Abstract
The taxonomic position of the green alga Capsosiphon fulvescens was first reported from Northern Europe and has since been reported from all over the world, including Korea. In Korea, C. fulvescens has been used as an essential edible economic alga for approximately 570 years, from the time of the Joseon Dynasty to the present, and is currently under development as a new aquaculture strain. Therefore, examining the taxonomic relationships between the European and Korean C. fulvescens is important. In this study, we analyzed nuclear 18S rDNA and ITS regions and compared them with the DNA sequences of authentic materials of North Atlantic C. fulvescens. Additionally, rbcL and tufA genes were sequenced to analyze genetic variations among populations. The results showed that the Korean and European C. fulvescens were different species. Moreover, the Korean C. fulvescens was distantly related to the North Atlantic C. fulvescens at the order level. Moreover, the Korean C. fulvescens formed a sister group with the North Pacific Pseudothrix borealis. Cryptic genetic diversity was observed at the intraspecific level among the Korean populations. These findings will help in tracing the origin of the Korean C. fulvescens and provide new genetic insights into this species.
1. Introduction
The aquaculture of green macroalgae has recently increased in Asia, and cultivation technologies are being developed for green algae that are putative strains for aquaculture [1]. Regarding the global production of green seaweed by marine aquaculture during the years 2014–2018, production of Ulva spp. in Korea, South Africa, and China accounted for 49% of the five-year average relative percentage by weight, whereas Codium fragile and Caulerpa spp. accounted for 21 and 5% of the total production, respectively. In Korea, Capsosiphon fulvescens (Ulvophyceae, Ulotrichales; Maesaengi (in Korean pronunciation)) accounted for 25% of the total production. The taxonomic position of C. fulvescens was first reported from Northern Europe (type locality: Sweden); however, this species is only cultivated in Korea in the North Pacific region [2].
In Korea, C. fulvescens has been used as an edible green alga for approximately 570 years, from the time of the Joseon Dynasty to the present. Taxonomically, C. fulvescens was first identified as Ulva fulvescens C. Agardh [3] and was later established as the genus Capsosiphon by Gobi [4]. Interestingly, 370 years before these modern taxonomic studies were conducted, C. fulvescens was recognized as a taxonomic entity in Korea (Joseon Dynasty) and was named “Maesani” in Korean (Sejong Sillok (1454) Volume 151, Geography of Jeolla-do [Veritable Records of the Joseon Dynasty; https://sillok.history.go.kr/intro/english.do accessed on 12 February 2025]) [5]. The physiological and ecological characteristics of this useful seaweed have been described in the Veritable Records of the Joseon Dynasty listed under the Memory of the World Programme (United Nations Educational, Scientific, and Cultural Organization [UNESCO]). This species was widely used as an edible seaweed during the time of the Joseon Dynasty (Revised and Expanded Edition of Survey of the Geography of Joseon, Volume 37, Jeolla-do; Lee Heng [6]; Jasaneobo, Jeong Yak Jeon [7]).
Four species have been recorded under the genus Capsosiphon: C. aureolus (C. Agardh) Gobi, C. aureus V.J. Chapman, C. fulvescens (C. Agardh) Setchell & N.L. Gardner, and C. groenlandicus (J. Agardh) K.L. Vinogradova [8]. Among these species, C. aureolus is currently regarded as a heterotypic synonym of C. fulvescens, and C. groenlandicus has been taxonomically changed into a type species of a new genus, Pseudothrix [9]. Therefore, C. fulvescens (C. Agardh) Setchell & N.L. Gardner [10] and C. aureus V.J. Chapman [11] are the only two species currently under the genus Capsosiphon. The presence of C. fulvescens has been recorded in Asia, Europe, the Arctic, and the sub-Antarctic islands [8]. From Korea [12,13,14] and Japan [15], only C. fulvescens has been reported. Morphological studies using an indoor culture method [16,17,18,19] were conducted to analyze the life cycle and reproductive cell characteristics of C. fulvescens while also assessing its taxonomic position at the order and family levels.
Molecular phylogenetic studies were conducted using the nuclear 18S rDNA and plastid ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit (rbcL) gene sequences and the plastid genome sequence of C. fulvescens; the phylogenetic relationships between C. fulvescens and other green algal species of the orders Ulotrichales and Ulvales were examined [20,21,22]. However, the plastid genome data of C. fulvescens were insufficient to resolve its taxonomic problems [22]. Thus, the taxonomic position of C. fulvescens remains controversial.
Conflicting results have been obtained using previously reported DNA sequences. The phylogenetic relationships between Ulvales and Ulotrichales were examined using 18S rDNA and rbcL DNA sequences [20]. The researchers used only the 18S rDNA sequence (GenBank accession number: AF499664) from the Atlantic C. fulvescens (source information: Monks Head Beach, Nova Scotia) to determine its phylogenetic relationship with other species belonging to Ulvales and Ulotrichales. The results of the study [20] were cited as important reference data in subsequent reports on molecular analyses [21,22]. However, in these subsequent studies [21,22], the molecular phylogeny was not analyzed, including that of the key reference of the 18S rDNA sequence from the European C. fulvescens [20]. Instead of the previously reported 18S rDNA sequence of North Atlantic C. fulvescens, ref. [20,21,22] used DNA sequences from the Korean samples. In the absence of a key reference 18S rDNA sequence, the phylogenetic relationships between C. fulvescens and other species belonging to Ulvales and Ulotrichales were analyzed and discussed in these studies [21,22].
Recently, novel DNA sequences (18S rDNA and internal transcribed spacer [ITS] regions) were reported [23] from unicellular marine coccoid green algae that had not been analyzed in previous molecular phylogenetic studies [20,21,22]. In this study, novel taxa of green algae, belonging to the orders Chlorocystidales and Sykidiales ord. nov., were included [23]. These novel sequences of the 18S rDNA and ITS regions can be used as novel DNA sequence references for the assessment of the taxonomic position of Korean C. fulvescens. In another study [9], Pseudothrix borealis, previously reported as C. groenlandicus from North Pacific Amaknak Island (Aleutian Islands, AK, USA), was identified as a new genus based on morphological characteristics and molecular data (DQ821514; 18S rDNA and ITS regions); in this study, the previously reported DNA sequences from the North Atlantic C. fulvescens were included (AF499664 [18S] and EU541503 [ITS1]).
In this study, we observed genetic differences between 18S rDNA and ITS sequences of the Korean and European C. fulvescens; therefore, we reexamined the taxonomic position of the Korean C. fulvescens using 18S rDNA and ITS sequences. Moreover, the results were compared with those obtained using the key reference DNA sequences from the European C. fulvescens [9,20]. We also determined the rbcL and translation elongation factor EF-Tu (tufA) gene sequences and examined intraspecific sequence variations for these two genes among Korean populations.
2. Materials and Methods
We examined field samples and herbarium specimens of the Korean C. fulvescens deposited at the National Institute of Biological Resources (NIBR) in Korea (Figure 1 and Figure 2; Table 1). Commercial goods in the public market were also analyzed. Field samples were collected and directly deposited onto silica gels for DNA analysis. We also used herbarium specimens collected from the Korean Peninsula. The advantage of using herbarium specimens was that samples from a wide range of collection sites could be analyzed, and the cryptic genetic features of Korean algal flora could be detected [24,25]. We examined the morphological characteristics of fresh field samples obtained from Korean individuals using a light microscope (Primostar 3; Zeiss, Germany; Figure 3).

Figure 1.
Aquaculture farm showing the cultivation of Capsosiphon fulvescens in Korea (Jangheung, Republic of Korea; 30 April 2014).

Figure 2.
A herbarium specimen (National Institute of Biological Resources [NIBR]) examined in this study (Wando, Republic of Korea, 15 March 2022; NIBR accession number: NIBRCL0000115865).

Table 1.
Sampling information and sequence data of Capsosiphon fulvescens from Korea. The 18S rDNA and tufA gene sequences were similar among samples (O); sequences that could not be determined are indicated using (-). Ribotypes of internal transcribed spacer (ITS) sequences (GenBank accession numbers: A (PQ557621), G (PQ557622), and R (PQ557623)) and haplotypes of ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit (rbcL) sequences are presented as A (PQ561384) or G (PQ561385). R indicates mixed bases (A/G) in the ITS sequence.
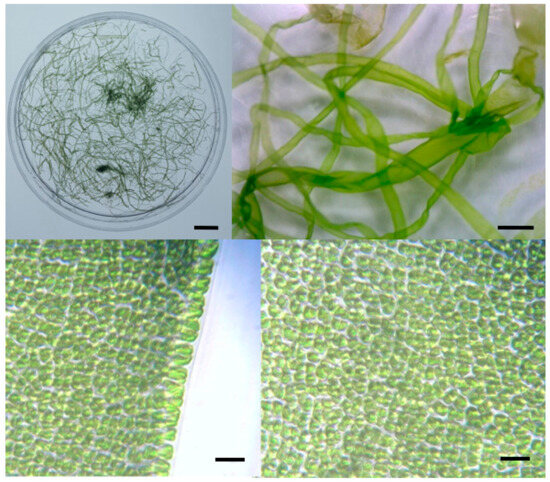
Figure 3.
Morphological features of vegetative cells of Korean Capsosiphon fulvescens. Scale bar: (1) 1 cm, (2) 1 mm, (3) 100 μm, and (4) 100 μm.
Total genomic DNA was extracted from a single strand of the filamentous plant body using the DNeasy Plant Mini Kit and TissueLyser LT (Qiagen, Hilden, Germany), according to the manufacturer’s instructions [24]. For the molecular phylogenetic analyses, we selected four DNA markers (nuclear DNA, ribosomal small subunit RNA gene [18S rDNA], ITS region, and plastid DNA: rbcL and tufA gene sequences). Nuclear DNA sequences (18S rDNA and ITS) were used as key references to determine the phylogenetic relationships between C. fulvescens and the related species belonging to Ulvales and Ulotrichales, based on the previously reported molecular phylogenetic studies [9,20].
Specific primer combinations were used to amplify the target regions (18S rDNA: A/SSU-inR1 [26]; ITS region: ITS1/ITS4 [27]; rbcL: RH1/rbc590 [20,28]; tufA: tufAF/tufAR [29]) using amfiXpand PCR Master Mix (GenDEPOT, Barker, TX, USA). The reaction conditions were as follows: 3 min at 95 °C followed by 40 cycles of 30 s at 94 °C, 30 s at 50 °C, and 1 min at 72 °C, with a final 7 min at 72 °C; the reactions were performed using a Thermal Cycler 9700 (Applied Biosystems, Foster City, CA, USA). The amplified PCR products were purified and sequenced in both directions (GenoTech, Daejeon, Republic of Korea). Chromatograms were assembled using Sequencher version 5.4.6 (Gene Codes, Ann Arbor, MI, USA), and the Basic Local Alignment Search Tool (BLAST; http://blast.ncbi.nlm.nih.gov/Blast.cgi accessed on 12 February 2025)as used to search for similarities between the identified and reference sequences. For molecular phylogenetic analysis, we aligned the sequences and inferred phylogenetic trees using the neighbor-joining and maximum likelihood methods using the MEGA program (version 6) [30]. The taxonomic position of Korean C. fulvescens in relation to European C. fulvescens remained consistent, despite model changes; therefore, we primarily represented the phylogenetic tree using the neighbor-joining method. Additionally, phylogenetic trees were analyzed using the maximum likelihood method (Figures S1 and S2). Pairwise distances were calculated using Kimura’s two-parameter model.
3. Results
In this study, we determined the 18S rDNA, ITS region, and rbcL and tufA gene sequences of the Korean samples of C. fulvescens (Table 1). The 18S rDNA sequences were isolated from nine samples of Korean C. fulvescens (5′-end partial sequences; 520 bp), and no intraspecific variation was found among the samples (GenBank accession number: PQ555428). A BLAST search revealed that our 18S rDNA sequences showed 100% similarity with the C. fulvescens sequences reported in previous studies (MG764430 in [22] and EU099917, EU099919, and EU099920 in [21]). A one-base difference was found between our 18S rDNA sequences and the EU099918 sequence (Jindo, Republic of Korea [21]). The species that was most closely related to the Korean C. fulvescens in terms of DNA sequence similarity (99.8%) was Pseudothrix borealis (DQ821514), which was previously identified as the North Pacific C. groenlandicus. The novel genus Pseudothrix was established in a later study [9]. In contrast, the Korean C. fulvescens was distantly related to the North Atlantic C. fulvescens (AF499664) reported in another study [20] (92.7% similarity). A BLAST analysis of the long DNA sequences obtained from the Korean C. fulvescens (EU099917–EU099920 and MG764430) revealed that these sequences showed 100% similarity with our sequences; the North Atlantic C. fulvescens (AF499664) showed 97% similarity (723/749) with our C. fulvescens samples. Phylogenetic analysis of 18S rDNA was conducted using long DNA sequences, including those from the Korean C. fulvescens (MG764430; Figure 4).
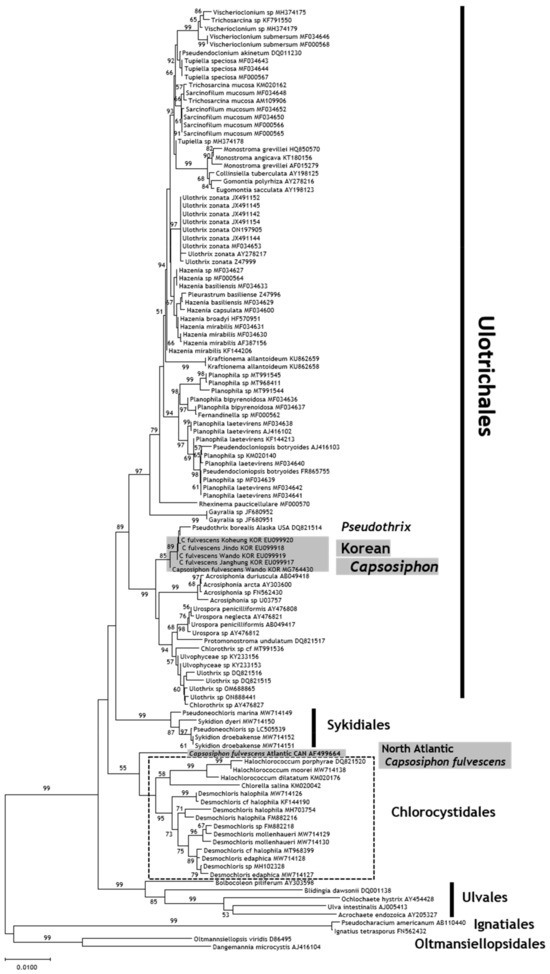
Figure 4.
Neighbor-joining tree constructed using sequences of the 18S rDNA region by applying Kimura’s two-parameter model. The bootstrap test was conducted with 2000 replicates.
The ITS sequences of the Korean C. fulvescens were determined; nine sequences (569 bp) were elucidated using field samples, herbarium specimens, and commercial goods available from public markets (Table 1). We found two ribotypes with a one-base difference among the Korean population (62nd position in the ITS1 sequence: A (GenBank accession number PQ557621) or G (PQ557622). In the case of commercial goods in the public market (Jangheung 1, Republic of Korea; PQ557623), the mixed base R (A/G) was found at the 62nd position. This indicates the presence of mixed strains in aquaculture farms of C. fulvescens. A similarity search revealed that the ITS sequences from the Korean samples of C. fulvescens were different (82% similarity) from those of the North Atlantic C. fulvescens (Kattegat, Denmark) [9]. In contrast, the Korean C. fulvescens ITS sequences showed the highest similarity (94.5%) with ITS sequences from Pseudothrix borealis (Capsosiphon groenlandicus DQ821514 in [9]) and less than 93.5% similarity with those from the other species belonging to Ulotrichales.
We also determined 12 sequences of rbcL (547 bp) using field samples, herbarium specimens, and commercial goods in the public market and six sequences (930 bp) of tufA using field samples (Table 1). The rbcL (G-type; PQ561385) and tufA sequences (PQ561386) showed 100% similarity to the previously reported sequences of the Korean C. fulvescens (MG727869 in [22]). The rbcL gene sequence from the Korean C. fulvescens showed 93.5% similarity with sequences reported from the other species of Ulotrichaceae deposited in GenBank (National Center for Biotechnology Information, NCBI); the tufA sequence showed less than 93% similarity to sequences reported from the other green algae deposited in GenBank (NCBI). Two haplotypes of the rbcL gene were found among the Korean populations (A (PQ561384) or G (PQ561385)) at the 418th position in the rbcL gene sequence), including the previously reported gene sequence from the Korean C. fulvescens (G in MG727869).
Molecular phylogenetic analyses were conducted using sequences of 18S rDNA and ITS regions (Figure 4 and Figure 5) because these were key reference DNA sequences obtained from the European C. fulvescens in previous studies [9,20]. The Korean C. fulvescens formed a separate group from the European C. fulvescens in the phylogenetic tree prepared using the 18S rDNA and ITS sequences. The Korean C. fulvescens formed a sister group with Pseudothrix borealis and was placed under the order Ulotrichales in the tree constructed using the 18S rDNA sequences (Figure 4); the Korean C. fulvescens clustered with Pseudothrix and Protomonostroma in the tree constructed using the ITS sequences (Figure 5). The European C. fulvescens formed a clade with the species belonging to Chlorocystidales and was distantly related to the Korean C. fulvescens. The North Pacific and North Atlantic C. fulvescens showed differences in their classification as green algae.
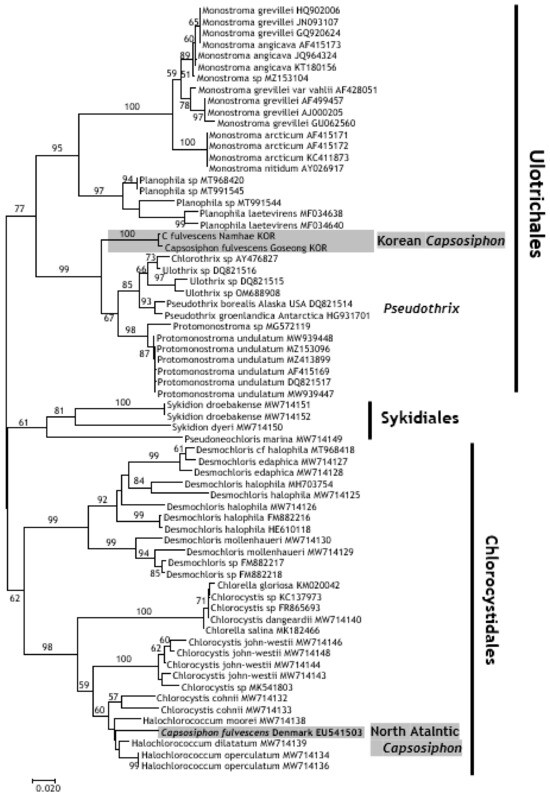
Figure 5.
Neighbor-joining tree constructed using sequences of the internal transcribed spacer (ITS) region by applying Kimura’s two-parameter model. The bootstrap test was conducted with 2000 replicates.
4. Discussion
The green alga C. fulvescens has been used as an essential edible economic alga in Korea since the time of the Joseon Dynasty (approximately 570 years ago). Its industrial use in Korea has predated modern taxonomic studies on this species in Europe by several hundred years [3,4]. This species has been used as an aquaculture species in Korea [2,31,32]. Currently, this species is important in aquaculture [1]. Although molecular phylogenetic studies have been conducted to determine the taxonomic relationships between C. fulvescens and other species of Ulotrichales and Ulvales, this topic remains debatable [20,21,22].
One of the causes for this confusion is the lack of genetic data required for conducting molecular phylogenetic studies [21,22]. In a report [20], the phylogenetic classifications reported in previous studies were summarized; the taxonomic relationships between the European C. fulvescens and other species belonging to Ulotrichales and Ulvales were also examined in that study using 18S rDNA and rbcL gene sequences. In particular, the researchers used only the 18S rDNA gene sequences to determine the taxonomic relationships among the species belonging to Ulotrichales and Ulvales. In another study [9], the 18S rDNA and ITS regions were examined using samples of the North Atlantic C. fulvescens and North Pacific Pseudothrix borealis (previously reported as C. groenlandicus). However, in two subsequent studies [21,22], these key reference DNA sequences [9,20] were not considered during the analysis of the taxonomic relationships between C. fulvescens and other Ulotrichales and Ulvales species. The authors deduced the phylogenetic relationships using only the Korean samples of C. fulvescens and did not analyze the ITS region sequences from those samples [21,22].
In this study, we determined the 18S rDNA and ITS sequences of the Korean C. fulvescens. The Korean C. fulvescens showed a high degree of sequence variation and a distant phylogenetic relationship with the European C. fulvescens (Figure 4 and Figure 5). Analysis of phylogenetic trees constructed using 18S rDNA and ITS sequences revealed that the Korean C. fulvescens was closely related to P. groenlandica (J. Agardh) Hanic & S.C. Lindstrom [9] (Ulotrichaceae, Ulotrichales). P. groenlandica was initially identified as C. groenlandicus from North Pacific Alaska and was taxonomically reexamined in a subsequent study [9]. In this report [9], the ITS sequence (EU541503) obtained from the North Atlantic C. fulvescens samples collected at Hirsholm, Kattegat, Denmark (by Ruth Nielsen; 1 September 1988) was published. The collection site was located near the type locality of C. fulvescens (Landskrona, Sweden, South of Kattegat). The ITS sequence of the North Atlantic C. fulvescens showed a high degree of sequence variation with that of the Korean C. fulvescens (82% similarity); the North Atlantic and Korean C. fulvescens formed distinct groups in the phylogenetic tree constructed using these ITS sequences (Figure 5).
The European C. fulvescens formed a cluster with the species putatively belonging to Chlorocystidales in the phylogenetic tree (Figure 4 and Figure 5). In contrast, the Korean C. fulvescens showed a close taxonomic relationship with the species belonging to Ulotrichales. In a report [23], novel DNA sequences (18S rDNA and ITS regions) from unicellular marine coccoid green algae belonging to the orders Chlorocystidales and Sykidiales nov. were published. These species were not included in previous molecular phylogenetic studies [21,22]. The plastid genome sequence data from C. fulvescens [22] were insufficient to determine the taxonomic position of C. fulvescens. However, data obtained from multiple taxa have been frequently used to resolve taxonomic problems (e.g., [33]). Therefore, we included the novel 18S rDNA and ITS sequences obtained from species belonging to Chlorocystidales and Sykidiales [23] in our analysis to determine the taxonomic positions of C. fulvescens from the North Pacific and North Atlantic regions (Figure 4 and Figure 5).
The results revealed that the North Pacific and North Atlantic C. fulvescens were separate taxonomic entities that were not closely related to each other. These species were placed in different taxonomic groups, putatively in Ulotrichales and Chlorocystidales, respectively (Figure 4 and Figure 5). Additionally, the results suggested that the Korean C. fulvescens should be placed in a new taxonomic group. In this study, we could not examine the type specimen of C. fulvescens and conducted a global sampling of C. fulvescens. Further studies including the type specimen and global samplings should be addressed for nomenclatural treatments.
In summary, we conducted a molecular phylogenetic analysis of the Korean C. fulvescens and examined the intraspecific DNA sequence variations. We believe that this genetic information will be useful for inferring the taxonomic relationships between C. fulvescens and other species belonging to Ulotrichales and Ulvales. These findings will help in tracing the origin of the North Atlantic C. fulvescens. Moreover, genetic variations in the ITS and rbcL sequences can be used to differentiate between various populations of the Korean C. fulvescens; this would also provide a helpful tool for the development of C. fulvescens as an aquaculture strain in Korea.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/d17020132/s1, Figure S1: Molecular phylogenetic analysis of 18S rDNA using the maximum-likelihood; Figure S2: Molecular phylogenetic analysis of the ITS region using the maximum-likelihood method.
Author Contributions
Conceptualization, S.-R.L.; methodology, E.-Y.L. and S.J.L.; software, S.-R.L.; validation, S.-R.L., E.-Y.L., and S.J.L.; formal analysis, E.-Y.L. and S.J.L.; investigation, S.-R.L., E.-Y.L., and S.J.L.; resources, S.J.L. and E.-Y.L.; writing—original draft preparation, S.-R.L.; writing—review and editing, E.-Y.L. and S.J.L.; visualization, E.-Y.L. and S.J.L.; supervision, S.-R.L.; project administration, S.J.L. and S.-R.L.; funding acquisition, E.-Y.L. and S.J.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by grants from the National Fishery Products Quality Management Service (Development of quarantine & disease control program for aquatic life: NFQS2025001) and the National Institute of Biological Resources (NIBR; grant number: NIBR202502103), funded by the Korea Ministry of Environment (MOE), Republic of Korea.
Data Availability Statement
Sequence data were deposited in GenBank under accession numbers (https://www.ncbi.nlm.nih.gov accessed on 12 February 2025).
Conflicts of Interest
The authors declare that there are no conflicts of interest.
References
- Moreira, A.; Cruz, S.; Marques, R.; Cartaxana, P. The underexplored potential of green macroalgae in aquaculture. Rev. Aquac. 2022, 14, 5–26. [Google Scholar] [CrossRef]
- Hwang, E.K.; Yi, Y.H.; Shin, W.J.; Sohn, C.H. Growth and maturation of a green alga, Capsosiphon fulvescens, as a new candidate for seaweed cultivation in Korea. In Proceedings of the 17th International Seaweed Symposium; Chapman, R.O., Anderson, R.J., Vreeland, V.J., Davison, I.R., Eds.; Oxford University Press: Oxford, UK, 2003; pp. 59–64. [Google Scholar]
- Agardh, C.A. Species Algarum Rite Cognitae, Cum Synonymis, Differentiis Specificis et Descriptionibus Succinctis; Volumen primum pars posterior; Ex officina Berlingiana: Lund, Sweden, 1823; pp. vii–viii, 399–531. [Google Scholar]
- Gobi, C. Berichte über die algologische Forschungen in finnischen Meerbusen im Sommer 1877 ausgeführt. Trudy Leningr. Obshch. Estest. 1879, 10, 83–92. [Google Scholar]
- National Institute of Korean History (NIKH). The Annals of the Joseon Dynasty. National Institute of Korean History. 2024. Available online: https://sillok.history.go.kr/id/kda_400070 (accessed on 31 October 2024).
- Lee, H. Revised and Expanded Edition of Survey of the Geography of Joseon. 1530. Available online: http://db.itkc.or.kr/inLink?DCI=ITKC_BT_B001A_0380_010_0040_2000_005_XML (accessed on 31 October 2024).
- Jeong, Y.J. Jasaneobo. 1814.
- Guiry, M.D.; Guiry, G.M. AlgaeBase. World-wide Electronic Publication, National University of Ireland, Galway. 2023. Available online: https://www.algaebase.org (accessed on 31 October 2024).
- Hanic, L.A.; Lindstrom, S.C. Life history and systematic studies of Pseudothrix borealis gen. et sp. nov. (= North Pacific Capsosiphon groenlandicus, Ulotrichaceae, Chlorophyta). Algae 2008, 23, 119–133. [Google Scholar] [CrossRef]
- Setchell, W.A.; Gardner, N.L. Phycological Contributions. I; University of California Publications in Botany: Berkeley, CA, USA, 1920; Volume 7, pp. 279–324, pls 21–31. [Google Scholar]
- Chapman, V.J. New entities in the Chlorophyceae of New Zealand. Trans. R. Soc. New Zealand 1952, 80, 47–58, ps 21, 22. [Google Scholar]
- Lee, I.K.; Kang, J.W. A check list of marine algae in Korea. Kor. J. Phycol. 1986, 1, 311–325. [Google Scholar]
- Lee, Y.; Kang, S. A Catalogue of the Seaweeds in Korea; Cheju National University Press: Jeju, Republic of Korea, 2001; pp. 1–662. [Google Scholar]
- Bae, E.H. Ulotrichales, Ulvales. In Algal Flora of Korea. Volume 1, Number 1. Chlorophyta: Ulvophyceae: Ulotrichales, Ulvales, Cladophorales, Bryopsidales. Marine Green Algae; Bae, E.H., Kim, H.S., Kwon, C.J., Hwang, I.K., Kim, G.H., Klochkova, T.A., Eds.; National Institute of Biological Resources: Incheon, Republic of Korea, 2010; pp. 7–52. [Google Scholar]
- Yoshida, T. Checklist of marine algae of Japan (Reviced in 2015). Jpn. J. Phycol. 2015, 63, 129. [Google Scholar]
- Bliding, C. A critical survey of European taxa in Ulvales, Part I. Capsosiphon Percursaria Blidingia Enteromorpha. Opera Bot. 1963, 8, 1–160. [Google Scholar]
- Migita, S. Life cycle of Capsosiphon fulvescens (C. agardh) Setchell and Gardner. Bull. Fac. Fish. Nagasaki Univ. 1967, 22, 21–31. [Google Scholar]
- Chihara, M. Developmental morphology and systematics of Capsosiphon fulvescens as found in Izu, Japan. Bull. Natl. Sci. Mus. 1967, 10, 163–170. [Google Scholar]
- Yoshida, K. Studies on germling development and life-history in Ulvaceae and Monostromaceae—Part I. Publ. Seto Mar. Biol. Lab. 1970, 17, 403–428. [Google Scholar] [CrossRef][Green Version]
- Hayden, H.S.; Waaland, J.R. Phylogenetic systematics of the Ulvaceae (Ulvales, Ulvophyceae) using chloroplast and nuclear DNA sequences. J. Phycol. 2002, 38, 1200–1212. [Google Scholar] [CrossRef]
- Sun, S.M.; Yang, S.H.; Golokhvast, K.S.; Le, B.; Chung, G. Reconstructing the phylogeny of Capsosiphon fulvescens (Ulotrichales, Chlorophyta) from Korea based on rbcL and 18S rDNA sequences. BioMed. Res. Int. 2016, 1, 1462916. [Google Scholar]
- Kim, D.; Lee, J.; Choi, J.W.; Yang, J.H.; Hwang, I.K.; Yoon, H.S. Flip-flop organization in the chloroplast genome of Capsosiphon fulvescens (Ulvophyceae, Chlorophyta). J. Phycol. 2019, 55, 214–223. [Google Scholar] [CrossRef] [PubMed]
- Darienko, T.; Rad-Menéndez, C.; Campbell, C.N.; Pröschold, T. Molecular phylogeny of unicellular marine coccoid green algae revealed new insights into the systematics of the Ulvophyceae (Chlorophyta). Microorganisms 2021, 9, 1586. [Google Scholar] [CrossRef]
- Lee, E.Y.; Lee, S.J.; Lee, S.R. A proposal for the lectotype designation of Ishige foliacea (Phaeophyceae, Ishigeaceae) using DNA barcoding. Diversity 2022, 14, 225. [Google Scholar] [CrossRef]
- Lee, S.J.; Choi, H.G.; Kim, J.H.; Lee, E.Y.; Lee, S.R. Genetic diversity of Cladophora oligocladoidea forming a bloom in the coastal area of Korea. Phycol. Res. 2023, 71, 77–80. [Google Scholar] [CrossRef]
- Lee, S.R.; Oak, J.H.; Chung, I.K.; Lee, J.A. Effective molecular examination of eukaryotic plankton species diversity in environmental seawater using environmental PCR, PCR-RFLP, and sequencing. J. Appl. Phycol. 2010, 22, 699–707. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- Manhart, J.R. Phylogenetic analysis of green plant rbcL sequences. Mol. Phylogenet. Evol. 1994, 3, 114–127. [Google Scholar] [CrossRef]
- Famà, P.; Wysor, B.; Kooistra, W.H.; Zuccarello, G.C. Molecular phylogeny of the genus Caulerpa (Caulerpales, Chlorophyta) inferred from chloroplast tufA gene1. J. Phycol. 2002, 38, 1040–1050. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Hwang, E.K.; Amano, H.; Park, C.S. Assessment of the nutritional value of Capsosiphon fulvescens (Chlorophyta): Developing a new species of marine macroalgae for cultivation in Korea. J. Appl. Phycol. 2008, 20, 147–151. [Google Scholar] [CrossRef]
- Hwang, E.K.; Park, C.S. Seaweed cultivation and utilization of Korea. Algae 2020, 35, 107–121. [Google Scholar] [CrossRef]
- Sorenson, M.D.; Oneal, E.; García-Moreno, J.; Mindell, D.P. More taxa, more characters: The hoatzin problem is still unresolved. Mol. Biol. Evol. 2003, 20, 1484–1498. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).