Abstract
Macrobrachium rosenbergii, commonly known as giant freshwater prawns (GFPs), is an economically and nutritionally important decapod crustacean species in China. Understanding the genetic diversity of selective breeding populations is crucial in breeding plans for selecting genetically diverse broodstocks and maintaining genetic diversity. The genetic structure of six breeding populations (Hefu (HF), Nantaihu No.2 (NTH), Jiaxin (JX), Shufeng (SF), Taiwan (TW), and Guangxi (GX)) of GFP in China was examined using 16 newly developed microsatellite loci and the mitochondrial control region (D-loop). The microsatellite data revealed that all 16 loci have high diversity, with all values of polymorphism information content (PIC) more than 0.5. The average expected heterozygosity (He, 0.89) and the number of alleles (Na, 18.25) of SF were the highest, followed by He (0.89) and Na (14.75) of the JX, and GX has the lowest He (0.83) and Na (11.31). The average PIC value for the six stocks ranged from 0.80 to 0.87. Pairwise comparisons revealed that Fst ranged from 0.03541 to 0.09637 and was significant (p < 0.05) between most populations, indicating from low to moderate genetic differentiation among the six populations. The D-loop analysis identified 114 variable sites and 29 haplotypes, with an average haplotype diversity (Hd) and nucleotide diversity (π) of 0.640 and 0.01247, respectively. Genetic differentiation among the six populations based on the D-loop was from moderate to high, with Fst values of 0.05603–0.80788, and all p < 0.05. This study demonstrates that selective breeding stocks of M. rosenbergii in China show moderate to high genetic diversity and have the potential for further selective breeding, providing a theoretical basis for conserving and utilizing M. rosenbergii genetic resources.
1. Introduction
Macrobrachium rosenbergii, giant freshwater prawn (GFP), is one of the most important farmed crustacean species owing to its delicious taste, rapid growth, short culture cycle, and high disease resistance [1]. This species is native to Southeast Asia and has been successfully introduced to several developed countries for commercial aquaculture [2]. Its global production increased to more than 337 thousand tons in 2022 [3]. In 1976, China (Mainland) introduced M. rosenbergii from Japan for the first time and continuously improved its culturing technology and farming scale. The GFP industry is an important component of Chinese aquaculture, providing major shrimp products to Chinese people and contributing significantly to exports and rural economic development. To date, China is the largest producer of M. rosenbergii globally, with a total production of 196,374 tons in 2023, according to the data of the China Fisheries Yearbook (2024) [4], contributing to over 50% of the global output [3].
The tremendous growth in the M. rosenbergii industry in China is attributable primarily to the continuous breeding of improved strains. Undoubtedly, the selective breeding program for M. rosenbergii has been well-established in China since its inception in 2006 [5], producing several viable new varieties, such as “South Taihu No. 2”, “South Taihu No. 3”, and “Shufeng No. 1”. Meanwhile, many notable M. rosenbergii hatcheries are working hard to develop new strains. Selective breeding is a key approach to enhancing the production traits of farmed animals. It has been successfully applied to improve the performance of numerous aquaculture species [6]. However, several significant problems are associated with breeding programs, including reduced genetic diversity, increased inbreeding levels, and reduced population size [7]. Over the past two decades, inbreeding depression has been reported to cause a severe decline in productivity in commercial aquaculture of M. rosenbergii in Mainland China [8], Thailand [9], India [10], and Vietnam [11]. At present, genetic deterioration in most hatchery stocks owing to inbreeding affects the quality of GFP seedlings, which significantly retards the sustainable development of the GFP farming industry in China [5]. Thus, a proper breeding program and strategy for conserving this species’ genetic diversity are urgently needed. Achieving this requires a good understanding of the genetic structure of the selective breeding populations.
In aquaculture, genetic structure and variability studies of selective breeding populations have been conducted extensively [12]. However, related research on M. rosenbergii is limited and is mainly concentrated in the breeding nucleus. For example, Sui et al. evaluated the genetic diversity, inbreeding levels, and effective population size of the M. rosenbergii breeding nucleus after eight generations of selection using a pedigree record [5]. Tang et al. analyzed the genetic diversity between two M. rosenbergii breeding strains (Shufeng nucleus breeding population and the introduced Zhengda population), including two hybrid populations generated from these two strains [12]. Their study revealed a moderate loss of genetic diversity with an increase in inbreeding levels among the four populations. Both studies emphasized the importance of conducting comprehensive studies on the genetic diversity of selective breeding populations of M. rosenbergii. Understanding the genetic diversity of species will help make informed choices in breeding plans regarding selecting genetically diverse brood stocks and maintaining genetic diversity in various stocks.
Molecular DNA markers have become indispensable and widely employed tools in population genetic research, offering effective means for assessing genetic diversity across a range of taxa. Several genetic diversity studies have been conducted on M. rosenbergii using allozymes [13], mtDNA [14], microsatellites [15,16,17,18], randomly amplified polymorphic DNA (RAPD) [19], and single-nucleotide polymorphisms (SNPs) [20]. In recent years, using multiple genetic markers to identify genetic variations has substantially aided population structure research and resource development in aquaculture. Many researchers have combined mitochondrial genes with microsatellite loci to conduct extensive research on the genetic diversity of various species, such as M. nipponense [21], M. rosenbergii [12], Misgurnu anguillicaudatus [22], Labeo rohita [23], and Oplegnathus fasciatus [24]. Combining these two markers can more comprehensively reflect the genetic diversity of an organism. Microsatellites are among the many marker systems that are highly dependable and reproducible for evaluating the genetic differences between and within populations. Mitochondrial DNA (mtDNA) has become an increasingly valuable molecular marker in genetic and evolutionary studies because of its unique properties (e.g., maternal inheritance, low rate of recombination, and higher mutation rate) compared to nuclear DNA. The control region (D-loop), which is a single non-coding segment in mtDNA, is highly polymorphic; thus, it is a powerful tool for defining the genetic structure of a population [25].
In this study, we investigated the genetic diversity of six selective breeding populations of M. rosenbergii, using microsatellite and mitochondrial D-loop as molecular markers. The results will provide details about the status of genetic diversity for M. rosenbergii selective breeding populations in China and serve as a theoretical foundation for conserving and utilizing M. rosenbergii genetic resources.
2. Materials and Methods
2.1. Sample Collection and Genomic DNA Extraction
In total, 202 samples were taken from six different prawn farms representing different breeding populations, among which three populations were collected from Huzhou city in Zhejiang province, including Hefu population (HF; 29 individuals), Nantaihu No. 2 population (NTH; 33 individuals), and Guangxi population (GX; 35 individuals); another two from Jiaxing city in Zhejiang province, including Jiaxing population (JX; 32 individuals) and Taiwan population (TW; 34 individuals); Shufeng population (SF; 39 individuals) was from Gaoyou city in Jiangsu province (Figure 1). Upon arrival at the laboratory, prawns were euthanized by thermal shock (at 14 °C for 20 min), and the muscle tissue was cut separately, stored in 95% ethanol, and frozen in a −20 °C freezer for subsequent DNA extraction. All animal experiments were conducted according to the Guide for Laboratory Animals developed by the Ministry of Science and Technology (Beijing, China) and approved by the Animal Care Committee of Huzhou University.
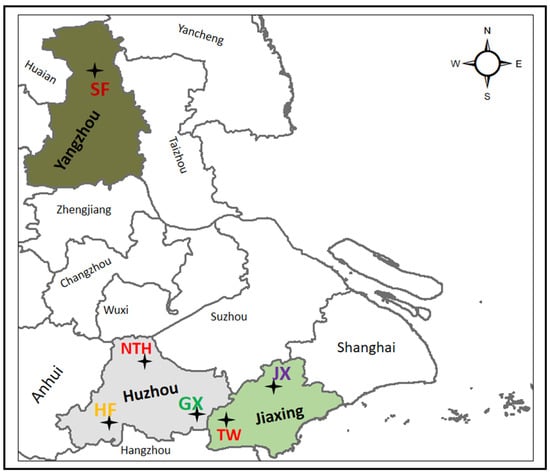
Figure 1.
Schematic map showing the six sampling sites in Zhejiang Province (Jiaxing and Huzhou) and Jiangsu Province (Gaoyou in Yangzhou). “HF” = Hefu population (29 individuals); “NTH” = Nantaihu No. 2 population (33 individuals); “GX” = Guangxi population (35 individuals); “JX” = Jiaxing population (32 individuals); “TW” = Taiwan population (34 individuals); “SF” = Shufeng population (39 individuals).
Approximately 25 mg of tissue was used for total genomic DNA extraction following the standard phenol-chloroform-isoamyl alcohol extraction and ethanol precipitation methods. The quality of the extracted DNA was checked by electrophoresis on a 1.5% agarose gel. The concentration was measured using a Nanodrop 2000 spectrometer (Thermo Fisher Scientific, Waltham, MA, USA), and DNA was diluted with sterilized ddH2O to 50 ng/μL for subsequent polymerase chain reaction (PCR).
2.2. mtDNA D-Loop Amplification and Sequencing
PCR was used to amplify D-loop segments. Primers were designed based on the complete mtDNA sequence of M. rosenbergii [25]. The forward primer was DL: 5′-CCTGAAACAGAGGAACAAGCAAG-3′, and the reverse primer was DH: 5′-TATTCGCCCTATCAAGACGACC-3′. The PCR was performed using a T100 thermal cycler (Bio-Rad, Hercules, CA, USA) in a reaction mixture of 50 μL, including 31 μL sterilized ddH2O, 5 μL 10× buffer, 4 μL dNTP (2.5 mM/L), 3 μL MgCl2 (25 mM/L), 1 µL of each primer (10 μM), 0.2 μL Taq DNA polymerase (5 U/μL) (Takara, Nojihigashi, Shiga, Japan), and 5 µL DNA. PCR was carried out with the following temperature profiles: initial denaturation at 94 °C for 3 min, 35 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 1 min, and a final elongation cycle at 72 °C for 5 min. Tianyi Huiyuan Biotechnology Co., Ltd. (Wuhan, China) purified and sequenced the amplified products.
2.3. Microsatellite Amplification and Genotyping
Sixteen pairs of microsatellite primers newly developed from the M. rosenbergii genome sequences were used to amplify the microsatellite loci (Table 1). PCR was performed in a final volume of 20 μL containing 10 μL 2× Taq Mix (Takara), 1 μL of each forward and reverse primer (10 μM), 7 μL sterilized ddH2O, and 1 μL DNA. PCR was carried out in a T100 thermal cycler (Bio-Rad, Hercules, CA, USA) using the following temperature profiles: an initial denaturation step at 94 °C for 5 min, followed by 35 cycles of denaturation at 94 °C for 30 s, annealing ranging from 52 °C to 60 °C (depending on the primers) for 30 s, and elongation at 72 °C for 45 s, and a final elongation step at 72 °C for 10 min. PCR products were visualized by gel electrophoresis on a 1.5% agarose gel under an illuminator (UV). Allele genotyping was resolved via capillary electrophoresis using an ABI 3730XL DNA analyzer (Applied Biosystems, Waltham, MA, USA). Fragment sizes were determined using GeneMarker V2.2.0. This work was performed by Tianyi Huiyuan Biotechnology Co., Ltd. (Wuhan, China).

Table 1.
The list of DNA microsatellite primer pairs designed for M. rosenbergii.
2.4. Microsatellite Data Analyses
For each of the six groups, the mean number of alleles (Na), observed heterozygosity (Ho), expected heterozygosity (He), and polymorphism information content (PIC) were calculated using POPGENE 1.31. GENEPOP 4.0 was used to identify possible deviations from Hardy–Weinberg equilibrium (HWE). Exact p-values were evaluated using the Markov chain method (dememorization = 10,000; batches = 100; iterations per batch = 5000). Bootstrapping with Arlequin 3.5 was used to calculate population pairwise Fst values and test for significance. Genetic differentiation was analyzed using Wright’s fixation index, that is, the acquisition of allele frequencies that differ across populations (Fis, Fit, Fst). A dendrogram based on the unweighted pair group method using arithmetical averages (UPGMA) was constructed using MEGA 11.0. STRUCTURE Ver. 2.3.4 was used to evaluate the most likely number of genetic clusters (K = 1–6). Before statistical analysis, genotype data at all loci were analyzed for data integrity using MicroChecker version 2.2.3. MicroChecker tests for null alleles, allelic dropout, and scoring errors due to stutter.
2.5. mtDNA D-Loop Sequence Alignment and Data Analyses
For the D-loop sequences of all samples, multiple alignment analysis was carried out using Clustal X version 2.1 software and manually corrected in SeaView version 4.0. The basic information of the sequence, such as sequence length, base composition, number of mutation sites, and number of simple information sites, was calculated using MEGA. The software DnaSP 5.0 and Arlequin were used to calculate the number of haplotypes (H), haplotype diversity (Hd), and nucleotide diversity (π). Arlequin was used to estimate various inter-population genetic differentiation indices (Fst). The phylogenetic relationships among the mtDNA D-loop haplotypes were reconstructed using the maximum likelihood method in MEGA, and bootstrap analyses (1000 replicates) were used to assess the relative robustness of the nodes. The median-joining method in POPART was used to construct the haplotype network.
3. Results
3.1. Microsatellite Polymorphism and Genetic Variation
All 202 samples from six populations were successfully genotyped by 16 microsatellite loci. The results showed that all 16 microsatellite loci were polymorphic in all six M. rosenbergii populations, and the levels of polymorphism varied depending on the locus (Table S1). The number of alleles per locus (Na), observed heterozygosity (Ho), and expected heterozygosity (He) varied widely among sampled populations (Table S1). The average Na (18.25) and He (0.89) of the SF were the highest, followed by Na (14.75) and He (0.89) of the JX, and GX had the lowest Na (11.31) and He (0.83). The average Ho ranged from 0.73 (GX) to 0.97 (SF), and the Ho of most microsatellite loci was lower than that of He, except for the SF. The average PIC values for the six stocks ranged from 0.80 (GX) to 0.87 (SF and JX). F-statistics for the 16 microsatellite loci among all six populations showed mean Fis, Fit, and Fst values of 0.0454, 0.1039, and 0.0612, respectively, and the number of migrants (Nm) among populations at different loci ranged from 2.0677 to 7.6788 with an average of 3.8320 (Table 2). The pairwise Fst values varied from 0.03541 (between JX and HF) to 0.09637 (between TW and GX) and were statistically significant (p < 0.05) between most populations except between JX and HF, JX and TW, JX and SF, HF and SF, and NTH and SF (Table 3). An unbiased estimation of HWE exact p values showed that, when the six stocks were treated as one population, no significant linkage disequilibrium among the loci was detected (p > 0.05), but, among 96 single population single locus detection data (6 populations × 16 primers), 35 showed significant deviation from HWE (p < 0.05; Table S1).

Table 2.
The values of F-statistics for all the loci across the six populations of M. rosenbergii based on sixteen microsatellite loci.

Table 3.
Genetic differentiation index (Fst) between populations based on SSR.
In the UPGMA dendrogram, the NTH and SF populations were grouped together and then clustered with GX, forming a clade. The JX and HF populations grouped, forming another clade in the median position, and the TW remained on an independent branch at the base of the dendrogram (Figure 2). The STRUCTURE software was used to cluster individuals into 2 ≤ K ≤ 6. The best K value is 3 when ΔK = m|L″ (K)|/s|L(K)| (Figure 3). The structure analysis results showed that all individuals were genetically mixed, especially in the HF population; individuals in other populations were slightly mixed, and the TW population was purer than the others. All studied individuals were divided into three groups. One group included all individuals with SF and HF, and most NTH individuals. Each group included all the TW and JX individuals. The GX population is almost a group; only a few individuals come from the NTH and HF.
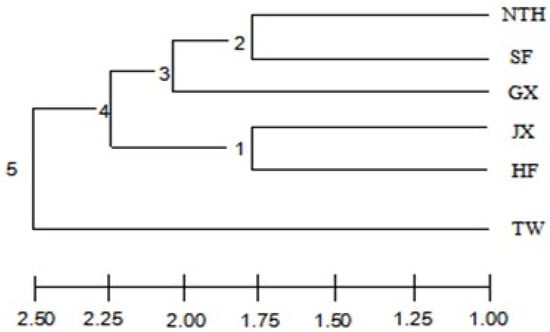
Figure 2.
Dendrogram based on genetic distance of microsatellites for six showing clusters from the six populations of M. rosenbergii.
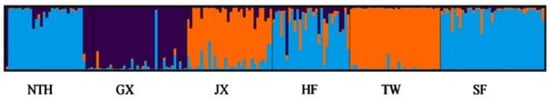
Figure 3.
Clustering diagrams for the six populations of M. resonbergii using STRUCTURE.
3.2. Genetic Diversity Based on D-Loop Sequences
The D-loop segment of 197 individuals from the six populations was successfully amplified and sequenced. After multiple sequence alignments and removal of inaccurate sequencing information at the beginning and end of the sequences, a dataset of 956 bp in total length was used for analysis, and 29 haplotypes were identified, defined by differences at 114 variable sites.
Five dominant haplotypes, including Hap9, Hap14, Hap16, Hap19, and Hap25, were found in most populations, whereas the others were found in only one or two (Table S2). Hap25 occurred in three populations, Hap9, Hap16, and Hap19, in four populations, and Hap14 in five. Hap14 was the most common haplotype, with a relative frequency of 25.35%. The combined relative frequency of the five dominant haplotypes was 65.91%, whereas the combined frequency of all remaining haplotypes was only 34.09%. The haplotype diversity (Hd) of the six groups ranges from 0.326 (TW) to 0.907 (HF), with an overall mean of 0.640 across all groups (Table 4). The group with the highest nucleotide diversity (π) was the SF population (0.02758), the following were NTH (0.01365) and JX (0.01303), the lowest was the GX population (π = 0.00398), and the average value of all populations was 0.01247 (Table 4). The Fst values among the six populations varied from 0.05603 (between JX and TW) to 0.80788 (between TW and SF). The differentiation between SF and other populations was much larger (all Fst > 0.5) than that of the other paired populations (Table 5), and the Fst values among all populations were statistically significant (p < 0.05). Maximum likelihood phylogenetic analysis clustered the 29 haplotypes into two distinct clades, Clade A and Clade B (Figure 4). There were 25 haplotypes in Clade A. The haplotype of the HF population was mainly clustered in Clade A and contained more JX and TW populations and a few haplotypes of the NTH and GX populations. Clade B contained four haplotypes, mainly with individuals from the JX and SF groups and only three from the NTH group (Figure 4 and Figure 5). The genetic distance of all pairwise haplotypes ranged from 0.001 to 0.014, and the average genetic distance between the two clades was 0.009.

Table 4.
Genetic diversity parameters based on D-loop sequences of each population of M. rosenbergii.

Table 5.
Genetic differentiation index (Fst) between populations based on D-loop.
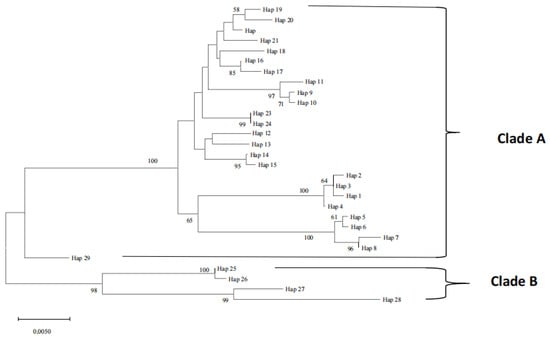
Figure 4.
The maximum likelihood phylogenetic tree of the entire D-loop for M. rosenbergii constructed by MEGA version 11.
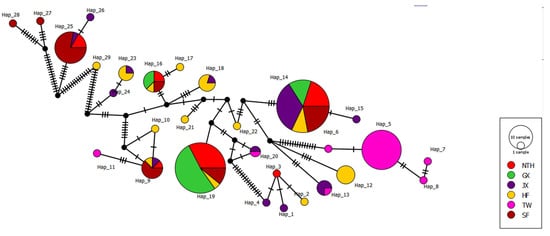
Figure 5.
The median-joining network based on haplotype frequencies of M. rosenbergii. The circle represents the haplotype; the circle size is proportional to the number of individuals in the haplotype. Each line in the network represents a single mutational change, and the branches are scaled to the number of polymorphic sites between each haplotype. Each black circle represents one missing haplotype.
4. Discussion
This study examined the genetic diversity of six selective breeding populations of M. rosenbergii in China using both microsatellite markers and mitochondrial DNA (D-loop gene) sequences. Genetic diversity is crucial for evolutionary potential and population fitness. Generally, higher genetic diversity enhances adaptive capacity and breeding potential, whereas lower diversity may compromise survival and increase extinction risk [26]. In the current study, 16 microsatellite loci were analyzed, all of which varied significantly in their degree of polymorphism, with the number of alleles detected at each locus ranging from 7 to 30, consistent with previous studies [13,27,28]. Polymorphic information content (PIC) measures genetic diversity within and between populations, with higher values indicating greater genetic richness and higher environmental adaptation [29]. All loci displayed high PIC values (>0.5), indicating highly polymorphic characteristics [30]. The high polymorphism (100%) found in all 16 microsatellite loci in this study was comparable to the findings of Ding et al. [27] for a cultured population of M. rosenbergii in China (100%). Furthermore, more alleles were observed in this study than in allozyme analysis [13] and RAPD analysis in wild and hatchery populations [19], demonstrating that microsatellites exhibit a greater level of polymorphism and are more suitable for population genetics studies than other traditional molecular markers.
The six M. rosenbergii populations in this study all showed high degrees of heterozygosity, with an average He ranging from 0.83 (GX) to 0.89 (SF and JX), indicating high genetic diversity. According to Qin et al., when heterozygosity is between 0.5 and 0.8, the population can be considered to have high diversity [31]. The high allelic diversity and heterozygosity of the selective breeding population suggest proper genetic management of the strains. Additionally, in this study, the mean Ho at all loci in all populations was less than the mean He, similar to the observations of Chareontawee et al. [9] and Ahmad et al. [13] in natural populations of M. rosenbergii in Thailand and Bangladesh, respectively. Deficiencies in heterozygosity may result from several factors, such as a reduction in effective breeding number, selection pressure at a particular locus, the Wahlund effect, and differences in the chances of mating among individuals [14]. Among these, a reduction in the breeding population is the most likely factor causing the loss of heterozygosity in M. rosenbergii selective breeding populations in China. Sample size easily influences observed heterozygosity, whereas effective heterozygosity can better reflect genetic diversity [31]. The genetic diversity of smaller populations declines faster than that of larger populations, and the loss of genetic variation results in the loss of fitness of a population. Similarly, heterozygous deletion increases homozygosity, the chance of a harmful allele’s homogeneous purity, and the chance of inbreeding decline, thereby reducing the adaptability of the species to environmental changes [32]. Therefore, in the absence of dominant exotic wild-type groups as breeding parents, selecting groups with higher heterozygosity for breeding can reduce the chance of inbreeding depression. The selection of cross-fertilization between groups with a large genetic differentiation index can be advantageous and slow the impact of inbreeding between breeding groups [15]. In most cases, selection and hatchery practices are the primary factors that reduce genetic variability in aquaculture populations. However, this study did not observe the adverse effects of selective breeding practices. To prevent inbreeding that might occur in later generations, breeders should maintain breeding records and pedigree information. Sui et al. constructed a pedigree of founders in a selection population of M. rosenbergii in China using 16 microsatellite loci [5]. Based on this information, they evaluated the genetic parameters and selection response of the harvest body weight of M. rosenbergii [33].
The D-loop analysis identified 114 variable sites and 29 haplotypes, with SF, HF, and JX showing the highest haplotype diversity (Hd > 0.5, π > 0.005), suggesting that they were highly genetically diverse. However, the GX and TW populations displayed lower diversity, highlighting the potential risks of inbreeding if not managed properly. Hd and π are important parameters for assessing genetic diversity [34]. Grant and Bowen determined that the critical value of Hd is 0.5 and that of π is 0.005, respectively. The greater the two values, the greater the degree of population diversity [35]. From the perspective of the haplotype number of each group, the HF group contained the largest number of haplotypes (13), followed by the JX group (11). Five dominant haplotypes were observed in most populations. Individuals with dominant haplotypes are generally adaptable and have larger breeding populations. In contrast, low-frequency haplotypes have smaller breeding populations owing to poor environmental adaptability, which is more likely to adversely affect the genetic diversity of the entire population [32]. The dominant haplotype reflects the breeding direction in the early artificial selection process, and individuals with high survival rates and excellent growth traits are retained in the directional selection process, so that the number of groups in the dominant haplotype gradually expands. In addition, haplotype clustering results showed that the haplotypes of the HF and JX groups had higher distributions in both branches, reflecting the rich genetic diversity of these two groups.
These results corroborate previous genetic diversity studies on M. rosenbergii using mtDNA in China, reinforcing the utility of mtDNA markers in aquaculture genetics. For instance, mtDNA COI and 16SrRNA fragments were used in a study on the genetic structure of four breeding populations of M. rosenbergii, which revealed that the populations showed low to moderate genetic diversity [5]. Recently, Jiang et al. employed mitochondrial sequences of the COI gene to study the genetic diversity of three populations of M. rosenbergii in China, and all populations showed high genetic diversity [18]. To our knowledge, the study of Yao et al. is the only study on the genetic diversity of the M. rosenbergii D-loop gene in China, which showed that the genetic diversity of the D-loop gene in M. rosenbergii is relatively rich. The deficiency is that they studied only a small number of samples, with a total of 16 samples for three populations [36]. The present study expanded the population and individuals, and the conclusions are consistent with theirs. An adequate sample size is a prerequisite for obtaining stable and reliable genetic results. The sample size in this study was relatively large, except for the TW population, which was less than the sample size requirement and could bias the diversity estimates. The sample size requirement of ≥30 individuals ensures statistical reliability while offering a more accurate representation of population genetic diversity. Larger sample sizes may be necessary, particularly for species with complex population structures or those subject to intensive artificial selection.
Comparatively, the mtDNA D-loop region and SSR results showed that the six M. rosenbergii breeding populations maintained substantial genetic diversity. The Na, He, and PIC values indicated that SF had the highest diversity, whereas GX demonstrated the least, a finding that aligns with the genetic diversity parameters Hd and π derived from the D-loop analysis. This suggests that the SF, HF, JX, and NTH groups possessed the highest diversity, starkly contrasting with GX, which remained the least diverse. Fst is an important indicator of genetic diversity among populations [32]. Different Fst values represent varying degrees of genetic differentiation: Fst < 0.05 indicates low genetic differentiation, 0.05 < Fst < 0.15 indicates moderate genetic differentiation, and Fst > 0.25 indicates high genetic differentiation [37]. The results of the SSR markers showed that the pairwise Fst among the six groups ranged from 0.03541 to 0.09637 (p < 0.05), indicating from low to moderate genetic differentiation among the six populations, while the Fst of the D-loop ranged from 0.05603 to 0.80788, indicating from moderate to high genetic differentiation. Moreover, based on SSR, the highest Fst value was found between TW and GX, and the lowest Fst value was found between HF and JX. However, based on the D-loop, the highest Fst value was obtained between SF and TW, and the lowest was obtained between JX and TW. The UPGMA trees based on the genetic distance of microsatellites clustered into two major groups (Figure 1), whereas the structure cluster analysis clustered into three major groups (Figure 2). The phylogenetic tree based on the D-loop haplotypes was clustered into two major groups and was not entirely consistent with the two clusters mentioned above.
The observed discrepancy between microsatellite and mitochondrial markers may arise from their limitations in detecting specific genetic variation, even though they both serve as valuable tools for genetic analysis. For instance, the mitochondrial D-loop gene exclusively reflects the maternal lineage, potentially biasing paternal or nuclear genomic variations. Additionally, microsatellites may exhibit homoplasy, where alleles of the same size are not identical by descent, complicating population genetic analyses. Future studies should leverage genomic approaches (e.g., SNP arrays or whole-genome sequencing) to provide a more detailed genetic structure that will aid in the breeding management practices of M. rosenbergi.
5. Conclusions
Overall, the analyses of this study confirmed that the genetic differentiation based on microsatellite markers and mitochondrial D-loop gene in all six populations was from moderate to high, among which four populations, including Shufeng, Hefu, Nantaihu No.2, and Jiaxing, have a higher degree of diversity and, hence, possess greater potential for long-term selective breeding. The results provide a theoretical basis for conserving and utilizing M. rosenbergii selective breeding strains in China. Much attention and precise knowledge of all the existing strains must be considered to design proper and effective conservation strategies. Therefore, urgent further studies with a higher number of microsatellites, another part of the mitochondrial genome, or involving all existing M. rosenbergii strains are recommended to ensure a better understanding of the detailed genetic structure of this important crustacean species.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/d17070437/s1, Table S1: Allele number (Na) observed heterozygosity (Ho) and Expected heterozygosity (He), polymorphism information content (PIC), and p value for testing Hardy-Weinberg equilibrium (P) in the six populations genetic diversity of M. rosenbergii; Table S2: Distribution of haplotypes in different populations of M. rosenbergii.
Author Contributions
Conceptualization, S.I.; methodology, S.I. and M.C.; software, S.I. and J.L.; validation, S.Y. and Z.X.; formal analysis, Q.T.; investigation, A.R.; resources, Y.D.; data curation, M.C.; writing—original draft preparation, S.I.; writing—review and editing, S.I. and Q.T.; visualization, Z.X.; supervision, G.Y.; funding acquisition, G.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the Key Scientific and Technological Grant of Zhejiang for Breeding New Agricultural (Aquaculture) Varieties (2021C02069-4-3) and the Earmarked Fund for CARS (CARS-48).
Institutional Review Board Statement
This study was conducted according to the Guide for Laboratory Animals developed by the Ministry of Science and Technology (Beijing, China). The experimental protocol for animal care and tissue collection was approved by Huzhou University (the ethical approval code: 20190625; approval date: 15 June 2019).
Data Availability Statement
The data are within the manuscript, with Supplementary Data provided separately. There is no additional dataset.
Acknowledgments
We are very grateful to the potential reviewers for their constructive and informative comments and corrections, which would help improve our manuscript.
Conflicts of Interest
Guoliang Yang, Zhenglong Xia and Miaoying Cai were employed by Jiangsu Shufeng Prawn Breeding Co., Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| GFP | Giant freshwater prawn |
| NTH | Nantaihu |
| HF | Hefu |
| GX | Guangxi |
| JX | Jiaxing |
| TW | Taiwan |
| SF | Shufeng |
| PCR | Polymerase chain reaction |
| DNA | Deoxyribonucleic acid |
| mt | Mitochondrial |
| RAPD | Randomly amplified polymorphic |
| SNP | Single nucleotide polymorphisms |
| SSR | Single sequence repeats |
| ddH2O | Double distilled water |
| μL | Microlitre |
| s | Second |
| min | Minute |
| ng | Nanogram |
| PIC | Polymorphic information content |
| Na | Mean number of alleles per locus |
| Ne | Effective number of alleles |
| He | Expected heterozygosity |
| Ho | Observed heterozygosity |
| HWE | Hardy–Weinberg equilibrium |
| N | Sample size |
| H | Haplotype number |
| Hd | Haplotype diversity |
| π | Nucleotide diversity |
| S | Number of polymorphic (segregating) sites |
| Fis | Inbreeding coefficient |
| Fit | Biological Fitness |
| Fst | Fixation index |
| Nm | Number of migrants |
| UPGMA | Unweighted pair group method with arithmetic |
| MEGA | Molecular evolutionary genetics analysis |
| A | Adenine |
| G | Guanine |
| C | Cytosine |
| T | Thymine |
References
- Xu, Y.; Feng, Y.; Zhang, S.; Yang, Z.; Xu, W.; Gong, J.; Gu, H. Effects of Macrobrachium nipponense on phytoplankton communities and water environmental factors in Macrobrachium rosenbergii culture. Aquac. Res. 2024, 2024, 4218312. [Google Scholar] [CrossRef]
- Tan, K.; Wang, W. The early life culture and gonadal development of giant freshwater prawn, Macrobrachium rosenbergii: A review. Aquaculture 2022, 559, 738357. [Google Scholar] [CrossRef]
- FAO (Food and Agriculture Organization of the United Nations). Yearbook of Fishery Statistics: Summary Tables; FAO: Rome, Italy, 2024. [Google Scholar]
- China Fishery Statistical Yearbook 2024; Fisheries Bureau Department of Agriculture of China: Beijing, China, 2024.
- Sui, J.; Luan, S.; Yang, G.; Chen, X.; Luo, K.; Gao, Q.; Wang, J.; Hu, H.; Kong, J. Genetic diversity and population structure of a giant freshwater prawn (Macrobrachium rosenbergii) breeding nucleus in China. Aquac. Res. 2018, 49, 2175–2183. [Google Scholar] [CrossRef]
- Boudry, P.; Allal, F.; Aslam, M.L.; Bargelloni, L.; Bean, T.P.; Brard-Fudulea, S.; Brieuc, M.S.O.; Calboli, F.C.F.; Gilbey, J.; Haffray, P.; et al. Current status and potential of genomic selection to improve selective breeding in the main aquaculture species of international council for the exploration of the sea (ICES) member countries. Aquac. Rep. 2021, 20, 100700. [Google Scholar] [CrossRef]
- Bijma, P.; Woolliams, J.A. Prediction of rates of inbreeding in populations selected on best linear unbiased prediction of breeding value. Genetics 2000, 156, 361–373. [Google Scholar] [CrossRef]
- Yu, L.; Zhu, X.; Liang, J.; Fan, J.; Chen, C. Analysis of genetic structure of wild and cultured giant freshwater prawn (Macrobrachium rosenbergii) using newly developed microsatellite. Front. Mar. Sci. 2019, 6, 323. [Google Scholar] [CrossRef]
- Chareontawee, K.; Poompuang, S.; Na-Nakorn, U.; Kamonrat, W. Genetic Diversity of hatchery stocks of giant freshwater prawn (Macrobrachium rosenbergii) in Thailand. Aquaculture 2007, 271, 121–129. [Google Scholar] [CrossRef]
- Pillai, B.R.; Ponzoni, R.W.; Das Mahapatra, K.; Panda, D. Genetic improvement of giant freshwater prawn Macrobrachium rosenbergii: A review of global status. Rev. Aquac. 2022, 14, 1285–1299. [Google Scholar] [CrossRef]
- Thanh, N.M.; Nguyen, N.H.; Ponzoni, R.W.; Vu, N.T.; Barnes, A.C.; Mather, P.B. Estimates of strain additive and non-additive genetic effects for growth traits in a diallel cross of three strains of giant freshwater prawn (Macrobrachium rosenbergii) in Vietnam. Aquaculture 2010, 299, 30–36. [Google Scholar] [CrossRef]
- Tang, Q.Y.; Xie, J.H.; Xia, Z.L.; Cai, M.Y.; Wu, Y.M.; Bai, L.H.; Du, H.K.; Li, J.F.; Yang, G.L. Genetic diversity of the breeding populations of giant freshwater prawn Macrobrachium rosenbergii. Acta Hydrobiol. Sin. 2020, 44, 1097–1104. [Google Scholar] [CrossRef]
- Ahammad, A.S.; Rahman, M.S.; Ahmed, M.B.U.; Rabbi, M.F.; Wahab, M.A. Morpho-genetic characterization of morphotypes of the giant freshwater prawn, Macrobrachium rosenbergii. Bangladesh J. Fish. 2019, 31, 31–40. [Google Scholar]
- Wahidah, W.; Omar, S.B.A.; Trijuno, D.D.; Nugroho, E.; Amrullah, A.; Khatimah, K. Genetic variation of giant freshwater prawns Macrobrachium rosenbergii wild population of South Sulawesi, Indonesia. Biodiversitas J. Biol. Divers. 2023, 24, 3081–3090. [Google Scholar] [CrossRef]
- Binur, R.; Pancoro, A. Inbreeding depression level of post-larvae freshwater prawn (Macrobrachium rosenbergii) from several hatcheries in Java, Indonesia. Biodiversitas J. Biol. Divers. 2017, 18, 609–618. [Google Scholar] [CrossRef]
- Lyu, M.; Huang, G.; Li, M.; Yang, Q.; Lu, X.; Gan, H.; Ruan, Z.; Huang, L.; Yang, Y.; Lu, T.; et al. Microsatellite analysis of genetic diversity of male giant freshwater prawn Macrobrachium rosenbergii with various morphotypes. Fish. Sci. 2019, 38, 355–360. [Google Scholar]
- Ge, J.; Xu, Z.; Huang, Y.; Lu, Q.; Pan, J.; Yang, J. Genetic variation in wild and cultured populations of the freshwater prawn, Macrobrachium nipponense, in China. J. World Aquac. Soc. 2011, 42, 504–511. [Google Scholar] [CrossRef]
- Jiang, F.; Dai, X.L. Analysis of the genetic diversity and sequence variation of mitochondrial COIgene from three populations of Macrobrachium rosenbergii. J. Fish. Res. 2023, 45, 8. [Google Scholar]
- Bala, B.; Mallik, M.; Saclain, S.; Islam, M.S. Genetic variation in wild and hatchery populations of giant freshwater prawn (Macrobrachium rosenbergii) revealed by randomly amplified polymorphic DNA markers. J. Genet. Eng. Biotechnol. 2017, 15, 23–30. [Google Scholar] [CrossRef]
- Agarwal, D.; Aich, N.; Pavan-Kumar, A.; Kumar, S.; Sabnis, S.; Joshi, C.G.; Koringa, P.; Pandya, D.; Patel, N.; Karnik, T.; et al. SNP mining in transcripts and concomitant estimation of genetic variation in Macrobrachium rosenbergii stocks. Conserv. Genet. Resour. 2016, 8, 159–168. [Google Scholar] [CrossRef]
- Chen, P.; Shih, C.; Chu, T.J.; Lee, Y.; Tzeng, T.D. Phylogeography and genetic structure of the oriental river prawn Macrobrachium nipponense (Crustacea: Decapoda: Palaemonidae) in East Asia. PLoS ONE 2017, 12, e0173490. [Google Scholar] [CrossRef]
- Ke, X.; Liu, J.; Gao, F.; Cao, J.; Liu, Z.; Lu, M. Analysis of genetic diversity among six dojo loach (Misgurnus anguillicaudatus) populations in the pearl river basin based on microsatellite and mitochondrial DNA markers. Aquac. Rep. 2022, 27, 101346. [Google Scholar] [CrossRef]
- Noorullah, M.; Zuberi, A.; Zaman, M.; Younas, W.; Hussain, S.; Kamran, M. Assessment of genetic diversity among wild and captive-bred Labeo rohita through microsatellite markers and mitochondrial DNA. Fish. Aquat. Sci. 2023, 26, 752–761. [Google Scholar] [CrossRef]
- Liu, S.; Yu, Q.; Chen, R.; Hu, W.; Yan, X.; Han, Q.; Xu, D.; Zhu, Q. Comparison of genetic diversity between hatchery-reared and wild rock bream (Oplegnathus fasciatus) based on microsatellite markers and mitochondrial COI sequences. Aquac. Res. 2024, 2024, 5570764. [Google Scholar] [CrossRef]
- Li, Y.; Song, J.; Shen, X.; Cai, Y.; Cheng, H.; Zhang, X.; Yan, B.; Chu, K.H. The First mitochondrial genome of Macrobrachium rosenbergii from China: Phylogeny and gene rearrangement within Caridea. Mitochondrial DNA Part B 2018, 4, 134–136. [Google Scholar] [CrossRef]
- Woodruff, D.S. Populations, Species, and conservation genetics. In Encyclopedia of Biodiversity; Elsevier: Amsterdam, The Netherlands, 2001; pp. 811–829. [Google Scholar] [CrossRef]
- Ding, Q.; Shi, M.; Ji, P.; Qin, L.; Gao, X.; Zhang, X.; Jiang, Q. Genetic diversity and differentiation of cultured Macrobrachium rosenbergii in China using newly developed microsatellite multiplex PCR panels. Aquac. Int. 2024, 32, 7895–7910. [Google Scholar] [CrossRef]
- Banu, M.R.; Siraj, S.S.; Christianus, A.; Ikhsan, N.F.M.; Rajaee, A.H. Genetic variation among different morphotypes of the male freshwater prawn Macrobrachium rosenbergii (De Man). Aquac. Rep. 2015, 1, 15–19. [Google Scholar] [CrossRef]
- Avval, S.E. Assessing Polymorphism Information Content (PIC) Using SSR molecular markers on local species of Citrullus Colocynthis. Case study: Iran, Sistan-Balouchestan Province. J. Mol. Biol. Res. 2017, 7, 42. [Google Scholar] [CrossRef]
- Zhang, H.-R.; Niu, S.-F.; Wu, R.-X.; Zhai, Y.; Tian, L.-T. Development and characterization of 26 polymorphic microsatellite markers in Lateolabrax maculatus and cross-species amplification for the phylogenetically related taxa. Biochem. Syst. Ecol. 2016, 66, 326–330. [Google Scholar] [CrossRef]
- Qin, Y.; Shi, G.; Sun, Y. Evaluation of genetic diversity in Pampus argenteus using SSR markers. Genet. Mol. Res. 2013, 12, 5833–5841. [Google Scholar] [CrossRef]
- Huang, X.; Yang, S.; Gong, J.; Zhao, Y.; Feng, Q.; Gong, H.; Li, W.; Zhan, Q.; Cheng, B.; Xia, J.; et al. Genomic analysis of hybrid rice varieties reveals numerous superior alleles that contribute to heterosis. Nat. Commun. 2015, 6, 6258. [Google Scholar] [CrossRef]
- Sui, J.; Luan, S.; Yang, G.; Xia, Z.; Luo, K.; Tang, Q.; Lu, X.; Meng, X.; Kong, J. Genetic parameters and selection response for the harvest body weight of the giant freshwater prawn (Macrobrachium rosenbergii) in a breeding program in China. PLoS ONE 2019, 14, e0218379. [Google Scholar] [CrossRef]
- Wu, J.; Wang, W.; Deng, D.; Zhang, K.; Peng, S.; Xu, X.; Zhang, Y.; Zhou, Z. Genetic diversity and phylogeography of Daphnia similoides sinensis located in the middle and lower reaches of the Yangtze river. Ecol. Evol. 2019, 9, 4362–4372. [Google Scholar] [CrossRef] [PubMed]
- Grant, W. Shallow population histories in deep evolutionary lineages of marine fishes: Insights from sardines and anchovies and lessons for conservation. J. Hered. 1998, 89, 415–426. [Google Scholar] [CrossRef]
- Yao, Q.; Yang, P.; Chen, L.Q.; Guo, H.; Zhang, H.; Yu, N.; Yang, G.L.; Wang, J.Y. Interspecific DNA sequence polymorphism in the mitochondrial D-loop gene from three populations of Macrobrachium rosenbergii. J. Fish. China 2007, 31, 18–22, (In Chinese with English abstract). [Google Scholar]
- Wright, S. Variability Within and Among Natural Populations. In Evolution and the Genetics of Populations: A Treatise in Four Volumes; The University of Chicago Press, Cop: Chicago, IL, USA; London, UK, 1984; Volume 4. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).