Abstract
In this manuscript we report on the regioselective electrophilic substitution at C(7) of 7-[2-deoxy-3,5-bis-O-(2-methylpropanoyl)-ß-D-erythro-pentofuranosyl]-2-(formylamino)-4-methoxy-7H-pyrrolo[2,3-d]pyrimidine (2) – a protected precursor of 7-deaza-2’-deoxyguanosine - using N,N-dimethyl-methyleniminium iodide (Eschenmoser’s salt) yielding the Mannich compound 4.
Most naturally occurring 7-deazapurine ribonucleosides [1,2] – both of the adenosine as well as of the guanosine type – carry substituents at the 7-position, and many of them which are found in tRNA represent Mannich bases such as the nucleoside “Q” {2-amino-5-(4,5-cis-dihydroxy-1-cyclopenten-3-yl-trans-aminomethyl-(7-ß-D-ribofuranosyl)-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, Queuosine, 1}. Many attempts have been made to direct a Mannich side chain regioselectively into position 7. In contrast to 7-deazaadenosine derivatives [3,4] this is not an easy task for 7-deazaguanosine precursors [5] and requires chemical detours [6]. For example, regioselective C(7) Mannich alkylation (morpholine, HCOH/HOAc) can be performed at 3’,5’-bis-O-toluoylated 6-methoxy-2-(methylthio)-7-deazapurine 2’-deoxy-ß-D-ribonucleoside, followed by a three-step aglycone conversion to 7-deazaguanine and deprotection of the glycone [7].
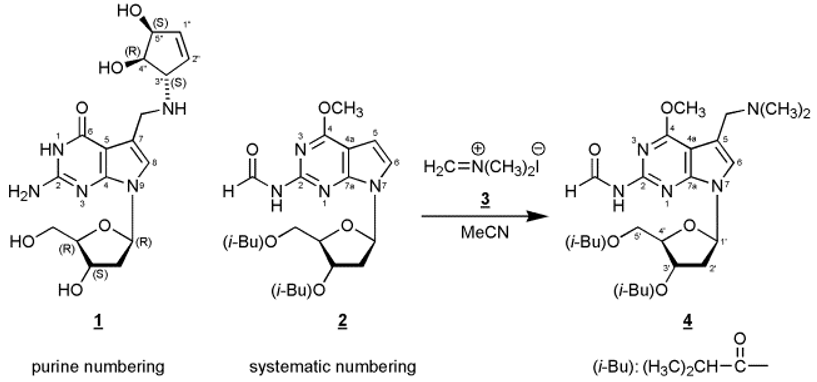
It has been reported for 7-deazaguanines that the position of electrophilic substitution strongly depends on the particular substituent pattern of the base as well as of the reaction conditions [8,9,10]. Of decisive importance is the observation that a free 2-amino group directs the electrophilic attack into the undesired 8-position (position 6 using systematic numbering) of the 7-deazaguanine moiety. This is the result of mesomeric stabilization of the σ-complex formed during electrophilic attack at the 8-position. On the other hand, 2-acylamino-7-deazaguanine derivatives form the desired 7-substituted compounds [8]
In this communication we disclose that reaction of the fully protected 7-deaza-2’-deoxy-7-deazaguanosine derivative 2 [8] with N,N-dimethyl-methyleniminium iodide (Eschenmoser’s salt) afforded the C(7) alkylated Mannich compound 4 in moderate yield. This reaction presents a new and alternative route to 7-deazapurine nucleosides with a Mannich side chain in position 7. The structure of 4 was unequivocally assigned by 1H- and 13C-NMR spectroscopy as well as by 1H-NMR NOE difference spectroscopy (see Experimental Procedure). 13C-NMR resonances were assigned applying DEPT-135 and [C,H]HETCOR spectra.
A further transformation of the tertiary amino group of 4 into other Mannich compounds can be accomplished after quarternization. Furthermore, the synthetic or enzymatic incorporation of Mannich bases derived from compounds such as compound 4 allows the introduction of reporter groups in a favourable position of a DNA molecule because the Mannich side chain protrudes into the major groove of a B-DNA double helix.
Experimental Procedure
7-[2-Deoxy-3,5-bis-O-(2-methylpropanoyl)-ß-D-erythro-pentofuranosyl]-5-(dimethylaminomethyl)-2-(formylamino)-4-methoxy-7H-pyrrolo[2,3-d]pyrimidine (4).
Compound 2 (305 mg, 0.7 mmol) was dissolved in acetonitrile (3 mL) and N,N-dimethyl-methyleniminium iodide (3, Eschenmoser’s Salz, 260 mg, 1.4 mmol) was added. After heating to 80°C for 24h, the reaction mixture was evaporated to a small volume, and compound 4 (67 mg, 20 %) was isolated as an amorphous solid by thick-layer chromatography (chloroform/methanol, 9:1, two developments, Rf, 0.4), elution from the silica gel with methanol and centrifugation. 1H-NMR ((D6)DMSO, 500.1 MHz): δ 10.73 (1H, d, HC=O, J = 9.9 Hz); 9.44 (1H, d, NH, J = 9.3 Hz); 7.31 (1H, s, H-C(6)); 6.49 (1H, dd, H-C(1’), J = 6.4, 8.0 Hz); 5.36 (1H, dd, H-C(3’), J = 2.8, 3.6 Hz); 4.28 - 4.17 (3H, m, H-C(4’), H2-C(5’)); 4.03 (3H, s, OCH3); 3.67 (2H, s, CH2); 2.86 (1H, m, iBu-CH); 2.63 – 2.57 (2H, m, H2-C(2’)); 2.47 (1H, m, iBu-CH); 2.28 (6H, s, N(CH3)2); 1.15 and 1.09 (12H, 4 iBu-CH3). 1H-NMR NOE Data ((D6)DMSO): proton irradiated: H-C(1’). NOE observed: H-C(6), 1.5 %; Hα-C(2’), 2.5 %; H-C(4’), 1.5 %. 13C-NMR ((D6)DMSO, 125.8 MHz): δ 175.6, 175.8 (2 C=O, iBu); 163.4 (C-4); 163.3 (C=O, formyl); 152.4 (C-7a); 152.0 (C-2); 121.6 (C-6); 111.4 (C-5); 101.5 (C-4a); 82.9 (C-1’); 81.1 (C-4’); 74.2 (C-3’); 63.6 (C-5’); 53.8 (CH2); 53.7 (OCH3); 44.1 and 33.1 (4 Me, iBu); 35.9 (C-2’); 18.6 (2 Me, N(CH3)2).
Supplementary materials
Supplementary File 1Supplementary File 2Supplementary File 3References and Notes
- Suhadolnik, R.J. Nucleosides As Biological Probes; John Wiley & Sons: New York, 1979; pp. 158–169. [Google Scholar]
- Limbach, P.A.; Crain, P.F.; McCloskey, J.A. Nucleic Acids Res 1994, 22, 2183.
- Watanabe, S.I.; Ueda, T. Nucleosides Nucleotides 1983, 2, 113.
- West, R.A. J. Org. Chem. 1961, 26, 4959. [CrossRef]
- Seela, F.; Lüpke, U. Chem. Ber. 1977, 110, 1462. [CrossRef]
- Seela, F.; Richter, R. Chem. Ber. 1978, 111, 2925.
- Seela, F.; Chen, Y.; Zulauf, M. Synthesis 1997, 1067.
- Ramzaeva, N.; Seela, F. Helv. Chim. Acta 1995, 78, 1083. [CrossRef]
- Akimoto, H.; Imamiya, E.; Hitaka, T.; Nomura, H.; Nishimura, S. J. Chem. Soc. Perkin Trans 1 1988, 1637.
- Benghiat, E.; Crooks, P.A. J. Heterocycl. Chem. 1983, 20, 1023. [CrossRef]
- #Purine numbering has been used within the General Part of the manuscript, systematic numbering within the Experimental Part.
© 2007 by MDPI (http://www.mdpi.org/). Reproduction is permitted for noncommercial purposes.