Abstract
A new species of Encotyllabe Diesing, 1850 (Monopisthocotylea: Capsalidae), Encotyllabe tantaliani n. sp., is described from the pharyngeal plates of the Lorna drum, Sciaena deliciosa (Tschudi, 1846) (Eupercaria: Sciaenidae), collected from two localities along the Peruvian coast. This new species was originally proposed as E. callaoensis Tantaleán, 1974, in an unpublished doctoral thesis, and is herein recognized as a nomen nudum under the International Code of Zoological Nomenclature (ICZN). Encotyllabe tantaliani n. sp. is distinguished from all known congeners by the following combination of morphological features: (1) an anteriorly tapering body proper, (2) slightly lobed testes markedly larger than the ovary, (3) vitelline follicles beginning at the level of the male copulatory organ (MCO) and absent from the regions of the reproductive organs, (4) a genital pore positioned posterolateral to the pharynx, and (5) an oblong-shaped MCO. Phylogenetic analysis based on cox1 sequence places E. tantaliani n. sp. in a clade with Encotyllabe percussa Morales-Ávila, Jufaili & Ogawa, 2024, a parasite of Lethrinus nebulosus (Forsskål, 1775) (Eupercaria: Lethrinidae) from the Arabian Gulf. Pairwise genetic distances support the distinctiveness of the new species from its closest congeners. Encotyllabe tantaliani n. sp. represents the first species of the genus described from a host belonging to the Sciaenidae host.
1. Introduction
The genus Encotyllabe Diesing, 1850 (Monopisthocotylea: Capsalidae), comprises a group of ectoparasitic monogeneans infecting marine teleost fish in tropical and subtropical waters of the Atlantic, Indian, and Pacific Oceans [1,2,3,4,5,6,7]. Although 27 nominal species have been assigned to Encotyllabe [8], recent morphological and molecular revisions have questioned the validity of several of them due to inadequate descriptions based on a limited number of specimens and the absence of morphometric and genetic data [4,6,7]. Currently, seven species are regarded as species inquirendae. Among these, Encotyllabe callaoensis Tantaleán, 1974, originally proposed in an unpublished doctoral thesis and never formally described, has been considered invalid [6,9]. As a result, only 19 species of Encotyllabe are currently recognized as valid [6,7,8].
The five most recently described species within this genus have been described using an integrative taxonomic approach that combines detailed morphological analysis with molecular data, particularly sequences of ribosomal and mitochondrial markers [4,6,7,10,11]. These include E. antofagastensis Sepúlveda, González & Oliva, 2014, and E. cheilodactyli Sepúlveda, González & Oliva, 2014, from Chilean waters [4], E. bifurcatum Taborda, Sepulveda, Luque, Escribano & Oliva, 2023, and E. parvum Taborda, Sepulveda, Luque, Escribano & Oliva, 2023, from Brazilian waters [6], and E. percussa Morales-Ávila, Jufaili, Ogawa, 2024, from the Arabian Sea and Gulf of Oman [7]. According to Taborda et al. [6], the most reliable diagnostic characteristics for species-level identification in Encotyllabe include overall body shape, relative size of several organs, relative position and shape of the testes, male copulatory organ (MCO) shape, extension and distribution of the vitellaria, the relative length of peduncle, and the size and shape of the anchors.
In this work, we describe E. tantaliani n. sp., a new capsalid species collected from the pharyngeal plates of Sciaena deliciosa (Tschudi, 1846) (Eupercaria: Sciaenidae), a demersal teleost of commercial importance along the Southeastern Pacific coast of Peru [12]. The new species is described based on an integrative approach combining detailed morphological data with molecular analysis of the mitochondrial cytochrome c oxidase subunit I (cox1) gene. Comparative assessments with closely related species and phylogenetic reconstruction are provided to support its taxonomic status and to contribute to the growing knowledge of the diversity and host associations of Encotyllabe in the Southeastern Pacific.
2. Materials and Methods
2.1. Sample Collection and Morphological Analysis
In January 2020 and February 2025, a total of 140 specimens of S. deliciosa were obtained by local fishermen using gillnets at two distinct localities along the Peruvian coast. Of these, 17 individuals were collected from the coastal zone of Puerto Santa Rosa (6°52′ S, 79°55′ W), Lambayeque Region, and 123 from the coastal zone of Lima (12°04′ S, 77°10′ W), Lima Region. The gills and pharyngeal plates were excised and placed in Petri dishes with seawater and examined for monogeneans with the aid of a stereomicroscope. Eleven monogeneans were observed alive on the pharyngeal plates, and seven were fixed in hot 4% formalin under slight coverslip pressure and subsequently preserved in 70% ethanol. Fixed specimens were stained with iron acetocarmine, clarified in Eugenol (4-allyl-2-ethoxyphenol), and mounted in Canada balsam. Two additional specimens were mounted directly in Gray and Wess medium for study of sclerotized structures [13]. The remaining two specimens were preserved in 96% ethanol for DNA extraction. Specimens were examined and photographed using a NikonTM Eclipse SI compound photomicroscope, and drawings were made with the aid of a drawing tube. All measurements are given in micrometers (µm), unless otherwise stated, and represent straight-line distances between the extreme points of the structures measured. Data are presented as the range, followed by the mean and number (n) of structures measured in parentheses. Body length represents the length of the body proper with the haptor. Fishes were identified employing the keys of Peruvian marine fishes of [12]. Type specimens were deposited in the Helminthological Collection of the Museum of Natural History at the San Marcos University (MUSM), Lima, Peru. Authorship of the new taxa is attributed to the first two and the last three authors following Recommendation 50.1 (Identity of authors) of the International Code of Zoological Nomenclature (ICZN). The type material of the Chauhanellus species described herein was deposited in the Helminthological Collection of the Museum of Natural History at the San Marcos University (MUSM), Peru.
2.2. Molecular Characterization and Phylogenetic Analyses
Genomic DNA was extracted using the TIANamp Genomic DNA Kit (TianGen, Beijing, China), according to the manufacturer’s recommendations. The mitochondrial DNA cytochrome c oxidase subunit I gene (cox1, partial fragment) was amplified using the primers ASmit1 (5′-TTTTT TGGGC ATCCT GAGGT TTAT-3′) and ASmit2 (5′-TAAAG AAAGA ACATA ATGAA AATG-3′) [6]. PCR assays were carried out in a total volume of 50 µL containing 25 µL of PCRBIO Taq Mix Red (PCR Biosystems), 1 µL of each primer, 16 µL of DNA sample, and ultrapure water to complete, using cycling parameters as previously described by these authors. The PCR products were purified with sparQ PureMag Beads (Quantabio, Beverly USA) following the manufacturer’s instructions and sequenced in Macrogen Inc. (Macrogen, Seoul, Republic of Korea) with the Sanger sequencing method. Sequences were edited and contigs were assembled using ProSeq 2.9 beta [14]. The National Center for Biotechnology Information (NCBI) sequence database (henceforth ‘GenBank’) was searched for similar sequences using BLAST 2.16.0 + (Basic Local Alignment Search Tool) [15]. Sequences generated in this study were aligned with selected sequences obtained from GenBank, using the software Clustal W 2.1 [16] (Table 1).

Table 1.
GenBank accession numbers for Encotyllabe species, host species, host family, locality, reference, and the species used as outgroup in the phylogenetic analysis.
Neobenedenia sp. (JQ782846) was used as an outgroup for the cox1 mDNA dataset. Sequence alignment was analyzed in JModelTest2 [18] to compare different models of DNA substitution in a hierarchical hypothesis-testing framework to select a base substitution model that best fits the data. The best model found was TrN+I+G, selected with the corrected Akaike information criterion (AICc; [19]). Next, the best model was implemented in MrBayes 3.2.7a [20] for Bayesian Inference analysis (BI). Unique random starting trees were used in the Metropolis-coupled MCMC [21]. The analysis was performed for a total of 5,000,000 generations. Visual inspection of log-likelihood scores against generation time indicated that the values reached a stable equilibrium before the 100,000th generation. Thus, a burn-in of 1000 samples was conducted; every 100th tree was sampled from the MCMC analysis, obtaining a total of 100,000 trees, and the tree topology represented the 50% majority rule consensus trees. Support for nodes in the BI tree topology was obtained by posterior probability. For the ML analysis, we used the default options in IQ-TREE run through the Cipress Science Gateway platform [22]. The robustness of the ML tree topology was assessed by bootstrap iterations of the observed data 1000 times. Phylogenetic trees were visualized and edited in Figtree 1.4.4 [23]. Pairwise genetic distances between the sequences of the cox1 gene were calculated in MEGA [24] using the Kimura 2-Parameter model [25].
3. Results
Capsalidae Baird, 1853
Encotyllabinae Monticelli, 1892
Encotyllabe tantaliani n. sp. Villena, Huerta, Cruces, Ñacari, and Chero
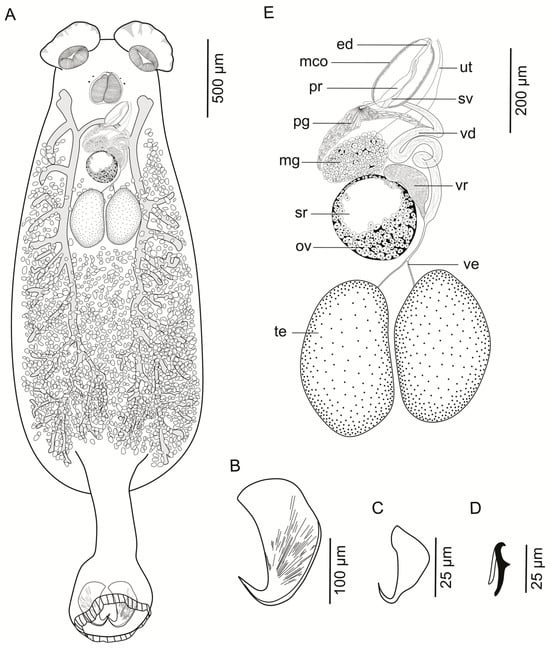
Figure 1.
Encotyllabe tantaliani n. sp. (A). Whole mount (ventral view). (B). Large anchor. (C). Small anchor. (D). Hook. (E). Reproductive system. Abbreviations: ed, ejaculatory duct; mco, male copulatory organ; mg, Mehlis’ gland; ov, ovary; pg, prostatic glands; pr, prostatic reservoir; sr, seminal receptacle; sv, seminal vesicle; te, testis; ut, uterus; ve, vas efferent; vd, vas deferens; and vr, vitelline reservoir.
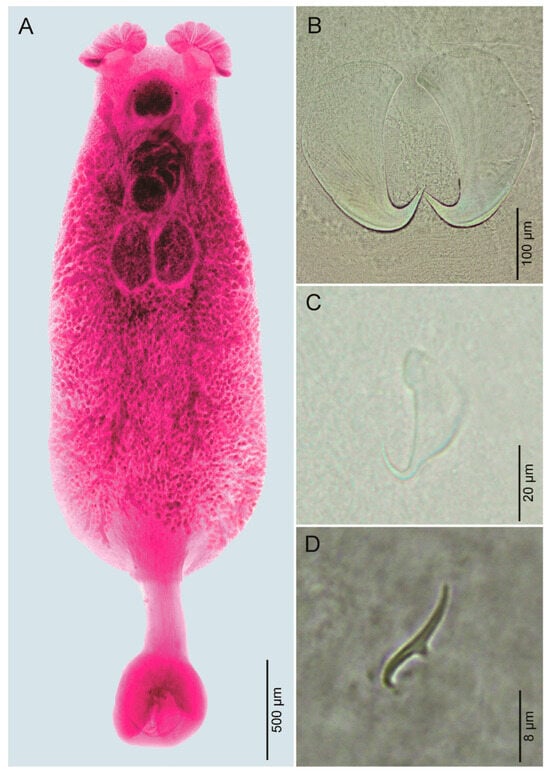
Figure 2.
Photomicrographs of Encotyllabe tantaliani n. sp. (A). Whole mount (ventral view). (B). Large anchor. (C). Small anchor. (D). Hook.
Syn. Encotyllabe callaoensis Tantaleán, 1974
Description (based on eight stained and mounted specimens): Body ellipsoidal, tapering anteriorly, rounded posteriorly, 1.97–3.11 (2.54; n = 8) mm long, with distinct ventral concavity; greatest width 0.73–1.50 (1.04; n = 8) mm, usually at level of posterior third of body. Tegument thin, surface smooth. Anterior attachment organs with two muscular suckers, 105–153 (129; n = 8) long, 138–221 (170; n = 8) wide, each partially surrounded by a fan-shaped incomplete membrane. Two pairs of eyespots are present at the level of the pharynx, equidistant. Pharynx eversible, pyriform, muscular, and glandular, 157–231 (195; n = 8) long, 150–276 (195; n = 8) wide; anterior margin folded into digitiform projections. Intestinal bifurcation postpharyngeal; intestinal ceca branched, extending to base of peduncle, not confluent posteriorly. Peduncle elongate, 521–879 (685; n = 7) long, 159–257 (225; n = 7) wide. Haptor bell-shaped, 416–566 (489; n = 8) in diameter, armed with one pair of large, broad anchors, one pair of small anchors, and 14 marginal hooks evenly distributed. Marginal membrane 25–72 (45; n = 8) long. Large anchor 222–327 (265; n = 8) long; small anchor 26–32 (28; n = 2) long; small-large anchor ratio 0.08–1.14:1; large-small anchor ratio 7–13:1; hooks, 8–14 (10; n= 8) long. Male copulatory organ (MCO) muscular, oblong, 175–250 (203; n = 8) long, 47–107 (78; n = 8) wide. Testes paired, juxtaposed, unequal in size, pre-equatorial, oval in shape; right testis 265–425 (338; n = 8) long, 126–251 (338; n = 8) wide, left testis 263–370 (316; n = 8) long, 134–234 (179; n = 8) wide; vas deferens sinistral, winding anteriorly, entering MCO at its base, dilated distally to form a fusiform seminal vesicle; prostatic duct joins ejaculatory duct and opens at tip of MCO; Goto’s glands not observed. Ovary oval, situated slightly lateral to vitelline reservoir, 124–232 (166; n = 8) long, 62–98 (77; n = 8) wide; vitelline reservoir sinistral; Mehlis’ glands conspicuous, surrounding ootype; uterus sinuous, extending anterolaterally along posterior margin of MCO. The genital pore is located on the left posterolateral of the pharynx. The vaginal aperture is situated on the left ventral side of the vitelline reservoir; vaginal duct not observed; seminal receptacle reniform, intraovarian. Vitelline follicles are dense, extending laterally and medially from the posterior third of the MCO to the base of the peduncle, not overlapping reproductive organs. Eggs not observed.
Type host. Sciaena deliciosa (Tschudi) (Perciformes: Sciaenidae), Lorna drum.
Site of infection. Pharyngeal plates.
Type locality. Puerto Santa Rosa (6°52′ S, 79°55′ W), Lambayeque Region, Peru, South America.
Other localities. Coastal zone of Lima (12°04′ S, 77°10′ W), Lima Region.
Specimens deposited. Holotype, MUSM XXXX; 5 paratype, MUSM XXXX.
Etymology. The new species is named in honor of Dr. Manuel Tantaleán Vidaurre (UNMSM), in recognition of his pioneering work on Peruvian monogeneans.
Zoobank registration: The Life Science Identifier (LSID) for Encotyllabe tantaliani n. sp. is urn:lsid:zoobank.org:act:EC5A5883-5387-4083-BD88-CFAFE2D54316.
Representative DNA sequences. PV637007 (cox1).
Differential diagnosis
Encotyllabe tantaliani n. sp. closely resembles E. antofagastensis, E. caballeroi Velasquez, 1977, E. chironemi Robinson, 1961, and E. parvum in possessing a proper body that tapers anteriorly. However, the new species differs from E. antofagastensis in having testes markedly larger than the ovary (vs. testes equal in size or slightly larger than the ovary in E. antofagastensis), vitelline follicles commencing at the level of the MCO base (vs beginning at the level of the pharynx base in E. antofagastensis), and an oblong MCO (vs curved distally in E. antofagastensis). Encotyllabe tantaliani n. sp. can be distinguished from E. caballeroi by the shape of the MCO (oblong vs. fusiform in E. caballeroi) and the position of the genital pore (posterolateral to the pharynx vs. directly posterior to the pharynx in E. caballeroi). The new species differs from E. chironemi in having pre-equatorial testes (vs. testes reaching or surpassing the midline of the body proper in E. chironemi) and by the distribution of the vitelline follicles, which are absent from the regions of the reproductive organs (vs. follicles extending into these regions in E. chironemi). Finally, E. tantaliani n. sp. differs from E. parvum in having testes much larger than the ovary (vs. testes much smaller than the ovary in E. parvum) (Table 2) and in the extent of the vitelline follicles (commencing at level of the MCO base in the new species vs. beginning at the pharynx level in E. parvum).

Table 2.
Morphometric comparison of the new Encotyllabe species and the most similar species (Encotyllabe antofagastensis, E. caballeroi, E. chironemi, and E. parvum). Range values are followed by mean values in parentheses.
Phylogenetic Relationships
One cox1 sequence was obtained from E. tantaleani n. sp. (334 bp in length). The final alignment of the cox1 gene dataset was 269 bp. The ML and BI analyses yielded identical topologies (Figure 3), revealing three main clades. The first clade included the undescribed species Encotyllabe sp. and E. vallei, with strong support (ML = 87%). The second clade comprised three subclades: E. antofagastensis and E. parvum formed a group sister to E. cheilodactyli (BI = 0.95; ML = 68%). This group, in turn, was sister to a third clade formed by E. percussa and E. tantaleani n. sp., although this relationship was weakly supported (BI = 0.68; ML = 63%). The genetic distance between E. tantaliani n. sp. and E. percussa was 8.7–9.1% (22–23 nucleotides of difference). The genetic divergence with congeneric species is shown in Table 3.
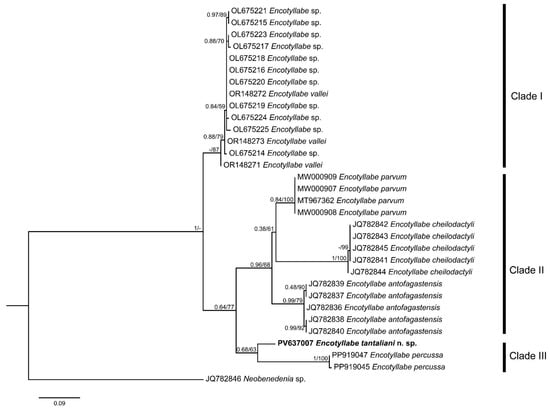
Figure 3.
Phylogenetic tree based on cox1 mDNA gene for Encotyllabe spp. inferred by Bayesian inference (BI) and maximum likelihood (ML). Numbers along branches indicate the bootstrap values obtained from the posterior probability support BI and ML (BI/ML).

Table 3.
Pairwise sequence divergences for cox1 mDNA (based on 269 bp) genes among species of Encotyllabe Clade 2 and 3 (Phylogenetic tree, Figure 3). Kimura 2-parameter distance is shown as percentage (above the diagonal) and the raw number of bp-pairwise differences below the diagonal. Due to the greater variability of the cox1 gene, the values are shown as a range.
4. Discussion
The species described herein as E. tantaliani n. sp. corresponds to E. callaoensis, a name originally proposed by Tantaleán [28] in his doctoral thesis based on specimens collected from the pharyngeal plates and gills of the sciaenid fishes Paralonchurus peruanus (Steindachner, 1875) and S. deliciosa in the coastal waters off Callao, central Peru. Although the thesis included a morphological description and the designation of a holotype and 11 paratypes, it was never formally published. According to the International Code of Zoological Nomenclature [29], a doctoral thesis does not constitute a valid publication unless formally issued with the intent of public and permanent scientific dissemination (Art. 8.1). While the description provided by Tantaleán (1974) meets the diagnostic requirements of Art. 13.1, the name remains unavailable due to the lack of a valid publication. The name E. callaoensis was later mentioned by Tantaleán et al. [30] in an academic textbook. However, this publication also does not meet the criteria for nomenclatural availability under the Code (Arts. 8.1.2, 8.4), as it lacks a formal type designation and does not provide a differential diagnosis. Nevertheless, Morales et al. [31] incorrectly treated E. callaoensis as a valid name. Consequently, E. callaoensis is regarded herein as a nomen nudum and holds no nomenclatural priority. A specimen labeled as E. callaoensis (MUSM 277), corresponding to the holotype designated in the original thesis, is currently housed at MUSM. However, this specimen cannot be considered a valid holotype because the name with which it is associated is unavailable. We herein provide a full morphological and molecular description of the species and formally name it E. tantaliani n. sp. Type specimens have been designated and deposited in the MUSM, a recognized institutional collection. The new name is proposed in accordance with the ICZN [29], thereby resolving the previous nomenclatural ambiguity associated with the unavailable name E. callaoensis.
Encotyllabe tantaliani n. sp. represents the first species of the genus described from a host belonging to the Sciaenidae (Table 4), expanding the known host range of Encotyllabe, which has been primarily associated with sparids, haemulids, carangids, cheilodactylids, and lethrinids (Table 4). Like other members of the genus, E. tantaleani n. sp. was found attached to the pharyngeal plates of its host, suggesting a conserved site specificity within the genus [1,7,11]. This preference for attachment to the pharyngeal plates may reflect ecological and functional adaptation for feeding in host species with benthic or demersal habits.

Table 4.
List of Encotyllabe species accepted as valid in this study, type host, host family, locality, and reference.
Host specificity in the genus Encotyllabe appears to be relatively high, with most species reported from a single host species and family [4,7,11]. This pattern suggests a narrow host range potentially shaped by long-term co-evolutionary processes and ecological compatibility between parasite and host. Nevertheless, the overall extent of host specificity in Encotyllabe remains poorly understood, especially in regions where parasitological sampling is limited. In the present study, E. tantaliani n. sp. was recovered exclusively from S. deliciosa; however, due to the lack of host diversity in the sampling effort, definitive conclusions regarding its host range cannot be made. Broader surveys that include sympatric fish species are required to determine whether this apparent specificity reflects true host restriction.
5. Conclusions
Encotyllabe tantaliani n. sp. is described as a new species of capsalid monogenean parasitizing the pharyngeal plates of S. deliciosa (Sciaenidae) off the coast of Peru, representing the first record of the genus Encotyllabe from a sciaenid host. The new species corresponds to E. callaoensis, a name originally proposed in an unpublished doctoral thesis by Tantaleán (1974), but herein recognized as a nomen nudum under the ICZN. The species is formally established based on an integrative approach combining detailed morphological data with molecular analysis of the mitochondrial cox1 gene. Phylogenetic analysis supports its placement within a clade comprising E. percussa, and pairwise genetic distances corroborate its distinctiveness from closely related congeners.
Author Contributions
Conceptualization, L.Ñ., C.L.C., A.M.-M. and J.D.C.; methodology, A.H., C.V., M.R., J.R. and K.M.; formal analysis, A.H., C.V., L.Ñ., C.L.C. and J.D.C.; data curation, A.H., C.V., M.R., J.R. and K.M.; writing—original draft prep, L.Ñ., A.M.-M. and J.D.C.; writing—review and editing, L.Ñ., A.M.-M., C.L.C. and J.D.C.; supervision, J.D.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Universidad Nacional Mayor de San Marcos—RR N° 004814-2025-R/UNMSM and project number B2510050v.
Institutional Review Board Statement
Ethical review and approval were waived for this study because the fish used were obtained from artisanal fisheries, which do not require special permits for collection.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Acknowledgments
We thank Lidia Sánchez (MUSM) for allowing us access to specimens under their care. The authors are grateful to the following people who helped to the collection of fishes in Peru: Alexander Reyes, Milagros Carrillo, and Cynthia E. Rodríguez, all from the National University Federico Villarreal (UNFV).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Khalil, L.F.; Abdul-Salam, J.B. The subfamily Encotyllabinae (Monogenea: Capsalidae) with the description of Alloencotyllabe caranxi n. g., n. sp. and Encotyllabe kuwaitensis n. sp. Syst. Parasitol. 1988, 11, 139–150. [Google Scholar] [CrossRef]
- Kardousha, M.M.; Al-Ansi, M.A.; Al-Khayat, J. Monogenea of the Arabian Gulf Fishes. 3. Encotyllabe spari and E. kuwaitensis (Capsalidae) from Qatari waters. Riv. Parassitol. 2002, 19, 227–235. [Google Scholar]
- Carvalho, A.R.; Luque, J.L. Three new species of monogeneans parasitic on Atlantic cutlassfish Trichiurus lepturus (Perciformes: Trichiuridae) from Southeastern Brazil. Acta Sci. Biol. Sci. 2012, 34, 359–365. [Google Scholar] [CrossRef]
- Sepúlveda, F.A.; González, M.T.; Oliva, M.E. Two new species of Encotyllabe (Monogenea: Capsalidae) based on morphometric and molecular evidence: Parasites of two inshore fish species of northern Chile. J. Parasitol. 2014, 100, 344–349. [Google Scholar] [CrossRef]
- Camargo, A.C.A.; Luque, J.L.; Santos, C.P. Mexicana rubra sp. nov. and Encotyllabe cf. spari off Rio de Janeiro. Helminthologia 2017, 54, 336–347. [Google Scholar] [CrossRef][Green Version]
- Taborda, N.; Sepulveda, F.A.; Luque, J.L.; Escribano, R.; Oliva, M.E. Two New Species of Encotyllabe (Monogenea: Capsalidae) from Brazil: Morphological and Molecular Evidence. Diversity 2023, 15, 706. [Google Scholar] [CrossRef]
- Morales-Ávila, J.R.; Al Jufaili, S.; Ogawa, K. Morpho-Molecular Characterization and Phylogenetic Relationships of Encotyllabe percussa n. sp. (Monogenea: Capsalidae) from the Spangled Emperor Lethrinus nebulosus (Teleostei, Lethrinidae). Syst. Parasitol. 2024, 101, 69. [Google Scholar] [CrossRef]
- World Register of Marine Species. Encotyllabe sp. (ID: 119265). 2023. Available online: https://www.marinespecies.org/aphia.php?p=taxdetails&id=119265 (accessed on 24 May 2025).
- Santillán, L.A.; Cruces, C.L.; Sáez, G.M.; Martínez-Rojas, R.; Mondragón-Martínez, A.; Murrieta Morey, G.A.; Quiñones, M.; Luque, J.L.; Chero, J.D. An Annotated Checklist of Monogeneans (Platyhelminthes, Monogenea) from Aquatic Vertebrates in Peru: A Review of Diversity, Hosts and Geographical Distribution. Animals 2024, 14, 1542. [Google Scholar] [CrossRef]
- Lablack, L.; Rima, M.; Georgieva, S.; Douniazed, M.; Kostadinova, A. Novel Molecular Data for Monogenean Parasites of Sparid Fishes in the Mediterranean and a Molecular Phylogeny of the Microcotylidae Taschenberg, 1879. Curr. Res. Parasitol. Vector-Borne Dis. 2021, 2, 100069. [Google Scholar] [CrossRef]
- Zedam, F.-Z.; Bouguerche, C.; Ahmed, M.; Tazerouti, F. Morphological and Molecular Characterization of Encotyllabe vallei Monticelli, 1907 (Monopisthocotylea, Monogenea) from the Gilthead Seabream Sparus aurata Linnaeus (Teleostei, Sparidae) from the Southwestern Mediterranean and Notes on Host Specificity of the Genus Encotyllabe Diesing, 1850. J. Helminthol. 2023, 97, e82. [Google Scholar]
- Chirichigno, N.; Vélez, J. Clave para Identificar los Peces Marinos del Perú; Instituto del Mar del Peru Special Publication: Callao, Peru, 1998; p. 500. [Google Scholar]
- Humason, G.L. Animal Tissue Techniques; W.H. Freeman: New York, NY, USA, 1979. [Google Scholar]
- Filatov, D.A. PROSEQ: A Software for Preparation and Evolutionary Analysis of DNA Sequence Data Sets. Mol. Ecol. Notes 2002, 2, 621–624. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X Version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Sepulveda, F.; González, M. Molecular and Morphological Analyses Reveal That the Pathogen Benedenia seriolae (Monogenea: Capsalidae) Is a Complex Species: Implications for Yellowtail Seriola spp. Aquaculture. Aquaculture 2013, 418–419, 94–100. [Google Scholar] [CrossRef]
- Santorum, J.M.; Darriba, D.; Taboada, G.L.; Posada, D. jmodeltest.org: Selection of Nucleotide Substitution Models on the Cloud. Bioinformatics 2014, 30, 1310–1311. [Google Scholar] [CrossRef]
- Posada, D.; Buckley, T.R. Model Selection and Model Averaging in Phylogenetics: Advantages of Akaike Information Criterion and Bayesian Approaches Over Likelihood Ratio Tests. Syst. Biol. 2004, 53, 793–808. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian Inference of Phylogenetic Trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Miller, J.C.; Tan, S.; Qiao, G.; Barlow, K.A.; Wang, J.; Xia, D.F.; Meng, X.; Paschon, D.E.; Leung, E.; Hinkley, S.J.; et al. A TALE Nuclease Architecture for Efficient Genome Editing. Nat. Biotechnol. 2011, 29, 143–148. [Google Scholar] [CrossRef]
- Rambaut, A. FigTree v1.4.4.; Institute of Evolutionary Biology: Edinburgh, UK, 2018. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis Across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Kimura, M. A Simple Method for Estimating Evolutionary Rates of Base Substitutions Through Comparative Studies of Nucleotide Sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Velasquez, C. A New Monogenetic Trematode, Encotyllabe caballeroi sp. nov. (Capsalidae) from a Marine Fish from the Philippines. Univ. Nac. Auton. De Mex. Inst. De Biol. Publicaciones Espec. 1977, 4, 117–120. [Google Scholar]
- Robinson, E.S. Some Monogenetic Trematodes from Marine Fishes of the Pacific. Trans. Am. Microsc. Soc. 1961, 80, 235–266. [Google Scholar] [CrossRef]
- Tantalean, M. Dos Nuevas Especies de Monogeneos Parásitos de Peces Comerciales del Mar Peruano. Biota 1974, 10, 235–242. [Google Scholar]
- International Commission on Zoological Nomenclature. Official Lists and Indexes of Names in Zoology; International Commission on Zoological Nomenclature: London, UK, 2012. [Google Scholar]
- Tantalean, M.; Carvajal, G.; Martinez, R.; Huiza, A. Helmintos Parásitos de Peces Marinos de la Costa Peruana. NCTL Ser. Div. Cient. 1982, 1, 40. [Google Scholar]
- Morales, E.; Sarmiento, L.; Sánchez, L.; Floríndez, D.; Lamas, G. Material Tipo de Helmintos en el Museo de Historia Natural, Universidad Nacional Mayor de San Marcos, (MUSM), Lima, Perú. Rev. Peru. De Biol. 2005, 12, 463–472. [Google Scholar] [CrossRef][Green Version]
- Williams, A.; Beverley-Burton, M. Redescription of Three Species of the Genus Encotyllabe (Capsalidae: Monogenea) from Fishes of the East Coast of Australia. Aust. J. Zool. 1989, 37, 45–53. [Google Scholar] [CrossRef]
- Noble, E.R. The Genus Encotyllabe (Class Trematoda) with a Description of a New Species. Trans. Am. Microsc. Soc. 1966, 85, 144–151. [Google Scholar] [CrossRef]
- Gupta, S.; Krishna, V. Encotyllabe punctatai sp.n., E. fotedari sp.n. and Neoencotyllabe muelleri g.n., sp.n. (Monogenea) from Marine Fishes. Helminthologia 1980, 17, 83–89. [Google Scholar]
- Monticelli, F.S. II Genere Encotyllabe Diesing; Serie VI, 4/001; Atti del Reale Istituto d’incoraggiamento di Napoli: Naples, Italy, 1907. [Google Scholar]
- Egorova, T.P. Recent composition of the subfamily Encotyllabinae (Monogenea: Capsalidae). Parazitologiya 2000, 34, 295–301. (In Russian) [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).