Abstract
Camellia sinensis cv. Xinyang10 is a nationally recognized elite tea cultivar selected from the Xinyang drought-resistant population, valued for its notable cold tolerance and broad adaptability. In this study, we present the first complete assembly and annotation of its mitogenome. The mitogenome features a multipartite structure, consisting of a circular chromosome (798,917 bp) and a linear chromosome (46,159 bp), harboring a total of 74 genes. We identified extensive repetitive sequences (244 simple sequence repeats and 998 long sequence repeats), 211 RNA editing sites, and 16,614 bp of chloroplast-derived DNA, indicating a highly dynamic genome. Positive selection was detected in nad1 and ccmFC. Phylogenetic analysis based on mitochondrial SNP markers placed C. sinensis Xinyang10 closest to C. sinensis var. pubilimba. Notably, a phylogeny reconstructed based on mitogenomic collinearity displayed a distinct geographical pattern, supporting the hypothesized westward-to-eastward migration route of tea plants from southwestern China. These findings provide valuable genomic resources and demonstrate the utility of the mitogenome in understanding the evolutionary history of tea plants.
1. Introduction
While animal mitogenomes are typically conserved and compact, plant mitogenomes are noted for their conspicuous structural intricacies, typically characterized by large genome sizes, frequent recombination events, a high proportion of repetitive sequences, and considerable variation in gene content and order [,]. Decoding the mitogenome is essential for understanding various biological processes in plants, including cellular energy metabolism [], male sterility mechanisms [,], stress responses [,,], phylogenetic relationships [], and adaptive evolution []. Evidence for positive selection acting on mitochondrial protein-coding genes (PCGs) is well-established in the literature [,], with adaptive evolution in these genes often linked to specific environmental adaptations or agronomic traits, such as cold tolerance [], drought resistance [], or reproductive development []. Additionally, the mitochondrial translation machinery demonstrates unique characteristics, including pronounced codon usage bias (CUB), which reflects the combined effects of genomic composition, mutational pressure, and natural selection, potentially influencing the efficiency and accuracy of gene expression []. Another key feature is the widespread occurrence of RNA editing, which fine-tunes gene function through post-transcriptional modifications—predominantly C-to-U conversions—and is indispensable for the functional maturation of mitochondrial genes []. Plant mitogenomes adopt various structural conformations, including single circular, branched, and linear multipartite molecules [], and may also coexist as mixed circular and linear forms []. However, the accurate resolution of plant mitogenomes has long been hindered by the prevalence of long repetitive sequences and highly dynamic rearrangement regions. Traditional short-read sequencing technologies struggle to span these repetitive elements, often resulting in highly fragmented assemblies that fail to accurately represent the complete genome architecture and rearrangement events. The emergence of long-read sequencing technologies (e.g., PacBio and Oxford Nanopore) represents a revolutionary shift for this field of study. With read lengths capable of spanning tens of kilobases, these technologies provide a powerful means to overcome these challenges, enabling complete and accurate assembly of complex plant mitogenomes as well as the resolution of structural variations such as repeat-mediated recombination.
The global tea industry, driven primarily by the cultivation of C. sinensis, represents a major agricultural sector worldwide, and its quality and adaptability have attracted widespread interest. Xinyang Maojian, one of China’s top ten famous teas, is highly appreciated by consumers for its appealing shape, abundant trichomes, high aroma, robust flavor, and strong sweet aftertaste. Xinyang, located in the Jiangbei tea-growing region, experiences relatively low annual precipitation and cold winters. Tea plants grown in this area have accumulated genetic adaptations to the local climate, resulting in unique physiological metabolism and harboring valuable genes related to stress resistance and flavor quality. These characteristics make it a valuable natural resource for tea plant breeding. C. sinensis cv. Xinyang10 is a cultivar developed through individual plant selection from the Xinyang population. It is a medium-sized plant with well-distributed and dense branches, elliptical and glossy leaves, light green buds, high fertility, uniform sprouting, and high yield. Its cold tolerance exceeds that of the Fuding Dabaicha cultivar, which itself is a nationally recognized, widely adaptable variety with medium-to-high cold tolerance []. While researchers have made considerable progress in deciphering the tea plant’s nuclear genome [,,,,], and a few mitogenomes of C. sinensis have been reported [,], along with a recently reported mitogenome of the related oil-tea camellia (C. oleifera) [], the mitogenome of C. sinensis cv. Xinyang10 remains poorly characterized. A comprehensive assembly and analysis of the complete mitogenome of this important cultivar will not only fill a critical gap in the organellar genomic information of this unique variety but also provide new insights into its biological underpinnings, such as stress resistance and cellular energy supply for flavor compound metabolism. Moreover, it will serve as a essential foundation for further comparative genomic studies.
Based on the aforementioned background and research gaps, this study aims to: (1) assemble and characterize the complete mitogenome of C. sinensis cv. Xinyang10, with a focus on its structural features including repeat sequences and gene order; (2) investigate potential variations at the mitogenome level that may be associated with cultivar differentiation, geographical adaptation, or agronomic traits; and (3) assess the utility of mitochondrial genomic information in elucidating the phylogenetic evolution of C. sinensis. To achieve these objectives, we employed PacBio long-read sequencing technology to generate high-fidelity data, followed by de novo assembly using efficient algorithms. We performed comprehensive genome annotation, repeat sequence analysis, codon usage bias assessment, and RNA editing site prediction with specialized bioinformatics tools. Furthermore, comparative genomic and phylogenomic approaches were applied to examine structural variations, conduct multi-genome alignments, and analyze selective pressures. Through this study, we aim to provide important foundational genomic resources for understanding the biological characteristics of C. sinensis cv. Xinyang10 and the evolutionary dynamics of tea plant mitogenomes, thereby providing insights into the origin and evolution of the tea plant.
2. Materials and Methods
2.1. Genome Sequencing, Assembly, and Annotation
Fresh and healthy leaves of C. sinensis cv. Xinyang10 were collected from the production base of Xinyang Xiangyun Tea Co., Ltd. (32.257296° N, 113.788973° E) in Shihe District, Xinyang City. Following collection, leaf samples were immediately flash-frozen in liquid nitrogen and stored at −80 °C at the Henan Provincial Key Laboratory of Tea Plant Biology. Prior to DNA extraction, the frozen leaves were ground to a fine powder. High-quality genomic DNA was then isolated using a modified CTAB protocol []. The extracted DNA was evaluated for integrity and size via 0.75% agarose gel electrophoresis. DNA purity was determined using a NanoDrop One spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Subsequently, the concentration was accurately quantified with a Qubit 3.0 Fluorometer (Life Technologies, Carlsbad, CA, USA). Approximately 8 μg of DNA was sheared to fragments of 15–20 kb using a Megaruptor 3 system, and the sheared product was purified using AMPure PB beads. An SMRTbell library was constructed with the Pacific Biosciences SMRTbell Express Template Prep Kit 2.0. The library was size-selected using the BluePippin™ system to enrich for fragments of ~15 kb, followed by primer annealing and polymerase binding using the DNA/Polymerase Binding Kit. After quality assessment and quantification, the SMRTbell template–polymerase complexes were loaded onto the PacBio Revio platform for sequencing. Raw subread data were saved in the XY10.bam file. Quality control metrics were generated using SMRTlink v12.0 (Pacific Biosciences, Menlo Park, CA, USA). HiFi reads were extracted from the XY10.bam file using samtools v1.19.2 [] and saved as XY10_hifi_reads.fastq.
The mitochondrial genome of C. sinensis cv. Xinyang10 was assembled using two distinct software tools. The first was PMAT v1.5.3 [], an efficient toolkit designed for de novo assembly of plant mitochondrial genomes using third-generation sequencing data (HiFi/CLR/ONT). The assembly was performed with the command: PMAT autoMito -i XY10_hifi.fastq -o./test_g3 -st hifi -g 3g -m. The second tool was oatk [], which specializes in de novo assembly of complex plant organelle genomes, particularly those containing long repetitive sequences, using PacBio HiFi data. The assembly command used was: oatk -k 1001 -c 7 -t 8 --nhmmscan nhmmscan -m embryophyta_mito.fam -o oatk.assembly XY10_hifi_reads.fastq. The resulting assembly graphs were visualized and manually adjusted using Bandage v.0.8.1 []. The final mitochondrial genome assembly was annotated using GeSeq [] (https://chlorobox.mpimp-golm.mpg.de/geseq.html, accessed on 25 March 2025) with default parameters. tRNA genes were further verified using tRNAscan-SE v.1.21 [], and start/stop codons of protein-coding genes were manually verified in BioEdit v7.2.1. The circular map of the mitochondrial genome of C. sinensis cv. Xinyang10 was generated using the Plant Mitochondrial Genomes Map (PMGmap) online tool [] (http://47.96.249.172:16086/home/, accessed on 21 May 2025).
2.2. Mitogenome Collinearity Analysis
Collinearity in mitochondrial genomes reflects the conservation of gene order or the occurrence of rearrangement events among species. The mitochondrial genome sequences of ten published C. sinensis cultivars were downloaded from the NCBI database (accessed on 12 February 2025). These included two small-leaf varieties (C. sinensis var. sinensis cv. Dahongpao (DHP, accession no. PP212895) and C. sinensis var. sinensis cv. Rougui (RG, accession no. PP212896)), four medium-leaf types (C. sinensis sp. Zhuyeqi (ZYQ, accession nos. PQ763484-PQ763490), C. sinensis sp. Baihaozao (BHZ, accession nos. PV340641-PV340651), C. sinensis cv. SRDMB (DMB, accession no. PP770440), and C. sinensis var. pubilimba (PUP, accession no. ON782577)), and four large-leaf Assam varieties (C. sinensis var. assamica (AOL, accession no. OL989850; AOM, accession no. OM809792; AMK, accession nos. MK574876 and MK574877; and AMH, accession no. MH376284)) [] (Table 1). It should be noted that a more recent mitogenome assembly (PV744423-PV744434) became publicly available after our data collection cutoff date and was therefore not included in this analysis. This more recent data will be incorporated in our future comparative studies.

Table 1.
Summary of the 11 assembled C. sinensis mitogenomes.
Pairwise comparisons of the 11 mitogenomes (including that of C. sinensis cv. Xinyang10) were performed using BLASTN. Homologous regions with a length greater than 500 bp and sequence similarity ≥ 70% were retained as conserved collinear blocks. The results were visualized using AliTV v1.0.6 [] to intuitively identify structural variations such as inversions, duplications, and deletions [].
2.3. Repetitive Sequence Analysis
Simple sequence repeats (SSRs) were identified using MISA [] (https://webblast.ipk-gatersleben.de/misa/, accessed on 5 July 2025) with the following parameters: mononucleotide: 10; dinucleotide: 5; trinucleotide: 4; tetranucleotide: 3; pentanucleotide: 3; and hexanucleotide: 3, and a maximum sequence length of 100 bp. REPuter [] (accessed on 5 July 2025) was employed to identify dispersed long sequence repeats (LSRs), including forward, reverse, complement, and palindromic types. The analysis parameters included a minimum repeat size of 30 bp and a Hamming distance of 3. The resulting repeat structures were then plotted using the Circos package within TBtools-II v2.360 [].
2.4. Intracellular Gene Transfer
Homologous DNA fragments between the chloroplast and mitochondrial genomes were identified using BLASTN v2.2.30 with an identity threshold of 70% and an E-value cutoff of 1 × 10−6 []. To mitigate the potential for redundant detection caused by the inverted repeat (IR) regions, only one copy of each IR was retained for analysis. The quadripartite regions of the chloroplast genome were identified using CPStools v3.0 [], and duplicate IR regions were removed using BioEdit v7.2.1 []. The complete chloroplast genome sequence of C. sinensis cv. Xinyang10 (accession no. MZ153237) was downloaded from NCBI []. All results were visualized using R v 4.3.
2.5. RNA Editing Prediction
The identification of RNA editing sites in the mitochondrial protein-coding genes (PCGs) of C. sinensis cv. Xinyang10 was performed using an integrated strategy combining RNA-Seq and Illumina genomic sequencing data. Raw RNA and DNA reads were first quality-trimmed using fastp with default parameters []. The processed RNA reads were then aligned to the XY10 mitochondrial reference sequence using HISAT2 v2.2.1 (in --dta mode) []. The resulting alignments were sorted with SAMtools v1.19.2 [], deduplicated using Picard, and supplemented with read group information. Variant calling was subsequently performed using GATK HaplotypeCaller v4.6.2.0 []. The same analytical pipeline was applied to the Illumina genomic reads to identify DNA-level SNPs. Candidate RNA editing sites were isolated by retaining only RNA-specific SNPs, which were extracted by excluding variants common to both RNA and DNA datasets using bcftools v1.22 [].
To further investigate the biological effects of RNA editing, the genomic coordinates and sequences of all PCGs were extracted from the reference annotation file (XY10.gb). A custom Python 3.11 pipeline was employed to precisely map each candidate editing site to its corresponding gene and codon position (i.e., 1st, 2nd, or 3rd nucleotide). By comparing the codon sequences before and after editing and translating them according to the mitochondrial genetic code, the resulting amino acid changes were systematically determined.
2.6. Codon Usage Analysis and Ka/Ks Estimation
Protein-coding genes (PCGs) from the C. sinensis cv. Xinyang10 mitochondrial genome were extracted using CPStools v3.0 []. To minimize bias, genes shorter than 300 bp were excluded from codon usage analysis. The relative synonymous codon usage (RSCU) values were calculated and visualized using the online bioinformatics platform (https://www.bioinformatics.com.cn, accessed on 5 June 2025), a publicly accessible resource for RSCU plotting. GC content at each codon position (1st, 2nd, and 3rd) was computed for every gene using the CPStools v3.0 []. ENC (effective number of codons) and GC content at the third synonymous codon position (GC3s) were assessed using CodonW v1.4.2 []. A parity rule 2 (PR2) plot was employed to analyze AT bias [A3/(A3 + T3)] and GC bias [G3/(G3 + C3)] at the third codon position across four-codon amino acids []. The ENC plot was used to evaluate the impact of mutational bias on codon usage: points lying on or near the expected curve indicate that codon usage is primarily constrained by mutation bias, whereas deviations suggest additional influences, such as natural selection [].
To investigate the strength and pattern of natural selection on the mitochondrial PCGs in C. sinensis cv. Xinyang10, shared protein-coding genes across the mitogenomes of C. sinensis cv. Xinyang10 and ten related tea cultivars were extracted using CPStools v3.0 []. The sequences were aligned with MAFFT v7.310 [], and the Ka/Ks ratio for each gene was computed using KaKs_Calculator v2.0. A Ka/Ks value > 1 indicates positive selection, =1 suggests neutral evolution, and <1 implies purifying selection []. The results were visualized as a heatmap using the ggbiplot package in R v4.3.
2.7. Phylogenetic Analysis
To evaluate the utility of the mitochondrial genome in reconstructing phylogenetic relationships and to elucidate the phylogenetic position of C. sinensis cv. Xinyang10 among other tea plants, we conducted a phylogenetic analysis using mitogenomic single nucleotide polymorphisms (SNPs). Published mitochondrial genomes from Ericales taxa were selected, including 23 species from Theaceae, five from Ericaceae, one from Pyrolaceae, one from Symplocaceae, three from Actinidiaceae, two from Primulaceae, and one from Ebenaceae. Nicotiana tabacum was used as the outgroup.
The mitochondrial genomes of the selected Ericales species were aligned to the N. tabacum reference mitogenome using Minimap2 v2.26 []. SNPs were called from the alignments using bcftools v1.22 [] with filtering thresholds set to DP ≥ 10 and QUAL ≥ 30, and were stored in VCF format. The SNP-based VCF files were converted into a multi-sequence FASTA alignment using bcftools consensus. Multiple sequence alignment was performed with MAFFT v7.310 [].
A maximum likelihood (ML) tree was reconstructed using raxmlHPC v8.2.12 [] under the GTRGAMMA model. We employed ModelFinder to select the optimal nucleotide substitution model under the Bayesian information criterion (BIC). Branch support was then evaluated with 1000 ultrafast bootstrap replicates. The resulting alignment was converted from FASTA to NEXUS format using MEGA v11 []. We employed MrBayes v3.2.7 [] for Bayesian inference (BI) of phylogeny under the GTR+G+I model. The analysis involved two independent runs of four Markov chains (one cold, three heated) for 10 million generations, sampling trees every 1000 generations. Following a burn-in of 2.5 million generations (25%), a consensus tree summarizing the remaining samples was generated, with nodal support given as posterior probabilities (PP).
3. Results
3.1. Mitogenome Assembly and Annotation
Sequencing on the PacBio Sequel II platform yielded 5,327,958 HiFi reads. These reads had a maximum length of 59,940 bp and an average length of 17,227 bp. The mitogenome of C. sinensis cv. Xinyang10 exhibited a multipartite conformation (Figure 1a). After manual resolution, the final mitogenome assembly consisted of two chromosomes: the first was circular with a length of 798,917 bp, and the second was linear with a length of 46,159 bp (Figure 1b). We have deposited the sequence in the NCBI database under the accession number PX398764 and PX398765. A total of 74 genes were annotated in the mitogenome of C. sinensis cv. Xinyang10, including 38 protein-coding genes (PCGs), 33 tRNA genes, and 3 rRNA genes. The overall GC content was 45.82% (Figure 1c). The distribution of introns among the protein-coding genes (PCGs) varied: the genes nad1, nad2, nad5, and nad7 contained the highest number, with four introns each. In contrast, a single intron was identified in ccmFC, rpl2, rps3, trnT-TGT, trnI-GAT, trnA-TGC, trnS-TGA, and trnT-GGT. The gene nad4 was unique in possessing two introns (Table 2). The total length of all 38 protein-coding genes was 33,367 bp. The longest gene was nad5 (2013 bp), while the shortest was sdh4 (93 bp), followed by atp9 (225 bp).
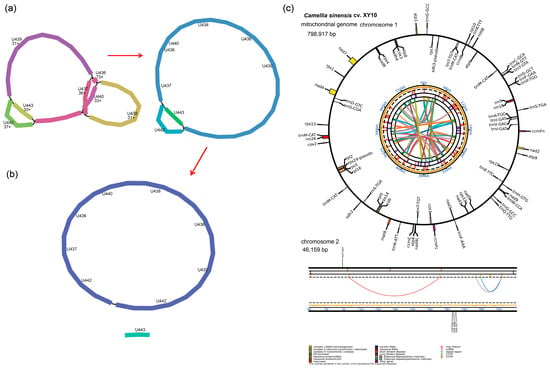
Figure 1.
Mitochondrial genome map of C. sinensis cv. Xinyang10. (a) Schematic of the preliminary assembly. (b) The master circle and linearized map, with numerically labeled contigs reflecting their connection order during the assembly process. (c) Annotation map of the mitochondrial genome.

Table 2.
Results of mitochondrial genome annotation in C. sinensis cv. Xinyang10.
3.2. Mitochondrial Genome Collinearity Between Diverse Tea Plants
A collinearity analysis between the C. sinensis cv. Xinyang10 mitogenome and ten other published sequences revealed multiple conserved genomic segments, indicating shared synteny among the 11 tea plants; however, the order of these blocks varied considerably, indicating extensive genomic rearrangements across different tea cultivars. Based on collinearity patterns, the large-leaf tea accession AMK from Yunnan Province, China, and AMH from Assam, India, formed the outermost branch (Figure 2). BHZ, AOL, and AOM from Hunan Province clustered together, while the remaining six accessions constituted a large inner clade. Within this inner group, C. sinensis cv. Xinyang10 from Henan Province and PUP from Jiangxi Province exhibited a sister relationship. Further inward, ZYQ from Hunan and DMB from Shangrao, Jiangxi were grouped together, and the innermost branch consisted of RG and DHP from Fujian Province.
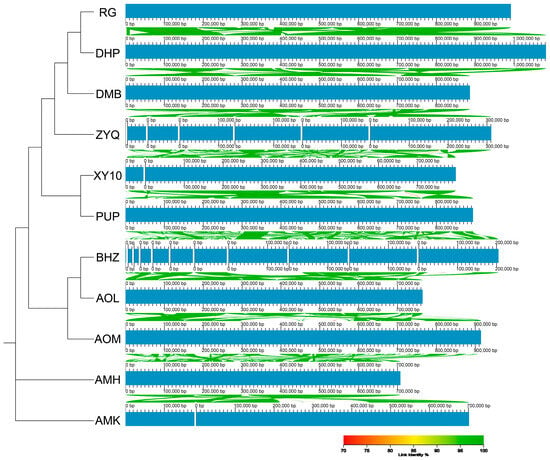
Figure 2.
Assessment of Synteny among Mitochondrial Genomesof Camellia sinensis cv. Xinyang10 and other 10 tea plants.
3.3. Repetitive Elements in the Mitochondrial Genome of C. sinensis cv. Xinyang10
A survey of the C. sinensis cv. Xinyang10 mitogenome revealed 244 simple sequence repeats (SSRs). The circular chromosome contained 231 SSRs, consisting of 28 mononucleotide, 62 dinucleotide, 28 trinucleotide, 98 tetranucleotide, 14 pentanucleotide, and 1 hexanucleotide repeats. The linear chromosome contained 13 SSRs, including 2 mononucleotide, 4 dinucleotide, 4 trinucleotide, 2 tetranucleotide, and 1 pentanucleotide repeat (Figure 3 and Table S1). The majority of SSRs were located in intergenic regions. SSRs were also detected within gene regions, including AT (in mttB), CTT (in rps2), AATG (in rps3), and CTAGT (in matR). Additionally, 998 dispersed long repeats (LSRs) with a length ≥ 30 bp were identified, comprising 451 forward repeats, 545 palindromic repeats, 1 reverse repeat, and 1 complementary repeat (Table S2).
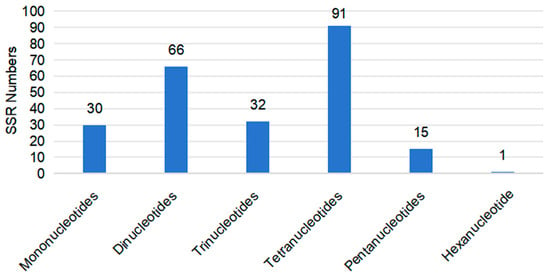
Figure 3.
The number of SSRs in the complete mitogenome of C. sinensis cv. Xinyang10.
3.4. Collinearity Analysis of the Mitochondrial GENOME and Chloroplast Genome of C. sinensis cv. Xinyang10
Analysis of sequence collinearity identified 23 homologous fragments, totaling 16,614 bp, that have been transferred from the chloroplast to the mitochondrial genome in C. sinensis cv. Xinyang10. These transferred sequences constitute 1.97% of the entire mitogenome length and were exclusively located on chromosome 1. Most homologous fragments ranged between 50 and 100 bp in length. The longest aligned region spanned 7731 bp with 99.47% sequence identity, mapping to positions 187,343–179,651 on mitochondrial chromosome 1 and 100,311–108,041 in the chloroplast genome. The shortest homologous fragment was 32 bp long with 100% identity, located at positions 762,361–762,394 on mitochondrial chromosome 1 and 88,505–88,538 in the chloroplast genome. Additionally, three perfectly matched regions (100% identity) were identified: one 1689 bp segment (mitochondrial positions 157,495–155,807) and one 42 bp segment (mitochondrial positions 186,712–186,671). Homologous regions containing repetitive sequences (SSRs or long repeats) constituted 90.4% (15,026 bp) of the total transferred sequences. All these fragments were located within the inverted repeat (IR) regions of the chloroplast genome. Furthermore, seven intact tRNA genes were detected in the homologous mitochondrial sequences: trnD-GUC, trnW-CCA, trnM-CAU, trnI-GAU, trnA-UGC, trnV-GAC, and trnN-GUU (Figure 4 and Table S3).
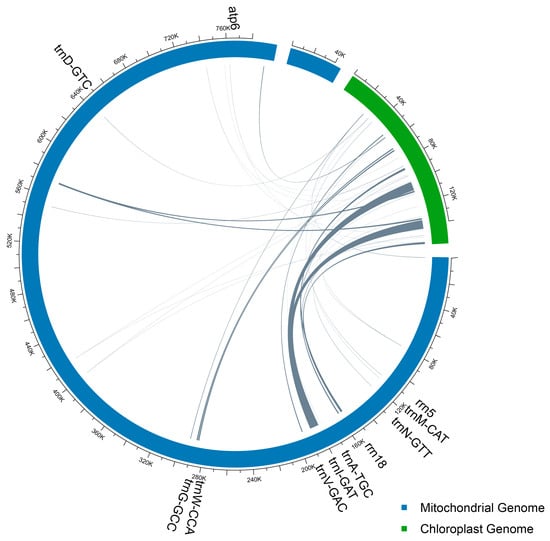
Figure 4.
Schematic for the chloroplast-to-mitochondrial gene transfer.
3.5. RNA Editing Serves to Modify the Physicochemical Properties of Encoded Amino Acids
A total of 211 RNA editing sites were identified in the mitogenome of C. sinensis cv. Xinyang10. Among these, 126 sites were successfully annotated within 25 protein-coding genes (PCGs), while the remaining 85 sites were located in non-coding regions. The top five genes with the highest number of editing events were cox2 (14 sites), cox1 (12 sites), atp6 and atp9 (11 sites each), and nad5 (8 sites) (Figure 5a). The two primary editing types were C-to-U and G-to-A, each accounting for approximately half of the total events. Notably, RNA editing was observed within the stop codon of both cox1 and atp9. The second codon position was the most frequent target for RNA editing, accounting for 61.1% of all sites, with the majority of remaining events occurring at the first position.
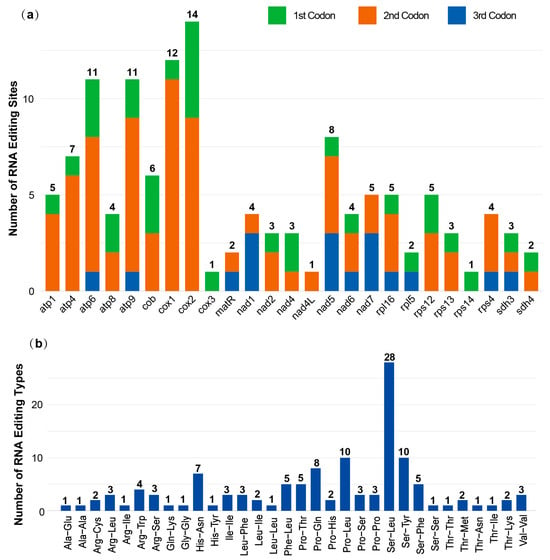
Figure 5.
RNA editing events in C. sinensis cv. Xinyang10. (a) Frequency of RNA editing sites per gene, (b) Impact of RNA editing on amino acid sequence.
The conversions of serine to leucine (28 events), proline to leucine (12 events), and serine to phenylalanine (10 events) were the three most common amino acid substitutions resulting from RNA editing. RNA editing altered the physicochemical properties of the encoded amino acids in 88.1% of the events, often involving changes in hydrophobicity. Specifically, 35% of the edited sites led to a change from hydrophilic to hydrophobic amino acids, while 15.1% resulted in a change from hydrophobic to hydrophilic residues (Figure 5b).
3.6. Codon Usage Bias in Mitochondrial Genome of C. sinensis cv. Xinyang10
A total of 9976 codons from 34 protein-coding genes in the complete mitogenome of C. sinensis cv. Xinyang10 were extracted and analyzed for codon usage bias. All 64 codons encoding 21 amino acids were identified. With the exception of methionine (ATG) and tryptophan (TGG), which showed no bias (RSCU = 1), all other amino acids exhibited codon usage preference (Figure 6 and Table S4). Twenty-nine codons had RSCU values greater than 1, indicating preferential usage. The alanine codon GCU showed the highest RSCU value (1.59), followed by the tyrosine codon UAU (1.54). In contrast, 33 codons had RSCU values less than 1, with the tyrosine codon UAC being the least preferred (RSCU = 0.47). Preferentially used codons (RSCU > 1) predominantly ended with A or U, while those with low usage preference (RSCU < 1) mainly ended with C or G. The Effective Number of Codons (ENC) plot was used to assess the major factors influencing codon usage bias. Most genes had ENC values below the expected curve, with only a few points lying near it (Figure S1). These findings suggest that natural selection, especially translational selection for optimizing highly expressed genes, along with other factors, were the predominant forces shaping codon usage bias, rather than mutation pressure.
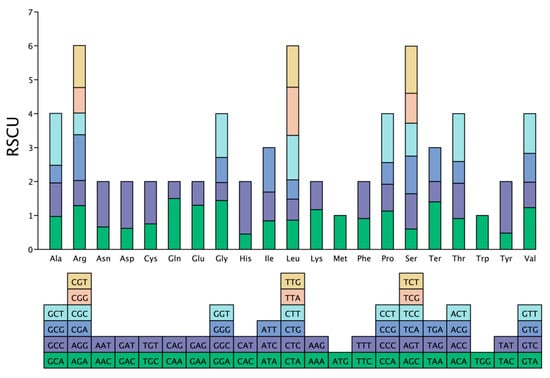
Figure 6.
Analysis of codon preference in mitochondrial protein-coding genes of C. sinensis cv. Xinyang10. The plot displays the relative synonymous codon usage (RSCU) values for codons corresponding to the 21 amino acids.
3.7. Assessing Selective Pressure in Tea Plants
Comparative analysis of the 11 tea plant mitogenomes revealed a core set of 29 shared protein-coding genes. Calculation of the Ka/Ks ratios for these orthologous genes between XY10 and the other ten cultivars revealed that the vast majority of genes had Ka/Ks values less than 0.5 or equal to 0 (Figure 7 and Table S5), indicating that these genes were subject to strong purifying selection. For example, cox2 exhibited signatures of purifying selection in all pairwise comparisons between XY10 and the other ten cultivars. Two genes were identified with Ka/Ks ratios greater than 1, suggesting positive selection. Specifically, the ccmFC gene in the comparison between XY10 and DMB had a Ka/Ks ratio of 1.83969, and the nad1 gene compared between XY10 and AMH showed a Ka/Ks ratio of 1.03976. These results indicate that ccmFC and nad1 are likely under positive selection.
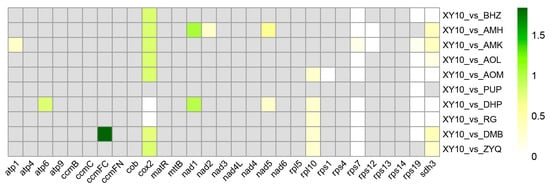
Figure 7.
Ka/Ks substitution rates of protein coding genes in C. sinensis cv. Xinyang10 and 10 other tea plants.
3.8. Phylogenetic Analysis
The phylogenetic trees constructed using both maximum likelihood (ML) and Bayesian inference (BI) exhibited strong congruence in their topologies. Nodal support values were generally higher in the Bayesian tree than in the ML tree (Figure 8). The phylogenetic results indicated that species within the family Theaceae formed a monophyletic clade with high support (BS/PP = 98/100), representing an inner branch within the order Ericales. Outside the Theaceae clade, the families Ericaceae, Pyrolaceae, Symplocaceae, and Actinidiaceae were successively positioned. The families Primulaceae and Ebenaceae formed a sister relationship, constituting the outermost branch of the Ericales phylogeny with support values of 79/92 (BS/PP).
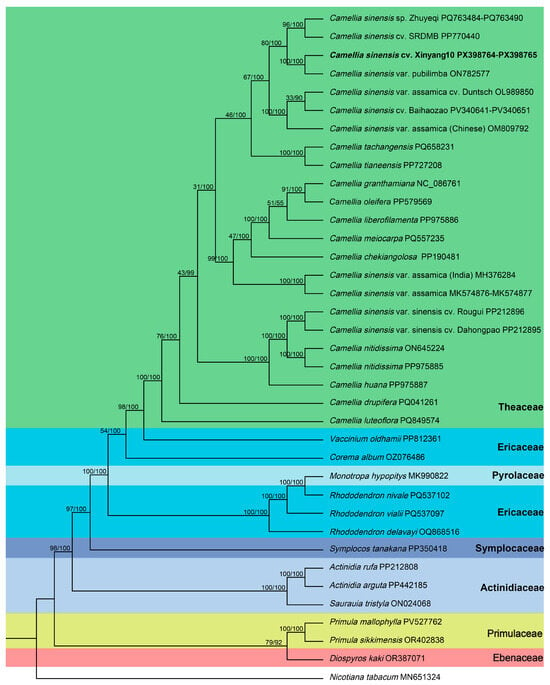
Figure 8.
Phylogenetic reconstruction of 35 species based on SNP data, inferred from both maximum likelihood (ML) and Bayesian inference (BI) methods. Nicotiana tabacum was used as the outgroup. Nodal support is indicated by ML bootstrap values and BI posterior probabilities. Different families within the order Ericales are highlighted in distinct colors, with family names indicated in the right side of the corresponding color blocks. Boldfaced species names in the phylogenetic tree represent the species sequenced in this study.
Within the Theaceae clade, internal nodes generally received lower support values than inter-familial branches. C. sinensis cv. Xinyang10 and C. sinensis var. pubilimba (PUP) formed a strongly supported clade (100/100). This clade was sister to a group comprising C. sinensis sp. Zhuyeqi (ZYQ) and C. sinensis cv. SRDMB (DMB), forming an internal Theaceae branch with support values of 80/100 (BS/PP). C. sinensis cv. Baihaozao (BHZ) clustered with two large-leaf tea accessions from China—C. sinensis var. assamica cv. Duntsch (AOL) and C. sinensis var. assamica (AOM)—and this group was sister to the aforementioned internal clade. Camellia tachangensis (from southwestern China) and Camellia tianeensis (from Guangxi) formed a distinct clade. The large-leaf tea accession AMK (from China) and AMH (from India) grouped together and were sister to a clade containing Camellia granthamiana, Camellia oleifera, Camellia liberofilamenta, Camellia meiocarpa, and Camellia chekiangolosa (from southwestern and southeastern China) with strong support (99/100). The small-leaf tea cultivars RG and DHP formed a clade sister to Camellia nitidissima, with Camellia huana positioned outside this sister group. Camellia luteoflora and Camellia drupifera were successively placed as the outermost lineages within Theaceae.
4. Discussion
4.1. Structure and Size of the C. sinensis cv. Xinyang10 Mitogenome
The size of plant mitogenomes varies dramatically across species. For instance, the mitogenome of Brassica rapa is only 219.78 kb [], while that of Cathaya argyrophylla reaches 18.99 Mb []. Differences in non-coding regions and the abundance of repetitive sequences are primary contributors to this observed genomic variation [,]. Among the reported mitogenomes in Ericales, AMH is the smallest (707,441 bp), and C. tianeensis is the largest (1,098,121 bp). The mitogenome of C. sinensis cv. Xinyang10 is 845,076 bp, which is comparable to other published tea plant mitogenomes such as DHP (1,082,025 bp), RG (991,788 bp), AOL (1,081,966 bp), AOM (914,855 bp), AMK (879,048 bp), AMH (707,441 bp), BHZ (909,843 bp), ZYQ (911,255 bp), DMB (886,354 bp), and PUP (707,441 bp). Plant mitogenomes are predominantly circular, though some exhibit complex structural conformations, including linear multipartite, branched, or mixed circular-linear architectures []. In this study, the mitogenome of C. sinensis cv. Xinyang10 consists of one circular and one linear DNA molecule, resembling the structure of AMK and showing the closest size similarity to it []. In contrast, the mitogenomes of ZYQ and DMB are linear and multipartite, comprising 7 and 11 linear DNA molecules, respectively [,].
Annotation of the C. sinensis cv. Xinyang10 mitogenome identified a complement of 74 genes, covering all genes found in other tea plant mitogenomes. Unlike other tea cultivars, all 30 protein-coding genes (PCGs) in XY10 were single-copy. In contrast, other tea mitogenomes contain at least one multi-copy PCG; for example, the nad5 gene in BHZ has three copies, and rps19 in ZYQ has two copies. The absence of duplicated PCGs in mitogenomes has also been observed in other species within Theaceae or even Ericales, such as in C. granthamiana [], Actinidia eriantha [], and Aegiceras corniculatum []. Consistent with other tea plant mitogenomes, eight tRNA genes in XY10 were multi-copy.
Genomic repeats are known to play crucial roles in intermolecular recombination and serve as important evidence for evolutionary and genetic characteristics [,,]. These repeats can promote structural variations that may influence gene function []. A total of 244 SSRs were identified in C. sinensis cv. Xinyang10 mitogenome, compared to 322 in DHP, 294 in RG, 316 in AOL, 260 in AOM, 250 in AMK, 201 in AMH and 264 in BHZ. SSR-containing genes in XY10 included mttB, matR, rps3, and rps1. The (CTT)4 motif in rps1 was also present in eight other tea cultivars but absent in ZYQ and BHZ [,]. Mutations or dysregulation of mttB and matR are associated with male sterility and represent key genes in Cytoplasmic Male Sterility (CMS) mechanism research for hybrid breeding []. As mitochondrial ribosomal subunit genes, rps1 and rps3 participate in the translation of mitochondrial mRNAs. Variations or deficiencies in these genes may reduce translational efficiency, affecting energy metabolism and fertility. SSR loci within these genes could serve as cultivar-specific markers linked to relevant quality traits.
In the long repeat (LSR) analysis, C. sinensis cv. Xinyang10 contained 998 LSRs—more than other reported tea cultivars DHP (747), RG (613), AOL (707), AOM (472), AMK (408), and AMH (298). By mediating genomic expansion, rearrangement, and evolutionary dynamics, long dispersed repeats are a key architectural feature of plant mitogenomes and a contributing factor to traits like cytoplasmic male sterility [,]. The abundance of LSRs in C. sinensis cv. Xinyang10 may contribute to seedling breeding and genetic improvement in tea plants.
During plant mitogenome evolution, frequent DNA transfer events [], abundant non-coding sequences [], and poorly conserved gene order with sparse distribution [] are common, even among close relatives [,]. Despite these structural variations, the length of homologous sequences between mitogenomes correlates with taxonomic relatedness, with closely related species sharing the largest homologous regions, including non-coding segments []. Collinearity analysis among the 11 tea plant mitogenomes revealed extensive rearrangements leading to substantial divergence in gene order. A phylogenetic tree constructed based on mitogenomic collinearity placed tea accessions from India and southwestern China at the basal positions, followed by those from Guizhou, Hunan, Jiangxi, and Henan in southern China, with accessions from Fujian in southeastern China forming the innermost clade. This result is consistent with the hypothesis proposed by Xia et al. []. Based on resequencing of 81 tea accessions from diverse origins, which suggested that tea plants originated in southwestern China and spread eastward. Our findings thus support the validity of collinearity-based phylogenetic reconstruction in mitogenomes.
4.2. Codon Usage Bias and Its Evolutionary Implications in the Mitogenome of C. sinensis cv. Xinyang10
Mitochondrial protein-coding genes (PCGs) play essential roles in oxidative phosphorylation, providing energy for cellular functions, and their evolutionary adaptations enable organisms to thrive in diverse environments. Furthermore, these genes influence cellular development and functions such as regulating apoptosis and oxidative stress responses []. Our analyses of RNA editing, codon usage bias, and selective pressures on mitochondrial PCGs collectively reveal their critical roles in bioenergetics, environmental adaptation, and cellular function. The number of RNA editing sites identified in the mitogenome of C. sinensis cv. Xinyang10 (330 sites) was lower than that in the other ten tea cultivars (ranging from 546 to 720). This discrepancy may be attributed to differences in detection methodologies. When comparing specific RNA editing events across all tea plants, C. sinensis cv. Xinyang10 exhibited a higher proportion of conversions to hydrophobic amino acids—such as serine to leucine—than other cultivars. Since increased hydrophobicity may enhance protein stability under low temperatures [], this bias toward hydrophobic conversions may be associated with the cold adaptation of C. sinensis cv. Xinyang10.
Codon usage bias is critical for determining protein structure, as well as for translational fidelity and efficiency. Analyzing this bias provides insights into the evolutionary adaptation of genomes [,]. We calculated GC content at different codon positions and the relative synonymous codon usage (RSCU) in the mitogenome of C. sinensis cv. Xinyang10 and compared these values with those of other tea plants. The overall GC content of all tea mitogenomes ranged between 45.62% and 45.82%. In all 11 Theaceae mitogenomes examined, including C. sinensis cv. Xinyang10, codons showed a pronounced bias toward A/T endings. These findings align with previous reports that dicot plants generally exhibit a preference for A/T-ending codons [].
The Ka/Ks ratio is an important metric for understanding species adaptation, identifying selective agents, assessing adaptive potential, and deciphering genetic and evolutionary processes []. Analysis of shared PCGs between the mitogenome of C. sinensis cv. Xinyang10 and ten other tea plants revealed Ka/Ks values greater than 1 for ccmFC and nad1, indicating that these genes have undergone positive selection during tea plant evolution. Notably, signals of positive selection in nad1 were also reported in a comparative analysis of six tea cultivars []. The nad1 gene encodes a subunit of nicotinamide adenine dinucleotide (NADH) dehydrogenase, which plays a crucial role in the initiation of ATP production. Studies of the Nicotiana tabacum mitogenome suggest that copy number variation and sequence changes in nad1 may lead to energy deficiency during pollen development []. As a perennial evergreen species that requires substantial energy to synthesize secondary metabolites (e.g., tea polyphenols and caffeine), the tea plant has high energy demands. Positive selection in nad1 may optimize the efficiency of the electron transport chain (ETC), thereby increasing ATP production to support rapid growth, stress tolerance, and the synthesis of bioactive compounds. Moreover, given that tea plants are cultivated across a range of environments from the tropics to temperate zones, positive selection in these genes may represent a “fine trade-off”—balancing the maintenance of sufficient energy output with minimizing reactive oxygen species (ROS) bursts under stress, thereby enhancing resistance to cold, drought, waterlogging, and other adversities. However, definitive establishment of causality between the observed positive selection in nad1 and cold stress adaptation in tea plants will require targeted functional validation through gene expression studies under controlled stress conditions and molecular genetics approaches. As such, nad1 represents a promising candidate gene for future studies on cold tolerance. It also serves as a valuable mitochondrial genetic marker for tea breeding. Notably, positive selection in nad1 has also been detected in the mitogenomes of the Hippophae genus [], Isopyrum anemonoides [], Fagopyrum dibotrys [], and Sapindus mukorossi [].
The ccmFC gene is involved in cytochrome c biogenesis. Cytochrome c is a key signaling molecule in mitochondria-mediated programmed cell death (PCD). Positive selection in ccmFC may affect the maturation and release of cytochrome c, thereby regulating PCD processes in tea plants. PCD is essential for plant development (e.g., tracheary element formation) and defense responses. Positive selection in ccmFC may enable tea plants to control PCD more precisely, facilitating efficient supportive tissue formation during development or triggering localized cell suicide in response to pathogen invasion to protect the whole plant. Positive selection in ccmFC has also been reported in the mitogenomes of Cinnamomum camphora [], Lindera aggregata [], Luffa cylindrica [], and Eichhornia crassipes [].
4.3. Phylogenetic Relationships Based on Mitochondrial Genomes
Clarifying the phylogenetic relationships of tea plants is crucial not only for revealing the genetic background of tea resources but also for conserving their diversity and understanding their adaptive evolution. Previous studies have largely relied on chloroplast and nuclear genomes to reconstruct phylogenetic relationships within the genus Camellia and even the family Theaceae. The broad evolutionary relationships among major lineages in Theaceae are relatively well resolved [,]. However, due to interspecific hybridization and polyploidization, relationships at the genus and species levels continue to be revised and refined []. For example, a phylogenetic analysis of 161 species from all four subgenera of Camellia based on four chloroplast regions (matK, rbcL, ycf1, and trnL-F) revealed that the genus can be divided into 13 clades (A–M). Species from the Subgenus Thea were distributed across multiple clades rather than forming a single monophyletic group []. Similarly, a phylogeny of 116 Camellia species constructed from 405 high-quality low-copy nuclear genes showed that most inferred relationships were strongly supported by morphological traits, but Section Thea was not monophyletic and instead clustered with Section Glaberrima [].
Owing to the scarcity of available mitogenome data within the Theaceae family and the fact that mitochondrial sequences have not traditionally been the marker of choice for plant phylogenetic reconstruction, studies on tea plant phylogeny based on mitogenomes are scarce. In the phylogenetic study of Ranunculales plants, Zhu et al. demonstrated that mitochondrial genes provide complementary and strongly supported evidence for resolving some controversial phylogenetic relationships within the order, confirming the significant potential of mitogenome sequences in reconstructing phylogenies of closely related taxa []. Li et al. used 573 SNPs from 20 mitogenomes to construct a phylogeny containing six tea accessions and found that the six C. sinensis samples clustered together, with C. sinensis var. assamica and C. sinensis var. sinensis forming distinct subclades []. In this study, we constructed a phylogenetic tree using 4537 SNPs from the mitogenomes of 36 Ericales species, including members of Theaceae. We found that C. sinensis samples did not cluster together; instead, they were intermingled with other Camellias. For instance, C. tachangensis and C. tianeensis clustered with both C. sinensis var. assamica and C. sinensis var. sinensis, while C. nitidissima grouped with two C. sinensis var. sinensis, RG and DHP. Similar findings were reported in a mitogenome-based phylogenetic study of oil-tea Camellias []. Phylogenies reconstructed using maximum likelihood (ML) and Bayesian inference (BI) methods based on protein-coding sequences from 16 complete mitochondrial and chloroplast genomes of Theaceae species (including C. sinensis) revealed incongruences between the mitochondrial and chloroplast trees. In the chloroplast tree, all tea accessions formed a single clade, whereas in the mitochondrial tree, three C. sinensis samples clustered together, and another grouped with C. nitidissima and C. gigantocarpa [].
Discrepancies between mitochondrial and chloroplast/nuclear phylogenies may arise from the distinct properties of mitogenomes, such as their tendency to retain ancestral polymorphisms or traces of introgression events []. Incomplete lineage sorting (ILS) may also contribute to these differences, leading to varying ancestral haplotypes among varieties. Furthermore, frequent hybridization within the genus Camellia—particularly between C. sinensis and closely related species such as C. taliensis and C. crassicolumna—may result in the “capture” of mitochondria across genetic backgrounds []. Tea cultivars may also originate from multiple independent domestication events or admixture between geographically distinct populations, leading to diverse mitogenomic profiles. Future studies should aim to sequence mitogenomes from a broader range of tea cultivars and other Camellia species. Integrating mitochondrial (mtDNA), chloroplast (cpDNA), and nuclear (nDNA) data in a phylogenomic framework will leverage the strengths of each genetic marker and yield a more robust and comprehensive understanding of tea plant evolutionary history [].
5. Conclusions
This study presents the complete mitochondrial genome of the elite tea cultivar C. sinensis cv. Xinyang10, revealing a multipartite architecture consisting of both circular and linear chromosomes. Thereby documenting the notable plasticity of mitogenome organization within C. sinensis, which ranges from single circular to multiple linear or mixed conformations. Our analysis further identified extensive genomic reorganization, inter-organellar sequence transfer, and lineage-specific evolutionary signatures.
Importantly, the phylogenetic pattern inferred from mitogenomic collinearity provides independent support for a postulated westward-to-eastward migration route of tea plants within China, offering novel insights into the species’ biogeographic history. The mitogenome likely harbors further valuable information regarding the origin and adaptive evolution of tea plants, yet unlocking this potential fully will require the inclusion of additional mitogenomes from geographically diverse local tea varieties to establish a more comprehensive comparative framework. Furthermore, the detection of positive selection in key mitochondrial genes (nad1 and ccmFC) identifies valuable candidate markers for mitochondrial-assisted breeding. While these genes represent promising targets for improving stress resilience, their specific functional roles in cold adaptation warrant further experimental validation. Collectively, this work not only enriches the genomic resources for tea plants but also demonstrates the utility of mitogenome data in reconstructing phytogeographic history and supporting the development of molecular breeding strategies.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/d17100705/s1, Table S1: The number of SSRs in the mitogenome of C. sinensis cv. Xinyang10; Table S2: The number of LSRs in mitogenome of C. sinensis cv. Xinyang10; Table S3: Sequence homology between the mitochondrial and chloroplast genomes of Camellia sinensis cv. Xinyang10; Table S4: RSCU values in Camellia sinensis cv. Xinyang10 mitogenome; Table S5: Ka/Ks substitution rates of protein coding genes in Camellia sinensis cv. Xinyang10 and other 10 tea plants. Figure S1: ENc-plot of C. sinensis cv. Xinyang10.
Author Contributions
Conceptualization, M.-H.Y. and W.T.; methodology, Y.-R.D.; software, M.-H.Y. and Y.-R.D.; validation, W.T., M.-H.Y. and Y.-R.D.; formal analysis, M.-H.Y., Y.-R.D., W.T., J.-M.S., G.-Q.P., L.-M.Y., W.-W.W. and T.-T.Z.; investigation, J.-M.S. and W.-W.W.; resources, G.-Q.P.; data curation, L.-M.Y.; writing—original draft preparation, Y.-R.D.; writing—review and editing, M.-H.Y.; visualization, M.-H.Y. and Y.-R.D.; supervision, W.T.; project administration, M.-H.Y. and W.T.; funding acquisition, M.-H.Y. and W.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (31800276), the Open Fund of Dabie Mountain Laboratory (DMLOF2024021), the Natural Science Foundation of Henan Province of China (252300420219), and the Nanhu Scholars Program for Young Scholars of XYNU granted to Ming-Hui Yan.
Data Availability Statement
To support the results, the mitochondrial genome data have been deposited in the NCBI database, with login accession numbers PX398764 and PX398765.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Wu, Z.Q.; Liao, X.Z.; Zhang, X.N.; Tembrock, L.R.; Broz, A. Genomic architectural variation of plant mitochondria—A review of multichromosomal structuring. J. Syst. Evol. 2022, 60, 160–168. [Google Scholar] [CrossRef]
- Morley, S.A.; Nielsen, B.L. Plant mitochondrial DNA. Front. Biosci. (Landmark Ed.) 2017, 22, 1023–1032. [Google Scholar]
- Jethva, J.; Schmidt, R.R.; Sauter, M.; Selinski, J. Try or Die: Dynamics of Plant Respiration and How to Survive Low Oxygen Conditions. Plants 2022, 11, 205. [Google Scholar] [CrossRef] [PubMed]
- Darracq, A.; Varré, J.S.; Maréchal-Drouard, L.; Courseaux, A.; Castric, V.; Saumitou-Laprade, P.; Oztas, S.; Lenoble, P.; Vacherie, B.; Barbe, V.; et al. Structural and content diversity of mitochondrial genome in beet: A comparative genomic analysis. Genome Biol. Evol. 2011, 3, 723–736. [Google Scholar] [CrossRef]
- Chen, Z.; Zhao, N.; Li, S.; Grover, C.E.; Nie, H.; Wendel, J.F.; Hua, J. Plant Mitochondrial Genome Evolution and Cytoplasmic Male Sterility. Crit. Rev. Plant Sci. 2017, 36, 55–69. [Google Scholar] [CrossRef]
- Liberatore, K.L.; Dukowic-Schulze, S.; Miller, M.E.; Chen, C.; Kianian, S.F. The role of mitochondria in plant development and stress tolerance. Free Radic. Biol. Med. 2016, 100, 238–256. [Google Scholar] [CrossRef]
- Quesada, V. The roles of mitochondrial transcription termination factors (MTERFs) in plants. Physiol. Plant. 2016, 157, 389–399. [Google Scholar] [CrossRef]
- de Cássia Monteiro-Batista, R.; Siqueira, J.A.; da Fonseca Pereira, P.; Barreto, P.; Feitosa-Araujo, E.; Araújo, W.L.; Nunes-Nesi, A. Potential roles of mitochondrial carrier proteins in plant responses to abiotic stress. J. Exp. Bot. 2025, eraf032. [Google Scholar] [CrossRef] [PubMed]
- Feng, G.; Jiao, Y.; Wang, Y.; He, D.; Liu, Q.; Linchen, R.; Gao, Y.; Wang, J.; Wang, X.; Huang, T.; et al. Complete mitochondrial genome assembly and comparative analysis of Fagopyrum dibotrys (Golden Buckwheat). BMC Plant Biol. 2025, 25, 985. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Yan, R.R.; Yang, C.Y.; Ling, L.Z.; Bai, X.X.; Ren, Q.F.; Hu, G.X. Mitochondrial genome analysis of the endangered Oreocharis esquirolii: Insights into evolutionary adaptation and conservation. BMC Plant Biol. 2025, 25, 827. [Google Scholar] [CrossRef]
- Xu, C.; Bi, W.; Ma, R.Y.; Li, P.R.; Liu, F.; Liu, Z.W. Assembly and comparative analysis of the complete mitochondrial of Spodiopogon sagittifolius, an endemic and protective species from Yunnan, China. BMC Plant Biol. 2025, 25, 373. [Google Scholar] [CrossRef] [PubMed]
- Gong, Y.; Luo, X.; Zhang, T.; Zhou, G.; Li, J.; Zhang, B.; Li, P.; Huang, H. Assembly and comparative analysis of the complete mitochondrial genome of white towel gourd (Luffa cylindrica). Genomics 2024, 116, 110859. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.; Zhang, Z.; Tso, N.; Zhang, S.; Chen, Y.; Shu, Q.; Li, J.; Liang, Z.; Wang, R.; Wang, J.; et al. Complete mitochondrial genome of Hippophae tibetana: Insights into adaptation to high-altitude environments. Front. Plant Sci. 2024, 15, 1449606. [Google Scholar] [CrossRef]
- Miao, X.; Yang, W.; Li, D.; Wang, A.; Li, J.; Deng, X.; He, L.; Niu, J. Assembly and comparative analysis of the complete mitochondrial and chloroplast genome of Cyperus stoloniferus (Cyperaceae), a coastal plant possessing saline-alkali tolerance. BMC Plant Biol. 2024, 24, 628. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liang, L.; Ran, J.; Yang, F.; Ran, M.; Yong, X.; Kong, C.; Tang, Y.; Li, H. Re-Sequencing the Mitochondrial Genome Unveils a Novel Isomeric Form of NWB CMS Line in Radish and Functional Verification of Its Candidate Sterile Gene. Horticulturae 2024, 10, 395. [Google Scholar] [CrossRef]
- Hao, J.; Liang, Y.; Wang, T.; Su, Y. Correlations of gene expression, codon usage bias, and evolutionary rates of the mitochondrial genome show tissue differentiation in Ophioglossum vulgatum. BMC Plant Biol. 2025, 25, 134. [Google Scholar] [CrossRef]
- Ramadan, A.; Alnufaei, A.A.; Fiaz, S.; Khan, T.K.; Hassan, S.M. Effect of salinity on ccmfn gene RNA editing of mitochondria in wild barley and uncommon types of RNA editing. Funct. Integr. Genom. 2023, 23, 50. [Google Scholar] [CrossRef]
- Bi, C.; Qu, Y.; Hou, J.; Wu, K.; Ye, N.; Yin, T. Deciphering the Multi-Chromosomal Mitochondrial Genome of Populus simonii. Front. Plant Sci. 2022, 13, 914635. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, J.; He, W.; Kan, S.; Liao, X.; Jordan, D.R.; Mace, E.S.; Tao, Y.; Cruickshank, A.W.; Klein, R.; et al. Variation in mitogenome structural conformation in wild and cultivated lineages of sorghum corresponds with domestication history and plastome evolution. BMC Plant Biol. 2023, 23, 91. [Google Scholar] [CrossRef]
- Guo, G.Y. Famous tea of China: Xinyangmaojian. J. Xinyang Agric. Coll. 1999, 9, 49–52. [Google Scholar]
- Chen, J.-D.; Zheng, C.; Ma, J.Q.; Jiang, C.K.; Ercisli, S.; Yao, M.Z.; Chen, L. The chromosome-scale genome reveals the evolution and diversification after the recent tetraploidization event in tea plant. Hortic. Res. 2020, 7, 63. [Google Scholar] [CrossRef]
- Chen, S.; Wang, P.; Kong, W.; Chai, K.; Zhang, S.; Yu, J.; Wang, Y.; Jiang, M.; Lei, W.; Chen, X.; et al. Gene mining and genomics-assisted breeding empowered by the pangenome of tea plant Camellia sinensis. Nat. Plants 2023, 9, 1986–1999. [Google Scholar] [CrossRef]
- Xia, E.; Tong, W.; Hou, Y.; An, Y.; Chen, L.; Wu, Q.; Liu, Y.; Yu, J.; Li, F.; Li, R.; et al. The Reference Genome of Tea Plant and Resequencing of 81 Diverse Accessions Provide Insights into Its Genome Evolution and Adaptation. Mol. Plant 2020, 13, 1013–1026. [Google Scholar] [CrossRef]
- Wang, X.; Feng, H.; Chang, Y.; Ma, C.; Wang, L.; Hao, X.; Li, A.l.; Cheng, H.; Wang, L.; Cui, P.; et al. Population sequencing enhances understanding of tea plant evolution. Nat. Commun. 2020, 11, 4447. [Google Scholar] [CrossRef]
- Kong, W.; Kong, X.; Xia, Z.; Li, X.; Wang, F.; Shan, R.; Chen, Z.; You, X.; Zhao, Y.; Hu, Y.; et al. Genomic analysis of 1,325 Camellia accessions sheds light on agronomic and metabolic traits for tea plant improvement. Nat. Genet. 2025, 57, 997–1007. [Google Scholar] [CrossRef]
- Li, L.; Li, X.; Liu, Y.; Li, J.; Zhen, X.; Huang, Y.; Ye, J.; Fan, L. Comparative analysis of the complete mitogenomes of Camellia sinensis var. sinensis and C. sinensis var. assamica provide insights into evolution and phylogeny relationship. Front Plant Sci. 2024, 15, 1396389. [Google Scholar]
- Zhang, F.; Li, W.; Gao, C.W.; Zhang, D.; Gao, L.Z. Deciphering tea tree chloroplast and mitochondrial genomes of Camellia sinensis var. assamica. Sci. Data 2019, 6, 209. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Yang, L.; Zhou, J.; Xiao, Z.; Lu, M.; Zeng, Y.; Tan, X. Mitochondrial genome study of Camellia oleifera revealed the tandem conserved gene cluster of nad5–nads in evolution. Front. Plant Sci. 2024, 15, 1396635. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Latif, A.; Osman, G. Comparison of three genomic DNA extraction methods to obtain high DNA quality from maize. Plant Methods 2017, 13, 1. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Bi, C.; Shen, F.; Han, F.; Qu, Y.; Hou, J.; Xu, K.; Xu, L.A.; He, W.; Wu, Z.; Yin, T. PMAT: An efficient plant mitogenome assembly toolkit using low-coverage HiFi sequencing data. Hortic. Res. 2024, 11, uhae023. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Brown, M.; Blaxter, M.; McCarthy, S.A.; Durbin, R. Oatk: A de novo assembly tool for complex plant organelle genomes. Genome Biol. 2025, 26, 235. [Google Scholar] [CrossRef]
- Wick, R.R.; Schultz, M.B.; Zobel, J.; Holt, K.E. Bandage: Interactive visualization of de novo genome assemblies. Bioinformatics 2015, 31, 3350–3352. [Google Scholar] [CrossRef]
- Tillich, M.; Lehwark, P.; Pellizzer, T.; Ulbricht-Jones, E.S.; Fischer, A.; Bock, R.; Greiner, S. GeSeq-versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017, 45, w6–w11. [Google Scholar] [CrossRef]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Chen, H.; Ni, Y.; Wu, B.; Li, J.; Burzyński, A.; Liu, C. Plant mitochondrial genome map (PMGmap): A software tool for the comprehensive visualization of coding, noncoding and genome features of plant mitochondrial genomes. Mol. Ecol. Resour. 2024, 24, e13952. [Google Scholar] [CrossRef]
- Ankenbrand, M.J.; Hohlfeld, S.; Hackl, T.; Förster, F. AliTV—Interactive visualization of whole genome comparisons. PeerJ Comput. Sci. 2017, 3, e116. [Google Scholar] [CrossRef]
- Zhang, H.; Meltzer, P.; Davis, S. RCircos: An R package for Circos 2D track plots. BMC Bioinform. 2013, 14, 244. [Google Scholar] [CrossRef]
- Beier, S.; Thiel, T.; Münch, T.; Scholz, U.; Mascher, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef]
- Kurtz, S.; Choudhuri, J.V.; Ohlebusch, E.; Schleiermacher, C.; Stoye, J.; Giegerich, R. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001, 29, 4633–4642. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Ye, W.; Zhang, Y.; Xu, Y. High speed BLASTN: An accelerated MegaBLAST search tool. Nucleic Acids Res. 2015, 43, 7762–7768. [Google Scholar] [CrossRef]
- Huang, L.; Yu, H.; Wang, Z.; Xu, W. CPStools: A package for analyzing chloroplast genome sequences. iMetaOmics 2024, 1, e25. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. In Proceedings of the Nucleic acids symposium series, Durham, NC, USA, 15–17 October 1999; pp. 95–98. [Google Scholar]
- Yan, M.H.; Liu, K.; Wang, M.; Lyu, Y.; Zhang, Q. Complete chloroplast genome of Camellia sinensis cv. Xinyang 10 and its phylogenetic evolution. Tea Sci. 2021, 41, 777–788. [Google Scholar]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Genovese, G.; Rockweiler, N.B.; Gorman, B.R.; Bigdeli, T.B.; Pato, M.T.; Pato, C.N.; Ichihara, K.; McCarroll, S.A. BCFtools/liftover: An accurate and comprehensive tool to convert genetic variants across genome assemblies. Bioinformatics 2024, 40, btae038. [Google Scholar] [CrossRef]
- Peden, J.F. Analysis of Codon Usage. Ph.D. Thesis, University of Nottingham, Nottingham, UK, 2000. Available online: https://codonw.sourceforge.net/JohnPedenThesisPressOpt_water.pdf (accessed on 9 October 2025).
- Huang, X.; Jiao, Y.; Guo, J.; Wang, Y.; Chu, G.; Wang, M. Analysis of codon usage patterns in Haloxylon ammodendron based on genomic and transcriptomic data. Gene 2022, 845, 146842. [Google Scholar] [CrossRef]
- Wright, F. The ‘effective number of codons’ used in a gene. Gene 1990, 87, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, Y.; Zhang, Z.; Zhu, J.; Yu, J. KaKs_Calculator 2.0: A toolkit incorporating gamma-series methods and sliding window strategies. Genom. Proteom. Bioinform. 2010, 8, 77–80. [Google Scholar] [CrossRef] [PubMed]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Xiao, W.; Wu, X.; Zhou, X.; Zhang, J.; Huang, J.; Dai, X.; Ren, H.; Xu, D. Assembly and comparative analysis of the first complete mitochondrial genome of zicaitai (Brassica rapa var. Purpuraria): Insights into its genetic architecture and evolutionary relationships. Front. Plant Sci. 2024, 15, 1475064. [Google Scholar] [CrossRef]
- Arimura, S.I. Fission and Fusion of Plant Mitochondria, and Genome Maintenance. Plant Physiol. 2018, 176, 152–161. [Google Scholar] [CrossRef]
- Gualberto, J.M.; Mileshina, D.; Wallet, C.; Niazi, A.K.; Weber-Lotfi, F.; Dietrich, A. The plant mitochondrial genome: Dynamics and maintenance. Biochimie 2014, 100, 107–120. [Google Scholar] [CrossRef]
- Gualberto, J.M.; Newton, K.J. Plant Mitochondrial Genomes: Dynamics and Mechanisms of Mutation. Annu. Rev. Plant Biol. 2017, 68, 225–252. [Google Scholar] [CrossRef]
- Kozik, A.; Rowan, B.A.; Lavelle, D.; Berke, L.; Schranz, M.E.; Michelmore, R.W.; Christensen, A.C. The alternative reality of plant mitochondrial DNA: One ring does not rule them all. PLoS Genet. 2019, 15, e1008373. [Google Scholar] [CrossRef]
- Zeng, W.J.; Zhu, Y.P.; Chen, J.X.; Li, H.Y.; Wang, S.H.; Gong, Y.H.; Chen, Z.Y. Analysis of Codon Usage Bias in Chloroplast and Mitochondrial Genomes of Camellia sinensis cv. ‘Zhuyeqi’. J. Tea Sci. 2025, 45, 201–218. [Google Scholar]
- Chen, Z.; Zhou, W.; Wang, Z.; Chen, Z.; You, X.; Gong, Y. Analysis of the Mitochondrial Genome of the Camellia sinensis cv. ‘Zhuyeqi’: Multichromosomal Structure, RNA Editing Sites, and Evolutionary Characterization. Front. Plant Sci. 2025, 16, 1644130. [Google Scholar] [CrossRef]
- Lu, C.; Gao, L.Z.; Zhang, Q.J. A high-quality genome assembly of the mitochondrial genome of the oil-tea tree Camellia gigantocarpa (Theaceae). Diversity 2022, 14, 850. [Google Scholar] [CrossRef]
- Wang, S.; Li, D.; Yao, X.; Song, Q.; Wang, Z.; Zhang, Q.; Zhong, C.; Liu, Y.; Huang, H. Evolution and Diversification of Kiwifruit Mitogenomes through Extensive Whole-Genome Rearrangement and Mosaic Loss of Intergenic Sequences in a Highly Variable Region. Genome Biol. Evol. 2019, 11, 1192–1206. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, S.; Zhang, Y. The complete mitochondrial genome of a mangrove plant: Aegiceras corniculatum and its phylogenetic implications. Mitochondrial DNA Part B 2020, 5, 1502–1503. [Google Scholar] [CrossRef]
- Ammiraju, J.S.; Zuccolo, A.; Yu, Y.; Song, X.; Piegu, B.; Chevalier, F.; Walling, J.G.; Ma, J.; Talag, J.; Brar, D.S.; et al. Evolutionary dynamics of an ancient retrotransposon family provides insights into evolution of genome size in the genus Oryza. Plant J. 2007, 52, 342–351. [Google Scholar] [CrossRef]
- Hu, J.; Gui, S.; Zhu, Z.; Wang, X.; Ke, W.; Ding, Y. Genome-Wide Identification of SSR and SNP Markers Based on Whole-Genome Re-Sequencing of a Thailand Wild Sacred Lotus (Nelumbo nucifera). PLoS ONE 2015, 10, e0143765. [Google Scholar] [CrossRef]
- Dong, S.; Zhao, C.; Chen, F.; Liu, Y.; Zhang, S.; Wu, H.; Zhang, L.; Liu, Y. The complete mitochondrial genome of the early flowering plant Nymphaea colorata is highly repetitive with low recombination. BMC Genom. 2018, 19, 614. [Google Scholar] [CrossRef]
- Yang, Z.; Ni, Y.; Lin, Z.; Yang, L.; Chen, G.; Nijiati, N.; Hu, Y.; Chen, X. De novo assembly of the complete mitochondrial genome of sweet potato (Ipomoea batatas [L.] Lam) revealed the existence of homologous conformations generated by the repeat-mediated recombination. BMC Plant Biol. 2022, 22, 285. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.Y.; Wang, Z.X.; Zhou, W.; Liu, S.J.; Xiao, Y.X.; Gong, Y.H. Complete sequencing of the mitochondrial genome of tea plant Camellia sinensis cv. ‘Baihaozao’: Multichromosomal structure, phylogenetic relationships, and adaptive evolutionary analysis. Front. Plant Sci. 2025, 16, 1604404. [Google Scholar] [CrossRef]
- Abdullah, M.; Sheraz, U.; Ain, A.T.; Nasir, B.; Hammad, S.; Shokat, S. Exploring the Strategies of Male Sterility for Hybrid Development in Hexaploid Wheat: Prevailing Methods and Potential Approaches. Rice 2025, 18, 53. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Ni, Y.; Zhang, X.; Li, J.; Chen, H.; Liu, C. The mitochondrial genomes of Panax notoginseng reveal recombination mediated by repeats associated with DNA replication. Int. J. Biol. Macromol. 2023, 252, 126359. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Kan, S.; Liao, X.; Zhou, J.; Tembrock, L.R.; Daniell, H.; Jin, S.; Wu, Z. Plant organellar genomes: Much done, much more to do. Trends Plant Sci. 2024, 29, 754–769. [Google Scholar] [CrossRef]
- Timmis, J.N.; Ayliffe, M.A.; Huang, C.Y.; Martin, W. Endosymbiotic gene transfer: Organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet. 2004, 5, 123–135. [Google Scholar] [CrossRef]
- Zandueta-Criado, A.; Bock, R. Surprising features of plastid ndhD transcripts: Addition of non-encoded nucleotides and polysome association of mRNAs with an unedited start codon. Nucleic Acids Res. 2004, 32, 542–550. [Google Scholar] [CrossRef]
- Kubo, T.; Nishizawa, S.; Sugawara, A.; Itchoda, N.; Estiati, A.; Mikami, T. The complete nucleotide sequence of the mitochondrial genome of sugar beet (Beta vulgaris L.) reveals a novel gene for tRNA(Cys)(GCA). Nucleic Acids Res. 2000, 28, 2571–2576. [Google Scholar] [CrossRef] [PubMed]
- Satoh, M.; Kubo, T.; Nishizawa, S.; Estiati, A.; Itchoda, N.; Mikami, T. The cytoplasmic male-sterile type and normal type mitochondrial genomes of sugar beet share the same complement of genes of known function but differ in the content of expressed ORFs. Mol. Genet. Genom. 2004, 272, 247–256. [Google Scholar] [CrossRef]
- Li, L.; Wang, B.; Liu, Y.; Qiu, Y.L. The complete mitochondrial genome sequence of the hornwort Megaceros aenigmaticus shows a mixed mode of conservative yet dynamic evolution in early land plant mitochondrial genomes. J. Mol. Evol. 2009, 68, 665–678. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, P.; Yi, B.; He, D. Current Advances in Plant Mitochondria: Application Revolution of Cytoplasmic Male Sterility. New Crops 2025, 3, 100081. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, N.; Ma, J.; Zhou, Y.; Wei, Q.; Tian, C.; Fang, Y.; Zhong, R.; Chen, G.; Zhang, S. Advances in cold-adapted enzymes derived from microorganisms. Front. Microbiol. 2023, 14, 1152847. [Google Scholar] [CrossRef]
- Hershberg, R.; Petrov, D.A. Selection on codon bias. Annu. Rev. Genet. 2008, 42, 287–299. [Google Scholar] [CrossRef]
- Tuller, T.; Waldman, Y.Y.; Kupiec, M.; Ruppin, E. Translation efficiency is determined by both codon bias and folding energy. Proc. Natl. Acad. Sci. USA 2010, 107, 3645–3650. [Google Scholar] [CrossRef]
- Mower, J.P. Variation in protein gene and intron content among land plant mitogenomes. Mitochondrion 2020, 53, 203–213. [Google Scholar] [CrossRef]
- Afzal Malik, W.; Afzal, M.; Chen, X.; Cui, R.; Lu, X.; Wang, S.; Wang, J.; Mahmood, I.; Ye, W. Systematic analysis and comparison of ABC proteins superfamily confer structural, functional and evolutionary insights into four cotton species. Ind. Crops Prod. 2022, 177, 114433. [Google Scholar] [CrossRef]
- He, D.; Li, Y.; Yuan, C.; Pei, X.; Damaris, R.N.; Yu, H.; Qian, B.; Liu, Y.; Yi, B.; Huang, C.; et al. Characterization of the CMS genetic regulation through comparative complete mitochondrial genome sequencing in Nicotiana tabacum. Plant Genome 2024, 17, e20409. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Lu, S.S.; Bi, Y.; Jiang, Y.M.; Feng, L.D.; He, J. Assembly and Analysis of the Mitochondrial Genome of Hippophae rhamnoides subsp. sinensis, an Important Ecological and Economic Forest Tree Species in China. Plants 2025, 14, 2170. [Google Scholar] [CrossRef] [PubMed]
- Yisilam, G.; Liu, Z.; Turdi, R.; Chu, Z.; Luo, W.; Tian, X. Assembly and comparative analysis of the complete mitochondrial genome of Isopyrum anemonoides (Ranunculaceae). PLoS ONE 2023, 18, e0286628. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, R.; Yun, Q.; Xu, Y.; Zhao, G.; Liu, J.; Shi, S.; Chen, Z.; Jia, L. Comprehensive analysis of complete mitochondrial genome of Sapindus mukorossi Gaertn.: An important industrial oil tree species in China. Ind. Crops Prod. 2021, 174, 114210. [Google Scholar] [CrossRef]
- Han, F.; Bi, C.; Zhao, Y.; Gao, M.; Wang, Y.; Chen, Y. Unraveling the complex evolutionary features of the Cinnamomum camphora mitochondrial genome. Plant Cell Rep. 2024, 43, 183. [Google Scholar] [CrossRef]
- Shi, Y.; Chen, Z.; Jiang, J.; Wu, W.; Yu, W.; Zhang, S.; Zeng, W. The assembly and comparative analysis of the first complete mitogenome of Lindera aggregata. Front. Plant Sci. 2024, 15, 1439245. [Google Scholar] [CrossRef]
- He, X.; Qian, Z.; Gichira, A.W.; Chen, J.; Li, Z. Assembly and comparative analysis of the first complete mitochondrial genome of the invasive water hyacinth, Eichhornia crassipes. Gene 2024, 914, 148416. [Google Scholar] [CrossRef]
- Yan, Y.; da Fonseca, R.R.; Rahbek, C.; Borregaard, M.K.; Davis, C.C. A new nuclear phylogeny of the tea family (Theaceae) unravels rapid radiations in genus Camellia. Mol. Phylogenet. Evol. 2024, 196, 108089. [Google Scholar] [CrossRef]
- Li, L.; Hu, Y.; He, M.; Zhang, B.; Wu, W.; Cai, P.; Huo, D.; Hong, Y. Comparative chloroplast genomes: Insights into the evolution of the chloroplast genome of Camellia sinensis and the phylogeny of Camellia. BMC Genom. 2021, 22, 138. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Folk, R.A.; Mo, Z.Y.; Peng, H.; Zhao, J.L.; Yang, S.X.; Yu, X.Q. Phylotranscriptomic analyses reveal deep gene tree discordance in Camellia (Theaceae). Mol. Phylogenet. Evol. 2023, 188, 107912. [Google Scholar] [CrossRef]
- Pang, Z.; Wang, Y.L.; Mantri, N.; Wang, Y.; Hua, X.J.; Quan, Y.P.; Zhou, X.; Jiang, Z.D.; Qi, Z.C.; Lu, H.F. Molecular phylogenetic relationships and taxonomy position of 161 Camellia species in China. Taiwania 2022, 67, 560–570. [Google Scholar]
- Wu, Q.; Tong, W.; Zhao, H.; Ge, R.; Li, R.; Huang, J.; Li, F.; Wang, Y.; Mallano, A.I.; Deng, W.; et al. Comparative transcriptomic analysis unveils the deep phylogeny and secondary metabolite evolution of 116 Camellia plants. Plant J. 2022, 111, 406–421. [Google Scholar] [CrossRef]
- Zhu, Z.J.; Qin, X.M.; Li, P.W.; Lu, Y.B.; Mo, X.Y.; Fang, Y.; Zhang, Q. Characterize the Complete Mitogenome of Semiaquilegia guangxiensis and Assess the Efficiency of the Mitochondrial Genes in Ranunculales Phylogeny. Ecol. Evol. 2025, 15, e71165. [Google Scholar] [CrossRef]
- Liang, H.; Qi, H.; Wang, C.; Wang, Y.; Liu, M.; Chen, J.; Sun, X.; Xia, T.; Feng, S.; Chen, C.; et al. Analysis of the complete mitogenomes of three high economic value tea plants (Tea-oil Camellia) provide insights into evolution and phylogeny relationship. Front. Plant Sci. 2025, 16, 1549185. [Google Scholar] [CrossRef] [PubMed]
- Zan, T.; He, Y.T.; Zhang, M.; Yonezawa, T.; Ma, H.; Zhao, Q.M.; Kuo, W.Y.; Zhang, W.J.; Huang, C.H. Phylogenomic analyses of Camellia support reticulate evolution among major clades. Mol. Phylogenet. Evol. 2023, 182, 107744. [Google Scholar] [CrossRef] [PubMed]
- Qu, X.J.; Zhang, X.J.; Cao, D.L.; Guo, X.X.; Mower, J.P.; Fan, S.J. Plastid and mitochondrial phylogenomics reveal correlated substitution rate variation in Koenigia (Polygonoideae, Polygonaceae) and a reduced plastome for Koenigia delicatula including loss of all ndh genes. Mol. Phylogenet. Evol. 2022, 174, 107544. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).