Abstract
Brain tumor detection in the initial stage is becoming an intricate task for clinicians worldwide. The diagnosis of brain tumor patients is rigorous in the later stages, which is a serious concern. Although there are related pragmatic clinical tools and multiple models based on machine learning (ML) for the effective diagnosis of patients, these models still provide less accuracy and take immense time for patient screening during the diagnosis process. Hence, there is still a need to develop a more precise model for more accurate screening of patients to detect brain tumors in the beginning stages and aid clinicians in diagnosis, making the brain tumor assessment more reliable. In this research, a performance analysis of the impact of different generative adversarial networks (GAN) on the early detection of brain tumors is presented. Based on it, a novel hybrid enhanced predictive convolution neural network (CNN) model using a hybrid GAN ensemble is proposed. Brain tumor image data is augmented using a GAN ensemble, which is fed for classification using a hybrid modulated CNN technique. The outcome is generated through a soft voting approach where the final prediction is based on the GAN, which computes the highest value for different performance metrics. This analysis demonstrated that evaluation with a progressive-growing generative adversarial network (PGGAN) architecture produced the best result. In the analysis, PGGAN outperformed others, computing the accuracy, precision, recall, F1-score, and negative predictive value (NPV) to be 98.85, 98.45%, 97.2%, 98.11%, and 98.09%, respectively. Additionally, a very low latency of 3.4 s is determined with PGGAN. The PGGAN model enhanced the overall performance of the identification of brain cell tissues in real time. Therefore, it may be inferred to suggest that brain tumor detection in patients using PGGAN augmentation with the proposed modulated CNN technique generates the optimum performance using the soft voting approach.
1. Introduction
Accurate diagnosis of brain tumors in the initial phase is very essential to averting the global death rate due to cancer-related illness [1]. Proper segmentation of brain tumors is indeed a key challenge and time-consuming procedure in the modern era due to the constantly increasing number of brain tumor patients all across the world [2]. To diagnose brain tumors accurately, radiologists adopt magnetic resonance (MR) imaging methods to visualize the inner shape of a patient’s brain securely and reduce the chances of any surgery [3,4]. MR imaging helps radiologists provide necessary data regarding a patient soft cell tissues that assist the doctors while handling cases related to the brain tumor in the real-time diagnosis of a patient [5,6,7]. Accurate segments of MR images are required to help doctors with the correct identification of brain tumors using multifarious computer-aided medical instruments. While the acquired MR images are being segmented accurately, the brain tumors may be categorized into two diverse categories, which are malignant (cancerous) and benign (non-cancerous) [8,9]. Benign (non-cancerous) brain tumors are further categorized into three diverse classes, which are meningiomas, gliomas, and pituitary tumors. The precise and timely identification of tissues is one of the major concerns and demands high focus. It is a tedious task because of the varied traits of the segmented brain tissues, such as the dimension and shape of the tissues as well as their accurate position with the intensity of the gray level [10,11,12]. Figure 1 illustrates the categorization of the brain tumor.
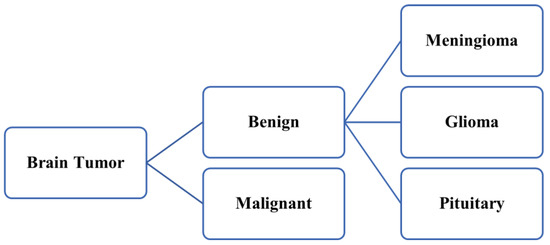
Figure 1.
Categorization of brain tumor.
Several brain tumor detection techniques aid in detecting brain tumors at early stages. These techniques may help clinicians continue a pragmatic diagnosis of patients as per the requirements. However, the majority of them have accuracy and latency limitations [13,14]. In addition, the data size used in the study is sparse and suffers from inadequate training [15]. The accurate detection and categorization of normal tissue as well as abnormal tissue is a major challenge. Brain tumor detection in the initial phase is essential for the effective diagnosis of the patients immediately. The main motivation to conduct this research was to overcome the major limitations of the existing work. Encouraged by the success of deep learning, various deep neural networks have proven to help overcome the limitations in the classification and detection of brain tumors. However, regardless of encouraging results, the scarcity of large volumes of data, generating realistic images completely different from the original ones, and the quality of synthesized images remain challenging issues to be addressed [16]. So, there is a need for a computationally interconnected advanced framework that can provide more reliability and accuracy with less response time delay so that it can be diagnosed with more precision. The initial diagnosis mainly emphasizes monitoring the symptoms of the patient facing any disorder related to brain cell tissues [17] Figure 2.
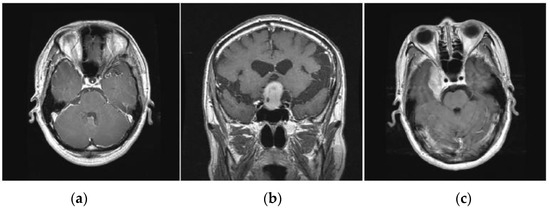
Figure 2.
Various types of brain tumors (a) Glioma (b) Pituitary (c) Meningioma.
Gliomas are positioned within the cerebral hemispheres; however, they may also appear in certain parts of the human brain. Pituitary tumors are positioned within the sella turcica, a minor depression within the skull that covers the pituitary gland. Meningiomas are positioned within the meninges, the membranes that cover the brain as well as the spinal cord.
Furthermore, such approaches can assist clinicians in the identification of brain cell tumors quickly and efficiently. This study aims to demonstrate both the strengths and major limitations of previously suggested brain tumor identification and categorization techniques as addressed in previous works. It also provides a comprehensive assessment of the examined literature analysis and reveals fresh investigation perspectives. Furthermore, this article proposes a new augmented modulated deep learning-based intelligent predictive model for brain tumor detection using a GAN ensemble.
Research Gap and Motivation
Existing clinical tools and ML-based models developed for brain tumor screening are limited in their potential to precisely identify varied-dimension tumors, particularly in the growing phase. This is due to the brain tumors diversity, variation in tumor development, and complex brain structure. Therefore, there must be developed an improvised model to detect brain tumors accurately.
The construction of an advanced and more robust predictive model for brain tumor identification utilizing the GAN ensemble approach has huge potential to address the limitations of existing approaches and improve the diagnosis process. GANs are a type of artificial intelligence (AI) technique that can be utilized to generate realistic images. By modulating the GANs along with ensemble learning, it is feasible to build a predictive model that is more precise and informative than conventional brain tumor screening approaches.
The construction of the proposed model would have a remarkable impact in the area of brain tumor diagnosis research. It would assist the clinical experts and allow accurate detection of brain tumors in the beginning stages, which may lead to enhanced treatment results. In addition to this, this proposed predictive model may be utilized to build novel diagnostic instruments and enhance the understanding of brain tumor development.
This research study has the potential to make a valuable contribution to the area of brain tumor research. The construction of this predictive model for brain tumor detection using a GAN ensemble would allow for early detection of tumors, which would lead to enhanced treatment results. In addition to this, the model may be utilized to build novel clinical instruments and enhance the understanding of brain tumor development.
The main contributions of the research study are as follows:
- In this research analysis, an integrated performance evaluation of different popular GAN approaches is done in the context of brain tumor symptom detection.
- This study proposes a novel augmented modulated deep learning-based advanced predictive model utilizing the voting-based GAN ensemble for the early detection of brain tumors.
- Enhanced hybrid CNN model with PGGAN architecture produced the best outcome recording, i.e., an optimum accuracy, precision, recall, F1-Score, and NPV of 98.85, 98.45%, 97.2%, 98.11%, and 98.09%, respectively. Additionally, the least time latency of 3.4 s is noted with the proposed hybrid model.
- The outcome of the implementation analysis through various performance parameters demonstrates that PGGAN is the most suitable augmented method for data generation with sufficient diversity in comparison with other GANs.
2. Literature Review and Background Study
Multifarious investigations have been conducted during the last decade for the early diagnosis of brain tumors, along with proper categorizations. However, the existing methods have various limitations due to several limitations, such as lower precision and accuracy in numerous cases.
2.1. Relevant Research in Context to Machine Learning (ML) for Brain Tumor Analysis
This section describes the existing research carried out in the field of early detection of diverse types of brain tumors by utilizing machine learning (ML) protocols in the real-time diagnosis of patients. Figure 3 depicts the sample depiction of brain tumor recognition utilizing conventional ML. In conventional ML, the first step is the pre-processing phase, which includes the utilization of the filter for removing unwanted noise for the picture contrast improvement and effective and easy identification of brain tumors.
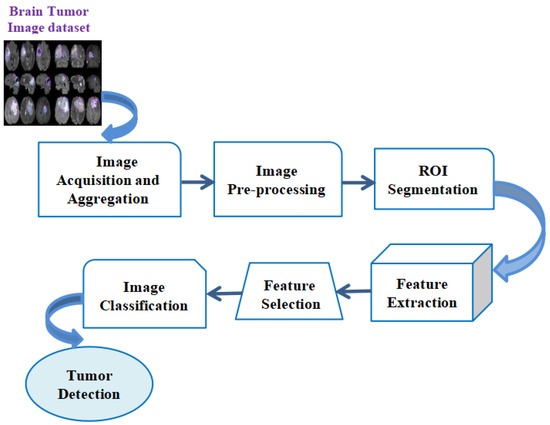
Figure 3.
Sample depiction of brain tumor recognition utilizing conventional Machine Learning.
In the initial pre-processing phase of the input pictures, the picture is first divided into multiple segments by utilizing segmentation methods such as the cluster-rooted scheme, thresholding, and edge-rooted split procedure. Later, the segmentation procedure is performed, and the multiple features extraction procedure is conducted to be rooted on the colored datasets, texture-based contrasts, and dimensions from a region of convergence (ROI). After that, multifarious principal features are to be evaluated by utilizing the feature-chosen method, namely the PCA (Principal Component Analysis) or the statistical examination. The selection of features is a procedure that aids in minimizing the input of a variable within the model by utilizing multiple relevant datasets and helps to eliminate the noise from the datasets. This is a very important procedure for the automated selection of some relevant features for the ML model based on these kinds of issues. Then, the chosen features are to be utilized to feed into the ML classifier, namely the neural network (NN) or the support vector machine (SVM). The chosen classifier may utilize the capture features vector with the destination class labels for determining the optimum boundary that separates every class. While the chosen ML-rooted classifier is to be trained, this could be utilized for the classification of novel, unknown datasets for the evaluation of the classes. The ML classifier would take the optimal features vector as a feed, and the outcome would be the object classes. Table 1 illustrates the methods utilized based on ML protocols for the detection of brain tumors.

Table 1.
Relevant Conventional ML methods for the detection of brain tumors.
2.2. Relevant Research in Context to Deep Learning for Brain Tumor Analysis
Proper brain tumor classification might, in turn, lead to the most suitable medical therapy and, as a result, increase human longevity significantly. Numerous existing approaches have been used to conduct a substantial study on accurate brain tumor tissue identification. However, current approaches for identifying brain tumors have several limitations, such as the lower accuracy of methods used in real-time recognition of MR images to identify and categorize brain tumors appropriately for early treatment of patients. The deep learning models are realistic and more pragmatic in terms of performance measures of various parameters, namely the sensitivity, specificity, or precision in real-time as compared to ML methods due to multifarious reasons, such as minimizing the subjective assessment for brain tumor identifications. Therefore, deep learning models are more secure and capable of training large datasets in real time.
CNN has been widely utilized, and the famous deep learning (DL) rooted prototypical utilized globally to segment and perform categorization procedures of the medicinal pictures. Figure 4 depicts the functional block diagram of brain tumor categorization utilizing the CNN technique. The CNN-rooted brain tumor categorization process is segregated into two diverse stages. One stage is known as the training stage, and another one is recognized as the testing stage. Each brain tumor picture is to be segregated into diverse classes by utilizing the names of the tags, like the tumor as well as without the tumor brain picture, etc. During the entire training stage, various kinds of processes are conducted, such as augmentation, picture categorization, picture accurate pre-processing, along with picture feature extraction to make the prediction prototypical. During the pre-processing stage, picture rescaling is to be employed to alter the picture dimension. Lastly, the CNN technique is to be utilized for automated categorization of the diverse kinds of brain tumors. Table 2 depicts the popular deep learning-based approaches used earlier for the recognition of diverse brain tumors.
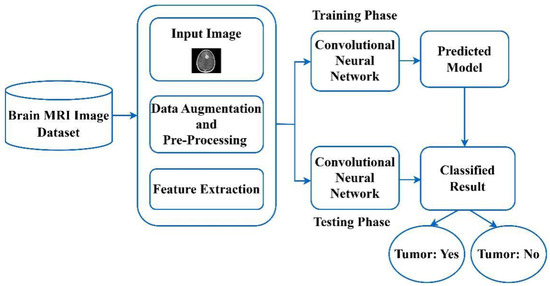
Figure 4.
A functional block diagram of brain tumor categorization utilizing the CNN method.

Table 2.
Popular Deep Learning based models for recognition of the diverse brain tumors.
This section provides a detailed procedure for our search strategy. In this study, the authors conducted an organized literature examination that aligned with the existing papers. For finding the pragmatically relevant work done by various authors, we performed our search utilizing diverse databases, which include PubMed, Scopus (SC), and WoS (Web of Science). The primary aim to perform the search properly was to recognize the most pragmatic studies among the GAN applications and diverse other recent functionality. We have searched the databases by utilizing the diverse kinds of specific synonyms of the major keywords, which are primarily related to the GAN. Further, we utilized various kinds of Boolean operatives, including the “GAN classification” OR “Types of GANs” OR “GAN categorization” OR “GAN for brain tumor” OR ‘GAN in the medical field’ AND ‘GAN for brain tissue recognition’. Furthermore, the WoS and Scopus databases have been fully utilized for identifying the multiple existing published papers by numerous authors in this field during the last decade all across the world. Figure 5 depicts the systematic procedure for relevant publication analysis.
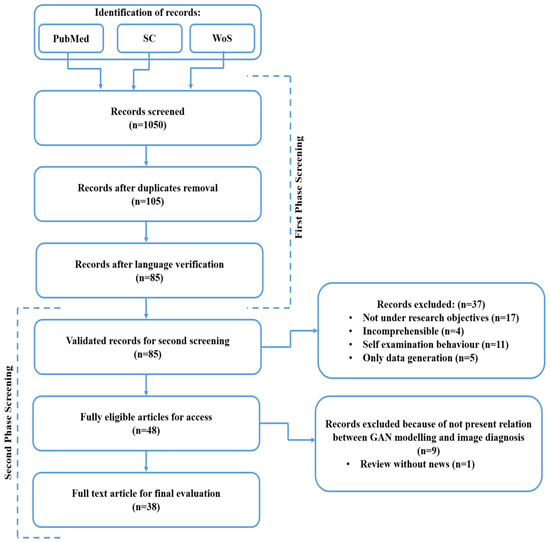
Figure 5.
A systematic procedure for relevant publications analysis.
In this research, a detailed analysis has been conducted utilizing the eligible research and review articles for an organized systematic review as well as a meta-analysis through PRISMA methodology. The PRISMA approach includes a set of algorithms for rational concepts and multiple longitudinal studies, which involve formatting and writing specifications. In this literature review, there is a three-step method that involves research question formation, guidelines to accept or discard the research, and a review of articles and online databases.
Figure 6 illustrates the analysis of articles used in the review process. This review involves 67% journal articles, 22% conference papers, and 11% book chapters in the initial screening and selection of the papers. The inclusion criteria are set on certain questionnaires, e.g., articles aimed at resolving issues with brain tumor identification utilizing AI and ML, search queries should involve the title of the scientific study, the abstract, and the entire body of reputed peer-reviewed journals or conferences. The exclusion criteria involve research articles published with identical findings, studies that are not relevant to brain tumor detection or segmentation, as well as scientific studies published in other languages rather than English.
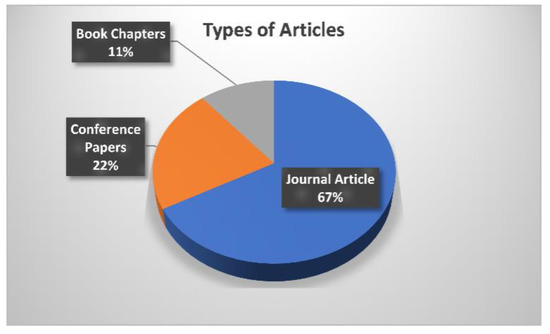
Figure 6.
Analysis of articles used in the review process.
In this study, GAN-based literature searching results offered 1050 diverse records, which are classified through the varied published articles that are associated with the main aim of this analysis. Initially, the recognition of the diverse records was done by utilizing the selected three databases, namely PubMed, SC (Scopus) as well as WoS. After, a total of 1050 screened records were obtained by using the databases. For accurate searching over the databases, we divided the entire screening procedure into two separate screenings, namely first-phase screening and second-phase screening. The records after the duplicate’s removal were identified (n = 105). The records after the language verification have been measured (n = 85). In the second phase screening procedure, validated records for the second screening were (n = 85), fully eligible articles for access were (n = 48) and a full-text article for the final evaluation was found (n = 38). Furthermore, the author filtered out the articles for review by considering the three classes of articles, such as original manuscripts, review manuscripts, and analytical manuscripts.
As observed from the existing literature survey, there exist research models and diverse approaches by different investigators in recent years for significant brain tumor categorization and identification that could be followed through diverse phases, namely prediction of the outcome, categorization, and diagnosis plan. However, such conventional developed models and screening approaches for brain tumor identification have certain restrictions in terms of accuracy in image categorization, increased execution time, and high computational overhead. Therefore, there is a need for the development of a new advanced deep learning-based intelligent predictive model for brain tumor detection, which can detect brain tumor symptoms with enhanced accuracy and minimal time delay, saving the time of clinicians in the real-time screening of the patients and perform diagnosis of patients in a more significant manner.
3. Working Principle of GAN
The tumor has been regarded as the most common cause of death across the globe. The broad diversity in sickness intensity, length of sickness, tumor position, numerous levels of susceptibility, or chemotherapeutic medicines might have been to blame for low tumor categorization. During the last few years, the proportion of individuals suffering from brain cell tissues has increased dramatically. Proper brain tumor categorization may lead to the best appropriate medicinal care and therefore considerably improve human longevity. The GAN (generative adversarial network) is a very powerful and correct approach that is capable of synthesizing novel MR images through the aid of latent vectors in real time. GAN is an ML-rooted framework that comprises a plurality of models such as generators and discriminators.
The GAN works primarily on the diverse three regulations; the beginner one is to make generative prototypical learning and datasets, which may be produced engaging numerous probabilistic illustrations. In the secondary stage, the model training process may be completed in any kind of inconsistent scenario. In the third phase, utilizing DLNN (Deep Learning rooted Neural Network) and utilizing artificial intelligence (AI) rooted protocol to train the entire system. Figure 7 depicts the overall working process of GAN. In the working of the GAN, a selected random input image is translated to the generator to map a noise sample to synthetic data and the synthetic sample images are to be generated. These synthetic sample images are to be used to apply to the discriminator where the obtained real sample images and synthetic sample images are compared and the discriminator separates the real data from the synthetic data in an accurate manner. Furthermore, based on the effective comparison this discriminator classifies the images into real and fake categories. Table 3 illustrates the types of GANs that form the GAN ensemble in our research. The GAN model architecture refers to the specified design of the model, which involves, layers types, the total number of layers used as well as the connection among different layers. The model training process means, the steps used in model training such as datasets utilized in model training, selection of the hyperparameters as well as specific stopping criteria utilized to stop the model training process, accordingly.
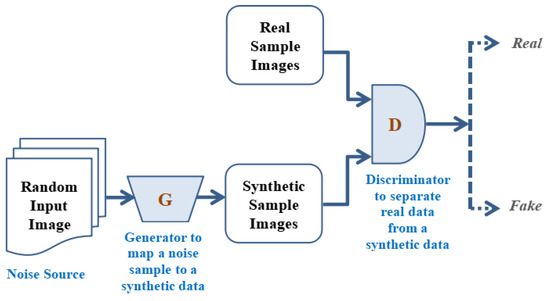
Figure 7.
Working principle of Generative adversarial Network.

Table 3.
Types of GANs that form GAN ensembles in our research [49].
The following is a systematic explanation of the GAN functioning.
Step 1: The operators have to be made utilizing the generator as a means of discriminative network commencing accurate datasets distribution in real-time.
Step 2: Now training the system such that the overall networks accountability level could be enlarged as well as entire discriminator networks may be misled employing creating such applicants, which are not fully combined for instance, which are a stagnant fragment of the distribution of the entire dataset.
Step 3: After that, the entire data could turn into personalizing training datasets for the entire discriminator in real time.
Step 4: To train the entire sampling data, which is accessible until the required accurateness is to be attained.
Step 5: To train the generator for producing the applicants while the entire discriminator is to be misled soon as this is fed with the arbitrary feed in it to perform the required processes in real-time.
Step 6: At last, back-propagation is to be applied for the entire generators along with the discriminator where in previous produces improved pictures, as well as later one, is to be accomplished at declining synthetic pictures.
4. Materials and Methods
This research analysis deals with the development of a novel hybrid and computationally augmented deep learning model for brain tumor detection in patients using GAN. This section discusses the dataset used in the study and the proposed framework for brain tumor assessment.
4.1. Dataset Used in the Study
In this work, the authors utilized available public data suggested by Cheng [60]. This dataset comprises overall 3064 CE-MR images, which include the three diverse kinds of brain tumors (for instance, the meningioma and glioma along with the pituitary) from 233 diversely ill candidates. The entire images utilized in the presented data were chosen in 2-Dimensional and not a single image was chosen in 3-Dimensional volume images in real-time performance measurement of the suggested model. In this investigation, authors encompassed all three of the accessible planes (for instance, the axial as well as coronal, along with the sagittal) images from the presented data. To balance the dataset, there has been selected oversampling technique. The oversampling technique duplicates minority class samples for matching multifarious majority class samples. It is achieved through duplicating samples, arbitrarily or by employing a method, namely the synthetic minority oversampling technique (SMOTE). Table 4 depicts the partition of the brain tumor dataset into training, validation, and test samples.

Table 4.
Partition of Brain tumor Dataset into training, validation, and test samples [60].
4.2. System Configuration
In this work, for experimentation work authors utilized the Ubuntu OS (Operating System) configuration: 18.04.2 LTS, which is maintained through the GeForce Nvidia Giga Texel Shader eXtreme (GTX) 1080 Graphics processing unit (GPU). We have written the entire codes using the PyCharm tool in Python, version v3.8.0 including numerous external libraries, for instance, Keras and TensorFlow, Matplotlib, and many more.
4.3. Proposed Methodology
Multifarious classes of GANs have diverse outcomes according to real-time implementations. The few commonly recognized classes of GAN are DCGAN (Deep Convolutional-Generative Adversarial Network), Conditional GAN, Cycle GAN, Info-GAN, and PG (progressive growing) GAN [48]. In this research work, the authors recognized PGGANs as one of the suitable approaches for the feature extractions of the brain tumor from multiple images as its accuracy, F1-score, and all other similar parameters are better in comparison to the other GANs. Figure 8 illustrates the proposed GAN ensemble Modulated CNN-based predictive model. The GAN ensemble used herein refers to a combined group of diverse GANs, which include PGGAN, Info GAN, Cycle GAN, DCGAN, and Conditional GAN. The key reason for the selection of the GAN ensemble approach in our study is to provide more stability to the model and to find a suitable GAN out of diverse selected GAN classes to detect and classify the brain tumor more accurately and effectively in real-time patients’ diagnosis and aids to the clinicians during the patients screening. The main reason to select the GAN ensemble in comparison to the single GAN is that the GAN ensemble is capable of pragmatic modeling the distribution of the normal dataset, thereby aiding in the detection of brain tumor anomalies in a significant manner.
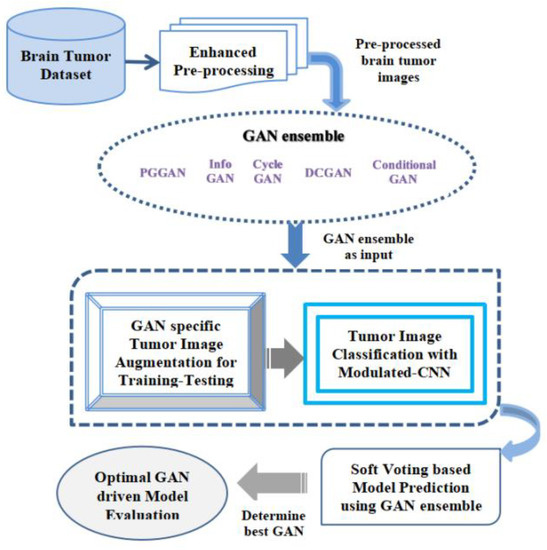
Figure 8.
Proposed GAN ensemble Hybrid CNN-based Predictive Model.
The explanation of the working procedure of the novel hybrid model is described in this section. The initial phase of the suggested model obtains the real-time MR image datasets of the various individuals for brain tumor detection effectively. Brain tumors are constantly becoming a serious concern around the world, as they are a group of irregular cells. These abnormal brain cell tissues are continuously becoming dangerous and cause deaths. For proper identification of the kind of brain tumors, namely the glioma, pituitary as well as meningioma, an accurate dataset sample was collected for accurate performance evaluation using the suggested GAN ensemble hybrid CNN-based predictive model. After obtaining the datasets, the enhanced pre-processing of the collected datasets samples is to be done efficiently with a high degree of precision. In this enhanced pre-processing phase, the entire acquired raw datasets are effectively transformed into clean datasets. The enhanced pre-processing procedure used here mainly consists of four separate phases, which include datasets quality assessment, cleansing of the obtained datasets, the transformation of the dataset, and minimization of the datasets. This image dataset pre-processing stage was applied for making the obtained datasets sample stable and highly reliable for suggested ensemble prediction analysis in real time. In this entire stage, numerous datasets pre-processing methods along with the accurate selection of diverse features of the pictures are to be done in a required manner constantly. The datasets pre-processing describes the translation of accessible datasets before this being fed in the suggested GAN ensemble Hybrid CNN-based predictive model. The authors primarily utilized methods, namely control missed values and obtained datasets’ accurate encoding. In the end, we opted dataset normalization technique to scale up the features in a required manner. After the pre-processing phase, the entire image datasets are to be translated towards the GAN ensemble in real time. Herein, multiple GANs are employed such as the PGGAN, Info GAN, Cycle GAN, DCGAN, and conditional GAN. The entire accessible image datasets were then processed through the chosen GAN ensemble for accurate performance evaluation. In this stage, the entire processed output from the GAN ensemble is further mapped for GAN-specific tumor image augmentation for performing the training and testing of entire accessible datasets for the proper brain cell tissue recognition in real time. This is a critical step, and a high degree of precision is required during the entire training and testing phase of the datasets. The datasets augmentation procedure in the proposed GAN ensemble Hybrid CNN-based predictive model helps in rising the datasets amount artificially as means of producing fresh dataset points through existing datasets. After the completion of the brain tumor picture augmentation that is training and testing, then the collected datasets are translated further for the brain cell tissues categorization procedure employing the modulated-CNN-based approach embedded within the suggested model, which provides more accurate outcomes in the image’s classification procedure. Once the categorization procedure is completed utilizing the suggested modulated-CNN-based approach, the received datasets are further input for the soft voting approach, which is based upon the suggested model prediction utilizing the GAN ensemble approach. In the soft computing approach used in the proposed model, the best GAN is chosen based upon various specified performance parameters such as accuracy, precision, as well as F1-Score, and Sensitivity, accurately with a high degree of precision for the optimal GAN-driven model evaluation in real-time. It is observed that PGGAN is computed to be the most promising GAN for fast, reliable, and accurate image classification during brain tumor categorization.
4.3.1. Pre-Processing Algorithm for the Skull Stripping
The obtained brain tumor dataset is translated to the image pre-processing segment. The pre-processing of the obtained dataset is to be done by utilizing the skull stripping along with the contrast enhancement. Table 5 illustrates the abbreviation used in the pre-processing algorithm for the Skull stripping.

Table 5.
Abbreviation used in the pre-processing algorithm for the Skull stripping.
The automated skull stripping of the MRI pictures has been recognized very important segment in MRI pictures’ efficient analysis. The accurateness of the human skull stripping protocol distresses multifarious apps, namely the tumor segmentation, pre-surgical arrangement, etc. This is a simple procedure for the removal of non-cerebral nerves, namely the skull and eyeball from MRI brain pictures. The pre-processing algorithm for the skull striping is illustrated below. In this algorithm, the MRI brain picture (IMR) denotes the input and the skull-stripped picture (IMRstripped) denotes the outcome. The detailed algorithm procedure is illustrated as follows. Initially, MR input images were binarized utilizing Otsu’s technique. This approach attains the threshold level which reduces in class-variation amongst two distinct categories. With the help of obtained binary images, the biggest connected component (CC) was received. The brain portion is the biggest CC within the obtained image. This biggest CC is further dilated by utilizing the 3 3 squared structured components to maintain the minimal brain data within the output images. Furthermore, the resultant image pixel was superimposed along with the accessed input picture to attain the stripped skull image, effectively.
Step 1: IMR ← Input MRI picture of a brain
Step 2: IMR bin ← Otsu (IMR)
Step 3: Conv ← Highest associated element (IMR bin)
Step 4: Conv’ ← Conv SEt1
Repeat
Step 5: Conv’Kt ← Conv’Kt−1 SEt2
Until
Conv’ Kt = Conv’ Kt−1
IMRIstripped ← Superimpose (Conv’ Kt)
4.3.2. Pre-Processing Algorithm for the MRI Picture Contrast Improvement
During the pre-processing stage of the MRI pictures, it is to be found that at certain times the contrast of the pictures is below a threshold limit. Table 6 illustrates the abbreviation used in the pre-processing algorithm for the MRI picture contrast improvement.

Table 6.
Abbreviation used in the pre-processing algorithm for the MRI picture contrast improvement.
For an effective visualization of the MRI pictures, the hardware, and software-level picture improvement, must be evaluated. The improvement of the MRI pictures contrast is a procedure for enhancing the visual elements of the received picture such that this could be appropriate for multifarious applications. The contrast improvement procedure provides additional clarity to the MRI pictures, which offers a solution for additional analysis and makes overall analysis much easier as well as fast in comparison to the other kinds of existing schemes. In this algorithm, the skull-stripped picture (IMRIstripped) is considered as an input and the contrast-improved picture (IMRIimproved) is the outcome. The key advantage of this specific pre-processing algorithm for the MRI picture contrast improvement rather than traditional methods is that it is faster, more accurate, and easy to implement and consumes very less time in comparison to the conventional approach. The main steps of this pre-processing algorithm are described as follows. In the beginning phase, the algorithm attained the complement of the applied stripped skull MRI image. Furthermore, the closing function has been applied over the complement picture as means of disk structured component. Later in the next phase, the complement is computed of the last outcome. Further, the mathematical function attains the variance among the resultant as well as the real picture. Lastly, the real picture has been added along with differentiated pictures for obtaining the contrast-improved images.
Step 1: IMRIstripped ← Skull stripping picture
Step 2: (Lt−1) IMRI
Step 3: Ct SEt2
Step 4: Dt ← (Lt−1) − Ct
Step 5: IMRIdiff ← IMRI − Dt
Step 6: IMRIimproved ← IMRIstripped + IMRIdiff
4.3.3. Classification Using Modulated-CNN
Figure 9 illustrates the block diagram of the suggested modulated CNN classifier. For the modulated CNN classifier, the utilized algorithm is given below. In this enhanced algorithm, we opted for Timage, labeled as an input, and the predicted outcome of the modulated CNN classifier is described through the output. Table 7 illustrates the abbreviation used in classification using Modulated-CNN. The entire steps of the suggested modulated CNN classification are illustrated as follows: In this classification process, the testing datasets are fed along with the Nk number datasets to the training network. Further, the predicted score is recorded for the testing process. The dataset Pt1 predicts through the model of modulated CNN. Later, the training begins using the classified data to compute the model outcome in terms of Malignant or Benign classification, effectively.
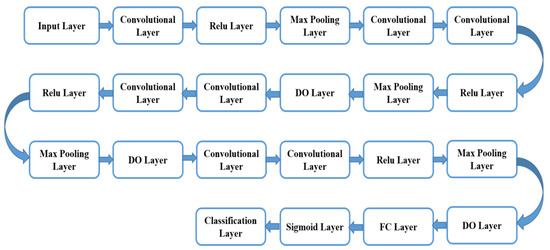
Figure 9.
Illustrates the block diagram of the suggested modulated CNN classifier.

Table 7.
Abbreviation used in Classification using Modulated-CNN.
Step 1: To Feed testing datasets along with the Nk number datasets to the training network
Step 2: To obtain the predicted scores
Step 3: For every testing, dataset Pt1 ← predicts via the model of modulated CNN
Step 4: Var X left ← 0 as well as Var X right ← 0
Step 5: if Pt1is found “Malignant” then
Step 6: Pt1 right ← Pt1 right +1
Step 7: Pt1 left ← Pt1 left +1
Step 8: if Pt1 right > Pt1 left
Step 9: Classification ← “Malignant”
Step 10: Else
Step 11: Classification ← “Benign”
Step 12: halt
In this work, the modulated CNN classifier has been built rooted on VGG-19, which includes seven convolutional layers; max-pooling layers were selected at four, as well as a full linked layer at one. The convolutional layer is indeed one of the key building elements of the suggested modulated CNN classifier. The modulated CNN approach is effective and beneficial to attain enhanced accuracy outcomes in comparison to conventional techniques.
In this research, the CNN classifier used is customized to attain more interpretability and accuracy. This classifier is selected and customized based on the complexity of the task as well as the available dataset size. In addition to this, various hyperparameters, namely learning rate, epoch, and batch size, were adjusted for optimal tuning in model training and testing.
This convolutional layer comprises multifarious filter parameters that require learning via appropriate training. The chosen filter dimensions are generally taken as small in comparison to the real picture. Each chosen filter convolves along with the picture and generates the map of activation. The max-pooling is a very significant operation that chooses the highest components via the feature map region enclosed through the filter. The fully linked layer aids in the compilation of the datasets extracted via the last layer to produce the end outcome. Furthermore, there have been used 4 ReLU (Rectified Linear Unit) layers in this suggested modulated CNN. The key benefit of the ReLU function utilization over the multiple activation functions is that there is not any requirement to activate the entire employed neuron at the same moment. The sigmoid layer has been utilized after the fully linked layer in place of softmax for the two-fold categorization. The sigmoid layer is employed as the second last layer the suggested modulated CNN classifier as it aids in classifier outcome alteration in the probability score. After every layer, a dropout (DO) layer has been installed for minimization of the overfitting [50]. This DO layer aids to ignore the arbitrarily chosen neuron during the training procedure. Through the simplification of the VGG-19 setup, the hyper-parameter turning procedure is more controllable. The VGG-19 network infrastructure is the CNN, which is to be trained on large-picture datasets through the ImageNet database. The VGG-19 network comprises 19 layers and may categorize the pictures into thousand diverse object classes, namely the keyboard, eraser, sharper, and many more. Consequently, this network infrastructure has learned opulent features illustration for multifarious image datasets. The total nineteen layers of the VGG-19 include three fully linked layers as well as 16 convolutional layers, which aid in the effective categorization of the pictures in diverse object groups. This approach is a suitable for picture categorization owing to the utilization of multifarious 3 × 3 filters inside every convolutional layer.
To enable clinicians to choose the appropriate kind of diagnosis as well as care strategy, properly forecast destiny, and effectively establish follow-up strategies, precision in categorizing tumor types is necessary. This investigation work has been carried out by utilizing the PGGAN architecture, which is rooted in the deep learning prototypical. This verification model comprises three diverse principal phases. The initial phase is known as the data pre-processing phase. The second phase is rooted in the deep learning prototypical to augment all the features of the feed in the picture accurately. The third phase of the illustrated model is the MR input image categorization phase. In the third phase, the VGG19 features extractor has been implemented for accurate and proper identification of all the features of the MR input images in real time. Figure 10 illustrates the structure of the PGGAN for 512 × 512-pixel MR picture in real-time training for the brain cell tissue generator.
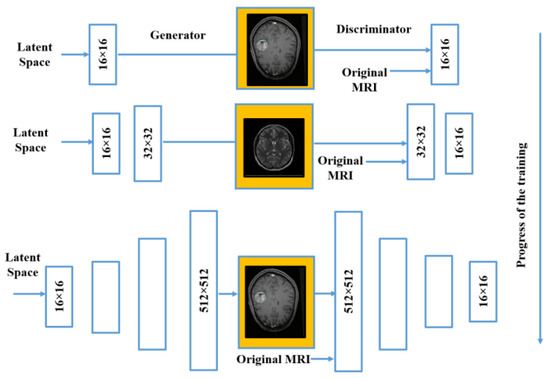
Figure 10.
PGGAN structure in real-time training for brain cell tissue generator.
5. Implementation Results and Discussion
Brain tumor cases are constantly increasing around the world. However, there are numerous clinical methods for determining the brain cell tissues in the diagnosis with the help of MR images taken by the radiologist. However, most of the existing methods have diverse limitations during early detection of the brain cell tissues, which is a huge problem and demands more attention towards novel research for correct recognition and segmentation of the brain tumor pragmatically. The authors of this paper present a hybrid and enhanced brain tumor assessment model based on the GAN ensemble and demonstrate PGGAN as the most promising approach for extracting characteristics of a brain tumor from real-time pictures.
Different evaluation indicators are utilized for validating the efficacy of the suggested methodology [49,50]. From the confusion matrix, four parameters are derived. True positives (TP) denote accurate predictions of the desired target affected by a brain tumor. True negatives (TN) represent accurate predictions of individuals not affected by a brain tumor. False positives (FP) highlight the inaccurate predictions of a normal individual shown as a brain tumor patient. False negatives (FN) depict inaccurate predictions of the desired target as normal individuals.
The accuracy rate denotes the proportion between accurate values under prediction and the cumulative values predicted, which is shown in Equation (1) as follows:
Precision represents the overall ratio of accurately predicted values to the total accurate values that includes both true and false predicted outcomes, which are denoted by Equation (2) as follows:
Recall illustrates the entire proportion of accurate likelihoods to the summation of accurate positively predicted values and inaccurate negatively predict outcomes, which are shown in Equation (3) as follows:
F-Score represents the weighted mean of the values generated from the computation of precision and recall metrics, which is shown in Equation (4) as follows:
The accuracy analysis of different kinds of GAN models is depicted in Figure 11. PGGAN achieved a 98.8% accuracy rate, which is higher than the other competing GANs. DCGAN with an accuracy of 97.24%, Cycle-GAN with an accuracy of 96.18%, info-GAN with an accuracy of 92.15%, and Conditional GAN with an accuracy of 90.24%. The reason for such high accuracy lies in the fact that PGGAN is very stable where the model size increases incrementally. The training and validation of the suggested GAN ensemble intelligent predictive model for brain tumor identification and categorization are done with higher precision to measure the model accuracy level on distinct GANs. The outcomes are recorded in real-time on the chosen datasets for model training and validation. The accuracy was measured on distinct images to identify the glioma, meningioma, and pituitary categories of the tumor. We selected three planes of the images, i.e., sagittal, coronal, and axial. For glioma, meningioma, and the pituitary, the total number of images were 1426, 708, and 930, respectively. The accuracy of the proposed model is measured very accurately as we segregated the entire chosen image datasets into three classes, i.e., training data, validation data, and testing data. The training data was considered at 60%, as well as validation data at 20% and the model testing data at 20%. The key advantage of the proposed GAN ensemble model is that it is capable of identifying and classifying the glioma, meningioma, and pituitary tumor from chosen images considering distinct planes in real-time, which is a significant advantage of the proposed GAN ensemble model over the existing models, which provides a lower accuracy level considering all the planes of the images.
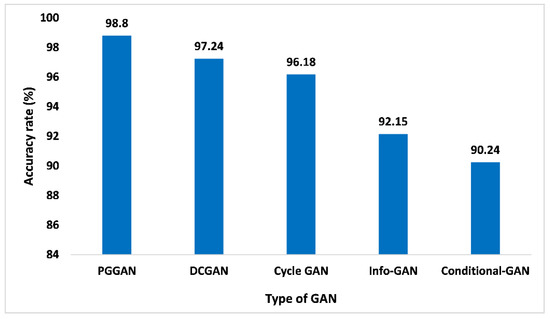
Figure 11.
Accuracy analysis for different types of GAN.
The precision matrices are a very important indicator for determining the proposed GAN ensemble model performance. The precision is referred to as overall true positive numbers divided by overall positive predictions, which means true positive numbers and false positive numbers added. When analyzing Figure 12, with 98.45% of precision, PGGAN attained the maximum, which is high compared to other GANs. While Precision values for DCGAN, Cycle-GAN, Info-GAN, and Conditional-GAN are 96.14%, 95.12%, 91.22%, and 88.02%, respectively. The incremental nature of PGGAN enhances the performance of the model. Thereby, the proposed GAN ensemble model offers a pragmatic and higher precision value on the PGGAN when compared with the other GAN approaches.
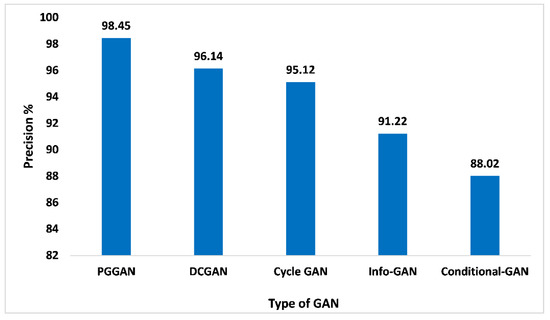
Figure 12.
Precision analysis for different types of GAN.
The recall performance matrices are not just restricted to binary categorization issues. Within the imbalanced categorization issue with the above two categories, the recall is evaluated as the addition of true positives across the overall categories divided by the addition of true positives and false negatives across every category. In addition to this, recall is indeed a very important metric that aids in quantifying the accurate positive numbers forecasted out of the entire forecast of positive. Similar to the precision matrices, which may comment over accurate forecasting of positives out of each forecasting of positives, the recall matrices may offer a sign of failed forecasting of positive. Figure 13 presents the outcome of PGGAN based on recall value is 97.2%, which is the maximum in comparison to DCGAN, Cycle-GAN, Info-GAN, and Conditional-GAN, which are 95.15%, 95.88%, 91.99%, and 90.14%, respectively. Thus, the proposed GAN ensemble model offers pragmatic advantages and a higher recall value when compared with other GANs.
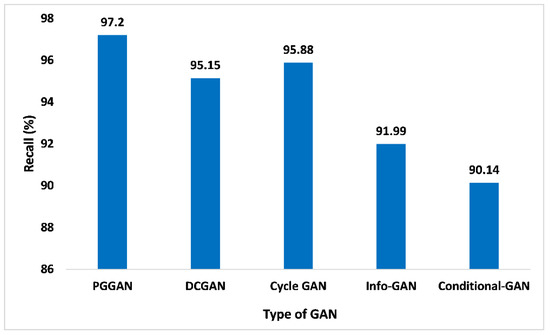
Figure 13.
Recall analysis for different types of GAN.
The F1-score is another significant performance matrix that combines the recall and precision of the classifier within one metric by considering the harmonic mean. This performance metric aids in comparing the performance of the proposed GAN ensemble model with distinct GANs. Similarly, performance analysis based on the F1-score is given in Figure 14. Compared to other GANs, PGGAN attained the maximum 98.11% F1 score. DCGAN, Cycle GAN, Info-GAN, and conditional-GAN attained the F1-score 96.01%, 96.03%, 91.77%, and 89.33%, respectively. Thus, the measured F1-score value of the proposed GAN ensemble model for PGGAN is improved and pragmatic when compared with the other GANs.
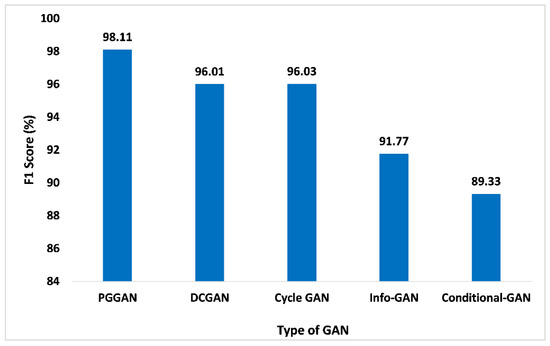
Figure 14.
F1-Score analysis for different types of GAN.
The NPV is described as the proportion of forecasted negatives that are true negatives. In addition to this, it demonstrates the likelihood that the forecasted negative is indeed the real negative. Figure 15 shows the NPV parameter of PGGAN is 98.09%, which is excellent among other competitive GANs. The NPV parameter values for DCGAN, Cycle-GAN, Info-GAN, and Conditional-GAN are 96.55%, 95.77%, 91.05%, and 89.09%, respectively. Thus, the proposed GAN ensemble model offers a higher NPV value over the PGGAN in comparison to the distinct other GANs such as the DCGAN and others.
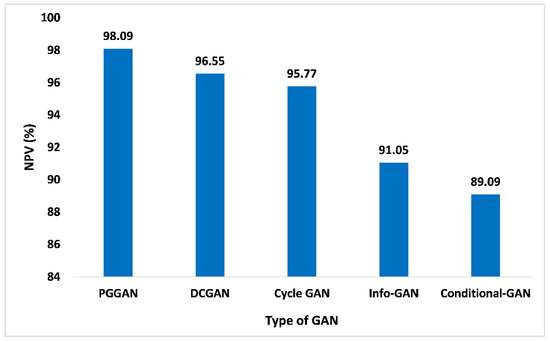
Figure 15.
NPV analysis for different types of GAN.
Figure 16 illustrates the execution time analysis for different types of GAN. The progressive growth of the generator and discriminator causes a lower execution time delay for PGGAN. The proposed GAN ensemble model consumes very minimal time on PGGAN, which is only 3.4 s, while the DCGAN, Cycle-GAN, Info-GAN, and Conditional-GAN consumes more time, which is 6.78 s, 9.42 s, 5.9 s, and 7.1 s, respectively, which is far greater in comparison to the PGGAN. Thereby, the proposed GAN ensemble model predicts that on PGGAN, the proposed model consumes very minimal time.
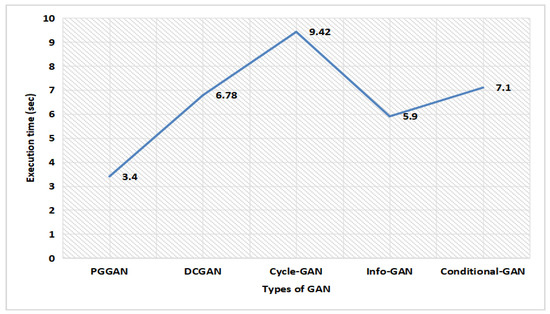
Figure 16.
Execution time analysis for different types of GAN.
Figure 17 illustrates the comparison of accuracy for different computational methods with PGGAN. The PGGAN has been globally recognized for the addition of the GAN training procedure, which can assist to train the multifarious generator prototype and may produce huge higher-quality pictures. It entails beginning with an extremely tiny picture as well as gradually accumulating stages to raise generator prototypical output dimensions and discriminator prototypical input dimensions until the required picture dimension is reached. Due to the diverse appearance and complexity of tumors, PGGAN provides satisfaction and high accuracy in the sense of tumor detection and will provide pre-treatment to the patients, so they can be cured. The proposed GAN ensemble model accuracy has been measured and compared with the other computational methods. In the analysis of proposed model accuracy with PGGAN and other methods, it has been found that the accuracy on the Bayesian approach [61], Capsule network [62], Feature extraction [63], CNN [64], Ensemble CNN [65], SVM [66], and Random Split-GAN [67] were 98.50%, 95.54%, 95.17%, 97.50%, 98.14%, 97.89%, and 96.0%, respectively. However, the proposed GAN ensemble model offers an accuracy level of 98.80% with the PGGAN, which is higher than the other computational methods.
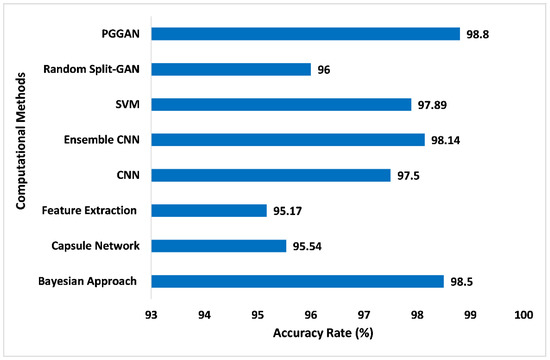
Figure 17.
Comparison of accuracy for different computational methods with PGGAN [61,62,63,64,65,66,67].
Table 8 presents the performance of the proposed model through various performance metrics such as accuracy, precision, recall, F1-score, and NPV with different portioning of training, testing, and validation ratio of the MRI brain tumor dataset. The best performance was observed for the ratio of 60:20:20 with scores of 98.8%, 98.45%, 97.2%, 98.11%, and, 98.09% for accuracy, precision, recall, F1-Score, and NPV, respectively.

Table 8.
Dataset portioning ratio (Training: Validation: Test samples) for different performance metrics.
Table 9 illustrates the performance metric evaluation of the proposed model for different diseases. The proposed model performance is computed for different disease classes and assessed through performance metrics such as accuracy, NPV, precision, recall, and F1-Score. The best optimal performance was observed for brain tumors with scores of 98.8%, 98.09%, 98.45%, 97.2%, and 98.11% for accuracy, NPV, precision, recall, and F1-Score, respectively.

Table 9.
Performance Metric Evaluation of the proposed model on different diseases.
Table 10 presents a cumulative analysis of performance metrics for different types of GANs. As observed from the table, the optimum outcome is generated using PGGAN. Here, 98.8%, 98.45%, 97.2%, 98.11%, and 98.09% are the recorded values for accuracy, precision, recall, F1-score, and NPV, respectively.

Table 10.
Cumulative analysis of performance metrics for different types of GANs.
In this experimental study design and implementation, a few of the identified insights into the limits and possible error sources of the proposed model for brain tumor detection have been identified. A few of the key limits and possible errors can involve the limited size of the datasets, challenges to managing the overfitting, etc. Although this proposed GAN ensemble model for brain tumor detection attains very optimized results, there can be limits to the research work and possible sources of the errors, which demand attention to resolve them. In addition to this, the generalizability of the proposed GAN ensemble model should be computed on larger datasets and in diverse clinical settings based on diverse tumor classes and sizes to determine its applicability beyond the input dataset it has been trained on.
6. Conclusions and Future Scope
Brain tumor has become one of the most prevalent causes of mortality all around the world. This research work designs a new hybrid augmentation-based modulated CNN architecture using a soft voting approach. GAN ensemble augmented brain MRI image data samples and the augmented dataset are fed into a modulated CNN model for the classification of images. PGGAN was found to be the most promising GAN technique, generating the most optimal values in performance metrics using a soft voting approach. The accuracy, precision, recall, F1-Score, and NPV were computed to be 98.8%, 98.45%, 97.2%, 98.11%, and 98.09%, respectively. It also recorded a very low execution time delay of only 3.4 s with PGGAN. The time required by the DCGAN, Cycle-GAN, Info-GAN, and Conditional-GAN is 6.78 s, 9.42 s, 5.9 s, and 7.1 s, respectively, which is higher in comparison to the PGGAN approach. As a result, a deep classification model using PGGAN with modulated CNN can be very effective in assisting medical professionals to assess brain tumor symptoms in patients.
Author Contributions
Conceptualization, S.S., S.M., B.P., A.K.B. and P.B.; methodology, S.S.; software, S.M.; validation, B.P., A.K.B. and P.B.; formal analysis, S.S. and S.M.; investigation, A.K.B.; resources, P.B.; data curation, S.S., S.M. and P.B.; writing—original draft preparation, S.S.; writing—review and editing, S.S., S.M. and A.K.B.; visualization, B.P.; supervision, S.M.; project administration, A.K.B.; funding acquisition, P.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Authors utilized publicly available datasets [60].
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kang, J.; Ullah, Z.; Gwak, J. MRI-Based Brain Tumor Classification Using Ensemble of Deep Features and Machine Learning Classifiers. Sensors 2021, 21, 2222. [Google Scholar] [CrossRef] [PubMed]
- Amin, J.; Sharif, M.; Haldorai, A.; Yasmin, M.; Nayak, R.S. Brain tumor detection and classification using machine learning: A comprehensive survey. Complex Intell. Syst. 2021, 8, 3161–3183. [Google Scholar] [CrossRef]
- Mishra, S.; Panda, A.; Tripathy, K.H. Implementation of re-sampling technique to handle skewed data in tumor prediction. J. Adv. Res. Dyn. Control Syst. 2018, 10, 526–530. [Google Scholar]
- Kumar, T.S.; Arun, C.; Ezhumalai, P. An approach for brain tumor detection using optimal feature selection and optimized deep belief network. Biomed. Signal Process. Control. 2022, 73, 103440. [Google Scholar] [CrossRef]
- Aastha; Mishra, S.; Mohanty, S. Integration of Machine Learning and IoT for Assisting Medical Experts in Brain Tumor Diagnosis. In Smart Healthcare Analytics: State of the Art; Springer: Singapore, 2022; pp. 133–164. [Google Scholar] [CrossRef]
- Ma, L.; Zhang, F. End-to-end predictive intelligence diagnosis in brain tumor using lightweight neural network. Appl. Soft Comput. 2022, 111, 107666. [Google Scholar] [CrossRef]
- Senan, E.M.; Jadhav, M.E.; Rassem, T.H.; Aljaloud, A.S.; Mohammed, B.A.; Al-Mekhlafi, Z.G. Early Diagnosis of Brain Tumour MRI Images Using Hybrid Techniques between Deep and Machine Learning. Comput. Math. Methods Med. 2022, 2022, 8330833. [Google Scholar] [CrossRef] [PubMed]
- Ramtekkar, P.K.; Pandey, A.; Pawar, M.K. Accurate detection of brain tumor using optimized feature selection based on deep learning techniques. Multimedia Tools Appl. 2023, 2, 1–31. [Google Scholar] [CrossRef]
- Haq, A.U.; Li, J.P.; Khan, S.; Alshara, M.A.; Alotaibi, R.M.; Mawuli, C. DACBT: Deep learning approach for classification of brain tumors using MRI data in IoT healthcare environment. Sci. Rep. 2022, 12, 15331. [Google Scholar] [CrossRef]
- Shelatkar, T.; Urvashi; Shorfuzzaman, M.; Alsufyani, A.; Lakshmanna, K. Diagnosis of Brain Tumor Using Light Weight Deep Learning Model with Fine-Tuning Approach. Comput. Math. Methods Med. 2022, 2022, 2858845. [Google Scholar] [CrossRef]
- Saeedi, S.; Rezayi, S.; Keshavarz, H.; Kalhori, S.R.N. MRI-based brain tumor detection using convolutional deep learning methods and chosen machine learning techniques. BMC Med. Informatics Decis. Mak. 2023, 23, 16. [Google Scholar] [CrossRef]
- Keerthana, A.; Kumar, B.K.; Akshaya, K.; Kamalraj, S. Brain Tumour Detection Using Machine Learning Algorithm. J. Physics: Conf. Ser. 2021, 1937, 012008. [Google Scholar] [CrossRef]
- Sarkar, A.; Maniruzzaman; Alahe, M.A.; Ahmad, M. An Effective and Novel Approach for Brain Tumor Classification Using AlexNet CNN Feature Extractor and Multiple Eminent Machine Learning Classifiers in MRIs. J. Sensors 2023, 2023, 1224619. [Google Scholar] [CrossRef]
- Akinyelu, A.A.; Zaccagna, F.; Grist, J.T.; Castelli, M.; Rundo, L. Brain Tumor Diagnosis Using Machine Learning, Convolutional Neural Networks, Capsule Neural Networks and Vision Transformers, Applied to MRI: A Survey. J. Imaging 2022, 8, 205. [Google Scholar] [CrossRef] [PubMed]
- Mahmud, I.; Mamun, M.; Abdelgawad, A. A Deep Analysis of Brain Tumor Detection from MR Images Using Deep Learning Networks. Algorithms 2023, 16, 176. [Google Scholar] [CrossRef]
- Mehnatkesh, H.; Jalali, S.M.J.; Khosravi, A.; Nahavandi, S. An intelligent driven deep residual learning framework for brain tumor classification using MRI images. Expert Syst. Appl. 2023, 213, 119087. [Google Scholar] [CrossRef]
- Methil, A.S. Brain Tumor Detection using Deep Learning and Image Processing. In Proceedings of the International Conference on Artificial Intelligence and Smart Systems, ICAIS 2021, Coimbatore, India, 25–27 March 2021. [Google Scholar]
- Shinde, A.S.; Mahendra, B.; Nejakar, S.; Herur, S.M.; Bhat, N. Performance analysis of machine learning algorithm of detection and classification of brain tumor using computer vision. Adv. Eng. Softw. 2022, 173, 103221. [Google Scholar] [CrossRef]
- Chen, B.; Zhang, L.; Chen, H.; Liang, K.; Chen, X. A novel extended Kalman filter with support vector machine based method for the automatic diagnosis and segmentation of brain tumors. Comput. Methods Programs Biomed. 2020, 200, 105797. [Google Scholar] [CrossRef] [PubMed]
- Islam, K.; Ali, S.; Miah, S.; Rahman, M.; Alam, S.; Hossain, M.A. Brain tumor detection in MR image using superpixels, principal component analysis and template based K-means clustering algorithm. Mach. Learn. Appl. 2021, 5, 100044. [Google Scholar] [CrossRef]
- Vankdothu, R.; Hameed, M.A. Brain tumor segmentation of MR images using SVM and fuzzy classifier in machine learning. Meas. Sensors 2022, 24, 100440. [Google Scholar] [CrossRef]
- Gajula, S.; Rajesh, V. An MRI brain tumour detection using logistic regression-based machine learning model. Int. J. Syst. Assur. Eng. Manag. 2022, 3, 1–11. [Google Scholar] [CrossRef]
- Saba, S.S.; Sreelakshmi, D.; Kumar, P.S.; Kumar, K.S.; Saba, S.R. Logistic regression machine learning algorithm on MRI brain image for fast and accurate diagnosis. Int. J. Sci. Technol. Res. 2020, 9, 7076–7081. [Google Scholar]
- Osman, A.F.I. Automated Brain Tumor Segmentation on Magnetic Resonance Images and Patient’s Overall Survival Prediction Using Support Vector Machines. In Proceedings of the Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries: Third International Workshop, BrainLes 2017, Quebec City, QC, Canada, 14 September 2017; Springer International Publishing: Cham, Switzerland, 2018; pp. 435–449. [Google Scholar]
- Felefly, T.; Achkar, S.; Lteif, T.; Azoury, F.; Khoury, C.; Sayah, R.; Barouky, J.; Farah, N.; Nasr, D.N.; Nasr, E. A Machine-Learning Model using MRI-based Radiomic Features to Predict Primary Site for Brain Metastases. Int. J. Radiat. Oncol. 2019, 105, E141. [Google Scholar] [CrossRef]
- Jeong, J.J.; Ji, B.; Lei, Y.; Wang, L.; Liu, T.; Ali, A.N.; Curran, W.J.; Mao, H.; Yang, X. Machine-learning based classification of glioblastoma using dynamic susceptibility enhanced MR image. In Proceedings of the Medical Imaging 2019: Biomedical Applications in Molecular, Structural, and Functional Imaging, San Diego, CA, USA, 16–21 February 2019; SPIE: San Diego, CA, USA, 2019. [Google Scholar] [CrossRef]
- Kiranmayee, B.V.; Rajinikanth, T.V.; Nagini, S. Enhancement of SVM based MRI Brain Image Classification using Pre-Processing Techniques. Indian J. Sci. Technol. 2016, 9, 1–7. [Google Scholar] [CrossRef]
- Sharif, M.; Amin, J.; Raza, M.; Anjum, M.A.; Afzal, H.; Shad, S.A. Brain tumor detection based on extreme learning. Neural Comput. Appl. 2020, 32, 15975–15987. [Google Scholar] [CrossRef]
- Anitha, R.; Raja, D.S.S. Development of computer-aided approach for brain tumor detection using random forest classifier. Int. J. Imaging Syst. Technol. 2018, 28, 48–53. [Google Scholar] [CrossRef]
- Asodekar, B.H.; Gore, S.A.; Thakare, A.D. Brain Tumor analysis Based on Shape Features of MRI using Machine Learning. In Proceedings of the 2019 5th International Conference on Computing, Communication, Control and Automation (ICCUBEA), Pune, India, 19–21 September 2019. [Google Scholar] [CrossRef]
- Gyorfi, A.; Csaholczi, S.; Fulop, T.; Kovacs, L.; Szilagyi, L. Brain Tumor Segmentation from Multi-Spectral Magnetic Resonance Image Data Using an Ensemble Learning Approach. In Proceedings of the IEEE International Conference on Systems, Man and Cybernetics, Toronto, ON, Canada, 11–14 October 2020. [Google Scholar] [CrossRef]
- Seere, S.K.H.; Karibasappa, K. Threshold Segmentation and Watershed Segmentation Algorithm for Brain Tumor Detection using Support Vector Machine. Eur. J. Eng. Res. Sci. 2020, 5, 516–519. [Google Scholar] [CrossRef]
- Kharat, K.; Kulkarni, P.P. Brain Tumor Classification Using Neural Network Based Methods. Int. J. Comput. Sci. Informatics 2012, 1, 2231–5292. [Google Scholar] [CrossRef]
- Qasem, S.N.; Nazar, A.; Qamar, A.; Shamshirband, S. A Learning Based Brain Tumor Detection System. Comput. Mater. Contin. 2019, 59, 713–727. [Google Scholar] [CrossRef]
- Selvapandian, A.; Manivannan, K. Performance analysis of meningioma brain tumor classifications based on gradient boosting classifier. Int. J. Imaging Syst. Technol. 2018, 28, 295–301. [Google Scholar] [CrossRef]
- Biradar, C. Measurement based Human Brain Tumor Recognition by Adapting Support Vector Machine. IOSR J. Eng. 2013, 3, 26–31. [Google Scholar] [CrossRef]
- Ajai, A.S.R.; Gopalan, S. Analysis of Active Contours Without Edge-Based Segmentation Technique for Brain Tumor Classification Using SVM and KNN Classifiers. In Advances in Communication Systems and Networks: Select Proceedings of ComNet 2019; Springer: Singapore, 2020. [Google Scholar] [CrossRef]
- Anantharajan, S.; Gunasekaran, S. Detection and Classification of MRI Brain Tumour Using GLCM and Enhanced K-NN. Comptes Rendus Acad. Bulg. Des Sci. 2021, 74, 260–268. [Google Scholar] [CrossRef]
- Rajagopal, R. Glioma brain tumor detection and segmentation using weighting random forest classifier with optimized ant colony features. Int. J. Imaging Syst. Technol. 2019, 29, 353–359. [Google Scholar] [CrossRef]
- Divyamary, D.; Gopika, S.; Pradeeba, S.; Bhuvaneswari, M. Brain Tumor Detection from MRI Images using Naive Classifier. In Proceedings of the 2020 6th International Conference on Advanced Computing and Communication Systems (ICACCS), Coimbatore, India, 6–7 March 2020. [Google Scholar] [CrossRef]
- Aswathy, A.L.; Chandra, S.S.V. Detection of Brain Tumor Abnormality from MRI FLAIR Images using Machine Learning Techniques. J. Inst. Eng. (India) Ser. B 2022, 103, 1097–1104. [Google Scholar] [CrossRef]
- Brindha, P.G.; Kavinraj, M.; Manivasakam, P.; Prasanth, P. Brain tumor detection from MRI images using deep learning techniques. IOP Conf. Series: Mater. Sci. Eng. 2021, 1055, 012115. [Google Scholar] [CrossRef]
- Kulkarni, S.M.; Sundari, G. A Framework for Brain Tumor Segmentation and Classification using Deep Learning Algorithm. Int. J. Adv. Comput. Sci. Appl. 2020, 11, 374–382. [Google Scholar] [CrossRef]
- Jia, Z.; Chen, D. Brain Tumor Identification and Classification of MRI images using deep learning techniques. IEEE Access 2020, 2, 1. [Google Scholar] [CrossRef]
- Suganya, M.; Sabitha, R.; Jasmine, J.A. Deep learning technique for brain tumor detection using medical image fusion. Int. J. Innov. Technol. Explor. Eng. 2019, 8, 867–870. [Google Scholar]
- Selvy, P.T.; Dharani, V.P.; Indhuja, A. Brain Tumour Detection Using Deep Learning Techniques. Int. J. Sci. Res. Comput. Sci. Eng. Inf. Technol. 2019, 169, 175. [Google Scholar] [CrossRef]
- Kumar, A.; Manikandan, R.; Rahim, R. A Study on Brain Tumor Detection and Segmentation Using Deep Learning Techniques. J. Comput. Theor. Nanosci. 2020, 17, 1925–1930. [Google Scholar] [CrossRef]
- Bathe, K.; Rana, V.; Singh, S.; Singh, V. Brain Tumor Detection Using Deep Learning Techniques. SSRN Electron. J. 2021, 4, 1–5. [Google Scholar] [CrossRef]
- Aamir, M.; Rahman, Z.; Dayo, Z.A.; Abro, W.A.; Uddin, M.I.; Khan, I.; Imran, A.S.; Ali, Z.; Ishfaq, M.; Guan, Y.; et al. A deep learning approach for brain tumor classification using MRI images. Comput. Electr. Eng. 2022, 101, 108105. [Google Scholar] [CrossRef]
- Sajjad, M.; Khan, S.; Muhammad, K.; Wu, W.; Ullah, A.; Baik, S.W. Multi-grade brain tumor classification using deep CNN with extensive data augmentation. J. Comput. Sci. 2019, 30, 174–182. [Google Scholar] [CrossRef]
- Yogananda, C.G.B.; Shah, B.R.; Vejdani-Jahromi, M.; Nalawade, S.S.; Murugesan, G.K.; Yu, F.F.; Pinho, M.C.; Wagner, B.C.; Emblem, K.E.; Bjørnerud, A.; et al. A Fully Automated Deep Learning Network for Brain Tumor Segmentation. Tomography 2020, 6, 186–193. [Google Scholar] [CrossRef] [PubMed]
- Noreen, N.; Palaniappan, S.; Qayyum, A.; Ahmad, I.; Imran, M.; Shoaib, M. A Deep Learning Model Based on Concatenation Approach for the Diagnosis of Brain Tumor. IEEE Access 2020, 8, 55135–55144. [Google Scholar] [CrossRef]
- Dipu, N.M.; Shohan, S.A.; Salam, K.M.A. Deep Learning Based Brain Tumor Detection and Classification. In Proceedings of the 2021 International Conference on Intelligent Technologies CONIT 2021, Hubli, India, 25–27 June 2021. [Google Scholar]
- Huang, H.; Yang, G.; Zhang, W.; Xu, X.; Yang, W.; Jiang, W.; Lai, X. A Deep Multi-Task Learning Framework for Brain Tumor Segmentation. Front. Oncol. 2021, 11, 690244. [Google Scholar] [CrossRef]
- Suganthe, R.C.; Revathi, G.; Monisha, S.; Pavithran, R. Deep learning based brain tumor classification using magnetic resonance imaging. J. Crit. Rev. 2020, 7, 347–350. [Google Scholar]
- Rehman, A.; Naz, S.; Razzak, M.I.; Akram, F.; Imran, M. A Deep Learning-Based Framework for Automatic Brain Tumors Classification Using Transfer Learning. Circuits Syst. Signal Process. 2020, 39, 757–775. [Google Scholar] [CrossRef]
- Shivdikar, A.; Shirke, M.; Vodnala, I.; Upadhaya, J. Brain Tumor Detection using Deep Learning. Int. J. Res. Appl. Sci. Eng. Technol. 2022, 10, 621–627. [Google Scholar] [CrossRef]
- Alsubai, S.; Khan, H.U.; Alqahtani, A.; Sha, M.; Abbas, S.; Mohammad, U.G. Ensemble deep learning for brain tumor detection. Front. Comput. Neurosci. 2022, 16, 1005617. [Google Scholar] [CrossRef]
- Alanazi, M.F.; Ali, M.U.; Hussain, S.J.; Zafar, A.; Mohatram, M.; Irfan, M.; AlRuwaili, R.; Alruwaili, M.; Ali, N.H.; Albarrak, A.M. Brain Tumor/Mass Classification Framework Using Magnetic-Resonance-Imaging-Based Isolated and Developed Transfer Deep-Learning Model. Sensors 2022, 22, 372. [Google Scholar] [CrossRef]
- Cheng, J. Brain Tumor Dataset. 2017. Available online: https://figshare.com/articles/dataset/brain_tumor_dataset/1512427/5 (accessed on 10 September 2017).
- Raja, P.S.; Rani, A.V. Brain tumor classification using a hybrid deep autoencoder with Bayesian fuzzy clustering-based segmentation approach. Biocybern. Biomed. Eng. 2020, 40, 440–453. [Google Scholar] [CrossRef]
- Adu, K.; Yu, Y.; Cai, J.; Tashi, N. Dilated Capsule Network for Brain Tumor Type Classification Via MRI Segmented Tumor Region. In Proceedings of the 2019 IEEE International Conference on Robotics and Biomimetics (ROBIO), Dali, China, 6–8 December 2019. [Google Scholar] [CrossRef]
- Vankdothu, R.; Hameed, M.A. Brain tumor MRI images identification and classification based on the recurrent convolutional neural network. Meas. Sensors 2022, 24, 100412. [Google Scholar] [CrossRef]
- Seetha, J.; Raja, S.S. Brain Tumor Classification Using Convolutional Neural Networks. Biomed. Pharmacol. J. 2018, 11, 1457–1461. [Google Scholar] [CrossRef]
- Younis, A.; Qiang, L.; Nyatega, C.O.; Adamu, M.J.; Kawuwa, H.B. Brain Tumor Analysis Using Deep Learning and VGG-16 Ensembling Learning Approaches. Appl. Sci. 2022, 12, 7282. [Google Scholar] [CrossRef]
- Sethy, P.K.; Behera, S.K. A data constrained approach for brain tumour detection using fused deep features and SVM. Multimedia Tools Appl. 2021, 80, 28745–28760. [Google Scholar] [CrossRef]
- Lamrani, D.; Cherradi, B.; El Gannour, O.; Bouqentar, M.A.; Bahatti, L. Brain Tumor Detection using MRI Images and Convolutional Neural Network. Int. J. Adv. Comput. Sci. Appl. 2022, 13, 452–460. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).