Evaluation of Odor Prediction Model Performance and Variable Importance according to Various Missing Imputation Methods
Abstract
1. Introduction
2. Materials and Methods
2.1. Material
2.1.1. Study Area
2.1.2. Data Sampling
2.2. Methods
2.2.1. Missing Imputation
- Single imputation
- Multiple imputations
- K-nearest neighbor (KNN) imputation
2.2.2. Data Preprocessing
- PCA
- PLS
2.2.3. Prediction Methods
- Multiple linear regression
- Support vector machine (SVM)
- Ensemble tree
- Random forest
- 2.
- Extremely randomized tree (Extra Tree)
- 3.
- XGBoost
- Deep neural network (DNN)
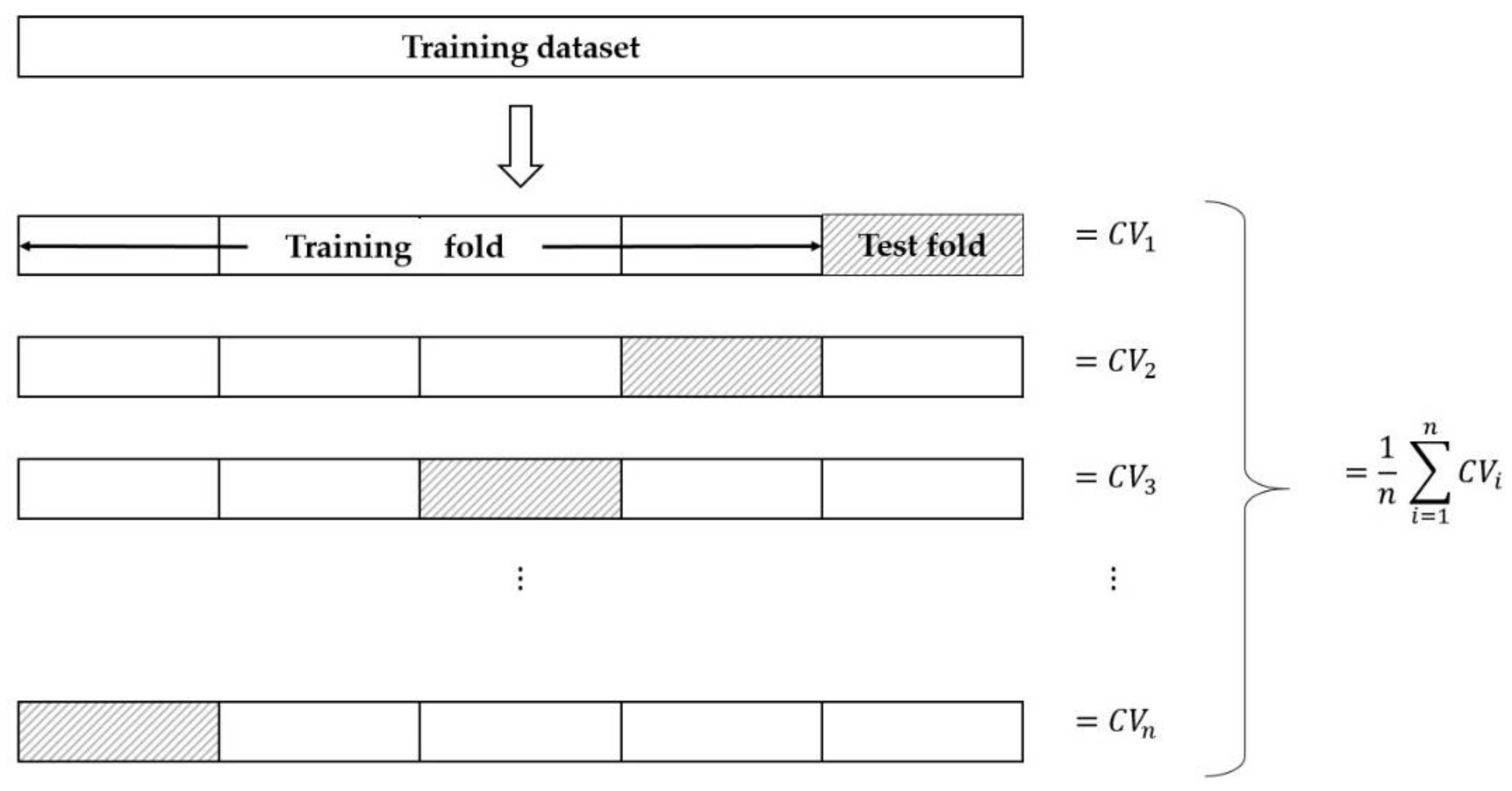
2.2.4. Leave-One-Out Cross-Validation (LOOCV)
3. Results
3.1. Proposed Method
3.2. Missing Imputation
3.3. Data Preprocessing
3.4. Prediction Method Performance
3.5. Variable Importance
4. Discussion and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wojnarowska, M.; Soltysik, M.; Turek, P.; Szakiel, J. Odour nuisance as a consequence of preparation for circular economy. Eur. Res. Stud. J. 2020, 23, 128–142. [Google Scholar] [CrossRef]
- Leníček, J.; Beneš, I.; Rychlíková, E.; Šubrt, D.; Řezníček, O.; Roubal, T.; Pinto, J.P. VOCs and odor episodes near the German–Czech border: Social participation, chemical analyses and health risk assessment. Int. J. Environ. Res. Public Health 2022, 19, 1296. [Google Scholar] [CrossRef] [PubMed]
- Byliński, H.; Gębicki, J.; Namieśnik, J. Evaluation of health hazard due to emission of volatile organic compounds from various processing units of wastewater treatment plant. Int. J. Environ. Res. Public Health 2019, 16, 1712. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.B.; Jeong, S.J. The relationship between odor unit and odorous compounds in control areas using multiple regression analysis. J. Environ. Health Sci. 2009, 35, 191–200. [Google Scholar] [CrossRef]
- Kim, J.B.; Jeong, S.J.; Song, I.S. The concentrations of sulfur compounds and sensation of odor in the residential area around Banwol-Sihwa industrial complex. J. Korean Soc. Atmos. Environ. 2007, 23, 147–157. [Google Scholar] [CrossRef]
- Rincón, C.A.; De Guardia, A.; Couvert, A.; Wolbert, D.; Le Roux, S.; Soutrel, I.; Nunes, G. Odor concentration (OC) prediction based on odor activity values (OAVs) during composting of solid wastes and digestates. Atmos. Environ. 2019, 201, 1–12. [Google Scholar] [CrossRef]
- Man, Z.; Dai, X.; Rong, L.; Kong, X.; Ying, S.; Xin, Y.; Liu, D. Evaluation of storage bags for odour sampling from intensive pig production measured by proton-transfer-reaction mass-spectrometry. Biosyst. Eng. 2020, 189, 48–59. [Google Scholar] [CrossRef]
- Hansen, M.J.; Jonassen, K.E.; Løkke, M.M.; Adamsen, A.P.S.; Feilberg, A. Multivariate prediction of odor from pig production based on in-situ measurement of odorants. Atmos. Environ. 2016, 135, 50–58. [Google Scholar] [CrossRef]
- Byliński, H.; Sobecki, A.; Gębicki, J. The use of artificial neural networks and decision trees to predict the degree of odor nuisance of post-digestion sludge in the sewage treatment plant process. Sustainability 2019, 11, 4407. [Google Scholar] [CrossRef]
- Yan, L.; Wu, C.; Liu, J. Visual analysis of odor interaction based on support vector regression method. Sensors 2020, 20, 1707. [Google Scholar] [CrossRef]
- Vigneau, E.; Courcoux, P.; Symoneaux, R.; Guérin, L.; Villière, A. Random forests: A machine learning methodology to highlight the volatile organic compounds involved in olfactory perception. Food Qual. Prefer. 2018, 68, 135–145. [Google Scholar] [CrossRef]
- Kang, J.H.; Song, J.; Yoo, S.S.; Lee, B.J.; Ji, H.W. Prediction of odor concentration emitted from wastewater treatment plant using an artificial neural network (ANN). Atmosphere 2020, 11, 784. [Google Scholar] [CrossRef]
- Hidayat, R.; Wang, Z.H. Odor classification in cattle ranch based on electronic nose. Int. J. Data Sci. 2021, 2, 104–111. [Google Scholar]
- Graham, J.W. Missing data analysis: Making it work in the real world. Annu. Rev. Psychol. 2009, 60, 549–576. [Google Scholar] [CrossRef]
- Dinh, D.T.; Huynh, V.N.; Sriboonchitta, S. Clustering mixed numerical and categorical data with missing values. Inf. Sci. 2021, 571, 418–442. [Google Scholar] [CrossRef]
- Li, X.; Wu, Z.; Zhao, Z.; Ding, F.; He, D. A mixed data clustering algorithm with noise-filtered distribution centroid and iterative weight adjustment strategy. Inf. Sci. 2021, 577, 697–721. [Google Scholar] [CrossRef]
- Jensen, B.B.; Jørgensen, H. Effect of dietary fiber on microbial activity and microbial gas production in various regions of the gastrointestinal tract of pigs. Appl. Environ. Microbiol. 1994, 60, 1897–1904. [Google Scholar] [CrossRef]
- Jang, Y.N.; Jung, M.W. Biochemical changes and biological origin of key odor compound generations in pig slurry during indoor storage periods: A pyrosequencing approach. BioMed Res. Int. 2018, 2018, 3503658. [Google Scholar] [CrossRef]
- Ministry of Environment (ME). Odor Management Manual; Ministry of Environment (ME): Sejong-si, Korea, 2012.
- Jang, Y.N.; Hwang, O.; Jung, M.W.; Ahn, B.K.; Kim, H.; Jo, G.; Yun, Y.M. Comprehensive analysis of microbial dynamics linked with the reduction of odorous compounds in a full-scale swine manure pit recharge system with recirculation of aerobically treated liquid fertilizer. Sci. Total Environ. 2021, 777, 146122. [Google Scholar] [CrossRef]
- Moriasi, D.N.; Gitau, M.W.; Pai, N.; Daggupati, P. Hydrologic and water quality models: Performance measures and evaluation criteria. Trans. ASABE 2015, 58, 1763–1785. [Google Scholar]
- Burnaev, E.; Vovk, V. Efficiency of conformalized ridge regression. In Proceedings of the Conference on Learning Theory, Barcelona, Spain, 13–15 June 2014; PMLR: USA, 2014; pp. 605–622. [Google Scholar]
- Geurts, P.; Ernst, D.; Wehenkel, L. Extremely randomized trees. Mach. Learn. 2006, 63, 3–42. [Google Scholar] [CrossRef]
- Schafer, J.L. Multiple imputation: A primer. Stat. Methods Med. Res. 1999, 8, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Li, J. K-nearest neighbor based missing data estimation algorithm in wireless sensor networks. Wirel. Sens. Netw. 2010, 2, 115. [Google Scholar] [CrossRef]
- Abdi, H.; Williams, L.J. Principal component analysis. Wiley Interdiscip. Rev. Comput. Statistics 2010, 2, 433–459. [Google Scholar] [CrossRef]
- Tobias, R.D. An introduction to partial least squares regression. In Proceedings of the Twentieth Annual SAS Users Group International Conference, Orlando, FL, USA, 2–5 April 1995; SAS Institute Inc.: Cary, NC, USA, 1995. [Google Scholar]
- Pradhan, A. Support vector machine-a survey. Int. J. Emerg. Technol. Adv. Eng. 2012, 2, 82–85. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Friedman, J.H. Greedy function approximation: A gradient boosting machine. Ann. Stat. 2001, 29, 1189–1232. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. Xgboost: A scalable tree boosting system. In Proceedings of the 22nd ACM Sigkdd International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
- Hecht-Nielsen, R. Theory of the backpropagation neural network. In Neural Networks for Perception; Academic Press: Cambridge, MA, USA, 1992; pp. 65–93. [Google Scholar]
- Kohavi, R. A study of cross-validation and bootstrap for accuracy estimation and model selection. In Proceedings of the International Joint Conference on Artificial Intelligence, San Francisco, CA, USA, 20–25 August 1995; pp. 1137–1145. [Google Scholar]
- Strobl, C.; Boulesteix, A.L.; Zeileis, A.; Hothorn, T. Bias in random forest variable importance measures: Illustrations, sources and a solution. BMC Bioinform. 2007, 8, 1–21. [Google Scholar] [CrossRef]
- Wei, P.; Lu, Z.; Song, J. Variable importance analysis: A comprehensive review. Reliab. Eng. Syst. Saf. 2015, 142, 399–432. [Google Scholar] [CrossRef]
- Faizin, R.N.; Riasetiawan, M.; Ashari, A. A Review of Missing Sensor Data Imputation Methods. In Proceedings of the 2019 5th International Conference on Science and Technology (ICST), Yogyakarta, Indonesia, 30–31 July 2019; pp. 1–6. [Google Scholar]
- Heskes, T.; Sijben, E.; Bucur, I.G.; Claassen, T. Causal shapley values: Exploiting causal knowledge to explain individual predictions of complex models. Adv. Neural Inf. Process. Syst. 2020, 33, 4778–4789. [Google Scholar]
- Lundberg, S.M.; Lee, S.I. A unified approach to interpreting model predictions. In Proceedings of the Advances in Neural Information Processing Systems 30, Long Beach, CA, USA, 4–9 December 2017. [Google Scholar]
- Das, A.; Rad, P. Opportunities and challenges in explainable artificial intelligence (xai): A survey. arXiv 2020, arXiv:2006.11371. [Google Scholar]
- Wojnarowska, M.; Ilba, M.; Szakiel, J.; Turek, P.; Sołtysik, M. Identifying the location of odour nuisance emitters using spatial GIS analyses. Chemosphere 2021, 263, 128252. [Google Scholar] [CrossRef] [PubMed]
- Velič, A.; Šurina, I.; Ház, A.; Vossen, F.; Ivanova, I.; Soldán, M. Modeling of Odor from a Particleboard Production Plant. J. Wood Chem. Technol. 2020, 40, 116–125. [Google Scholar] [CrossRef]













| Class | Variable | Sampling | Analytical Instrument | Analytical Conditions |
|---|---|---|---|---|
| Response Variable | Complex odor | Lung Sampler and Polyester Aluminum bag (10 L) | Air dilution method, Korea | |
| Explanatory Variable | Ammonia | Solution Absorption | UV/vis (Shimadzu) | Wavelength range 640 nm |
| Sulfur Compounds | Lung Sampler and Polyester Aluminum bag (10 L) | GC/PFPD (456-GC, Scion instruments) | Column: CP-Sil 5CB (60 m × 0.32 mm × 5 μm) Oven Condition: 60 °C (3 min) → (8 °C/min) → 160 °C (9 min) | |
| VOCs | Tenax TA tube adsorption | GC/FID (CP-3800, Varian) | Column: DB-WAX (30 m × 0.25 mm × 0.25 μm) Oven Condition: 40 °C → (8 °C/min) → 150 °C → (10 °C/min) → 230 °C |
| Class | Name (Abbreviation) | Unit | Missing/Total | Type | ||
|---|---|---|---|---|---|---|
| Response variables | Complex Odor (OC) | 0/57 | Float64 | |||
| Explanatory variables | Type | Name (abbreviation) | Unit | MDL * | Missing/Total | Type |
| Ammonia | Ammonia (NH3) | ppm | 0.08 | 0/57 | Float64 | |
| Sulfur compounds | Hydrogen sulfide (H2S) | ppb | 0.06 | 0/57 | Float64 | |
| Methyl mercaptan (MM) | 0.07 | 31/57 | Float64 | |||
| Dimethyl sulfide (DMS) | 0.08 | 36/57 | Float64 | |||
| Dimethyl disulfide (DMDS) | 0.05 | 52/57 | Float64 | |||
| Volatile Organic Compounds (VOCs) | Acetic acid (ACA) | ppb | 0.07 | 0/57 | Float64 | |
| Propanoic acid (PPA) | 0.34 | 1/57 | Float64 | |||
| Isobutyric acid (IBA) | 0.52 | 1/57 | Float64 | |||
| Normality butyric acid (BTA) | 0.96 | 1/57 | Float64 | |||
| Isovaleric acid (IVA) | 0.49 | 0/57 | Float64 | |||
| Normality valeric acid (VLA) | 0.53 | 1/57 | Float64 | |||
| Phenol (Ph) | ppb | 0.09 | 2/57 | Float64 | ||
| P-Cresol (p-C) | 0.06 | 1/57 | Float64 | |||
| Indole (ID) | ppb | 0.40 | 21/57 | Float64 | ||
| Skatole (SK) | 0.38 | 8/57 | Float64 | |||
| General Notations | |
| n | Number of data |
| i-th observation value of response variable | |
| Imputation parameter | |
| m | Number of imputation datasets in multiple imputations |
| k | Number of neighbors in K-nearest neighbor imputation |
| Data preprocessing | |
| p | Number of variables |
| k | Number of principal components in principal components analysis |
| i-th principal component in principal components analysis | |
| T | The latent variable made by partial least square |
| Prediction methods | |
| p-th regression coefficient in multiple regression | |
| i-th unexplained error in multiple regression | |
| i-th observation value of p-th explanatory variable in multiple regression | |
| k | Number of decision tree in random forest |
| i-th observation value of explanatory variable | |
| Leave-One-Out Cross-Validation | |
| k | Number of fold in LOOCV |
| i-th cross-validation result value | |
| Rank | R2 | MAE | Method | ||
|---|---|---|---|---|---|
| Imputation | Data Preprocessing | Prediction Method | |||
| 1 | 0.376 | 729.06 | Multivariate imputation using Bayesian ridge | Standardization | Extra Tree |
| 2 | 0.283 | 849.32 | KNN imputation | PCA | XGBoost |
| 3 | 0.281 | 822.88 | KNN imputation | PCA | Random forest |
| 4 | 0.267 | 783.74 | Multivariate imputation using Extra Tree | Standardization | Extra Tree |
| 5 | 0.238 | 811.28 | KNN imputation | Standardization | Extra Tree |
| Rank | R2 | MAE | Method | ||
|---|---|---|---|---|---|
| Imputation | Data Preprocessing | Prediction Method | |||
| 1 | 0.315 | 939.80 | Simple imputation using median | Standardization | Extra Tree |
| 2 | 0.311 | 964.68 | Simple imputation using mean | Standardization | Extra Tree |
| 3 | 0.311 | 982.80 | Multivariate imputation using Bayesian ridge | Standardization | Extra Tree |
| 4 | 0.305 | 961.72 | KNN imputation | Standardization | Extra Tree |
| 5 | 0.268 | 959.85 | Multivariate imputation using Bayesian ridge | PCA | Extra Tree |
| Univariate Imputation | Multivariate Imputation | Multiple Imputation | KNN Imputation | ||||
|---|---|---|---|---|---|---|---|
| Mean | Median | Bayesian Ridge | Extra Tree | Bayesian Ridge | Gaussian Process Regression | ||
| Ammonia | 0.314 | 0.314 | 0.314 | 0.314 | 0.314 | 0.314 | 0.314 |
| Hydrogen sulfide | −0.020 | −0.020 | −0.020 | −0.020 | −0.020 | −0.020 | −0.020 |
| Methyl mercaptan | 0.272 | 0.361 | 0.357 | 0.465 | 0.335 | 0.420 | 0.535 |
| Dimethyl sulfide | −0.173 | −0.125 | −0.217 | −0.026 | −0.141 | −0.116 | −0.184 |
| Acetic acid | 0.331 | 0.331 | 0.331 | 0.331 | 0.331 | 0.331 | 0.331 |
| Propanoic acid | 0.218 | 0.218 | 0.225 | 0.220 | 0.226 | 0.227 | 0.223 |
| Isobutyric acid | 0.203 | 0.204 | 0.211 | 0.208 | 0.212 | 0.213 | 0.209 |
| Normality butyric acid | 0.174 | 0.177 | 0.181 | 0.180 | 0.181 | 0.184 | 0.180 |
| Isovaleric acid | 0.147 | 0.147 | 0.147 | 0.147 | 0.147 | 0.147 | 0.147 |
| Normality valeric acid | 0.154 | 0.159 | 0.163 | 0.163 | 0.163 | 0.163 | 0.163 |
| Phenol | 0.208 | 0.214 | 0.225 | 0.225 | 0.221 | 0.224 | 0.223 |
| P-Cresol | 0.322 | 0.325 | 0.334 | 0.333 | 0.332 | 0.334 | 0.333 |
| Indole | 0.220 | 0.237 | 0.289 | 0.256 | 0.195 | 0.279 | 0.258 |
| Skatole | 0.150 | 0.175 | 0.204 | 0.200 | 0.171 | 0.225 | 0.178 |
| Loading | |||||
|---|---|---|---|---|---|
| PC1 | PC2 | PC3 | PC4 | ||
| Ammonia | Ammonia | 0.080 | −0.672 | 0.000 | −0.310 |
| Sulfur compounds | Hydrogen sulfide | 0.038 | −0.062 | 0.949 | −0.114 |
| Methyl mercaptan | 0.045 | −0.300 | 0.086 | 0.762 | |
| Dimethyl sulfide | 0.092 | 0.448 | 0.144 | −0.220 | |
| Volatile Organic Compounds (VOCs) | Acetic acid | 0.263 | −0.248 | −0.148 | −0.383 |
| Propanoic acid | 0.331 | 0.075 | −0.062 | −0.085 | |
| Isobutyric acid | 0.337 | 0.043 | −0.064 | −0.076 | |
| Normality butyric acid | 0.294 | 0.251 | 0.054 | −0.054 | |
| Isovaleric acid | 0.328 | 0.179 | 0.003 | 0.121 | |
| Normality valeric acid | 0.321 | 0.149 | −0.019 | 0.173 | |
| Phenol | 0.328 | −0.036 | −0.094 | 0.104 | |
| P-Cresol | 0.326 | −0.179 | 0.034 | −0.074 | |
| Indole | 0.298 | −0.183 | 0.165 | 0.093 | |
| Skatole | 0.300 | −0.003 | 0.034 | 0.174 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, D.-H.; Woo, S.-E.; Jung, M.-W.; Heo, T.-Y. Evaluation of Odor Prediction Model Performance and Variable Importance according to Various Missing Imputation Methods. Appl. Sci. 2022, 12, 2826. https://doi.org/10.3390/app12062826
Lee D-H, Woo S-E, Jung M-W, Heo T-Y. Evaluation of Odor Prediction Model Performance and Variable Importance according to Various Missing Imputation Methods. Applied Sciences. 2022; 12(6):2826. https://doi.org/10.3390/app12062826
Chicago/Turabian StyleLee, Do-Hyun, Saem-Ee Woo, Min-Woong Jung, and Tae-Young Heo. 2022. "Evaluation of Odor Prediction Model Performance and Variable Importance according to Various Missing Imputation Methods" Applied Sciences 12, no. 6: 2826. https://doi.org/10.3390/app12062826
APA StyleLee, D.-H., Woo, S.-E., Jung, M.-W., & Heo, T.-Y. (2022). Evaluation of Odor Prediction Model Performance and Variable Importance according to Various Missing Imputation Methods. Applied Sciences, 12(6), 2826. https://doi.org/10.3390/app12062826






