Abstract
We used a multi-locus phylogenetic approach (i.e., combining both mitochondrial and nuclear DNA fragments) to address some long-standing taxonomic inconsistencies within the diverse fish clade of Combtooth Blennies (Blenniidae—unranked clade Almadablennius). The obtained phylogenetic trees revealed some major inconsistencies in the current taxonomy of Parablennini, such as the paraphyletic status of the Salaria and Parablennius genera, casting some doubt regarding their actual phylogenetic relationship. Furthermore, a scarce-to-absent genetic differentiation was observed among the three species belonging to the genus Chasmodes. This study provides an updated taxonomy and phylogeny of the former genus Salaria, ascribing some species to the new genus Salariopsis gen. nov., and emphasizes the need for a revision of the genus Parablennius.
1. Introduction
Combtooth blennies (Blenniidae Rafinesque 1810; herein, blennies) are a diverse clade (>400 species) of nearshore, cryptobenthic fishes that inhabit temperate and tropical marine environments and inland water bodies worldwide [1]. The first major revisions of blenny taxonomy since Norman [2], subdivided the family into six tribes [3,4], each of which was characterized by generic revisions based on morphological characters (Salariini, [5], Omobranchini [6], Phenablenniini [4]; Nemophini [7], and Parablenniini and Blenniini [8,9,10]). The specific membership of these tribes, and generic boundaries within, have remained relatively stable, with the exception of Parablenniini and Blenniini, which have been the subject of great disagreement since Zander [9] rejected the new genera set by Bath [8] (See Table 1 in [11] for history of generic revisions).
Our understanding of the taxonomy of this group has changed significantly with the advent of molecular systematics. In particular, the Almadablennius clade (Parablenniini + Blenniini [12]) has received much attention since Almada et al. [11] published a phylogeny where Blenniini sensu Williams [13] was nested within Parablennini sensu Williams [13] and the genus Lipophrys was paraphyletic, demonstrating that the available taxonomy was inconsistent with phylogeny. Subsequent efforts to investigate the relationships and clarify taxonomy within the Almadablennius clade (e.g., [12,14,15,16,17]), have led to useful taxonomic changes, such as Microlipophyrs being split from Lipophrys (e.g., [15]) and the resolution of species membership within Blenniini and Parablenniini. Despite these updates and multiple lines of evidence suggesting problems, the taxonomy of the Almadablennius clade remains unresolved (e.g., paraphyly of Parablennius, Hypleurochilus and Salaria, and deep split between Mediterranean and Atlantic specimens of Scartella cristata; see Hundt and Simons [16]). In the light of these problems, we re-examined the phylogenetic relationships of the Almadablennius clade using partial sequences of two nuclear and two mitochondrial loci with the explicit aim of testing the monophyly and revising the taxonomy of the genus Salaria.
2. Methods
A total of 49 specimens of blennies belonging to 32 morphospecies were collected in the field or obtained as gifts from the colleagues listed in the acknowledgment section (Table 1). Specimens were fixed in 96% ethanol in situ and identified in the laboratory, using the most updated morphological identification keys [18,19,20].

Table 1.
List of species sampled, catalog number, locality, and GenBank accession number for molecular loci sampled. Novel GenBank accession numbers are reported in bold. * Salariopsis gen. nov. Roman numbers in brackets refer to analysed specimens shown in Figure 1.
Total genomic DNA was extracted from muscle or fin clips using a Qiagen DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA, USA), according to manufacturer suggested protocol. Polymerase chain reaction (PCR) was used to amplify fragments of two nuDNA exons (ectodermal-neural cortex 1-like protein, Enc1, and the cardiac muscle myosin heavy chain 6 alpha, myh6) and two mtDNA fragments (16S ribosomal RNA, 16S, and the control region, D-loop). PCR reactions contained 1.5 μL template DNA, 2.75 μL water, 6.25 μL GoTaqR Green Master Mix (Promega, Madison, WI), with 1.0 μL of each primer (10μM) (see [12,21,22] for the primer pairs used for the different loci). Exonuclease 1 and shrimp alkaline phosphatase were added to PCR products for enzymatic purification at manufacturer-suggested thermal profiles. Automated Sanger sequencing of purified PCR products was performed using ABI Prism R BigDye Terminator v. 3.1 chemistry (Applied Biosystems, Foster City, CA, USA) at the Biomedical Genomics Center DNA Sequencing and Analysis Facility at the University of Minnesota, USA. Complementary heavy and light strands were aligned into contiguous sequences (contigs) and edited in Geneious v. 6.1.8 (Biomatters Ltd., Auckland, New Zealand). Alignments were visually inspected for potential misalignments and, when appropriate, verified by checking amino acid translations.
All sequences were aligned with the software MEGAX [23], using the ClustalW method [24]. All novel sequences were deposited in GenBank (see Table 1 for their Accession Numbers). The alignment of the novel fragments and those downloaded from GenBank were trimmed to fragments of 801 bp (Enc1) 754 bp (myh6) 517 bp (16S) 308 bp (D-loop), respectively. In addition, publicly available sequences belonging to the study taxa and the outgroups Diademichthys lineatus (Sauvage, 1883) (Gobiesocidae) and Enneapterygius minutus (Günther, 1877) (Tripterygiidae) were downloaded from GenBank and included in the analyses (see Table 1 for their GenBank Accession Number, AN).
In order to test whether the mitochondrial and nuclear fragments could be combined for joint analyses, the incongruence length difference test (ILD, [25]) as implemented in PAUP* v. 4.0b10 [26] was used. According to Cunningham [27], if p > 0.01, pooling the data improves the phylogenetic accuracy, and thus it is admissible to merge the tested datasets into a single matrix. This condition was fulfilled both for the concatenation of all the genetic markers analysed in the frame of this study (p = 1). Therefore, the fragments of both the mtDNA and nuDNA loci were concatenated in a single, partitioned dataset. The best evolutionary model for each locus was selected among models analysed by MrBayes v. 3.2.6 [28] using Bayesian model choice criteria (nst = mixed, rates = gamma). The phylogenetic analyses of the partitioned concatenated dataset, including the fragments of the amplified DNA loci, were conducted using Bayesian Inference (BI) and Maximum Likelihood (ML) framework in the software package MrBayes and PhyMl v. 3 [29], respectively. Bootstrap values [30] were calculated with 1000 replicates in the ML trees, whereas the node posterior probability values were reported in the BI tree. In the BI analyses, two independent Markov Chain Monte Carlo analyses were performed with 1 million generations (temp.: 0.2; default priors). Trees and parameter values were sampled every 100 generations, with the result of 10,000 trees for each analysis. Convergence of chains was assessed to ensure proper mixing (Effective Sample Size, ESS, greater than 200 in all the analyses performed). The initial 25% of trees were discarded as “burn-in”.
3. Results
All phylogenetic analyses based on the concatenated DNA dataset were congruent and nodes were well-supported. Most of the genera included in the analyses proved to be monophyletic, with the noteworthy exception of Salaria Forsskål, 1775 and Parablennius Miranda Ribeiro, 1915, which were paraphyletic.
The Parablenniini are separated from Blenniini by a cladogenetic event, with an uncorrected p-distance between the two tribes of 15.5% (Figure 1). There are two well-supported major subclades within Parablenniini: a clade that includes the investigated freshwater Salaria species (see Figure 1, clade “I”), and a clade that includes the remaining analysed ingroup taxa (see Figure 1, clade “II”). Within clade “II”, the genus Parablennius is split into two different subclades; one subclade includes Parablennius intermedius, P. tasmanianus, P. yatabei, P. incognitus, P. zvonimiri, P. salensis, P. pilicornis, P. rouxi, P. tentacularis and the genus Hypleurochilus Gill, 1861 (subclade “IIA”, see Figure 1); the second subclade includes the rest of the analysed Parablennius species (i.e., P. gattorugine, P. ruber, P. parvicornis, and P. sanguinolentus) along with representatives of the genera Chasmodes Valenciennes, 1836, Hypsoblennius Gill, 1861, Scartella Jordan, 1886 and Salaria (subclade “IIB”, see Figure 1). The uncorrected p-distance between the two subclades (i.e., “IIA” and “IIB”) is 13.4%.
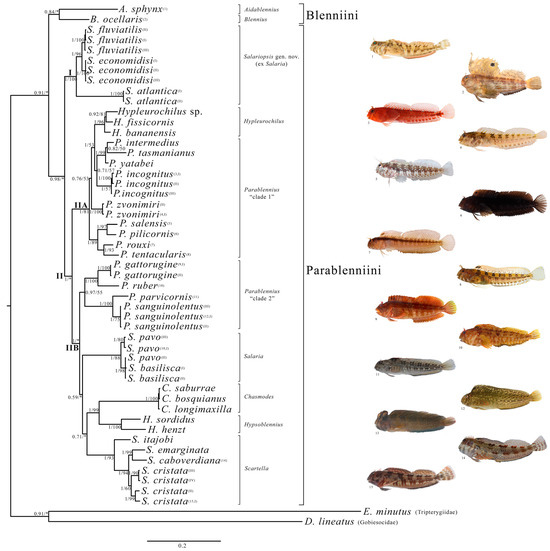
Figure 1.
Bayesian phylogram of the studied Blenniidae samples based on the concatenated mtDNA and nuDNA dataset. Node statistical support is reported as nodal posterior probabilities (Bayesian Inference of phylogeny, BI)/bootstrap values (maximum likelihood, ML). Asterisks indicate a bootstrap support value lower than 50. Square brackets group the samples according to the current taxonomy. Arabic numbers in brackets refer to the blennies’ images attached next to the phylogram. Roman numbers in brackets refer to specimens listed in Table 1. (I), freshwater Salariopsis gen. nov. (ex Salaria) clade; (II), marine Parablenniini clade; (IIA), Hypleurochilus and Parablennius “clade 1” subclades; (IIB), Parablennius “clade 2”, Salaria, Chasmodes, Hypsoblennius and Scartella subclades.
4. Discussion
The phylogenetic trees obtained in the present study highlight some important inconsistencies in the current taxonomy of Parablenniini: (i) the genera Parablennius and Salaria are paraphyletic; (ii) some alleged Parablennius species cluster with the genus Hypleurochilus; (iii) a scarce-to-absent genetic differentiation was observed between the three species belonging to the genus Chasmodes. Our study strongly supports prior findings which suggested a sharp differentiation between the marine and freshwater species currently ascribed to the genus Salaria (e.g., [12,17]). To date, three species are formally described within the freshwater clade of Salaria: the widespread S. fluviatilis; S. economidisi, endemic to Lake Trichonis (Greece), and S. atlantica, endemic to Morocco. Moreover, a further undescribed taxon of putative species rank occurs in the Middle East (see [31,32]).
This group of freshwater blennies is deeply divergent from its alleged marine congeneric taxa, by an extent much greater than that reported by Doadrio et al. [33], Hundt et al. [12] and Vecchioni et al. [17], thus stressing the inappropriateness of their current generic assignment. Even though some studies (e.g., [34]) found some clear osteological differences between S. pavo and S. fluviatilis, to date, the absence of morphological synapomorphies is a recurrent issue [35]). Based on these results, the taxonomical status of the freshwater species currently ascribed to the genus Salaria must be revised. Considering that the type taxon of the genus Salaria is S. basilisca (Valenciennes, 1836) (see also [36]), the species of the marine clade belong to Salaria s.s.. Conversely, no genus-level epithet is available for the divergent freshwater clade currently ascribed to “Salaria”. We propose the new genus Salariopsis, which includes the species Salariopsis fluviatilis, S. economidisi and S. atlantica.
5. Systematics
Family: Blenniidae Rafinesque, 1910
Genus: Salariopsis new genus (Zoobank link LSID: http://zoobank.org/urn:lsod:zoobank.org:pub:1884E670-F6E7-48F8-AF7F-19380579DB8)
Type species of the genus: Salariopsis fluviatilis (Asso, 1801)
Synonyms: none
Etymology: By adding the suffix—opsis, from the ancient Greek ὄψῐς (view, appearance), to the epithet “Salaria”, we want to highlight its apparent, but misleading, morphological similarity to the blenniid genus Salaria Forsskål, 1775
Morphological diagnosis: Fishes of the genus Salariopsis and Salaria have many overlapping meristic counts. However, Salariopsis possess fewer soft dorsal and anal fin elements than Salaria. In fact, Salariopsis has 16–17 dorsal and 16–19 anal fin rays, whereas Salaria has 22–25 and 23–28 fin rays, respectively (see Table 2).

Table 2.
Meristic data compiled from literature for comparison of fin element counts. Superscripts indicate source: a Bath [8], b Kottelat [37], c Doadrio et al. [33], and d Tiralongo [20]. Presence of two spines in the pelvic fins of Salaria atlantica could not be confirmed. Tiralongo [20] added two new observations from S. basilisca: a specimen with 28 anal fin rays and another with 2 pelvic fin rays.
The novel data used in this study provided results in accordance with previous molecular studies of the Almadablennius clade (e.g., [11,12,16,17,35]): the genus Parablennius s.l. proved to be paraphyletic, supporting the likely presence of at least two distinct and distantly related genera currently joined together within this name. Furthermore, the genus Hypleurochilus s.l. was nested within Parablennius (Figure 1, subclade “IIA”). Considering that these two genera share similar morphological features [35] and that the phylogenetic relationships are not in accordance with the current systematics, the taxonomic status of these two genera should be reassessed.
The remaining results largely agree with previous phylogenetic studies and taxonomy, while also providing direction for future studies of speciation and phylogeography. For example, a clade containing Chasmodes, Scartella, and Hypsoblennius was recovered, similar to previous studies (e.g., [16]).
The genus Chasmodes includes three species, Chasmodes saburrae, C. bosquianus and C. longimaxilla. Recently, Javonillo and Harold [38] highlighted the existence of a scarce interspecific divergence among the species of this genus, and their sister group relationship with a clade including the genera Scartella, Hypsoblennius and Hypleurochilus based on 12S mitochondrial DNA sequences. Our results are partially in contrast to those reported by Javonillo and Harold [38]. In fact, even if we detected a very low interspecific divergence (mean uncorrected p-distance about 0.22%) among the Chasmodes species and a sister group relationship between Chasmodes and the genera Scartella and Hypsoblennius, we did not observe the same phylogenetic relationship with the Hypleurochilus taxa (see Figure 1). This is probably due to our richer sampling effort, which includes more species than those investigated by Javonillo and Harold [38]. The scarce differentiation detected between the Chasmodes species is possibly related to their recent origin linked to sea-level fluctuations, as proposed by Javonillo and Harold [38]. However, bearing in mind that these species have a different ecology, phenotypic plasticity, i.e., an adaptive response to different local habitats and ecology, might be playing a major role in driving the diversification of the three Chasmodes lineages and might be accountable for their morphological variations.
Finally, our phylogenetic analyses confirm the monophyly of the genus Scartella, as already proposed by other authors [12,35,39], finding a sister clade relationship of this genus with the clade that includes Chasmodes spp. and Hypsoblennius spp. Within the Scartella clade, an uncorrected p-distance of 5.23% separating the Mediterranean versus the Atlantic specimens of S. cristata (see Table 1) suggests the possible presence of well-characterised parapatric lineages within this species, whose taxonomical rank should be the object of dedicated research.
Author Contributions
Conceptualization, P.J.H., M.A. and A.M.S.; methodology, P.J.H., A.C.C., F.M. and L.V.; writing—original draft preparation, L.V. and P.J.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available in GenBank (see Table 1 for their Accession Numbers).
Acknowledgments
We wish to thank Stamatis Zogaris (Hellenic Centre for Marine Research, Greece) for the help he provided us with the collection of Greek samples of Salariopsis economidisi (Hellenic Ministry of Environment and Energy licence no. 173241/1497/27-8-2018). Samuel P. Iglésias (Muséum National d’Histoire Naturelle, France) and Francesco Tiralongo (Ente Fauna Marina Mediterranea, Italy) kindly provided blenniid photos included in Figure 1.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Fricke, R.; Eschmeyer, W.N.; Fong, J.D. Species by Family/Subfamily. 2018. Available online: http://researcharchive.calacademy.org/research/ichthyology/catalog/SpeciesByFamily.asp (accessed on 10 December 2021).
- Norman, J.R. Notes on the blennioid fishes. I. A provisional synopsis of the genera of the family Blenniidae. Ann. Mag. Nat. Hist. 1943, 11, 793–812. [Google Scholar] [CrossRef]
- Springer, V.G. Osteology and classification of the fishes of the family Blenniidae. Bull. Am. Mus. Nat. 1968, 284, 1–85. [Google Scholar]
- Springer, V.G.; Smith-Vaniz, W.F. A new tribe (Phenablenniini) and genus (Phenablennius) of blenniid fishes based on Petroscirtes heyligeri Bleeker. Copeia 1972, 1, 64–71. [Google Scholar] [CrossRef]
- Smith-Vaniz, W.F.; Springer, V.G. Synopsis of the tribe Salariini, with description of five new genera and three new species (Pisces: Blenniidae). Smithson. Cont. Zool. 1971, 73, 1–72. [Google Scholar] [CrossRef][Green Version]
- Springer, V.G. Synopsis of the tribe Omobranchini with descriptions of three new genera and two new species (Pisces: Blenniidae). Smithson. Cont. Zool. 1972, 130, 1–31. [Google Scholar] [CrossRef]
- Smith-Vaniz, W.F. The saber-toothed blennies, tribe Nemophini (Pisces, Blenniidae). Monogr. Acad. Nat. Sci. Phila. 1976, 19, 1–196. [Google Scholar]
- Bath, H. Revision der Blenniini (Pisces: Blenniidae). Senckenb. Biol. 1977, 57, 167–234. [Google Scholar]
- Zander, C.D. Kritische Anmerkungen zur “Revision der Blenniini (Pisces: Blenniidae)” von H. Bath (1977). J. Zool. Syst. Evol. Res. 1978, 16, 290–296. [Google Scholar] [CrossRef]
- Bock, M.; Zander, C.D. Osteological characters as tool for blenniid taxonomy—A generic revision of European Blenniidae (Percomorphi; Pisces). J. Zool. Syst. Evol. Res. 1986, 24, 138–143. [Google Scholar] [CrossRef]
- Almada, F.; Almada, V.C.; Guillemaud, T.; Wirtz, P. Phylogenetic relationships of the north-eastern Atlantic and Mediterranean blenniids. Zool. J. Linn. Soc. 2005, 86, 283–295. [Google Scholar] [CrossRef]
- Hundt, P.J.; Iglésias, S.P.; Hoey, A.S.; Simons, A.M. A multilocus molecular phylogeny of combtooth blennies (Percomorpha: Blennioidei: Blenniidae): Multiple invasions of intertidal habitats. Mol. Phylogenetics Evol. 2014, 70, 47–56. [Google Scholar] [CrossRef]
- Williams, J.T. Phylogenetic relationships and revision of the blenniid fish genus Scartichthys. Smithson. Contrib. Zool. 1990, 492, 1–30. [Google Scholar] [CrossRef]
- Almada, V.C.; Robalo, J.I.; Levy, A.; Freyhof, J.; Bernardi, G.; Doadrio, I. Phylogenetic analysis of Peri-Mediterranean blennies of the genus Salaria: Molecular insights on the colonization of freshwaters. Mol. Phylogenetics Evol. 2009, 52, 424–431. [Google Scholar] [CrossRef] [PubMed]
- Levy, A.; Wirtz, P.; Floeter, S.R.; Almada, V.C. The Lusitania Province as a center of diversification: The phylogeny of the genus Microlipophrys (Pisces: Blenniidae). Mol. Phylogenetics Evol. 2011, 58, 409–413. [Google Scholar] [CrossRef] [PubMed]
- Hundt, P.J.; Simons, A.M. Extreme dentition does not prevent diet and tooth diversification within combtooth blennies (Ovalentaria: Blenniidae). Evolution 2018, 72, 930–943. [Google Scholar] [CrossRef]
- Vecchioni, L.; Marrone, F.; Belaiba, E.; Tiralongo, F.; Bahri-Sfar, L.; Arculeo, M. The DNA barcoding of Mediterranean combtooth blennies suggests the paraphyly of some taxa (Perciformes. Blenniidae). J. Fish Biol. 2019, 94, 339–344. [Google Scholar] [CrossRef]
- Nakabo, T. (Ed.) Fishes of Japan with Pictorial Keys to the Species, 3rd ed.; Tokai University Press: Tokyo, Japan, 2013. [Google Scholar]
- Iglésias, S.P. Actinopterygians from the North-Eastern Atlantic and the Mediterranean (A Natural Classification Based on Collection Specimens, with DNA Barcodes and Standardized Photographs; Provisional Version 08, 1 April 2012; MNHN: Paris, France, 2013; Volume I (plates), 245p, Available online: http://www.mnhn.fr/iccanam (accessed on 16 March 2021).
- Tiralongo, F. Blennies of the Mediterranean Sea; Amazon Fulfillment: Poland, 2020. [Google Scholar]
- Ostellari, L.; Bargelloni, L.; Penzo, E.; Patarnello, P.; Patarnello, T. Optimization of single-strand conformation polymorphism and sequence analysis of the mitochondrial control region in Pagellus bogaraveo (Sparidae. Teleostei): Rationalized tools in fish population biology. Anim. Genet. 1996, 27, 423–427. [Google Scholar] [CrossRef]
- Li, C.; Orti, G.; Zhang, G.; Lu, G. A practical approach to phylogenomics: The phylogeny of ray-finned fish (Actinopterygii) as a case study. BMC Ecol. Evol. 2007, 7, 1–11. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting. position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Farris, J.S.; Kallersjo, M.; Kluge, A.G.; Bult, C. Testing significance of incongruence. Cladistics 1995, 10, 315–319. [Google Scholar] [CrossRef]
- Swofford, D.L. Phylogenetic Analysis Using Parsimony (*and Other Methods); Version 4; Sinauer Associate: Sunderland, MA, USA, 2004. [Google Scholar]
- Cunningham, C.W. Can three incongruence tests predict when data should be combined? Mol. Biol. Evol. 1997, 14, 733–740. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; Van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes v. 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Gascuel, O. A simple. fast. and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Confidence-limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Belaiba, E.; Marrone, F.; Vecchioni, L.; Bahri-Sfar, L.; Arculeo, M. An exhaustive phylogeny of the combtooth blenny genus Salaria (Pisces. Blenniidae) shows introgressive hybridization and lack of reciprocal mtDNA monophyly between the marine species Salaria basilisca and Salaria pavo. Mol. Phylogenetics Evol. 2019, 135, 210–221. [Google Scholar] [CrossRef]
- Wagner, M.; Zogaris, S.; Berrebi, P.; Freyhof, J.; Koblmüller, S.; Magnan, P.; Laporte, M. Diversity and biogeography of Mediterranean freshwater blennies (Blenniidae, Salaria). Divers. Distrib. 2021, 27, 1832–1847. [Google Scholar] [CrossRef]
- Doadrio, I.; Perea, S.; Yahyaoui, A. A new species of the genus Salaria Forsskål, 1775 (Actinopterygii, Blennidae) in Morocco. Graellsia 2011, 67, 151–173. [Google Scholar] [CrossRef]
- Ferrito, V.; Mauceri, A.; Minniti, F.; Isaja, M.; Maisano, M.; Tigano, C. Comparative morphological studies of the neurocranium and the gills of two species of blennies living in different habitats. Acta Histochem. 2007, 109, 428–436. [Google Scholar] [CrossRef] [PubMed]
- Levy, A.; von der Heyden, S.; Floeter, S.R.; Bernardi, G.; Almada, V.C. Phylogeny of Parablennius Miranda Ribeiro, 1915 reveals a paraphyletic genus and recent Indo-Pacific diversification from an Atlantic ancestor. Mol. Phylogenetics Evol. 2013, 67, 1–8. [Google Scholar] [CrossRef]
- Krupp, F.; Schneider, W. The Fishes of the Jordan River Drainage Basin and Azraq Oasis; Pro Entomologia c/o Natural History Museum: Basel, Switzerland, 1989. [Google Scholar]
- Kottelat, M. Salaria economidisi, a new species of freshwater fish from Lake Trichonis, Greece, with comments on variation in S. fluviatilis (Teleostei: Blenniidae). Rev. Suisse Zool. 2004, 111, 121–137. [Google Scholar] [CrossRef]
- Javonillo, R.; Harold, A.S. A systematic review of the genus Chasmodes (Teleostei: Perciformes: Blenniidae). Zootaxa 2010, 2558, 1–16. [Google Scholar] [CrossRef]
- Araujo, G.S.; Vilasboa, A.; Britto, M.R.; Bernardi, G.; von der Heyden, S.; Levy, A.; Floeter, S.R. Phylogeny of the comb-tooth blenny genus Scartella (Blenniiformes: Blenniidae) reveals several cryptic lineages and a trans-Atlantic relationship. Zool. J. Linn. Soc. 2020, 190, 54–64. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).