedt, K..V.; et al. Hunting monolignol transporters: Membrane proteomics and biochemical transport assays with membrane vesicles of Norway spruce. J. Exp. Bot. 2020, 71, 6379–6395. [Google Scholar] [CrossRef]
- Zhao, Q.; Nakashima, J.; Chen, F.; Yin, Y.; Fu, C.; Yun, J.; Shao, H.; Wang, X.; Wang, Z.-Y.; Dixon, R.A. LACCASE is necessary and nonredundant with PEROXIDASE for lignin polymerization during vascular development in Arabidopsis. Plant Cell 2013, 25, 3976–3987. [Google Scholar] [CrossRef] [Green Version]
- Perkins, M.L.; Schuetz, M.; Unda, F.; Smith, R.A.; Sibout, R.; Hoffmann, N.J.; Wong, D.C.J.; Castellarin, S.D.; Mansfield, S.D.; Samuels, L. Dwarfism of high-monolignol Arabidopsis plants is rescued by ectopic LACCASE overexpression. Plant Direct 2020, 4, e00265. [Google Scholar] [CrossRef] [PubMed]
- Herrero, J.; Fernández-Pérez, F.; Yebra, T.; Novo-Uzal, E.; Pomar, F.; Pedreño, M.Á.; Cuello, J.; Guéra, A.; Esteban-Carrasco, A.; Zapata, J.M. Bioinformatic and functional characterization of the basic peroxidase 72 from Arabidopsis thaliana involved in lignin biosynthesis. Planta 2013, 237, 1599–1612. [Google Scholar] [CrossRef] [PubMed]
- Herrero, J.; Esteban-Carrasco, A.; Zapata, J.M. Looking for Arabidopsis thaliana peroxidases involved in lignin biosynthesis. Plant Physiol. Biochem. 2013, 67, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Le Roy, J.; Huss, B.; Creach, A.; Hawkins, S.; Neutelings, G. Glycosylation is a major regulator of phenylpropanoid availability and biological activity in plants. Front. Plant Sci. 2016, 7, 735. [Google Scholar] [CrossRef] [Green Version]
- Samanta, A.; Das, G.; Das, K.S. Roles of flavonoids in plants. Int. J. Pharm. Sci. Technol. 2011, 6, 12–35. [Google Scholar]
- Behr, M.; Neutelings, G.; El Jaziri, M.; Baucher, M. You want it sweeter: How glycosylation affects plant response to oxidative stress. Front. Plant Sci. 2020, 11, 571399. [Google Scholar] [CrossRef]
- Rauf, A.; Imran, M.; Abu-Izneid, T.; Iahtisham-Ul-Haq; Patel, S.; Pan, X.; Naz, S.; Silva, S.A.; Saeed, F.; Suleria, R.H.A. Proanthocyanidins: A comprehensive review. Biomed. Pharmacother. 2019, 116, 108999. [Google Scholar] [CrossRef]
- Dixon, R.A.; Sarnala, S. Proanthocyanidin biosynthesis—A matter of protection. Plant Physiol. 2020, 184, 579–591. [Google Scholar] [CrossRef]
- Pang, Y.; Cheng, X.; Huhman, D.V.; Ma, J.; Peel, G.J.; Yonekura-Sakakibara, K.; Saito, K.; Shen, G.; Sumner, L.W.; Tang, Y.; et al. Medicago glucosyltransferase UGT72L1: Potential roles in proanthocyanidin biosynthesis. Planta 2013, 238, 139–154. [Google Scholar] [CrossRef]
- Zhao, J.; Dixon, R.A. MATE transporters facilitate vacuolar uptake of epicatechin 3′-O-glucoside for proanthocyanidin biosynthesis in Medicago truncatula and Arabidopsis. Plant Cell 2009, 21, 2323–2340. [Google Scholar] [CrossRef] [Green Version]
- Rehman, H.M.; Nawaz, M.A.; Shah, Z.H.; Ludwig-Müller, J.; Chung, G.; Ahmad, M.Q.; Yang, S.H.; Lee, S.I. Comparative genomic and transcriptomic analyses of Family-1 UDP glycosyltransferase in three Brassica species and Arabidopsis indicates stress-responsive regulation. Nature 2018, 8, 1875. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gourlay, G.; Constabel, C.P. Condensed tannins are inducible antioxidants and protect hybrid poplar against oxidative stress. Tree Physiol. 2019, 39, 345–355. [Google Scholar] [CrossRef] [PubMed]
- Yin, R.; Han, K.; Heller, W.; Albert, A.; Dobrev, P.I.; Zažímalová, E.; Schäffner, A.R. Kaempferol 3-O-rhamnoside-7-O-rhamnoside is an endogenous flavonol inhibitor of polar auxin transport in Arabidopsis shoots. New Phytol. 2014, 201, 466–475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.; Li, C.Y.; Qi, Y.; Park, S.; Gibson, S.I. SIS8, a putative mitogen-activated protein kinase kinase kinase, regulates sugar-resistant seedling development in Arabidopsis. Plant J. 2014, 77, 577–588. [Google Scholar] [CrossRef] [Green Version]
- Feucht, W.; Dithmar, H.; Polster, J. Nuclei of tea flowers as targets for flavanols. Plant Biol. 2004, 6, 696–701. [Google Scholar] [CrossRef]
- Feucht, W.; Treutter, D.; Polster, J. Flavanol binding of nuclei from tree species. Plant Cell Rep. 2004, 22, 430–436. [Google Scholar] [CrossRef]
- Saslowsky, D.E.; Warek, U.; Winkel, B.S.J. Nuclear localization of flavonoid enzymes in Arabidopsis. J. Biol. Chem. 2005, 280, 23735–23740. [Google Scholar] [CrossRef] [Green Version]
- Hernández, I.; Alegre, L.; Van Breusegem, F.; Munné-Bosch, S. How relevant are flavonoids as antioxidants in plants? Trends Plant Sci. 2009, 14, 125–132. [Google Scholar] [CrossRef]
- Feucht, W.; Schmid, M.; Treutter, D. DNA and flavonoids leach out from active nuclei of Taxus and Tsuga after extreme climate stresses. Plants 2015, 4, 710–727. [Google Scholar] [CrossRef]
- Inoue, S.; Morita, R.; Kuwata, K.; Kunieda, T.; Ueda, H.; Hara-Nishimura, I.; Minami, Y. Tissue-specific and intracellular localization of indican synthase from Polygonum tinctorium. Plant Physiol. Biochem. 2018, 132, 138–144. [Google Scholar] [CrossRef]
- Cheng, X.; Muhammad, A.; Li, G.; Zhang, J.; Cheng, J.; Qiu, J.; Jiang, T.; Jin, Q.; Cai, Y.; Lin, Y. Family-1 UDP glycosyltransferases in pear (Pyrus bretschneideri): Molecular identification, phylogenomic characterization and expression profiling during stone cell formation. Mol. Biol. Rep. 2019, 46, 2153–2175. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.P.; Liu, B.; Sun, Y.; Chiang, V.L.; Sederoff, R.R. Enzyme-enzyme interactions in monolignol biosynthesis. Front. Plant Sci. 2019, 9, 1942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bassard, J.E.; Richert, L.; Geerinck, J.; Renault, H.; Duval, F.; Ullmann, P.; Schmitt, M.; Meyer, E.; Mutterer, J.; Boerjan, W.; et al. Protein-protein and protein-membrane associations in the lignin pathway. Plant Cell 2012, 24, 4465–4482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saunders, J.A.; McClure, J.W. The distribution of flavonoids in chloroplasts of twenty five species of vascular plants. Phytochemistry 1976, 15, 809–810. [Google Scholar] [CrossRef]
- Zaprometov, M.N.; Nikolaeva, T.N. Chloroplasts isolated from kidney bean leaves are capable of phenolic compound biosynthesis. Russ. J. Plant Physiol. 2003, 50, 623–626. [Google Scholar] [CrossRef]
- Liu, Y.; Gao, L.; Xia, T.; Zhao, L. Investigation of the site-specific accumulation of catechins in the tea plant (Camellia sinensis (L.) O. Kuntze) via vanillin-HCl staining. J. Agric. Food Chem. 2009, 57, 10371–10376. [Google Scholar] [CrossRef]
- Khorobrykh, S.; Havurinne, V.; Mattila, H.; Tyystjärvi, E. Oxygen and ROS in photosynthesis. Plants 2020, 9, 91. [Google Scholar] [CrossRef] [Green Version]
- Nagaoka, K.; Hanioka, N.; Ikushiro, S.; Yamano, S.; Narimatsu, S. The effects of N-glycosylation on the glucuronidation of zidovudine and morphine by UGT2B7 expressed in HEK293 cells. Drug Metab. Pharmacokinet. 2012, 27, 388–397. [Google Scholar] [CrossRef]
- Huang, F.C.; Giri, A.; Daniilidis, M.; Sun, G.; Härtl, K.; Hoffmann, T.; Schwab, W. Structural and functional analysis of UGT92G6 suggests an evolutionary link between mono- and disaccharide glycoside-forming transferases. Plant Cell Physiol. 2018, 59, 857–870. [Google Scholar] [CrossRef]
- Chen, Q.; Liu, X.; Hu, Y.; Wang, Y.; Sun, B.; Chen, T.; Luo, Y.; Zhang, Y.; Li, M.; Liu, Z.; et al. Broaden the sugar donor selectivity of blackberry glycosyltransferase UGT78H2 through residual substitutions. Int. J. Biol. Macromol. 2021, 166, 277–287. [Google Scholar] [CrossRef]
- Tiwari, P.; Sangwan, R.S.; Sangwan, N.S. Plant secondary metabolism linked glycosyltransferases: An update on expanding knowledge and scopes. Biotechnol. Adv. 2016, 34, 714–739. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.Y.; Nam, K.H.; Kim, A.R.; Park, J.; Yoo, E.S.; Baik, K.U.; Yu, Y.H.; Park, M.H. In-vitro and in-vivo immunomodulatory effects of syringin. J. Pharm. Pharmacol. 2001, 53, 1287–1294. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Shin, K.M.; Park, H.J.; Jung, H.J.; Kim, H.J.; Lee, Y.S.; Rew, J.H.; Lee, K.T. Anti-inflammatory and antinociceptive effects of sinapyl alcohol and its glucoside syringin. Planta Med. 2004, 70, 1027–1032. [Google Scholar] [CrossRef] [PubMed]
- Niu, H.S.; Liu, I.M.; Cheng, J.T.; Lin, C.L.; Hsu, F.L. Hypoglycemic effect of syringin from Eleutherococcus senticosus in streptozotocin-induced diabetic rats. Planta Med. 2008, 74, 109–113. [Google Scholar] [CrossRef]
- Chu, Y.; Kwon, T.; Nam, J. Enzymatic and metabolic engineering for efficient production of syringin, sinapyl alcohol 4-O-glucoside, in Arabidopsis thaliana. Phytochemistry 2014, 102, 55–63. [Google Scholar] [CrossRef]
- Panche, A.N.; Diwan, A.D.; Chandra, S.R. Flavonoids: An overview. J. Nutr. Sci. 2016, 5, e47. [Google Scholar] [CrossRef] [Green Version]
- Hsu, C.P.; Shih, Y.T.; Lin, B.R.; Chiu, C.F.; Lin, C.C. Inhibitory effect and mechanisms of an anthocyanins- and anthocyanidins-rich extract from purple-shoot tea on colorectal carcinoma cell proliferation. J. Agric. Food Chem. 2012, 60, 3686–3692. [Google Scholar] [CrossRef]
- Vanholme, B.; Desmet, T.; Ronsse, F.; Rabaey, K.; Van Breusegem, F.; De Mey, M.; Soetaert, W.; Boerjan, W. Towards a carbon-negative sustainable bio-based economy. Front. Plant Sci. 2013, 4, 174. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).