Abstract
A new species, Robertgurneya jejuensis sp. nov., was described from sandy sediment samples collected at a depth of 25 m on Mun Island, Jeju, in June 2018. The new species is morphologically similar to Robertgurneya similis similis (Scott A., 1896) and Robertgurneya donghaensis Bang, 2021; this is the second record of the genus Robertgurneya in South Korea. The morphological characteristics of the similis group within the genus Robertgurneya, to which the new species is ascribed, are summarized here. Furthermore, an identification key is provided based on the summary. Molecular identification of the collected specimens, based on the nuclear 18S ribosomal RNA (18S rRNA) and the mitochondrial cytochrome oxidase subunit I fragment, was obtained. Finally, a phylogenetic tree was constructed to present the position of Robertgurneya within the Miraciidae family based on 18S rRNA sequences which is relatively conserved. As a result, the relationship with sister genera morphologically similar to Robertgurneya was also molecularly confirmed.
1. Introduction
Miraciidae Dana, 1846, is a large family of benthic copepods, Harpacticoida [1]. It includes three subfamilies, 64 valid genera, and 506 species (WoRMS Editorial Board 2020), most of which were transferred from the former Diosaccidae family by Willen in 2002 [2]. In South Korea, 20 genera and 36 miraciid species identified at the species level have been reported to date, e.g., [3,4,5,6,7,8].
Sediment samples were collected from the sandy sediment in the subtidal zone at 25 m to investigate meiofauna. As a result, we collected a new species of Robertgurneya Apostolov & Marinov, 1988, on Mun Island, Jeju. The new species, Robertgurneya jejuensis sp. nov., is morphologically similar to the type species of Robertgurneya similis similis (Scott A., 1896) and the latest species, Robertgurneya donghaensis Bang, 2021. Morphological differences between these species and the new species were found in the number of setae in the mouth, the length ratio of A1 segment, the difference in type and relative length of setae, and the presence or absence of ornamentation on appendages. Currently, 19 valid species (and subspecies) exist worldwide in Robertgurneya; this is the second record of Robertgurneya in South Korea after the report of R. donghaensis [9].
The type species and diagnosis of genus Robertgurneya were established in 1988 [10] and the genus was formally named according to the International Commission on Zoological Nomenclature; however, this genus was first proposed by Lang in 1944 [11]. He divided the genus into two species groups and designated the type of each group as follows: similis group (type: Stenhelia simulans Norman & Scott T., 1905) and spinulosus group (type: Amphiascus spinulosus Sars G.O., 1911). Lang (1948) [12] also commented on dividing these two groups into separate genera [13]. Gómez (2020) [14] supported this incomplete opinion by proposing the new genus Robertgurneyella Gómez, 2020, setting Amphiascus spinulosus Sars G.O., 1911 as the type species and suggested that the original spinulosus group was renamed the rostrata group.
This study summarizes the contents of comparing the morphological characteristics between the constituent species of the similis group to which the new species belongs. The identification key is presented based on this. In addition, the gene sequences obtained in this study are the first reported sequences in Robertgurneya. Finally, a phylogenetic analysis was conducted to investigate the gene sequence-based phylogenetic relationship between Robertgurneya and other genera in Miraciidae uploaded to NCBI.
2. Materials and Methods
2.1. Specimen Collection and Observation
Coarse sandy sediment samples from 25 m deep were collected from Mun Island, Jeju, South Korea (33°13′38.4″ N 126°33′49.3″ E) (Figure 1) by SCUBA diving in June 2018. Sandy sediments were collected by using acrylic corers and these were filtered through a 38 μm sieve and fixed with 99% ethanol.
Figure 1.
Map of sample locality.
Harpacticoids were identified following Huys et al. (1996) [15] and Wells (2007) [16] under a compound microscope at 400–1000× magnification. All drawings were prepared using a drawing tube on an Olympus BX51 differential interference contrast microscope (Olympus, Tokyo, Japan). Scanning electron micrographs were obtained using a Hitachi S-3400N scanning electron microscope (SEM) at Chung-Ang University. The descriptive terminology followed that of Huys et al. (1996) [15]. Abbreviations used in the text are as follows: A1, antennule; A2, antenna; benp, baseoendopod; ae, aesthetasc; exp, exopod; enp, endopod; P1–P6, first to sixth thoracopod; exp (enp)-1 (2, 3) denotes the proximal (middle, distal) segment of a three-segmented ramus. Specimens were deposited in the collection of the National Institute of Biological Resources (NIBR), voucher code: NIBRIV0000901874 - NIBRIV0000901877.
To compare the morphology more accurately with a domestically reported species (Robertgurneya donghaensis Bang, 2021), which is very similar in morphology to a new species, the paratypes stored in the NIBR were observed.
2.2. DNA Extraction and Amplification
For DNA extraction and amplification, the two specimens, holotype and allotype, were transferred to ultrapure water for 1 h to remove the ethanol. The specimens were then prepared for non-destructive DNA extraction in worm lysis buffer [17]. Specimens were placed in tubes containing 25 μL lysis buffer and placed in a Takara thermocycler (Takara, Otsu, Shiga, Japan) with the following settings: 65 °C for 15 min, 95 °C for 20 min, and 15 °C for 2 min. The specimens were then maintained for morphological identification and described after genetic confirmation. Unpurified total genomic DNA was stored at −20 °C for long-term storage. Fragments from two genes, the nuclear 18S ribosomal RNA (18S rRNA) and mitochondrial cytochrome oxidase subunit I (mtCOI) genes, were amplified using a polymerase chain reaction (PCR) premix (BIONEER Co., Labopass, Korea), 2 μL of primers, 3 μL of genomic DNA as a template, and 5 μL of ultra-pure water.
The PCR primers that were used were 18S-F1, 18S-F3, 18S-R7, and 18S-R9 [18] for 18S ribosomal DNA. The amplification protocol consisted of an initial denaturation at 94 °C for 5 min, followed by 33 cycles of denaturation at 94 °C for 30 s, annealing at 47 °C for 30 s, extension at 72 °C for 1 min, and a final extension at 72 °C for 10 min.
For mtCOI, the LCO1490 and Cop-COI-2189R primer sets were used. The amplification protocol consisted of an initial denaturation at 94 °C for 5 min, followed by 40 cycles of denaturation at 94 °C for 1 min, annealing at 45 °C for 2 min, extension at 72 °C for 3 min; and a final extension at 72 °C for 10 min [19].
Successful amplification was confirmed by electrophoresis on 1% agarose gel. The PCR products were sent to Macrogen (Seoul, Korea) for purification and DNA sequencing. DNA was sequenced on an ABI automatic capillary sequencer, using the same sets of primers as those used for amplification. All the obtained sequences were visualized using Finch TV version 1.4.0 (https://digitalworldbiology.com/FinchTV; Geospiza Inc., Denver, CO, USA, accessed on 13 August 2018). The quality of each sequence was evaluated, and low-resolution peaks were checked by comparing the forward and reverse strands. Basic Local Alignment Search Tool (BLAST) [20] searches revealed that the obtained sequences were of copepod origin and not contaminants.
2.3. Phylogenetic Analyses
Evolutionary analyses were performed using MEGA version 11 [21]. The sequences were aligned using the multiple sequence comparison by log-expectation (MUSCLE) method. Phylogenetic trees were constructed using neighbor-joining (NJ) and maximum likelihood (ML) methods. For the ML tree, the K2 + G + I model was selected according to the model test in MEGA version 11. Pairwise distance was computed in the same software, using the p-distance. The analysis of 18S rRNA involved 14 nucleotide sequences, including the sequences secured in this study. Among the sequences of the two individuals obtained in this study, the sequence with the longer length was included in the phylogenetic analysis. The sequences of other miraciid species and outgroups were obtained from NCBI (Table 1). Among the data uploaded to NCBI, it was determined that the sequences (350−450 bp) that were too short were not suitable for use in this study; therefore, only those that satisfy a length of 1600 bp or longer were used.

Table 1.
GenBank numbers of 18S rRNA sequences used in phylogenetic analyses in this study.
3. Results
3.1. Systematics
Class: Copepoda H. Milne-Edwards, 1840
Order: Harpacticoida Sars G.O., 1913
Family: Miraciidae Dana, 1846
Genus: Robertgurneya Apostolov & Marinov, 1988
Robertgurneya jejuensis sp. nov.
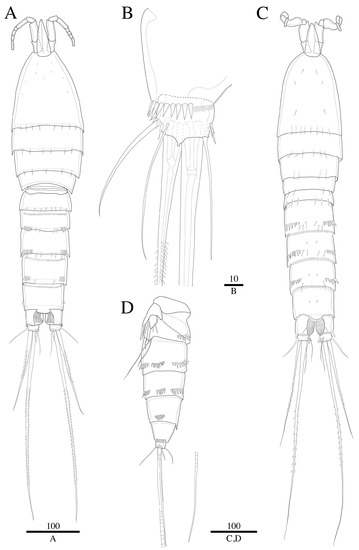
Figure 2.
Robertgurneya jejuensis sp. nov. Female. (A) Habitus, dorsal. (B) Caudal ramus, ventral. Male. (C) Habitus, dorsal. (D) Urosome, lateral. Scale bars: 100 μm (A,C,D); 10 μm (B).
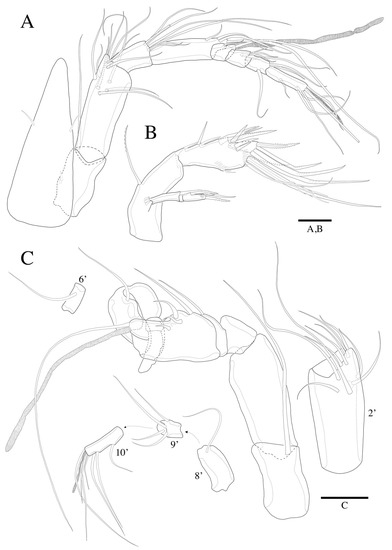
Figure 3.
Robertgurneya jejuensis sp. nov. Female. (A) A1 and rostrum, dorsal. (B) A2. Male. (C) A1 and segments. Scale bars: 20 μm.
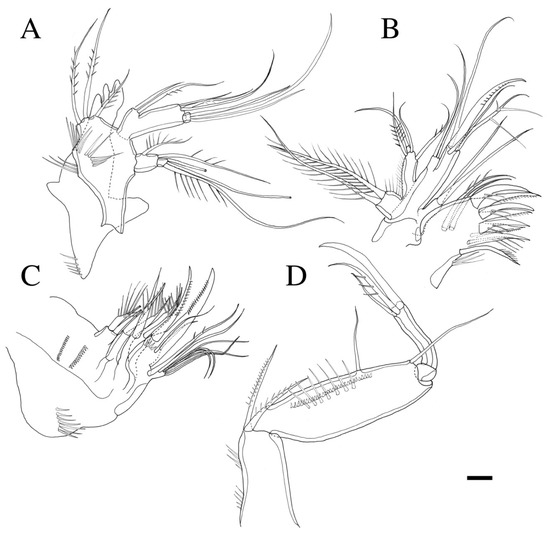
Figure 4.
Robertgurneya jejuensis sp. nov. (A) Mandible. (B) Maxillule. (C) Maxilla. (D) Maxilliped. Scale bar: 10 μm.
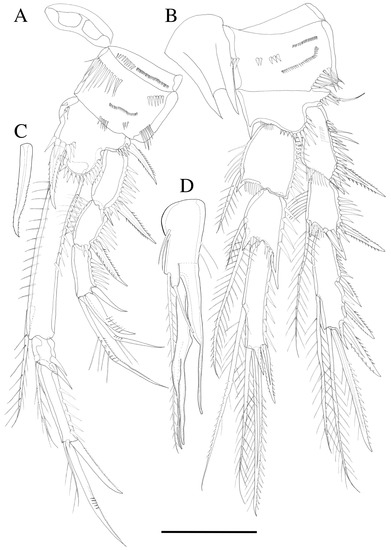
Figure 5.
Robertgurneya jejuensis sp. nov. Female. (A) P1. (B) P2. Male. (C) Inner spine of P1 basis. (D) P2 enp-2. Scale bar: 50 μm.
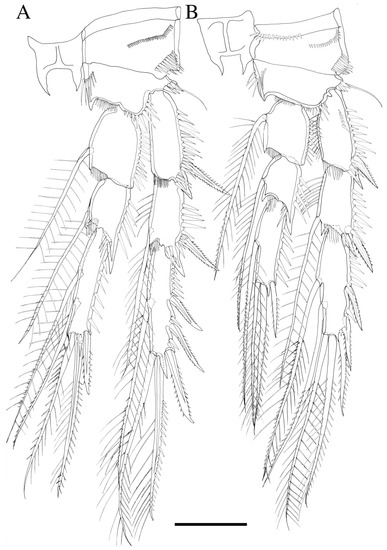
Figure 6.
Robertgurneya jejuensis sp. nov. (A) P3. (B) P4. Scale bar: 50 μm.
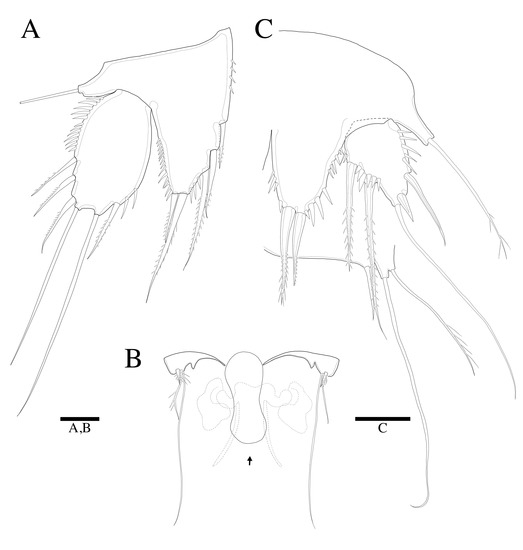
Figure 7.
Robertgurneya jejuensis sp. nov. Female. (A) P5. (B) Genital field and P6 (arrow: epicopulatory bulb). Male. (C) P5 and P6. Scale bars: 20 μm.
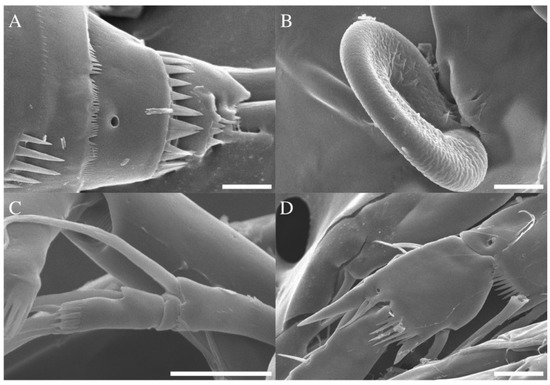
Figure 8.
Robertgurneya jejuensis sp. nov. Female. SEM photographs. (A) Caudal ramus, lateral. (B) Epicopulatory bulb in genital field. (C) A2 exopod. (D) P2 segments with distal ornamentation. Scale bars: 10 μm.
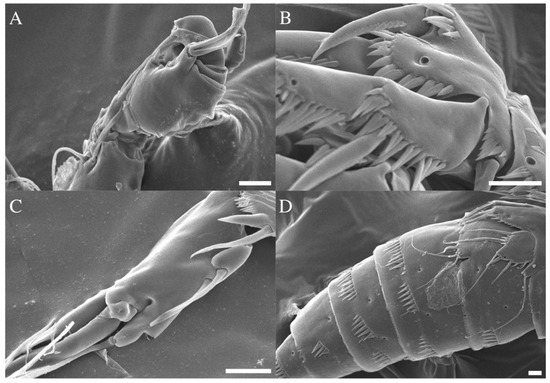
Figure 9.
Robertgurneya jejuensis sp. nov. Male. SEM photographs. (A) A1. (B) P1 basis. (C) P2 enp-2. (D) Urosome, lateral. Scale bars: 10 μm.
ZooBank Registration LSID
Urn: lsid:zoobank.org:act:799CAA2D-009C-4A82-950E-44E4476DE931
Type locality: 33°13′38.4″ N 126°33′49.3″ E; Hangaechang, Mun Island, Seogwipo-si, Jeju Island, Korea; collected on 18 June 2018, by Wonchoel Lee and Raehyuk Jeong. Sediment type: coarse sand; depth: 25 m; water temperature: 19 °C.
Material examined: Holotype; 1♀(NIBRIV0000901874) on 1 slide. Allotype; 1♂(NIBRIV0000901875) on 1 slide. Paratype; 2♀♀(NIBRIV0000901876) dissected on 20 slides, 2♀♀ and 2♂♂ on a SEM stub (NIBRIV0000901877).
Etymology: The species name was given as “jejuensis” because the species was discovered from Jeju Island, South Korea.
DNA-barcode: Sequences were submitted to GenBank. Genbank accession number: 18S rRNA (1755 base pairs); OP798781/mtCOI (531 base pairs); OP797675. The pairwise distance based on mtCOI gene data of holotype and allotype specimens was 0.008 (standard error: 0.004), confirming that female and male individuals are of the same species.
3.2. Description
3.2.1. Description of Female
Total body length 875 µm (specimens of holotype and paratype, n = 3) (Figure 2A); body cylindrical, slightly tapering behind. Rostrum prominent, pointing upward, tapering distally, reaching mid-length of second antennular segment, defined at base as in Figure 2A and Figure 3A; with two small sensilla. Cephalothorax bell-shaped.
Caudal ramus (Figure 2B and Figure 8A). Approximately 0.7 times as long as greatest width, ventral margin armed with pore; each ramus armed with seven setae; caudal terminal setae pinnate, other setae all naked.
A1 (Figure 3A). Eight-segmented, slender; seg-2 about 1.7 times as long as seg-1; seg-4 2 times as long as seg-3. Armature formula (segment-[number of seta/setae]): 1-[1], 2-[11], 3-[7], 4-[4 + ae], 5-[2], 6-[4], 7-[3], 8-[4 + acrothek]. Setae all naked. Aesthetasc on seg-4 fused basally with adjacent seta and about 1.5 times as long as distal four segments combined. Apical acrothek consisting of aesthetasc and two bare setae.
A2 (Figure 3B). Endopod two-segmented, enp-2 with nine setae. Exopod three-segmented (Figure 8C), exp-1 with pinnate seta; exp-2 without seta and small; exp-3 with one seta laterally and two setae distally.
Mandible (Figure 4A). Gnathobase bearing chitinous projection on ventral surface; palp with three pinnate setae and setule rows; exopod two-segmented, exp-1 with two unipinnate setae laterally and distally, exp-2 with two bare setae and an unipinnate seta; endopod two-segmented, enp-1 with two proximal and three distal setae, enp-2 with three bare setae.
Maxillule (Figure 4B). Praecoxal arthrite bearing six elements distally with a pinnate seta, and three bare setae on surface; coxa with two setae; basis with six setae; endopod bearing four setae; exopod bearing two pinnate setae.
Maxilla (Figure 4C). Syncoxa with three endites; proximal and middle endites with two spines, respectively; distal endite with two pinnate spines and one naked seta; allobasis transformed to curved claw bearing pinnate spine and two thin bare setae on surface; endopod with seven setae.
Maxilliped (Figure 4D). Subchelate; syncoxa with two pinnate setae; basis with small spinules along inner margin and a row of fine spinules on surface, 3.5 times as long as broad, bearing two bare setae; endopod elongate, with a strong claw, a bare seta and an unipinnate seta.
P1 (Figure 5A). Basis inner margin with a bipinnate spine inner distally; exopod three-segmented; exp-1 inner margin naked, exp-1 and exp-2 with a bipinnate outer spine and ornamented with coarse spinules along outer margin, exp-2 inner margin with fine spinules without seta, exp-3 inner margin naked, exp-3 with two outer spines and a long geniculate seta and a relatively short, geniculate setae distally; endopod three-segmented; enp-1 longer than exopod, about 6.5 times longer than wide with one inner pinnate seta distally, inner and outer margin ornamented with fine spinules; enp-2 small, ornamented with coarse spinules along outer margin, with inner pinnate seta; enp-3 about three times longer than enp-2, outer margin ornamented with coarse spinules, bearing an unipinnate seta at inner distal corner, a spine and a geniculate seta apically.
P2 (Figure 5B). Rami three-segmented; basis with naked seta and inner margin with fine spinules; exp-1 without inner seta, hyaline frills on inner distal margin (Figure 8D), exp-2 with one inner pinnate seta at distal, both exp-1 and exp-2 with a bipinnate outer spine, inner margin ornamented and outer margin ornamented with coarse spinules, exp-3 with a plumose inner seta, a long plumose seta at inner terminal, a long seta with plumose inner side and pinnate outer side at outer terminal and three outer spines, proximal outer margin ornamented with coarse spinules; endopod about as long as exopod, enp-1 with a plumose inner seta, inner margin with setules; enp-2 with two inner setae; enp-3 with a plumose inner seta, two setae distally and a spine at outer distal corner; endopod segments outer margin ornamented with small spinules.
P3 (Figure 6A). Rami three-segmented; exp-1 without inner seta; exp-2 with a plumose inner seta at distal, both exp-1 and exp-2 hyaline frills on inner distal margin and outer margin ornamented with coarse spinules; exp-3 with a plumose inner seta, a long seta at inner terminal, a long seta with plumose inner side and pinnate outer side at outer terminal and three outer pinnate spines, proximal outer margin ornamented with coarse spinules; endopod shorter than exopod, both enp-1 and enp-2 with a plumose inner seta and hyaline frills on inner distal margin, enp-3 with two plumose inner setae, two setae distally, and a spine at outer distal corner, endopod segments outer margin ornamented with coarse spinules.
P4 (Figure 6B). Rami three-segmented; basis with bare outer seta; exp-1 posterior surface ornamented with coarse spinules, exp-1 without inner seta, both exp-1 and exp-2 inner margin ornamented with fine spinules, hyaline frills on inner distal margin and outer margin ornamented with coarse spinules; exp-3 with two long inner setae, a long seta at inner terminal, a long seta with plumose inner side and pinnate outer side at outer terminal and three outer spines, proximal outer margin ornamented with coarse spinules; endopod shorter than exopod, both enp-1 and enp-2 with one inner seta and hyaline frills on inner distal margin, enp-3 with one inner seta, two setae distally and a spine at outer distal corner, endopod segments outer margin ornamented with coarse spinules.
Armature formulae are shown in Table 2.

Table 2.
Armature formulae of legs 1–4.
P5 (Figure 7A). Baseoendopod and exopod distinct, baseoendopod with naked outer basal seta; endopodal lobe bearing three inner spines and two bipinnate distal setae; exopod oval, elongated, 1.7 times longer than wide, with spinules along outer margins, two fine spinules on inner distal margin, bearing a bipinnate inner seta, two apical long bare setae and three unipinnate outer setae.
3.2.2. Description of Male
General shape of body (Figure 2C) same as in the female. Urosome somites (Figure 2C,D and Figure 9D) with several rows of spinule on dorsal, ventral, and lateral surface. Sexual dimorphism shown in antennule, inner projection on P1 basis, endopod of P2, P5, and P6.
A1 (Figure 3C and Figure 9A). Subchirocer. Ten-segmented. The fifth segment swollen. Armature formula: 1-[1], 2-[10], 3-[8], 4-[1], 5-[4 + (1 + ae)], 6-[1], 7-[1], 8-[1], 9-[4], 10-[5 + acrothek]. Setae all naked.
P2 endopod (Figure 5D and Figure 9C). Two-segmented; enp-1 with inner pinnate seta; enp-2 modified, bearing two short naked inner setae and one long pinnate seta, terminal seta, and two outer distal spiniform projection.
P5 (Figure 7C and Figure 9D). Baseoendopod and exopod distinct; endopodal lobe ornamented with spinules at outer margin, longer than exopod, armatured with two pinnate spines. Exopod with five setae in total, including two pinnate inner setae covered with four setules on base of setae, long bare distal seta and two bare outer setae, inner margin and outer margin ornamented with spinules.
4. Discussion
Following the identification keys to harpacticoid species [12,25], this copepod is identifiable as R. similis similis (Scott A. 1896). However, the new species and R. similis similis were distinguished by a combination of the following morphological characteristics: (1) the length of the secondary segment of A1 (longer in the new species), (2) the type of inner setae on P2 enp-2 (plumose in the new species), (3) the inner margin of P4 exp-2 (with long setules on the inner margin in the new species), (4) the type and length of P5 setae (with longer distal setae on P5 exp and benp in the new species), and (5) the ornamentation and distance between P5 benp distal setae and inner setae (wider distance and setules in the new species).
The new species described here also has morphological features similar to those of the most recently reported Korean species, R. donghaensis Bang 2021 [9]. However, there were obvious differences in some features (Table 3). The morphological differences between the two species are noticeable in the number of setae in the mouth part, the length ratio of the A1 segments, the difference in setae length, and the presence or absence of ornamentations on the appendages and body surfaces. The male tends to be relatively less decorated in the new species.

Table 3.
Morphological comparison of new species and R. donghaensis.
Gómez (2020) suggested a key to Robertgurneya species [14]. After this, as two new species of the similis group were additionally reported, the updated identification key in this study includes 11 species of the similis group. This group is a set of species within the genus that have in common that P2 enp-2 has two inner setae. Table S1 compares several morphological features of female congeners in the similis group and new species. Because these species are generally similar in morphological characteristics, minor features were selected for the comparison. Based on the contents of Table S1, the following identification key was organized.
Key to the Species of the Robertgurneya similis group:
- P1 enp-2 without inner seta …………………………………………………................R. remaneiP1 enp-2 with inner seta ……………………………………………………………....…........... 2
- P2 enp-1 without inner seta; P3 enp-3 with one inner seta; female P5 setae benp: exp = 4:5 ……………………………………………………………………………....................... R. brevipesP2 enp-1 with inner seta; P3 enp-3 with two inner setae; female P5 setae benp: exp = 5:6…………………………………………………………………………………………............... 3
- P5 exp l/b = 1.5 …………………………………………………………………………..… ..........4P5 exp l/b > 1.5 ………………………………………………………………………..….............. 7
- P1 enp-1/exp = 1; P1 enp-1 inner seta reaching 2/3 of enp-3……………….......... R. hopkinsiP1 enp-1 > exp; P1 enp-1 inner seta reaching the middle of enp-3 ……………………....... 5
- P5 exp distal margin swollen between setae II and III in both sexes …………...... R. smithiP5 exp distal margin normal, oval-shaped ………………………………………..….…........ 6
- P1 basis inner spine of male reaching tip of P1 exp ……………………....... R. falklandiensisP1 basis inner spine of male reaching P1 exp-2 …………………………….…........ R. diversa
- Male P5 exp with six setae ……………………………………………………......... R. simulansMale P5 exp with five setae …………………………………………………………..….......... 8
- Female P5 exp l/b = 2; P5 benp without ornamentation between seta II and III …........... 9Female P5 exp l/b < 2; P5 benp with ornamentation between seta II and III ……….…………………………………………………………….…....................... R. jejuensis sp. nov.
- P1 exp/enp-1 = 2/3; female P5 exp distal margin swollen………...…...…...... R. donghaensisP1 exp/enp-1 = 4/5; female P5 exp distal margin normal, oval-shaped ……….……........ 10
- A1 first/second segment length ratio = 0.38; female P5 exp outer setae short and bulbous; P5 benp distal setae length ratio I/II = 1/4...…………….............… R. similis bulbamphiascoidesA1 first/second segment length ratio = 1; female P5 exp outer setae slender and pinnate; P5 benign distal setae length ratio I/II = 1 …………………………………....... R. similis similis
When identification was based on another identification key [26], this species was morphologically closely identified to Amphiascoides subdebilis (Willey, 1935). The morphological characteristics of the species that differed from this species are P2 exp-3 without inner seta, P5 exp with five setae, and A1 with stubby segments.
In this study, a phylogenetic study was conducted on genera within Miraciidae based on the nuclear 18S rRNA fragment sequences, which is highly conserved. To make the most of the miraciid sequences from the NCBI database, the analysis was performed using only the 18S rRNA, which had the most diverse genera uploaded. When comparing the new species with other miraciids using 18S rRNA sequences, it appeared similar to Amphiascoides on the phylogenetic tree (Figure 10). Indeed, many Amphiascoides species have undergone genus changes. Eight species were transferred to Robertgurneya [13], and nine were transferred to Paramphiascella [12]. Therefore, it can be inferred that these three genera are closely related, both morphologically and molecularly. Meanwhile, Gómez et al. (2021) conducted a phylogenetic analysis based on morphological features and confirmed that Robertgurneya, Amphiascoides, and Paramphiascella formed the closest clade in the phylogenetic tree [27]. Therefore, the phylogenetic tree in this study provides genetic support for the results of Gómez et al. (2021) based on morphological characteristics. It is, therefore, necessary to conduct a morphological and molecularly integrated review of these three genera in the future. The mtCOI sequence (NCBI no. OP797675) obtained in this study will be useful as basic data for future research.
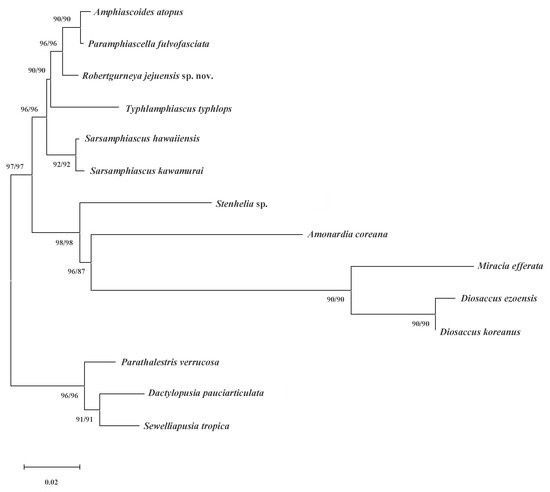
Figure 10.
Maximum likelihood phylogenetic tree including nine genera of Miraciidae and outgroups based on 18S rRNA sequences. Tree shows bootstrap values (ML/NJ, %) together in one tree because the topologies of the two trees created in this study were the same.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/d14121127/s1, Table S1: Morphological comparison of new species and congeners in similis group.
Author Contributions
Conceptualization, W.L. and J.Y.; formal analysis, W.L. and J.Y.; investigation, W.L. and J.Y.; resources, W.L.; writing—original draft preparation, W.L. and J.Y.; writing—review and editing, W.L. and J.Y.; visualization, J.Y.; project administration, W.L.; funding acquisition, W.L. and J.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, grant number 2021R1I1A2043807, 2022R1I1A1A01069134, and supported by a grant from the National Institute of Biological Resources (NIBR202203105), funded by the Ministry of Environment (MOE) of the Republic of Korea.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The voucher specimens of the species examined in the present study were deposited to the National Institute of Biological Resources (NIBR).
Acknowledgments
The authors would like to express gratitude to anonymous reviewers and the Biodiversity laboratory (Hanyang University, Korea) members.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Boxshall, G.A.; Halsey, S.H. An Introduction to Copepod Diversity; Ray Society: London, UK, 2004. [Google Scholar]
- Willen, E. Notes on the Systematic Position of the Stenheliinae (Copepoda, Harpacticoida) Within the Thalestridimorpha and Description of Two New Species from Motupore Island, Papua New Guinea. Cah. Biol. Mar. 2002, 43, 27–42. [Google Scholar]
- Song, S.J.; Yun, S.G.; Chang, C.Y. New Records on Three Harpacticoid Copepods Associated with Marine Macroalgae in Korea. Fish. Aquat. Sci. 1999, 2, 189–198. [Google Scholar]
- Song, S.J.; Rho, H.S.; Kim, W. A New Species of Amonardia (Copepoda: Harpacticoida: Miraciidae) from the Cultivated Brown Alga, Undaria pinnatifida. Integr. Biosci. 2007, 11, 69–77. [Google Scholar] [CrossRef][Green Version]
- Chang, C.Y. Three Miraciid Copepods (Harpacticoida, Miraciidae) from South Korea. Anim. Syst. Evol. Divers. 2009, 25, 215–225. [Google Scholar] [CrossRef][Green Version]
- Nam, E.; Lee, W. First Record of the Genus Sinamphiascus (Copepoda: Harpacticoida) from Korean Waters. J. Species Res. 2012, 1, 44–55. [Google Scholar] [CrossRef]
- Karanovic, T.; Kim, K. New Insights into Polyphyly of the Harpacticoid Genus Delavalia (Crustacea, Copepoda) Through Morphological and Molecular Study of an Unprecedented Diversity of Sympatric Species in a Small South Korean Bay. Zootaxa 2014, 3783, 1–96. [Google Scholar] [CrossRef][Green Version]
- Karanovic, T. Cladistic and Quantitative Shape Analyses of Five New Syntopic Sarsamphiascus (Copepoda, Harpacticoida): Problems and Solutions for Diosaccin Systematics and Taxonomy. Syst. Biodivers. 2020, 18, 810–833. [Google Scholar] [CrossRef]
- Bang, H.W. A New Species of the Genus Robertgurneya (Copepoda: Harpacticoida: Miraciidae) from the East Sea of Korea. Korean J. Environ. Biol. 2021, 39, 590–603. [Google Scholar] [CrossRef]
- Apostolov, A.M.; Marinov, T.M. Copepoda, Harpacticoida (morski kharpaktikoidi). Izd Vo Na Bŭlgarskata Akad. a Na Nauk. 1988, 18, 1–384. [Google Scholar]
- Lang, K. Monographie der Harpacticiden (Vorläufige Mitteilung); Almqvist & Wiksellls Boktryckeri: Upssala, Sweden, 1944. [Google Scholar]
- Lang, K. Monographie der Harpacticiden; Håkan Ohlsson: Lund, Sweden, 1948; Volume 2, pp. 1–1682. [Google Scholar]
- Huys, R. Unresolved Cases of Type Fixation, Synonymy and Homonymy in Harpacticoid Copepod Nomenclature (Crustacea: Copepoda). Zootaxa 2009, 2183, 1–99. [Google Scholar] [CrossRef]
- Gómez, S. A New Species of Robertgurneya Apostolov Marinov, 1988, with an Illustrated Record of R. rostrata (Gurney, 1927), an Amended Genus Diagnosis and Comments on R. Soyeri (Apostolov, 1974) and R. spinulosa (Sars, 1911) (Harpacticoida: Miraciidae). Zootaxa 2020, 4861, 4861. [Google Scholar] [CrossRef]
- Huys, R.; Gee, J.M.; Moore, C.G.; Hamond, R. Marine and Brackish Water Harpacticoid Copepods. Published for the Linnean Society of London and the Estuarine and Coastal Sciences Association by Field Studies Council. Sypopses Br. Fauna (New Ser.) London U.K. 1996, 51, 1–352. [Google Scholar]
- Wells, J.B.J. An Annotated Checklist and Keys to the Species of Copepoda Harpacticoida (Crustacea). Zootaxa 2007, 1568, 1–872. [Google Scholar] [CrossRef]
- Williams, B.D.; Schrank, B.; Huynh, C.; Shownkeen, R.; Waterston, R.H. A Genetic Mapping System in Caenorhabditis elegans Based on Polymorphic Sequence-Tagged Sites. Genetics 1992, 131, 609–624. [Google Scholar] [CrossRef]
- Yamaguchi, S.; Endo, K. Molecular Phylogeny of Ostracoda (Crustacea) Inferred from 18S Ribosomal DNA Sequences: Implication for Its Origin and Diversification. Mar. Biol. 2003, 143, 23–38. [Google Scholar] [CrossRef]
- Bucklin, A.; Ortman, B.D.; Jennings, R.M.; Nigro, L.M.; Sweetman, C.J.; Copley, N.J.; Sutton, T.; Wiebe, P.H.A. “Rosetta Stone” for Metazoan Zooplankton: DNA Barcode Analysis of Species Diversity of the Sargasso Sea (Northwest Atlantic Ocean). Deep Sea Res. II 2010, 57, 2234–2247. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Huys, R.; Mackenzie-Dodds, J.; Llewellyn-Hughes, J. Cancrincolidae (Copepoda, Harpacticoida) Associated with Land Crabs: A Semiterrestrial Leaf of the Ameirid Tree. Mol. Phylogenet. Evol. 2009, 51, 143–156. [Google Scholar] [CrossRef]
- Yeom, J.; Lee, W. A New Species of the Genus Sarsamphiascus Huys, 2009 (Copepoda: Harpacticoida: Miraciidae) from a Sublittoral Zone of Hawaii. PeerJ 2020, 8, e8506. [Google Scholar] [CrossRef]
- Lim, B.J.; Bang, H.W.; Moon, H.; Back, J. Integrative Description of Diosaccus koreanus sp. nov.(Hexanauplia, Harpacticoida, Miraciidae) and Integrative Information on Further Korean Species. ZooKeys 2020, 927, 1–35. [Google Scholar] [CrossRef]
- Wells, J.B.J. Keys to Aid in the Identification of Marine Harpacticoid Copepods; Department of Zoology, University of Aberdeen: Aberdeen, Scotland, 1976; pp. 1–215. [Google Scholar]
- Lang, K. Copepoda Harpacticoida from the Californian Pacific Coast. K. Sven. Vetensk. Handl. 1965, 10, 1–560. [Google Scholar]
- Gómez, S.; Corgosinho, P.H.C.; Rivera-Sánchez, K.I. Proposal of New Genera and Species of the Subfamily Diosaccinae (Copepoda: Harpacticoida: Miraciidae). Eur. J. Taxon. 2021, 759, 1–62. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).