Abstract
Colorectal cancer (CRC) is one of the leading causes of mortality worldwide, with rising cases in individuals under 50 years old, classified as early-onset CRC (EO-CRC). EO-CRC is characterized by having clinical features related to a worse prognosis and outcome. This underscores the critical need for early detection biomarkers. ncRNAs emerge as potential biomarkers for diagnosis, prognosis, and treatment response in other types of cancers. Sequencing data from the NCBI Bioproject PRJNA787417 were analyzed to identify differentially expressed miRNAs in early- and late-onset colorectal cancer (EO-CRC and LO-CRC). Differential expressions were assessed with a log fold change threshold of 1 and an adjusted p-value of 0.05. Predicted mRNA targets were identified via ENCORI and analyzed for pathway enrichment using the SHINYGO algorithm. RNA-seq analysis identified a 25-ncRNA EO-CRC signature, including hsa-miR-195 (downregulated) and hsa-miR-549a (upregulated), with enrichment analyses suggesting associations with MAPK, PI3K, VEGF, and KRAS pathways commonly linked to angiogenesis, migration, and invasion. This preliminary report highlights a 25-gene deregulated signature in EO-CRC, in which hsa-miR-195 and hsa-miR-549a emerge as biomarkers of clinical relevance, regulating key genes involved in angiogenesis, migration, and invasion. Their dysregulation could contribute to the aggressive clinical features and poor outcomes observed in EO-CRC.
1. Introduction
Colorectal cancer (CRC) is one of the leading causes of morbidity and mortality worldwide, with a rising incidence in adults under 50 years old, classified as early-onset CRC (EO-CRC). It is projected that the incidence of colorectal cancer (CRC) among young adults in the United States will rise by 1.1% annually for colon tumors and 1.7% for rectal tumors, resulting in 10% of all colon cancers and 22% of all rectal cancers being diagnosed in this demographic by 2030 [1]. EO-CRC more commonly presents distinct clinical features, such as left-sided and rectal distribution, high proportion of mucinous and signet ring cells, poorer cell differentiation, and peritoneal metastasis [2,3]. These aggressive features are linked to a markedly lower survival rate in this population. In 2023, the Gastroenterology Department at Mexico’s National Cancer Institute reported that 30% of deaths caused by colorectal cancer occurred among younger patients [4], highlighting the necessity of identifying molecular biomarkers for early detection in this population.
Several germline and somatic mutations have been identified in CRC patients, with APC, MSH6, KRAS, TP53, BRAF, and SMAD4 being among the most prevalent genetic alterations. However, although EO-CRC displays more aggressive clinicopathological features than cases diagnosed at older ages, current evidence indicates that their somatic and germline mutational profiles are largely similar between them and therefore do not adequately explain the differences in tumor behavior [5]. This provides an opportunity to investigate additional molecular mechanisms and to identify novel non-genetic biomarkers that may better explain EO-CRC biology.
Previous studies have demonstrated that non-coding RNAs (ncRNAs), specifically microRNAs (miRNAs) and long non-coding RNAs (lncRNAs), regulate key molecular processes and exhibit differential expressions in cancer patients. Consequently, ncRNAs have been proposed as potential biomarkers for diagnosis, prognosis, and treatment response in several types of cancer. For prostate cancer, PCA3 is the first lncRNA approved by the FDA (Food and Drug Administration) as a diagnostic biomarker more specific than the prostatic antigen [6]. In the EO-CRC context, several miRNA signatures have been proposed. For instance, hsa-miR-193a-5p, hsa-miR-210, hsa-miR-513a-5p, and hsa-miR-628-3p have been shown to diagnose colorectal cancer in young adults with high precision through liquid biopsy, with their expression decreasing after surgery—underscoring their diagnostic and prognostic potential [7]. These findings emphasize the need to develop additional biomarkers. Therefore, the aim of this preliminary report is to present early bioinformatic evidence supporting a 25-gene deregulated signature in EO-CRC, from which hsa-miR-195 and hsa-miR-549a showed particularly relevant clinical associations, as recognized at the ESMO Gastrointestinal Congress 2025 [8]. By sharing these initial results, we seek to emphasize the potential impact of these molecules and encourage their further investigation.
2. Results
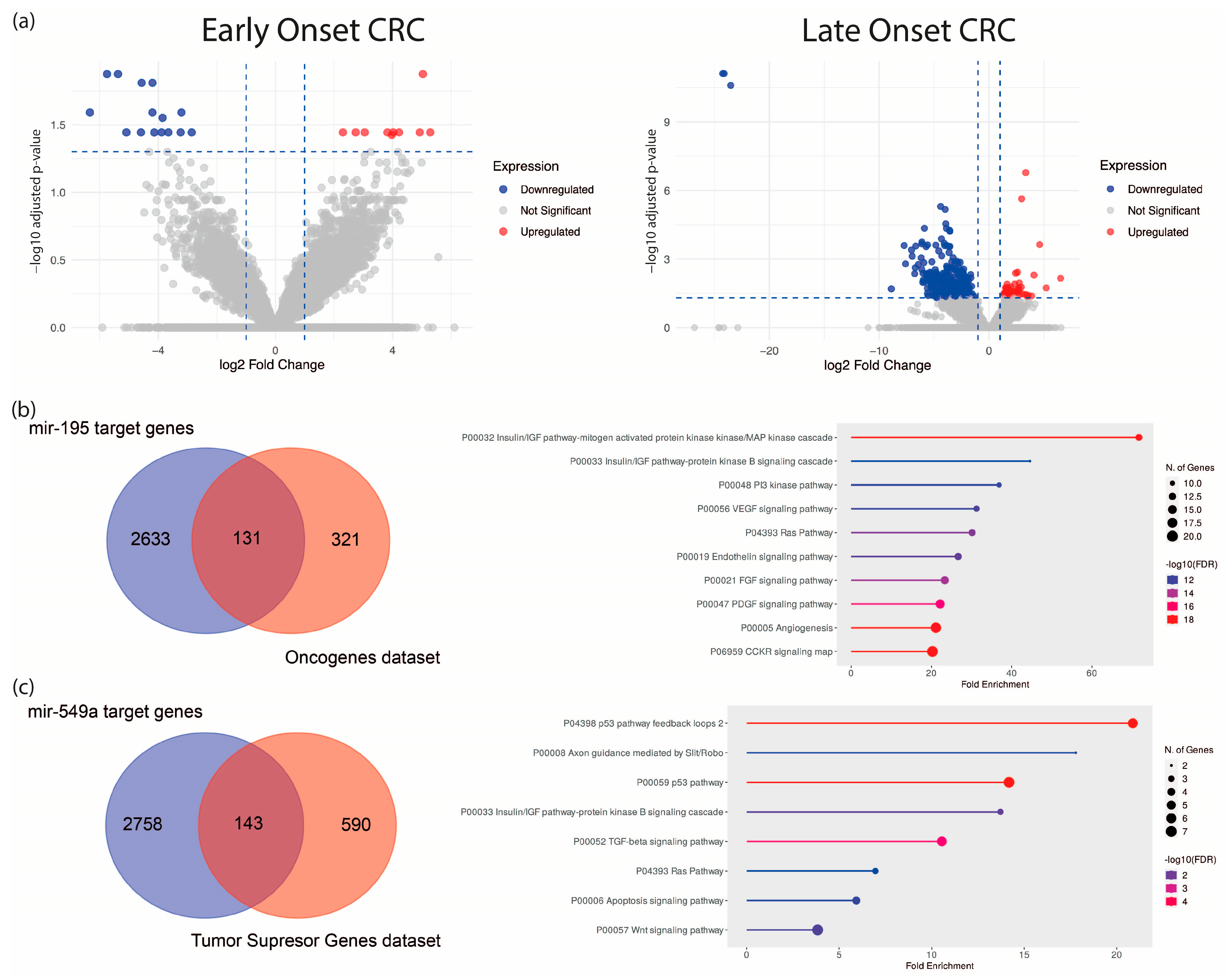
To establish a specific molecular signature of EO-CRC, we analyzed an RNA-seq dataset from eight EO-CRC and five LO-CRC patients (Bioproject PRJNA787417). Differential expression analysis revealed 25 dysregulated ncRNA genes in EO-CRC and 451 in LO-CRC (normalized to their respective age-matched healthy control groups), suggesting distinct ncRNA expression profiles between both groups, with greater dysregulation in older patients (Figure 1a, Table 1 and Table 2, and Supplementary Table S1). To refine the EO-CRC-specific signature, we used a Venn diagram to distinguish ncRNAs unique to EO-CRC from those shared with LO-CRC. This analysis identified 21 ncRNAs exclusive to EO-CRC and four shared between both groups, which are hsa-miR-195, hsa-miR-549a, GAS5, and a novel transcript sense-intronic to GK. Notably, hsa-miR-195 was downregulated, while hsa-miR-549a was upregulated. Among the identified signatures, hsa-miR-195 and hsa-miR-549a emerged as the only miRNAs, and their shared dysregulation across EO-CRC and LO-CRC suggests that they could represent conserved regulatory molecules potentially involved in early colorectal tumorigenesis across age groups.
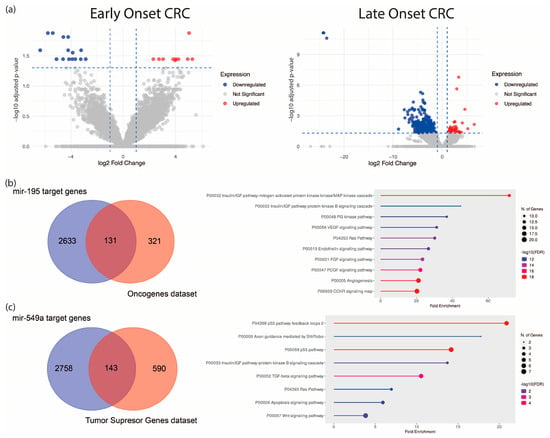
Figure 1.
Differential expression analysis of ncRNAs in EO-CRC and LO-CRC. (a) Volcano plot of dysregulated ncRNAs in both groups. (b) Predicted hsa-miR-195 targets intersecting with tumor suppressor genes and their enriched pathways. (c) Predicted hsa-miR-549a targets intersecting with oncogenes and associated oncogenic pathways.

Table 1.
Up- and downregulated ncRNAs in EO-CRC. Values are expressed in log2 FoldChange. In red, miRNAs shared between EO-CRC and LO-CRC are highlighted.

Table 2.
Up- and downregulated ncRNAs in LO-CRC. Summary of the 10 most overexpressed and downregulated ncRNAs in LO-CRC patients. The complete list comprising all 451 ncRNAs is provided in the Supplementary Materials.
To explore their functional relevance, we predicted hsa-miR-195 and hsa-miR-549a target genes using the ENCORI algorithm, identifying 2764 and 2901 predicted targets, respectively. These were cross-referenced via a Venn diagram with oncogene and tumor suppressor gene datasets (OncoKB™ Cancer Gene List), respectively. The resulting genes were subjected to gene set enrichment analysis, which implied an association with major oncogenic pathways, including MAPK, PI3K, VEGF, and KRAS (Figure 1b,c).
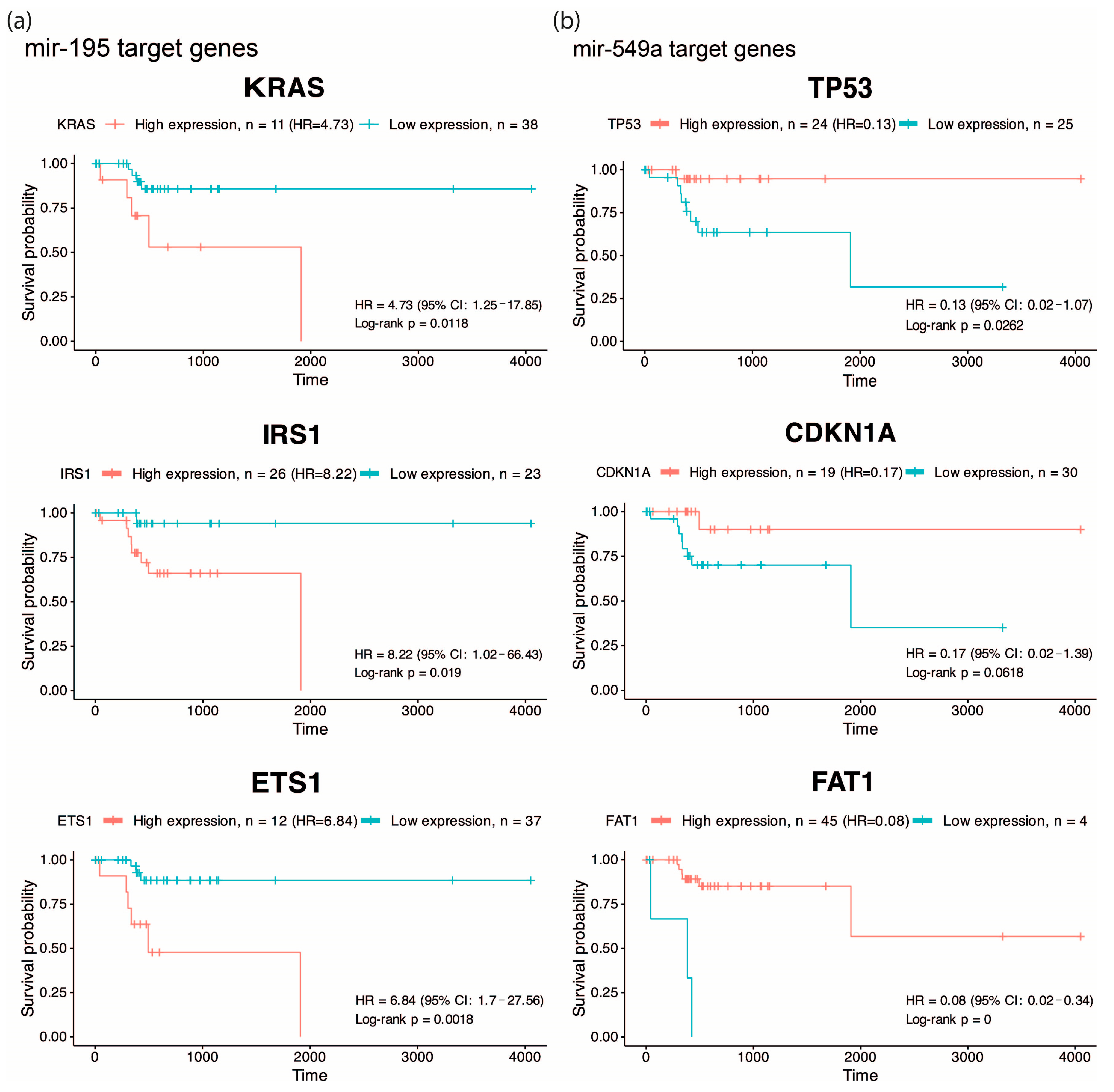
One of the most important features of EO-CRC is its aggressive clinical presentation at diagnosis, frequently associated with peritoneal metastasis. Accordingly, we focused on pathways related to angiogenesis, migration and cellular invasion, and by overall survival analysis, we found that high expression of KRAS, IRS1, ETS1 (targets of hsa-miR-195, Figure 2a) and low expression of TP53, CDKN1A and FAT1 (targets of hsa-miR-549a, Figure 2b) were associated with low survival (HR: 4.73, 8.22, 6.84, 0.13, 0.17 and 0.08), suggesting that hsa-miR-195 and hsa-miR-549a target genes may serve as potential prognostic biomarkers in CRC. Altogether, these findings highlight hsa-miR-195 and hsa-miR-549a as promising regulators of key oncogenic pathways in CRC.
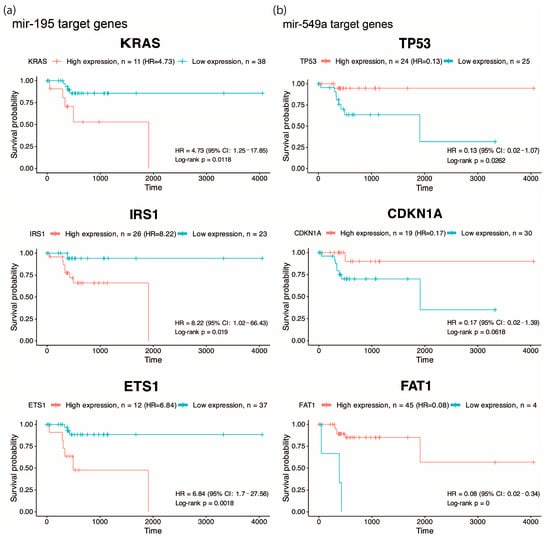
Figure 2.
Overall survival analysis of predicted targets. Kaplan–Meier curves showing overall survival according to the expression of KRAS, IRS1, and ETS1 (predicted targets of hsa-miR-195) (a), and TP53, CDKN1A, and FAT1 (predicted targets of hsa-miR-549a) (b) in EO-CRC patients, highlighting their association with poor prognosis..
3. Discussion
The present study provides an initial characterization of ncRNA dysregulation in EO-CRC based on a focused, well-defined cohort, identifying molecules with potential clinical relevance that warrant further evaluation in larger and independent populations. In this context, our results highlight miR-195 and miR-549a as clinically relevant biomarkers in EO-CRC. Consistent with previous reports, miR-195 downregulation has been associated with CRC progression, in part through the dysregulation of oncogenic pathways such as Wnt/β-catenin signaling, a molecular axis frequently altered in early-onset disease [9,10,11]. Other studies have indicated that Kras and PIK3 signaling pathways are essential to EO-CRC establishment [5,12], which are predicted to be targeted genes of miR-195 (Figure 1b,c). In the KrasG12D-Cre mouse model, miR-195 levels were reduced at 10 weeks of age; however, at 30 weeks of age, miR-195 restored basal levels, suggesting that downregulation of this miRNA and Kras mutation could be necessary to allow early tumor initiation [13]. In addition, in endothelial cells, breast cancer, and metabolic disease, the miR-195/IRS1 axis is described as a regulator of PI3K/AKT pathways, promoting tumor growth and tissue remodeling when miR-195 is downregulated [14,15,16]. For instance, in gastric cancer, miR-195-5p was shown to suppress ETS1, a known oncogenic factor whose elevated expression promotes proliferation and invasion [17]. Furthermore, there is evidence about miR-195’s relevance in clinical studies. Its low expression is associated with poor outcomes in 140 CRC formalin-fixed paraffin-embedded samples [18]; further, in CRC plasma, the miR-195 levels were significantly lower than in healthy control plasma, emerging as a potential CRC biomarker [19,20].
On the other hand, miR-549a is poorly studied in cancer and other diseases; it has been investigated for its function as a circulating diagnostic biomarker for multiple sclerosis, but without significant results [21]. Additionally, it has been studied as a prognostic/predictive biomarker in locally advanced adenocarcinomas of the gastroesophageal junction, where its overexpression was correlated with a positive therapy response [22]. Meanwhile, an advanced breast cancer clinical study indicated that miR-549a overexpression was negatively associated with clinical outcome in 59 fulvestrant-treated patients [23]. Specifically, in CRC, there is only one study that reveals that elevated levels of miR-549a in CRC tissue and plasma were associated with the invasion of tumor depth (p = 0.044), and according to the results of ROC analysis, miR-549a may serve to diagnose CRC [24]. Although miR-549a has been less explored in colorectal cancer, one of its predicted targets, FAT1, emerges as a molecule of relevance. In our analysis, low FAT1 expression was strongly associated with reduced overall survival, suggesting a potential tumor-suppressive role in EO-CRC (Figure 2b). This observation aligns with genomic studies reporting that FAT1 frequently harbors mutations in EO-CRC, including cohorts of African, Asian, and Western patients [25,26,27,28]. Given that loss-of-function mutations in FAT1 can impair its regulatory activity, the recurrent alterations described in these studies raise the possibility that genomic disruption of FAT1 may contribute to its reduced expression, thereby amplifying the deleterious effects predicted from miR-549a-mediated regulation.
Sequencing genomic studies indicate that the mutational landscape of colorectal cancer (encompassing genes such as APC, MSH6, KRAS, TP53, BRAF, and SMAD4) does not substantially differ between EO-CRC and LO-CRC [5]. This has raised growing interest in non-genetic regulation, including epigenetic and post-transcriptional mechanisms, as potential contributors to the aggressive clinical phenotype of EO-CRC. In this context, reduced miR-195 expression in CRC has been reported to be regulated through competing endogenous RNA mechanisms involving several lncRNAs (SNHG1, AFAP1-AS1, and PEG13) as well as circRNAs (circSEMA5A, circ_0060927, and circRASSF2) [11,29,30,31]. Moreover, growing evidence suggests that environmental exposures may contribute to EO-CRC through microbiome-associated and epigenetic mechanisms capable of modulating ncRNA regulatory networks, highlighting the importance of future investigations aimed at elucidating how microbiome alterations influence EO-CRC development in young populations [32].
4. Materials and Methods
4.1. ncRNA Identification
To identify differentially expressed ncRNAs between patients with EO-CRC and late-onset colorectal cancer LO-CRC, sequencing data were accessed from the NCBI bioproject PRJNA787417 [33]. The data set included samples from seven young healthy patients (ages 32–47), eight young patients with colorectal cancer (ages 33–47), five late healthy patients (ages 60–72), and five patients with late colorectal cancer (ages 60–64). Raw sequencing data were downloaded in FASTQ format using the Sequence Read Archive (SRA) with applied trimming and filtering. Contaminating 3′ adapter sequences (TGGAATTCTCGGGTGCCAAGG) were removed using Cutadapt (v.5.0) within the SystemPipe framework parameters, m, q20, and trim-n, ensuring high-confidence reads that are suitable for the downstream analysis. Sequence alignment was performed with Bowtie2 (Jhons Hopkins University, Baltimore, MD, USA, v.2.5.4) against the ENSEMBL Index of non-coding RNAs, corresponding to Homo_sapiens.GRCh38.113.chr.gtf, with the parameters-verysensitive and -N1 (allows maximum 1 mistmatch). The resulting SAM files were converted to BAM using Rsamtools (v.2.20.0), and alignment counts (excluding genomic mapping) were generated with this same package. Finally, differential expression analysis was performed using DESeq2 (v.1.44.0), comparing CRC patients with age-matched healthy controls (EO-CRC vs. young healthy controls, and LO-CRC vs. late healthy individuals), the threshold considered for differential expression was log2 fold change > 1 or <−1, and an adjusted p-value of <0.05. Based on these results, we selected only miR-195 and miR-549a for subsequent analysis.
4.2. mRNA Target Prediction and Pathway Enrichment Analysis
To predict the mRNA targets of miR-195 and miR-549a, we employed the ENCORI (Yang Lab, Xiamen, China, starBase v2.0) algorithm, and as selection criteria, the interactions of miRNA-target were performed by multiple target-predicting programs (PITA, miRanda, PicTar, and TargetScan). Due to their level of expression, we considered miR-195 as a tumor suppressor miR and miR-549a as an oncomiR. Thus, by a Venn diagram, we compared the predicted target genes for each miRNA with an oncogene and tumor suppressor gene dataset to obtain the curated target genes, which were loaded into Shiny GO (South Dakota State University, Brookings, SD, USA, v.0.85.1) gene-set enrichment tool using a 0.05 p-value cutoff of FDR.
4.3. Survival Analysis
Clinical and RNAseq data corresponding to the TCGA colon and rectal adenocarcinoma cohorts were downloaded using the Bioconductor TCGAbiolinks package (v.2.32.0). A total of 49 colorectal cancer patients aged ≤ 50 years of age, with complete clinical and expression data, were included in the survival analysis. The median follow-up duration was 457 days (range: 0–4051 days), during which nine death events were recorded. The expression of each gene was evaluated as a continuous variable and divided into high and low expression groups using the survminer package (v.0.5.1). Kaplan–Meier curves were compared between groups using a log-rank test using the R (RStudio Inc., Boston, MA, US, v.4.0.1+748) survival package (v.3.8.3).
5. Conclusions
This preliminary analysis reveals a 25-ncRNA signature in EO-CRC and identifies miR-195 and miR-549a as pivotal molecules with potential clinical relevance. The biological evidence supporting miR-195 loss and miR-549a overexpression provides a plausible molecular framework for the aggressive behavior observed in young patients. These findings, presented at the ESMO Gastrointestinal Congress 2025, highlight the promise of these miRNAs as candidate non-invasive biomarkers, whose clinical utility will require further validation in independent cohorts and liquid biopsy-based studies.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms27031379/s1.
Author Contributions
Conceptualization, J.C.-H., F.R.-I., G.C.-R., and Y.S.-P.; Methodology, F.R.-I., B.C.-L. and A.X.-M.; Software, F.R.-I.; Formal analysis, F.R.-I.; Investigation, J.C.-H., C.P.-P., and E.O.M.-S.; Writing—review and editing, J.C.-H., J.A.M.-G., and C.M.G.-C.; Visualization, G.C.-R., C.M.G.-C., and Y.S.-P.; Supervision, B.C.-L. and J.A.M.-G.; Funding acquisition, J.C.-H. and Y.S.-P. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Secretaria de Ciencia, Humanidades, Tecnología e Innovación (SECIHTI), México (grant number: CBF-2025-I-1783).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in this study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Acknowledgments
F.R.-I. is grateful to SECIHTI for a graduate student scholarship (fellowship 4018214).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Harrold, E.; Latham, A.; Pemmaraju, N.; Lieu, C.H. Early-Onset GI Cancers: Rising Trends, Genetic Risks, Novel Strategies, and Special Considerations. Am. Soc. Clin. Oncol. Educ. Book 2023, 43, e398068. [Google Scholar] [CrossRef]
- Wu, C.W.K.; Lui, R.N. Early-Onset Colorectal Cancer: Current Insights and Future Directions. World J. Gastrointest. Oncol. 2022, 14, 230–241. [Google Scholar] [CrossRef]
- Weinberg, B.A.; Marshall, J.L. Colon Cancer in Young Adults: Trends and Their Implications. Curr. Oncol. Rep. 2019, 21, 3. [Google Scholar] [CrossRef]
- Carbajal, B.; Coronel-Hernández, J.; Herrera, M.; Ruiz-Garcia, E.; Miyagui-Adame, S.M.; Diaz-Romero, C.; Osiris, E.; Esponda-Mendoza, P.M.; Carlos, P.; Calderillo-Ruiz, G. Age as a Predictor of Overall Survival in Colorectal Cancer. Diagnostic 2024, 14, 2550. [Google Scholar] [CrossRef] [PubMed]
- Storandt, M.H.; Shi, Q.; Eng, C.; Lieu, C.; George, T.; Stoppler, M.C.; Mauer, E.; Yilma, B.; Fragkogianni, S.; Teslow, E.A.; et al. Genomic Landscapes of Early-Onset Versus Average-Onset Colorectal Cancer Populations. Cancers 2025, 17, 836. [Google Scholar] [CrossRef] [PubMed]
- Shu, X.; Wang, J.; Cai, W.; Lin, S.; Li, J.; Ma, X.; Ying, Y.; Wang, Y.S.T.; Wang, X.; Chen, H.; et al. Clinical Assessment of Urinary Prostate Cancer Antigen 3 in Chinese Population: A Large-Scale, Prospective and Multicenter Study. World J. Surg. Oncol. 2024, 22, 355. [Google Scholar] [CrossRef]
- Nakamura, K.; Hernández, G.; Sharma, G.G.; Wada, Y.; Banwait, J.K.; González, N.; Perea, J.; Balaguer, F.; Takamaru, H.; Saito, Y.; et al. A Liquid Biopsy Signature for the Detection of Patients With Early-Onset Colorectal Cancer. Gastroenterology 2022, 163, 1242–1251.e2. [Google Scholar] [CrossRef]
- Coronel-Hernández, J.; Ruiz, G.C.; Rodríguez-Izquierdo, F.; Muñoz, A.X.; Carbajal-López, B.; Trujano-Camacho, S.; Pérez-Yépez, E.A.; Pérez-Plasencia, C. MiR-195 and MiR-549a as Potential Biomarkers for Early-Onset Colorectal Cancer. Ann. Oncol. 2025, 36, S41. [Google Scholar] [CrossRef]
- Li, B.; Wang, S.; Wang, S. MiR-195 Suppresses Colon Cancer Proliferation and Metastasis by Targeting WNT3A. Mol. Genet. Genom. 2018, 293, 1245–1253. [Google Scholar] [CrossRef]
- Zhang, X.; Xu, J.; Jiang, T.; Liu, G.; Wang, D.; Lu, Y. MicroRNA-195 Suppresses Colorectal Cancer Cells Proliferation via Targeting FGF2 and Regulating Wnt/β-Catenin Pathway. Am. J. Cancer Res. 2016, 6, 2631–2640. [Google Scholar]
- Yang, L.; Bi, T.; Zhou, S.; Lan, Y.; Zhang, R. CircRASSF2 Facilitates the Proliferation and Metastasis of Colorectal Cancer by Mediating the Activity of Wnt/β-Catenin Signaling Pathway by Regulating the MiR-195-5p/FZD4 Axis. Anti-Cancer Drugs 2021, 32, 919–929. [Google Scholar] [CrossRef]
- Kirzin, S.; Marisa, L.; Guimbaud, R.; De Reynies, A.; Legrain, M.; Laurent-Puig, P.; Cordelier, P.; Pradère, B.; Bonnet, D.; Meggetto, F.; et al. Sporadic Early-Onset Colorectal Cancer Is a Specific Sub-Type of Cancer: A Morphological, Molecular and Genetics Study. PLoS ONE 2014, 9, e103159. [Google Scholar] [CrossRef]
- Rachagani, S.; Macha, M.A.; Menning, M.S.; Dey, P.; Pai, P.; Smith, L.M.; Mo, Y.Y.; Batra, S.K. Changes in MicroRNA (MiRNA) Expression during Pancreatic Cancer Development and Progression in a Genetically Engineered KrasG12D;Pdx1-Cre Mouse (KC) Model. Oncotarget 2015, 6, 40295–40309. [Google Scholar] [CrossRef]
- Huang, Q.; Zhou, H.; Yu, S. Long Non-coding RNA PEG13 Regulates Endothelial Cell Senescence through the MicroRNA-195/IRS1 Axis. Exp. Ther. Med. 2023, 26, 584. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, X.; Zou, C.; Kung, H.F.; Lin, M.C.; Dress, A.; Wardle, F.; Jiang, B.H.; Lai, L. MiR-195 Inhibits Tumor Growth and Angiogenesis through Modulating IRS1 in Breast Cancer. Biomed. Pharmacother. 2016, 80, 95–101. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Jeong, W.T.; So, Y.S.; Lim, H.B.; Lee, J. Taxifolin and Sorghum Ethanol Extract Protect against Hepatic Insulin Resistance via the MiR-195/IRS1/PI3K/AKT and AMPK Signalling Pathways. Antioxidants 2021, 10, 1331. [Google Scholar] [CrossRef]
- Sui, H.; Zhu, C.; Li, Z.; Yang, J. Propofol Suppresses Gastric Cancer Tumorigenesis by Modulating the Circular RNA-PVT1/MiR-195-5p/E26 Oncogene Homolog 1 Axis. Oncol. Rep. 2020, 44, 1736–1746. [Google Scholar] [CrossRef]
- Bayat, A.; Raad, M.; Sharafshah, A.; Ahmadvand, M.; Aminian, H. Identification of MiR-195-5p as a Novel Prognostic Biomarker for Colorectal Cancer. Mol. Biol. Rep. 2022, 49, 6453–6457. [Google Scholar] [CrossRef]
- Al-Sheikh, Y.A.; Ghneim, H.K.; Softa, K.I.; Al-Jobran, A.A.; Al-Obeed, O.; Mohamed, M.A.V.; Abdulla, M.; Aboul-Soud, M.A.M. Expression Profiling of Selected MicroRNA Signatures in Plasma and Tissues of Saudi Colorectal Cancer Patients by QPCR. Oncol. Lett. 2016, 11, 1406–1412. [Google Scholar] [CrossRef] [PubMed]
- Wada, Y.; Shimada, M.; Murano, T.; Takamaru, H.; Morine, Y.; Ikemoto, T.; Saito, Y.; Balaguer, F.; Bujanda, L.; Pellise, M.; et al. A Liquid Biopsy Assay for Noninvasive Identification of Lymph Node Metastases in T1 Colorectal Cancer. Gastroenterology 2021, 161, 151–162.e1. [Google Scholar] [CrossRef]
- Montazeri, M.; Eskandari, N.; Mansouri, R. Evaluation of the Expressed MiR-129 and MiR-549a in Patients with Multiple Sclerosis. Adv. Biomed. Res. 2021, 10, 48. [Google Scholar] [CrossRef] [PubMed]
- Odenthal, M.; Hee, J.; Gockel, I.; Sisic, L.; Schmitz, J.; Stoecklein, N.H.; Driemel, C.; Möhlendick, B.; Schmidt, T.; Knoefel, W.T.; et al. Serum MicroRNA Profiles as Prognostic/Predictive Markers in the Multimodality Therapy of Locally Advanced Adenocarcinomas of the Gastroesophageal Junction. Int. J. Cancer 2015, 137, 230–237. [Google Scholar] [CrossRef]
- Torrisi, R.; Vaira, V.; Giordano, L.; Destro, A.; Basilico, V.; Mazzara, S.; Salvini, P.; Gaudioso, G.; Fernandes, B.; Rudini, N.; et al. Predictors of Fulvestrant Long-Term Benefit in Hormone Receptor-Positive/HER2 Negative Advanced Breast Cancer. Sci. Rep. 2022, 12, 12789. [Google Scholar] [CrossRef]
- Akbar, S.; Mashreghi, S.; Kalani, M.R.; Valanik, A.; Ahmadi, F.; Aalikhani, M.; Bazi, Z. Blood MiRNAs MiR-549a, MiR-552, and MiR-592 Serve as Potential Disease-Specific Panels to Diagnose Colorectal Cancer. Heliyon 2024, 10, e28492. [Google Scholar] [CrossRef]
- Yildiz, S.; Chambuso, R.; Rebello, G.; Ramesar, R. High Burden of Variants of Uncertain Significance in Early-Onset Colorectal Cancer among Indigenous African Patients: A Call for Global Research Equity in Cancer Genetics. Mol. Biol. Rep. 2025, 52, 684. [Google Scholar] [CrossRef]
- Tang, J.; Peng, W.; Tian, C.; Zhang, Y.; Ji, D.; Wang, L.; Jin, K.; Wang, F.; Shao, Y.; Wang, X.; et al. Molecular Characteristics of Early-Onset Compared with Late-Onset Colorectal Cancer: A Case Controlled Study. Int. J. Surg. 2024, 110, 4559–4570. [Google Scholar] [CrossRef]
- Holowatyj, A.N.; Wen, W.; Gibbs, T.; Seagle, H.M.; Keller, S.R.; Velez Edwards, D.R.; Washington, M.K.; Eng, C.; Perea, J.; Zheng, W.; et al. Racial/Ethnic and Sex Differences in Somatic Cancer Gene Mutations among Patients with Early-Onset Colorectal Cancer. Cancer Discov. 2023, 13, 570–579. [Google Scholar] [CrossRef]
- Li, P.; Meng, Q.; Xue, Y.; Teng, Z.; Chen, H.; Zhang, J.; Xu, Y.; Wang, S.; Yu, R.; Ou, Q.; et al. Comprehensive Genomic Profiling of Colorectal Cancer Patients Reveals Differences in Mutational Landscapes among Clinical and Pathological Subgroups. Front. Oncol. 2022, 12, 1000146. [Google Scholar] [CrossRef]
- Bai, J.; Xu, J.; Zhao, J.; Zhang, R. LncRNA SNHG1 Cooperated with MiR-497/MiR-195-5p to Modify Epithelial–Mesenchymal Transition Underlying Colorectal Cancer Exacerbation. J. Cell. Physiol. 2020, 235, 1453–1468. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhu, Z.; Hou, X.; Sun, Y. LncRNA AFAP1-AS1 Promotes the Progression of Colorectal Cancer through MiR-195-5p and WISP1. J. Oncol. 2021, 2021, 6242798. [Google Scholar] [CrossRef] [PubMed]
- Ye, Q.; Liu, S.; Lin, S.; Xie, W. Circular RNA CircSEMA5A Facilitates Colorectal Cancer Development by Regulating MicroRNA-195-5p to Target CCNE1 Axis. Cell. Signal. 2023, 107, 110649. [Google Scholar] [CrossRef] [PubMed]
- Xie, S.; Zhi, Y.; Badehnoosh, B. Microbial Modulators of the Epigenome: Probiotic Regulation of MiRNAs and LncRNAs in Health and Disease and Preventive Medicine. Gut Pathog. 2026, 18, 4. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Wu, W.; Chang, W.; Wu, R.; Sun, X.; Wu, H.; Liu, Z. MiR-31-5p-DMD Axis as a Novel Biomarker for Predicting the Development and Prognosis of Sporadic Early-onset Colorectal Cancer. Oncol. Lett. 2022, 23, 157. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.