Abstract
During forest vegetation rehabilitation, changes in aboveground litter and underground root inputs affect soil microbial communities. Clarifying the effects of forest ecosystem carbon inputs on soil microbial community structure can provide a theoretical basis for the microbial driving mechanism of soil fertility evolution and ecosystem rehabilitation of vegetation rehabilitation in degraded red soil. Our research focuses on a Schima superba pure forest recovered from eroded and degraded red soil in a subtropical region. Five treatments were set as follows: control treatment (CT), mycorrhiza (M), root + mycorrhiza (RM), litter + root + mycorrhiza (LRM), and double litter + root + mycorrhiza (DLRM). We used Illumina HiSeq technology to study the effects of different organic carbon inputs on soil microbial community structure. The results showed that all organic carbon input treatments reduced the total population of soil bacteria by 55–79%; M, RM, and DLRM treatments increased the quantity of operational taxonomic units (OTUs) by 25–37%, ACE index by 25–34%, and Chao1 index by 28–39%. Acidobacteria, Proteobacteria, and Actinobacteria were the dominant bacteriophyta in the Schima superba pure forest soil. The relative abundance of Alphaproteobacteria decreased by 55% under LRM treatment, and Thermoleophilia increased by 81% under M treatment. The dominant fungal phyla were Basidiomycota and Ascomycota. RM, LRM, and DLRM treatments reduced the relative abundance of Sordariomycetes by 46–64% and increased the relative abundance of Mortierellomycetes by 251–615%. The order of effects of different organic carbon inputs on the bacterial community composition at the phylum level was LRM > RM > M > DLRM and that on the fungal community composition was DLRM > LRM and RM > M. Alphaproteobacteria, Thermoleophilia, Sordariomycetes, and Mortierellomycetes were the main microbial groups affected by changes in organic carbon inputs. Soil organic carbon and total nitrogen were the key factors affecting the change of Mortierellomycetes. The bacterial community mainly affected the activity of soil acid invertase, while the fungal community affected the activities of various enzymes, with positive or negative effects. We concluded that the organic carbon inputs changed the species and quantity of soil microorganisms in the Schima superba forest, and the influence of organic carbon input on the fungal community structure was greater than that of bacteria.
1. Introduction
Soil microorganisms are one of the essential components of soil [1]. Microorganisms regulate the dynamic balance of various biochemical and physiological processes in soil, participate in the process of material transformation and energy flow, and play a crucial role in maintaining soil ecological function and stability [2,3]. Microbial community structure is the main factor driving ecosystem processes, and soil microorganisms can adapt to changes in the soil environment through changes in their community composition [4,5]. Therefore, microorganisms are crucial to examine better and are indicators of ecosystem productivity and soil quality [6,7].
Self-fertilization—litter, fine root, and mycorrhiza—supplements the organic carbon in forestland soils, which differs from the artificial addition of organic matter that agricultural soils experience. Litter input is the primary way for plants to return soil nutrients and is also considered an essential substrate for soil microbial metabolism in forest ecosystems [8]. Soil microbial communities mediate the decomposition process of litter, which improves the availability of soil carbon and nitrogen nutrients and causes changes in the microbial community structure [9]. Compared with boreal temperate forests, soil microorganisms in tropical and subtropical forests are more sensitive to changes in litter input, which is related to litter residence time [10]. Litter quantity is one of the main factors controlling soil microbial community structure [11], but there is still controversy about the effects of litter addition and removal. Lu et al. [12] showed that the contents of soil bacteria and fungi increased with litter quantity. Li et al. [13] showed that adding litter significantly increased fungi content and reduced Gram-positive bacteria content in the incubation experiment, and removing litter reduced fungal biomass [14]. However, Sayer [15] showed that the addition of litter did not cause a corresponding increase in the abundance of fungi. These results may be affected by by factors such as the chemical quality of litter, the length of test period, zonal differences, and having a certain complexity, so more field test data are needed to support or verify these results.
The input of underground roots facilitates the distribution of photosynthates from aboveground to underground. Root input also releases root exudates such as polysaccharides, organic acids, and amino acids into the soil, thus affecting the microbial activity and biomass [16]. Bluhm et al. [8] studied the response of soil microorganisms by cutting off the input of root-derived resources and found that microorganisms not only depended on the decomposition resources of litter but also largely depended on the root-derived resources, indicating that changes in underground root input are also one of the important factors affecting soil microbial community structure. Mycorrhiza is a symbiont of some fungi in soil and plant roots. Mycorrhizal fungi—such as arbuscular mycorrhizal fungi—can interact with soil microbial communities, producing a variety of exudates (e.g., glomalin, soil-related proteins and polysaccharides), affecting the decomposition of organic matter, and changing the composition of bacterial and fungal communities [17,18]. In relatively poor soil with low carbon input of litter, the effect of mycorrhizal fungi on plants is more prominent, and plants can regulate the mycorrhizal fungi community by secreting effective carbon sources from roots [19]. However, the impact of mycorrhiza input on microbial community structure is often rarely considered.
Studies on detritus input and removal treatment (DIRT) mainly focus on the carbon cycle-related indicators such as soil respiration and soil organic carbon, and most of them only focus on the single impact of litter or root input on organic carbon. However, the comprehensive effects of three different input carbon sources of litter, root, and mycorrhiza are still unclear. More specifically, the relationships between the three carbon inputs and soil microbial community structure after vegetation rehabilitation in degraded red soil is rarely examined. In the past, the importance of microorganisms was underestimated because many microbial species were undiscovered or unculturable [20]. In recent years, with the diversification of analysis methods, the research on soil microorganisms has become more accurate. For example, high-throughput sequencing technology can truly reveal the species composition and diversity of microbial communities, but it is relatively less applied in DIRT-related research. In addition, although soil enzyme activities can represent microbial activities to some extent, little is known about which microbial groups affect which soil enzyme activities.
Schima superba is one of the most widely distributed and dominant tree species in subtropical evergreen broad-leaved forests and has universal applicability and a typical role in ecological rehabilitation in the red soil region of China. Therefore, our research object is the Schima superba forest rehabilitated from the eroded and degraded red soil in the subtropical region of China. Illumina HiSeqc sequencing technology allows us to study the changes in soil microbial community structure and alpha diversity under different organic carbon input treatments (Control treatment (CT), mycorrhiza (M), root + mycorrhiza (RM), litter + root + mycorrhiza (LRM), and double litter + root + mycorrhiza (DLRM)). We investigated the relationships between microbial community, soil chemical properties, and enzyme activity. The aims of this study were to: (1) reveal the response of soil bacterial and fungal community structure to the changes in organic carbon input; (2) determine the critical soil factors affecting the soil microbial community structure of the artificial rehabilitated forest; and (3) identify the significant microbial communities closely related to soil specific enzyme activities. At the same time, we put forward two hypotheses: (1) organic carbon input (litter, root, and/or mycorrhiza) will increase the population of soil microorganisms compared with the treatment without organic carbon input, and (2) in the low pH environment of red soil, organic carbon input may make acid-resistant fungi more sensitive, so the influence of organic carbon input on the fungal community structure is greater than that of bacteria. This study has important significance for understanding and recognizing soil quality improvement and the maintenance of ecological function in subtropical forests.
2. Materials and Methods
2.1. Site Description
Our research was conducted in the Luoxi township (26°44′ N, 115°04′ E), Taihe County, Jiangxi Province, China. This region has an average altitude of 80 m and a subtropical humid monsoon climate—an 18.6 °C average annual temperature and 1726 mm average annual precipitation [21]. The original evergreen broad-leaved forest was transformed into grassland in the 1960s due to long-term degradation—woodcutting and stump digging—and grows upon red soil developed from quaternary red clay [21]. In 1991, the Jiangxi Agricultural University and Taihe Forestry Bureau selected coniferous and broad-leaved tree species such as Pinus elliottii, Pinus massoniana, Liquidambar fomosana, and Schima superba for forest rehabilitation. The vegetation recovery area in this region reached 133 ha, forming a typical forest landscape in the hilly region of southern China.
2.2. Experimental Design and Sample Collection
In December 2018, we randomly selected three independent Schima superba pure forests planted 27 years ago, and we set a standard plot of 20 m × 20 m in each forest stand. We used a random block method for our experimental design. Five treatments were established in each standard plot (see Table 1, Appendix A Figure A1 for details). (1) CT: trenches of 15 cm wide and 50 cm deep were dug around the treatment plot, and plasterboards were used to isolate the plot from the surrounding soil to prevent roots and hyphae from growing into the plot. After the aboveground litter was removed from the soil surface, a rectangular nylon mesh frame of 1 m × 0.5 m × 0.6 m with a pore diameter of 1 mm (no nylon mesh at the bottom) was placed to avoid external litter entry. (2) M: the ditches around the treatment plot were 50 cm deep, and the roots were isolated by nylon mesh with a pore diameter of 37 μm, allowing hyphae to enter the plot. After the aboveground litter was removed from the soil surface, a rectangular nylon mesh frame of 1 m × 0.5 m × 0.6 m with a pore diameter of 1 mm (no nylon mesh) at the bottom was placed to avoid external litter entry. (3) RM: after the aboveground litter was removed from the soil surface, a rectangular nylon mesh frame of 1 m × 0.5 m × 0.6 m with a pore diameter of 1 mm (no nylon mesh at the bottom) was placed to block the aboveground litter, and the aboveground litter was collected monthly. (4) LRM: undisturbed soil, not treated. (5) DLRM: a rectangular nylon mesh frame of 1 m × 0.5 m × 0.6 m with a pore diameter of 1 mm and no nylon mesh at the top and bottom was placed, and the aboveground litter collected from the nylon mesh frame in RM treatment was placed evenly in this treatment every month. Each treatment was replicated five times, with each treatment area being 0.5 × 1 m2. According to our examination, Schima superba forests have a stem density of 2175 stems ha−1, canopy coverage of 94%, mean tree height of 8.31 m, and mean tree diameter at breast height of 11.03 cm.

Table 1.
List of abbreviations.
In August 2020, we used a small soil shovel to collect 0–10 cm topsoil from each treatment in each standard plot. Five samples from the same treatment in the same standard plot were combined to form one composite sample. We took 15 soil samples from 3 standard plots to the laboratory in an incubator on ice. After removing plant residues, gravel, and other sundries, we divided each soil sample into two parts by the quartering method. We put one part through a 2 mm sieve, freeze-dried it, and stored it in a freezer at −20 °C to determine the microbial community structure. The other was dried naturally, ground, and screened to determine soil chemical properties and enzyme activities.
2.3. Determination of Soil Chemical Properties and Enzyme Activities
We determined soil pH in the mixed solution with a soil/water ratio of 1:2.5 (w/v). We used the dichromate oxidation and Kjeldahl methods to measure soil organic carbon (SOC) and total nitrogen (TN), respectively [22]. The ammonium fluoride–hydrochloric acid extraction method was used to determine available phosphorus (AP), and we used the ammonium acetate extraction method to determine available potassium (AK) [22]. We determined soil sucrase (SC), soil urease (UE), soil acid phosphatase (ACP), soil acid invertase (AI), and soil polyphenol oxidase (PPO) activities using a soil sucrase kit (BC0240), soil urease kit (BC0120), soil acid phosphatase kit (BC0140), soil acid invertase kit (BC3075), and soil polyphenol oxidase kit (BC0110) from Solarbio Science & Technology Co. (Beijing, China), in U/g [23,24].
2.4. Soil DNA Extraction, PCR Amplification, and Sequence Data Analysis
We extracted soil DNA using a metagenomic DNA Extraction Kit (GENErary) and detected DNA integrity by 1% agarose gel electrophoresis. We used a real-time fluorescence quantitative method for PCR amplification of the V3+V4 region of bacteria 16S rRNA and the ITS1 region of fungi. The bacterial primers were 338f (ACTCCTACGGGAGGCAGCAG) and 806r (GGACTACHVGGGTWTCTAAT) [25]. The fungal primers were ITS1F (CTTGGTCATTTAGAGGAAGTAA) and ITS2 (GCTGCGTTCTTCATCGATGC) [25]. Illumina HiSeq 2500 sequencing platform and paired-end (PE) sequencing method in Beijing Baimaike Biotechnology Co., Ltd. (Beijing, China) carried out the DNA extraction, PCR amplification, and library construction.
Trimmomatic V0.33 software filtered the raw reads obtained by sequencing. Then, Cutadapt 1.9.1 software identified and removed primer sequences, and we obtained high-quality reads without primer sequences. We acquired clean reads by using Flash V1.2.7 software overlap to splice high-quality reads of samples. We then identified and removed the chimeric sequences using Uchime V4.2 software to obtain effective reads. Usearch software clustered reads at a similarity level of 97%, which obtained operational taxonomic units (OTUs). We used alpha index analysis software QIIME2 to calculate species richness index (ACE and Chao1) and species diversity index (Simpson and Shannon), and we calculated the relative abundance of bacteria and fungi at different classification levels.
2.5. Statistical Analysis
A one-way ANOVA analyzed the effects of different treatments on soil properties, microbial population, and alpha diversity index. We used Duncan’s method to detect the differences among treatments, and we set the significance level to p = 0.05. We log10 transformed the OTUs of soil bacteria and fungi for normalization. We drew Venn diagrams of soil bacteria and fungi OTUs under different treatments using R 4.0.5 software and Origin 2018 software (OriginLab, Northampton, MA, USA) to draw relative abundance diagrams of dominant bacteria and fungi species at the phylum level. We used Canoco 5 software (Microcomputer Power, Ithaca, NY, USA) to draw the redundancy analysis (RDA) diagram of soil bacterial and fungal communities of bacteria and fungi class levels with a greater than 1% relative abundance. We used Spearman correlation analysis to examine the relationships among soil bacterial and fungal communities, alpha diversity, soil chemical properties, and enzyme activities—all analyses were conducted using SPSS 19.0 (IBM, Armonk, NY, USA).
3. Results
3.1. Effects of Different Treatments on Soil Chemical Properties and Enzyme Activities
Compared with the CT treatment, the DLRM treatment increased the SOC and TN contents by 52% and 30% (p < 0.05), respectively. At the same time, there were no significant differences in soil pH value, AP, and AK contents among the different treatments (p > 0.05, Table 2). RM, LRM, and DLRM reduced the activities of soil PPO by 34%, 49%, and 62% (p < 0.05), respectively, while there were no significant differences in soil SC, UE, and ACP activities among different treatments (p > 0.05).

Table 2.
Soil chemical properties and enzymatic activity of different treatments.
3.2. Effects of Different Treatments on Soil Microbial OTUs and Diversity
The coverage of both bacterial and fungal libraries was above 99.5%, indicating that the sequencing data could represent the actual situation of soil microbial communities under different treatments. However, a small number of microbial species were still undiscovered. We detected 412,446 bacterial gene sequences and 424,230 fungal gene sequences in 15 soil samples from 5 treatments, including 1109 bacterial OTUs and 1004 fungal OTUs. Compared with CT treatment, M, RM, LRM, and DLRM treatments reduced the total population of soil bacteria by 55%, 79%, 67%, and 56% (p < 0.05), respectively. At the same time, there was no significant difference in the population of soil fungi among treatments (p > 0.05, Table 3). The effect of different treatments on fungal OTUs was greater than that of bacteria OTUs, and the M, RM, and DLRM treatments increased fungal OTUs by 37%, 32%, and 25% (p < 0.05), respectively. There were 781 bacteria OTUs and 415 fungi OTUs in common among the 5 treatments (Figure 1). We found 12 unique bacteria OTUs and 18 unique fungi OTUs under the CT treatment, 42 unique bacteria OTUs and 9 unique fungi OTUs under the M treatment, 13 unique bacteria OTUs and 51 unique fungi OTUs under the RM treatment, 5 unique bacteria OTUs and 21 unique fungi OTUs under the LRM treatment, and 16 unique bacteria OTUs and 25 unique fungi OTUs under the DLRM treatment.

Table 3.
Soil microbial population, OTUs, and alpha diversity in different treatments.
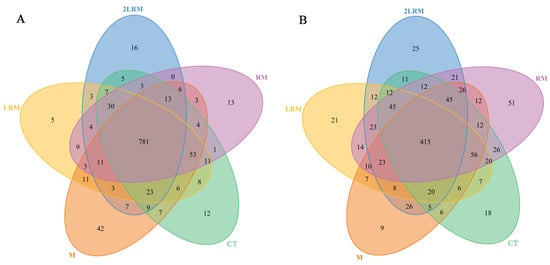
Figure 1.
Venn diagram of soil bacteria (A) and fungi (B) in different treatments.
Compared with the CT treatment, the LRM treatment reduced the Shannon diversity index of soil bacteria by 8% (p < 0.05), while the richness index (ACE and Chao1) and Simpson index showed no significant difference among all treatments (p > 0.05, Table 3). The M, RM, and DLRM treatments significantly increased the soil fungal richness index—the ACE index increased by 34%, 29%, and 25%, and the Chao1 index increased by 39%, 32%, and 28% (p < 0.05), respectively. The M treatment increased the Shannon diversity index of soil fungi by 30% (p < 0.05), while there were no significant differences in the Simpson index among all treatments (p > 0.05). Therefore, the effect of different treatments on the fungal richness index was greater than that of bacteria.
3.3. Effects of Different Treatments on Soil Microbial Community Composition and Structure
We detected 22 bacterial phyla in the soil samples of the 5 treatments. The top ten bacteria phyla in relative abundance were Acidobacteria (35%), Proteobacteria (30%), Actinobacteria (17%), WPS−2 (4.6%), Chloroflexi (3.4%), Verrucomicrobia (3.2%), Planctomycetes (Planctomycetes, 2.3%), Patescibacteria (1.8%), Bacteroidetes (0.72%), and Gemmatimonadetes (0.54%), as shown in Figure 2A. Among them, the dominant bacteria phyla were Acidobacteria, Proteobacteria, and Actinobacteria, which presented a relative abundance of 82% in total. Compared with the CT treatment, the LRM treatment reduced the relative abundance of Proteobacteria by 43% (p < 0.05). The dominant bacteria classes were Acidobacteriia (35%), Alphaproteobacteria (22%), Actinobacteria (6.8%), Gammaproteobacteria (6.7%), and Thermoleophilia (5.9%), accounting for 76% of the total relative abundance (Table 4). The LRM treatment reduced the relative abundance of Alphaproteobacteria by 55% (p < 0.05), while the M treatment increased the relative abundance of Thermoleophilia by 81% (p < 0.05).
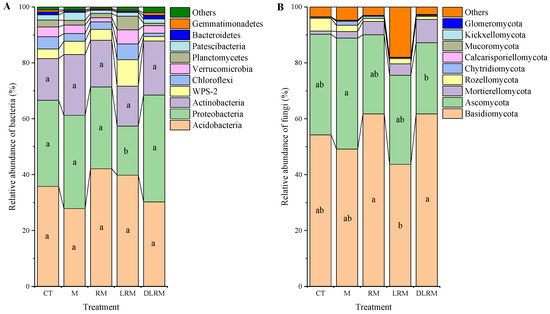
Figure 2.
Relative abundance of major soil bacteria (A) and fungi (B) at phylum level in different treatments. Different lowercase letters indicate significant difference among treatments (p < 0.05).

Table 4.
Relative abundance of major soil bacteria and fungi at class level in different treatments (%).
For fungi, we detected the nine phyla shown in Figure 2B—Basidiomycota (54%), Ascomycota (32%), Mortierellomycota (4.2%), Rozellomycota (2.1%), Chytridiomycot (0.68%), Calcarisporiellomycota (0.15%), Mucoromycota (0.07%), Kickxellomycota (0.04%), and Glomeromycota (0.01%). Basidiomycota and Ascomycota were the dominant fungi phyla, accounting for 86% of the total relative abundance. Compared with the CT treatment, the relative abundance of Basidiomycota and Ascomycota in other treatments did not change significantly. The dominant fungi classes were Agaricomycetes (47%), Sordariomycetes (10%), and Leotiomycetes (9.7%), which presented a relative abundance of 67% in total (Table 4). RM, LRM, and 2LRM treatments decreased the relative abundance of Sordariomycetes (46–64%, p < 0.05) but increased the relative abundance of Mortierellomycetes (251–615%, p < 0.05).
3.4. Effects of Soil Properties on Microbial Community Composition
RDA analysis based on microbial OTUs at the class level and soil chemical properties showed that soil chemical properties had a specific impact on microbial community structure. For bacterial communities, the first and second axes explained 50% and 12% of variation, respectively, and the two axes jointly explained 62% of variation (Figure 3A). Uncultured_bacterium_p_wps−2 and Planctomycetacia were positively correlated with soil AI, while Alphaproteobacteria, Thermoleophilia, and Deltaproteobacteria were negatively correlated with soil AI (Table 5). The first and second axes explained 31% and 19% of the variation for soil fungal communities, respectively. The two axes jointly explained 50% of the variation (Figure 3B). Sordariomycetes and Leotiomycetes were positively correlated with soil PPO. Mortierellomycetes were positively correlated with soil SOC, TN, and SC but negatively correlated with soil PPO. Dothideomycetes were negatively correlated with soil AI. Rozellomycotina_cls_Incertae_sedis was negatively correlated with soil UE (Table 5). In addition, among bacteria classes, Ktedonobacteria was positively correlated with the species richness index, while Alphaproteobacteria, Actinobacteria, Acidimicrobiia, and Saccharimonadia were positively correlated with the species diversity index. Among fungi classes, Sordariomycetes, Eurotiomycetes, and Dothideomycetes were also positively correlated with the species diversity index (Table 5).
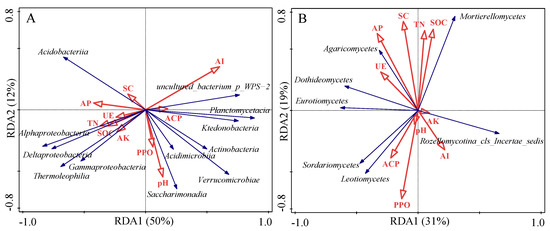
Figure 3.
Redundancy analysis (RDA) of soil bacteria (A) and fungi (B) at class level and soil chemical properties and enzymatic activity. pH, soil potential of hydrogen potential of hydrogen; SOC, soil organic carbon; TN, total nitrogen; AP, available phosphorus; AK, available potassium; SC, sucrase; UE, urease; ACP, acid phosphatase; AI, acid invertase; PPO, polyphenol oxidase.

Table 5.
Relationships between microbial community at class level and alpha diversity, soil chemical properties, and enzymatic activity.
4. Discussion
In our study, the comparison of RM, LRM, and DLRM treatments showed that the population of soil bacteria and fungi increased with litter input but did not reach a statistically significant level (Table 3). Nadelhoffer et al. [26] also observed no change in bacterial or fungal biomass in the soil of a deciduous site after the addition of aboveground litter. However, Lu et al. [12] found that litter addition increased soil bacterial and fungal contents, and litter could significantly change the soil microbial community through incubation experiments. Notably, unlike the study by Lu et al. [12], which added Chinese fir litter after crushing, the decomposition of Schima superba litter may be slower under natural conditions. Our study also found that compared with the CT treatment, each organic carbon input treatment greatly reduced the soil bacteria population but increased the soil fungi population, although the increase was not statistically significant, which was inconsistent with our first hypothesis. Bacteria have long been thought to benefit from labile carbon input. However, recent studies have shown that fungi also benefit from labile carbon inputs and may utilize them more quickly than bacteria [27,28]. Conversely, removing litter, roots, and mycorrhiza means a lack of labile organic carbon input and leads to carbon limitation. Carbon constraint increases physiological stress on microbial communities, with bacteria and fungi competing for similar resources [14]. Elser [29] believed that microbes change their substances and community structure to cope with environmental changes when there is an insufficient carbon source supply. Trenching causes bacteria to experience extraordinary stress [8], so bacteria strengthen the use of alternative resources by increasing their population. Another reason may be related to the soil water content since the addition of litter impedes the infiltration of rainwater. Roots also increase the absorption of water, resulting in a decline in soil moisture, which in turn affects microbial activity [16,30]. Of course, this needs to be verified because of the lack of soil moisture monitoring. Brant et al. [14] showed that rootless plots usually contain larger quantities of actinomycete biomarkers and lower amounts of fungal biomarkers. In our study, the isolation of roots also reduced the population of soil fungi. However, it did not reach a statistically significant level, which may be related to the duration of the treatments. We waited less than 2 years from the establishment of treatments to sampling. The treatments had existed for 5 years in the study by Brant et al. [14], and the change in soil microbial community was relatively noticeable.
Soil microbial community structure is vital in maintaining forest ecosystem structure and function. Many studies have documented that the aboveground and underground carbon input changes can affect the composition of soil microbial communities [31,32]. In our study, the proportion of OTUs shared by bacteria and fungi in different treatments was 70% and 41%, respectively (Figure 1). A considerable proportion of unique microbial OTUs (especially fungi) indicated that carbon input changed the species and quantity of soil microorganisms in Schima superba pure forest, and different input modes had different effects. Wan et al. [33] studied the soil microbial communities of two subtropical plantations (Cunninghamia lanceolata and Mytilaria laosensis), and the results showed that the change of carbon input from aboveground litter had a greater impact on soil microbial communities than that from underground roots. Brant et al. [14] and Wang et al. [32] found that root-derived carbon had a greater impact on soil microbial community composition in temperate and subtropical coniferous forests through DIRT experiments. Unfortunately, we did not consider the treatment with root input only in the design. Instead, we considered the combined effect of mycorrhizal input, both root and mycorrhizal input, and simultaneous input with litter. We found that the order of effects of different organic carbon inputs on the bacterial community structure at the phylum level was LRM > RM > M > DLRM, and that the order of effects for the fungal community structure was DLRM > LRM and RM > M (Figure 2; Appendix A Table A1). Among the main microbial communities at the phylum and class levels studied, different organic carbon inputs significantly affected two bacterial phyla, seven bacterial classes, five fungal phyla, and eight fungal classes (Figure 2; Table 4). Meanwhile, the inputs of mycorrhiza, root + mycorrhiza, and double litter + root + mycorrhiza significantly increased the soil fungi’s richness index (Chao1and ACE) and OTUs. In contrast, the richness index and OTUs of soil bacteria were not statistically significant among treatments (Table 3). These results indicated that different organic carbon inputs had a greater impact on the fungal community than the bacterial community. Thus, this supports our second hypothesis. The reason may be that most bacteria are more suitable for growth in neutral or alkaline environments, and the acidic conditions of red soil restrict the growth and development of soil bacteria. At the same time, acid-resistant fungi that can grow normally in acidic soil can replace bacteria to decompose organic matter. As a result, fungi are more sensitive to organic carbon inputs. Liu et al. [34] also showed that soil pH was the main factor affecting the composition and diversity of the microbial community. For example, pH influences the form of soil compounds, thus affecting the absorption of nutrients by microorganisms. The different responses of bacteria and fungi to different inputs of organic carbon indicate that they occupy different ecological niches. Bacteria are mainly affected by soil nutrients and root exudates (such as organic acids, amino acids, and polysaccharides), while fungi obtain nutrients mainly through decomposition of aboveground and underground litter such as fallen branches, leaves, and root cortex tissues [35].
In addition, litter significantly affects the microbial community structure through the availability of nutrients produced by its different chemical components and the unique soil microenvironment the litter creates [36,37]. The relative abundance of microbial communities may be altered by changes in resource quality due to differences in aboveground and belowground litter quality, with potentially important effects on the metabolic functions of microbial communities. Meanwhile, the production of aboveground litter and the growth of underground roots have strong seasonality. However, this study only discussed the changes in microbial communities in a single sampling after adding and removing litter, roots, and mycorrhiza in Schima superba forests. Moreover, this study used plasterboards to isolate roots and mycorrhiza, potentially blocking the lateral migration process of soil water and nutrients. The limitations of this method might have an impact on the soil microbial community. Therefore, one μm nylon mesh should be considered in a follow-up study, which can prevent the growth of roots and hyphae and ensure the migration of water and air. At the same time, considering the effects of different organic carbon input treatments on soil temperature, moisture, soil respiration, and carbon storage, the dynamics of soil microorganisms in different vegetation rehabilitation types of forestlands can be carried out to explore further the evolution of soil fertility and ecological function in eroded and degraded forestland.
The dominant bacterial phyla in the soil of the Schima superba forest were Acidobacteria, Proteobacteria, and Actinobacteria, among which the relative abundance of Acidobacteria was the highest (Figure 2A), which was consistent with the results of Sang et al. [38] in subtropical forests. Acidobacteria are acidophilic and oligotrophic, can be distributed in various habitats, and are comparable in quantity to Proteobacteria. The bacteria in this phylum can degrade plant residue polymers, participate in the iron cycle, have photosynthetic capacity, and participate in the metabolism of single-carbon compounds [39,40,41,42,43]. Low pH environments can promote the abundance of Acidobacteria [35]. Sun et al. [44] found that the top three dominant bacterial groups in the relative abundance of boreal forest soil were also the bacterial phyla mentioned above. However, the relative abundance of Proteobacteria was higher than that of Acidobacteria, which was related to the high soil nutrient content. Proteobacteria are the most abundant phylum among bacteria, most of which are facultative or obligate anaerobic microorganisms, and many groups of this phylum can fix nitrogen in soil [25]. Previous studies have shown that the relative abundance of Proteobacteria increases with the increase of organic matter and is higher in nutrient-rich soils [45,46]. However, in our study, the LRM treatment reduced the relative abundance of soil Alphaproteobacteria (the main class of Proteobacteria), mainly because organic carbon input was more conducive to the growth and utilization of fungi. Different treatments increased the relative abundance of related fungal phyla (Figure 2; Table A1). In the bacterial classes with high relative abundance, the M treatment significantly increased the relative abundance of Thermoleophilia (Table 4). Thermoleophilia belongs to Actinomycetes and copiotrophic microbiological groups, which are more inclined to use labile carbon [47,48]. Mycelium in mycorrhiza can produce a variety of exudates [17], providing more labile carbon sources, which are beneficial to the growth of microorganisms.
The dominant fungi phyla of Schima superba were Basidiomycota and Ascomycota (Figure 2B), which was similar to the results of Tedersoo et al. [49] on soil fungi in terrestrial ecosystems. Basidiomycota are the most common type of fungi, which are widely distributed, diverse and abundant, and are positively correlated with highly stable organic components such as cellulose and lignin [50]. Schima superba is an arbuscular mycorrhizal species whose mycorrhiza cannot decompose organic matter. Schima superba can only stimulate the decomposition of saprophytic fungi by providing photosynthate to saprophytic fungi [51]. Ascomycetes are mostly saprophytic fungi, which can decompose refractory organic matter [52]. The RM, LRM, and 2LRM treatments significantly reduced the relative abundance of Sordariomycetes and increased the relative abundance of Mortierellomycetes (Table 4), which benefited from the increase of carbon input and supported the growth of many microorganisms. The leaching and decomposition of fresh litter releases a large amount of labile carbon, providing many carbon sources for microorganisms [51]. Since Mortierellomycota has high utilization efficiency for simple carbohydrates and is positively correlated with the content of active carbon components in soil [53], the relative abundance of Mortierellomycetes is improved.
Soil microbial community composition is directly and indirectly affected by soil properties. RDA analysis results in this study showed that soil pH, SOC, TN, AP, and AK contents and enzyme activities explained 62% and 50% of the variation in soil bacterial and fungal communities, respectively (Figure 3). Our results indicate that in addition to the above soil properties, the variation in soil microbial community composition was also affected by other environmental factors, e.g., temperature, soil pores, texture, aggregates, protozoa [54,55,56]. Liu et al. [57] found that Actinobacteria and Deltaproteobacteria were significantly correlated with SOC. However, similar results were not observed in this study, which is consistent with the results of Zhong et al. [58] and Zeng et al. [59]. We also found that other soil chemical properties had no significant effect on soil bacterial classes (Table 5), indicating that bacteria in this area were less sensitive to these soil chemical properties. Different from bacterial classes, Mortierellomycetes were significantly positively correlated with SOC and TN, indicating that SOC and TN were the key soil factors affecting the change in Mortierellomycetes.
Soil enzymes, mainly produced by microorganisms, plant roots, and animals, participate in the synthesis or decomposition of soil organic matter and other nutrients. Enzymes directly or indirectly affect soil biochemical reactions with soil bacteria and fungi. In this study, RM, LRM, and DLRM treatments all reduced the activity of soil polyphenol oxidase, mainly because the inputs of litter, roots, and mycorrhiza increased available carbon sources. Microorganisms did not need to secrete more polyphenol oxidase to promote the release of nutrient elements from lignin, phenols, and other refractory components of organic matter. Here we also illuminated which microbial groups affect soil enzyme activity: uncultured_bacterium_p_WPS−2 and Planctomycetacia, Alphaproteobacteria, Thermoleophilia, and Deltaproteobacteria affected soil acid sucrase activity in positive or negative directions. Mortierellomycetes affected the activities of soil sucrase and polyphenol oxidase. Mortierellomycetes was positively correlated with soil sucrase activity, indicating that the existence of Mortierellomycetes, a fungus group, promoted the hydrolysis of sucrose into monosaccharides and increased soil carbon availability, thus contributing to the direct uptake and utilization of organisms. In contrast, the presence of Rozellomycotina_cls_Incertae_sedis inhibited soil urease activity and reduced soil nitrogen availability. In addition, the correlation between soil microorganisms and alpha diversity showed that there were more microbial species in some classes of bacteria and fungi (such as Actinobacteria, Ktedonobacteria, and Sordariomycetes) which caused a significant increase in species richness or diversity indices.
5. Conclusions
The main bacterial groups affected by changes of organic carbon inputs were Alphaproteobacteria and Thermoleophilia, while the main fungal groups affected were Sordariomycetes and Mortierellomycetes, and the fungal community was more sensitive to carbon input than the bacterial community. Organic carbon input significantly reduced the population of soil bacteria. The inputs of mycorrhiza, root + mycorrhiza, and double litter + root + mycorrhiza significantly increased soil fungi’s OTUs and richness index. Nevertheless, the bacteria population was much larger than the fungi population under different treatments, indicating that organic carbon input did not change the dominance of bacteria in this region. The order of effects of different organic carbon inputs on the bacterial community composition at the phylum level was LRM > RM > M > DLRM, and that on the fungal community composition was DLRM > LRM and RM > M. Whether the effects of carbon inputs on soil microbial community composition change with the increase of treatment time needs to be further demonstrated by long-term experimental field data. Soil organic carbon and total nitrogen were the key soil factors affecting the change of Mortierellomycetes, indicating that organic carbon input could affect microbial communities by changing soil chemical properties. The bacterial community mainly affected the activity of soil acid invertase, while the fungal community affected the activities of various enzymes. Here, we identified the significant microbial groups closely related to soil specific enzyme activities. These findings will help to further understand the important role of microorganisms in soil quality improvement and ecosystem rehabilitation.
Author Contributions
Conceptualization and methodology, R.H.; formal analysis and investigation, L.Z. and G.H.; data collection and curation, J.W., H.G., L.L. and M.Y.; writing—original draft preparation, L.Z.; writing—review and editing, R.H. and J.W.; funding acquisition, L.Z., G.H. and R.H.; resources, Y.L. and X.Z.; supervision, R.H. All authors have read and agreed to the published version of the manuscript.
Funding
The Science and Technology Research Project of Education Department of Jiangxi Province, China (Nos. GJJ190974, GJJ190945), the Natural Science Foundation of Jiangxi Province, China (No. 20202BABL213045), and the National Natural Science Foundation of China (No. 31660192).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets used or analyzed during the current study are available from the corresponding author on reasonable request.
Acknowledgments
We would like to thank Hongyu Wan and Haoze Wu for help in the field.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A
Figure A1.
Locations of the plots and treatments of Schima superba forests. CT, control treatment; M, mycorrhiza; RM, root + mycorrhiza; LRM, litter + root + mycorrhiza; DLRM, double litter + root + mycorrhiza.

Table A1.
Relative abundance of major soil bacteria and fungi at class level in different treatments (%).
Table A1.
Relative abundance of major soil bacteria and fungi at class level in different treatments (%).
| Type | Phylum | CT | M | RM | LRM | DLRM |
|---|---|---|---|---|---|---|
| Bacteria | Acidobacteria | 36 a | 28 a | 42 a | 40 a | 30 a |
| Proteobacteria | 31 a | 33 a | 29 a | 17.5 b | 38 a | |
| Actinobacteria | 15 a | 22 a | 17 a | 14 a | 19 a | |
| WPS−2 | 3.4 a | 4.7 a | 3.9 a | 9.5 a | 1.6 a | |
| Chloroflexi | 4.4 a | 2.8 a | 2.7 a | 5.7 a | 1.2 a | |
| Verrucomicrobia | 3.5 a | 3.1 a | 1.5 a | 5.0 a | 2.7 a | |
| Planctomycetes | 2.4 a | 1.7 a | 1.5 a | 4.9 a | 0.88 a | |
| Patescibacteria | 1.9 ab | 2.8 a | 1.2 b | 1.4 b | 1.5 b | |
| Bacteroidetes | 1.1 a | 0.55 b | 0.28 b | 0.41 b | 1.3 a | |
| Gemmatimonadetes | 0.70 ab | 0.35 b | 0.40 b | 0.33 b | 0.94 a | |
| Fungi | Basidiomycota | 54 ab | 49 ab | 62 a | 44 b | 62 a |
| Ascomycota | 36 ab | 40 a | 28 ab | 32 ab | 25 b | |
| Mortierellomycota | 1.2 d | 2.4 cd | 4.7 b | 4.1 bc | 8.4 a | |
| Rozellomycota | 4.5 a | 2.2 a | 1.0 a | 1.8 a | 1.0 a | |
| Chytridiomycota | 0.40 c | 1.4 a | 0.87 b | 0.35 c | 0.36 c | |
| Calcarisporiellomycota | 0.09 b | 0.26 a | 0.10 b | 0.06 b | 0.25 a | |
| Mucoromycota | 0.05 a | 0.05 a | 0.06 a | 0.11 a | 0.07 a | |
| Kickxellomycota | 0.01 b | 0.03 ab | 0.08 a | 0.01 b | 0.05 ab | |
| Glomeromycota | 0.004 b | 0.03 a | 0.005 b | 0.004 b | 0.002 b |
The data in the table are mean values (n = 3), and different lowercase letters indicate significant difference among treatments (p < 0.05).
References
- Pankhurst, C.E.; Ophel-Keller, K.; Doube, B.M.; Gupta, V.V.S.R. Biodiversity of soil microbial communities in agricultural systems. Biodivers. Conserv. 1996, 5, 197–209. [Google Scholar] [CrossRef]
- Navarrete, A.A.; Tsai, S.M.; Mendes, L.W.; Faust, K.; de Hollander, M.; Cassman, N.A.; Raes, J.; van Veen, J.A.; Kuramae, E.E. Soil microbiome responses to the short-term effects of Amazonian deforestation. Mol. Ecol. 2015, 24, 2433–2448. [Google Scholar] [CrossRef] [PubMed]
- Averill, C.; Hawkes, C.V. Ectomycorrhizal fungi slow soil carbon cycling. Ecol. Lett. 2016, 19, 937–947. [Google Scholar] [CrossRef] [PubMed]
- Van der Heijden, M.G.A.; Bardgett, R.D.; van Straalen, N.M. The unseen majority: Soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol. Lett. 2008, 11, 296–310. [Google Scholar] [CrossRef]
- Luo, C.Y.; Zhang, B.X.; Liu, J.; Wang, X.X.; Han, F.P.; Zhou, J.H. Effects of different Ages of Robinia pseudoacacia plantations on soil physiochemical properties and microbial communities. Sustainability 2020, 12, 9161. [Google Scholar] [CrossRef]
- Kennedy, A.C.; Smith, K.L. Soil microbial diversity and the sustainability of agricultural soils. Plant Soil 1995, 170, 75–86. [Google Scholar] [CrossRef]
- Felske, A.; Wolterink, A.; van Lis, R.; de Vos, W.M.; Akkermans, A.D.L. Response of a soil bacterial community to grassland succession as monitored by 16S rRNA levels of the predominant ribotypes. Appl. Environ. Microbiol. 2000, 66, 3998–4003. [Google Scholar] [CrossRef]
- Bluhm, S.L.; Eitzinger, B.; Ferlian, O.; Bluhm, C.; Schröter, K.; Pena, R.; Maraun, M.; Scheu, S. Deprivation of root-derived resources affects microbial biomass but not community structure in litter and soil. PLoS ONE 2019, 14, e0214233. [Google Scholar] [CrossRef]
- Nevins, C.J.; Nakatsu, C.; Armstrong, S. Characterization of microbial community response to cover crop residue decomposition. Soil Biol. Biochem. 2018, 127, 39–49. [Google Scholar] [CrossRef]
- Xu, S.; Liu, L.; Sayer, E.J. Variability of aboveground litter inputs alters soil physicochemical and biological processes: A meta-analysis of litterfall-manipulation experiments. Biogeosciences 2013, 10, 5245–5272. [Google Scholar] [CrossRef]
- Brant, J.B.; Sulzman, E.W.; Myrold, D.D. Microbial community utilization of added carbon substrates in response to long-term carbon input manipulation. Soil Biol. Biochem. 2006, 38, 2219–2232. [Google Scholar] [CrossRef]
- Lu, X.R.; Yin, Y.; Feng, J.X.; Ma, H.L.; Gao, R.; Yin, Y.F. Effects of Chinese fir litter and its biochar amendment on soil microbial community structure. Acta Sci. Circumstantiae 2019, 39, 3090–3098. (In Chinese) [Google Scholar]
- Li, Y.; Zhou, C.F.; Qiu, Y.X.; Tigabu, M.; Ma, X.Q. Effects of biochar and litter on carbon and nitrogen mineralization and soil microbial community structure in a China fir plantation. J. For. Res. 2019, 30, 1913–1923. [Google Scholar] [CrossRef]
- Brant, J.B.; Myrold, D.D.; Sulzman, E.W. Root controls on soil microbial community structure in forest soils. Oecologia 2006, 148, 650–659. [Google Scholar] [CrossRef] [PubMed]
- Sayer, E.J. Using experimental manipulation to assess the roles of leaf litter in the functioning of forest ecosystems. Biol. Rev. 2006, 81, 1–31. [Google Scholar] [CrossRef]
- He, K.Y.; Shen, Y.W.; Feng, J.G.; Han, M.G.; Zhou, Y.Q.; Zhu, B. Effects of altered plant detritus input on soil respiration and its temperature sensitivity in a Pinus sylvestris var. mongolica Plantation. Acta Sci. Nat. Univ. Pekin. 2021, 57, 361–370. (In Chinese) [Google Scholar]
- Caesar-TonThat, A.J.; Espeland, E.; Sainju, U.M.; Lartey, R.T.; Gaskin, J.F. Effectsof Agaricus lilaceps fairy rings on soil aggregation and microbial communitystructure in relation to growth stimulation of western wheatgrass (Pascopyrumsmithii) in eastern Montana rangeland. Microb. Ecol. 2013, 66, 120–131. [Google Scholar] [CrossRef]
- Xu, J.; Liu, S.; Song, S.; Guo, H.; Tang, J.; Yong, J.W.; Ma, Y.; Chen, X. Arbuscular mycorrhizal fungi influence decomposition and the associated soil microbial community under different soil phosphorus availability. Soil Biol. Biochem. 2018, 120, 181–190. [Google Scholar] [CrossRef]
- Kaiser, C.; Fuchslueger, L.; Koranda, M.; Gorfer, M.; Stange, C.F.; Kitzler, B.; Rasche, F.; Strauss, J.; Sessitsch, A.; Zechmeister-Boltenstern, S.; et al. Plants control the seasonal dynamics of microbial N cycling in a beech forest soil by belowground Callocation. Pedobiologia 2011, 92, 1036–1051. [Google Scholar]
- Tedersoo, L.; Nilsson, R.H.; Abarenkov, K.; Jairus, T.; Sadam, A.; Saar, I.; Bahram, M.; Bechem, E.; Chuyong, G.; Kõljalg, U. 454 Pyrosequencing and Sanger sequencing of tropical mycorrhizal fungi provide similar results but reveal substantial methodological biases. New Phytol. 2010, 188, 291–301. [Google Scholar] [CrossRef]
- Wei, X.; Li, Q.; Liu, Y.; Liu, S.; Guo, X.; Zhang, L.; Niu, D.; Zhang, W. Restoring ecosystem carbon sequestration through afforestation: A sub-tropic restoration case study. Forest Ecol. Manag. 2013, 300, 60–67. [Google Scholar] [CrossRef]
- Sparks, D.L.; Page, A.L.; Helmke, P.A.; Loeppert, R.H.; Soltanpour, P.N.; Tabatabai, M.A.; Johnston, C.T.; Sumner, M.E. Methods of Soil Analysis. Part 3: Chemical Methods; Soil Science Society of America Inc.: Madison, WI, USA, 1996. [Google Scholar]
- Hou, Q.; Wang, W.; Yang, Y.; Hu, J.; Bian, C.; Jin, L.; Li, G.; Xiong, X. Rhizosphere microbial diversity and community dynamics during potato cultivation. Eur. J. Soil Biol. 2020, 98, 103176. [Google Scholar] [CrossRef]
- Liu, B.; Wang, S.; Wang, J.; Zhang, X.; Shen, Z.; Shi, L.; Chen, Y. The great potential for phytoremediation of abandoned tailings pond using ectomycorrhizal Pinus sylvestris. Sci. Total Environ. 2020, 719, 137475. [Google Scholar] [CrossRef]
- Ren, Q.; Yuan, J.; Wang, J.; Liu, X.; Ma, S.; Zhou, L.; Miao, L.; Zhang, J. Water level has higher influence on soil organic carbon and microbial community in Poyang Lake wetland than vegetation type. Microorganisms 2022, 10, 131. [Google Scholar] [CrossRef]
- Nadelhoffer, K.J.; Boone, R.D.; Bowden, R.D.; Canary, J.D.; Kaye, J.; Micks, P.; Ricca, A.; Aitkenhead, J.A.; Lajtha, K.; McDowell, W.H. The DIRT experiment: Litter and root influences on forest soil organic matter stocks and function. In Forest Landscape Dynamics in New England: Ecosystem Structure and Function as a Consequence of 5000 Years of Change; Foster, D., Aber, J., Eds.; Oxford University Press: New York, NY, USA, 2004. [Google Scholar]
- Lemanski, K.; Scheu, S. Incorporation of 13C labelled glucose into soil microorganisms of grassland: Effects of fertilizer addition and plant functional group composition. Soil Biol. Biochem. 2014, 69, 38–45. [Google Scholar] [CrossRef]
- Rousk, J.; Hill, P.W.; Jones, D.L. Priming of the decomposition of ageing soil organic matter: Concentration dependence and microbial control. Funct. Ecol. 2015, 29, 285–296. [Google Scholar] [CrossRef]
- Elser, J.J.; Sterner, R.W.; Gorokhova, E.A.; Fagan, W.F.; Markow, T.A.; Cotner, J.B.; Harrison, J.F.; Hobbie, S.E.; Odell, G.M.; Weider, L.W. Biological stoichiometry from genes to ecosystems. Ecol. Lett. 2008, 3, 540–550. [Google Scholar] [CrossRef]
- DeAngelis, K.M.; Pold, G.; Topçuoğlu, B.D.; van Diepen, L.T.; Varney, R.M.; Blanchard, J.L.; Melillo, J.; Frey, S.D. Long-term forest soil warming alters microbial communities in temperate forest soils. Front. Microbiol. 2015, 6, 104. [Google Scholar] [CrossRef]
- Feng, W.T.; Zou, X.M.; Schaefer, D. Above- and belowground carbon inputs affect seasonal variations of soil microbial biomass in a subtropical monsoon forest of southwest China. Soil Biol. Biochem. 2009, 41, 978–983. [Google Scholar] [CrossRef]
- Wang, Q.K.; He, T.X.; Wang, S.L.; Wang, S.L.; Liu, L. Carbon input manipulation affects soil respiration and microbial community composition in a subtropical coniferous forest. Agric. Forest Meteorol. 2013, 178–179, 152–160. [Google Scholar] [CrossRef]
- Wan, X.H.; Huang, Z.Q.; He, Z.M.; Yu, Z.P.; Wang, M.H.; Liu, R.Q.; Zheng, L.J. Changes of above- and belowground carbon input affected soil microbial biomass and community composition in two tree species plantations in subtropical China. Acta Ecol. Sin. 2016, 36, 3582–3590. (In Chinese) [Google Scholar]
- Liu, J.; Liu, M.; Wu, M.; Jiang, C.; Chen, X.; Cai, Z.; Wang, B.; Zhang, J.; Zhang, T.; Li, Z. Soil pH rather than nutrients drive changes in microbial community following long-term fertilization in acidic Ultisols of southern China. J. Soil Sediments 2018, 18, 1853–1864. [Google Scholar] [CrossRef]
- Liu, G.Y.; Chen, L.L.; Shi, X.R.; Yuan, L.Y.; Lock, T.R.; Kallenbach, R.L. Changes in rhizosphere bacterial and fungal community composition with vegetation restoration in planted forests. Land Degrad. Dev. 2019, 30, 1147–1157. [Google Scholar] [CrossRef]
- Luo, Z.K.; Feng, W.T.; Luo, Y.Q.; Baldock, J.; Wang, E.L. Soil organic carbon dynamics jointly controlled by climate, carbon inputs, soil properties and soil carbon fractions. Glob. Chang. Biol. 2017, 23, 4430–4439. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Wang, L.; Hu, Y.; Tsang, Y.F.; Zhang, Y.; Wu, J.; Fu, X.; Sun, Y. Plant litter composition selects different soil microbial structures and in turn drives different litter decomposition pattern and soil carbon sequestration capability. Geoderma 2018, 319, 194–203. [Google Scholar] [CrossRef]
- Sang, R.; Li, S.; Huang, X.; Liu, W.; Liang, X.; Su, J. Effects of soil properties and plant diversity on soil microbial community composition and diversity during secondary succession. Forests 2021, 12, 805. [Google Scholar] [CrossRef]
- Hugenholtz, P.; Goebel, B.M.; Pace, N.R. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 1998, 180, 4765–4774. [Google Scholar] [CrossRef]
- Barns, S.M.; Takala, S.L.; Kuske, C.R. Wide distribution and diversity of members of the bacterial kingdom Acidobacterium in the environment. Appl. Environ. Microb. 1999, 65, 1731–1737. [Google Scholar] [CrossRef]
- Bryant, D.A.; Costas, A.M.; Maresca, J.A.; Chew, A.G.; Klatt, C.G.; Bateson, M.M.; Tallon, L.J.; Hostetler, J.; Nelson, W.C.; Heidelberg, J.F.; et al. Candidatus Chloracidobacterium thermophilum: An aerobic phototrophic acidobacterium. Science 2007, 317, 523–526. [Google Scholar] [CrossRef]
- Lu, S.P.; Gischkat, S.; Reiche, M.; Akob, D.M.; Hallberg, K.B.; Kusel, K. Ecophysiology of Fe-cycling bacteria in acidic sediments. Appl. Environ. Microb. 2010, 76, 8174–8183. [Google Scholar] [CrossRef]
- Pankratov, T.A.; Ivanova, A.O.; Dedysh, S.N.; Liesack, W. Bacterial populations and environmental factors controlling cellulose degradation in an acidic Sphagnum peat. Environ. Microbiol. 2011, 13, 1800–1814. [Google Scholar] [CrossRef]
- Sun, H.; Terhonena, E.; Koskinenb, K.; Paulinb, L.; Kasanena, R.; Asiegbu, F.O. Bacterial diversity and community structure along different peat soils in boreal forest. Appl. Soil Ecol. 2014, 74, 37–45. [Google Scholar] [CrossRef]
- McCaig, A.E.; Glover, L.A.; Prosser, J.I. Molecular analysis of bacterial community structure and diversity in unimproved and improved upland grass pastures. Appl. Environ. Microbiol. 1999, 65, 1721–1730. [Google Scholar] [CrossRef]
- Navarrete, I.A.; Tsutsuki, K. Land-use impact on soil carbon, nitrogen, neutral sugar composition and related chemical properties in a degraded Ultisol in Leyte, Philippines. Soil Sci. Plant Nutr. 2008, 54, 321–331. [Google Scholar] [CrossRef]
- Eilers, K.G.; Lauber, C.L.; Knight, R.; Fierer, N. Shifts in bacterial community structure associated with inputs of low molecular weight carbon compounds to soil. Soil Biol. Biochem. 2010, 42, 896–903. [Google Scholar] [CrossRef]
- Wang, H.; Liu, S.; Zhang, X.; Mao, Q.; Li, X.; You, Y.; Wang, J.; Zheng, M.; Zhang, W.; Lu, X.; et al. Nitrogen addition reduces soil bacterial richness, while phosphorus addition alters community composition in an old-growth N-rich tropical forest in southern China. Soil Biol. Biochem. 2018, 127, 22–30. [Google Scholar] [CrossRef]
- Tedersoo, L.; Bahram, M.; Põlme, S.; Kõljalg, U.; Yorou, N.S.; Wijesundera, R.; Ruiz, L.V.; Vasco-Palacios, A.M.; Thu, P.Q.; Suija, A.; et al. Global diversity and geography of soil fungis. Science 2014, 346, 1256688. [Google Scholar] [CrossRef]
- Lopes, A.R.; Manaia, C.M.; Nunes, O.C. Bacterial community variations in an alfalfa-rice rotation system revealed by 16S rRNA gene 454-pyrosequencing. FEMS Microbiol. Ecol. 2014, 87, 650–663. [Google Scholar] [CrossRef]
- Garcia-Oliva, F.; Sveshtarova, B.; Oliva, M. Seasonal effects on soil organic carbon dynamics in a tropical deciduous forest ecosystem in westernMexico. J. Trop. Ecol. 2003, 19, 179–188. [Google Scholar] [CrossRef]
- Beimforde, C.; Feldberg, K.; Nylinder, S.; Rikkinen, J.; Tuovila, H.; Dörfelt, H.; Gube, M.; Jackson, D.J.; Reitner, J.; Seyfullah, L.; et al. Estimating the phanerozoic history of the ascomycota lineages: Combining fossil and molecular data. Mol. Phylogenet. Evol. 2014, 78, 386–398. [Google Scholar] [CrossRef]
- Wang, Y.; Yu, Z.; Li, Y.; Wang, G.; Liu, J.; Liu, J.; Liu, X.; Jin, J. Microbial association with the dynamics of particulate organic carbon in response to the amendment of elevated CO2-derived wheat residue into a Mollisol. Sci. Total Environ. 2017, 607–608, 972–981. [Google Scholar] [CrossRef] [PubMed]
- Chau, J.F.; Bagtzoglou, A.C.; Willig, M.R. The effect of soil texture on richness and diversity of bacterial communities. Environ. Forensics 2011, 12, 333–341. [Google Scholar] [CrossRef]
- Naether, A.; Foesel, B.U.; Naegele, V.; Wüst, P.K.; Weinert, J.; Bonkowski, M.; Alt, F.; Oelmann, Y.; Polle, A.; Lohaus, G.; et al. Environmental factors affect acidobacterial communities below the subgroup level in grassland and forest soils. Appl. Environ. Microbiol. 2012, 78, 7398–7406. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Liu, N.; Zhang, Y. Soil aggregates regulate the impact of soil bacterial and fungal communities on soil respiration. Geoderma 2019, 337, 444–452. [Google Scholar] [CrossRef]
- Liu, J.; Sui, Y.; Yu, Z.; Shi, Y.; Chu, H.; Jin, J.; Liu, X.; Wang, G. High throughput sequencing analysis of biogeographical distribution of bacterial communities in the black soils of northeast China. Soil Biol. Biochem. 2014, 70, 113–122. [Google Scholar] [CrossRef]
- Zhong, Y.; Yan, W.; Shangguan, Z. Impact of long-term N additions upon coupling between soil microbial community structure and activity, and nutrient-use efficiencies. Soil Biol. Biochem. 2015, 91, 151–159. [Google Scholar] [CrossRef]
- Zeng, Q.; Liu, Y.; An, S. Impact of litter quantity on the soil bacteria community during the decomposition of Quercus wutaishanica litter. PeerJ 2017, 5, e3777. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).