Determination of Differential miRNA Expression Profile in People with Noise-Induced Hearing Loss
Abstract
1. Introduction
1.1. Noise-Induced Hearing Loss (NIHL) and Its Clinical Importance
1.2. Molecular Mechanisms in Cochlear Injury
1.3. Role of MicroRNAs (miRNAs) in Cochlear Pathology
1.4. miRNA Profiling in NIHL
1.5. Study Objective
2. Results
2.1. Sociodemographic and Audiometric Data in Noise-Induced Hearing Loss
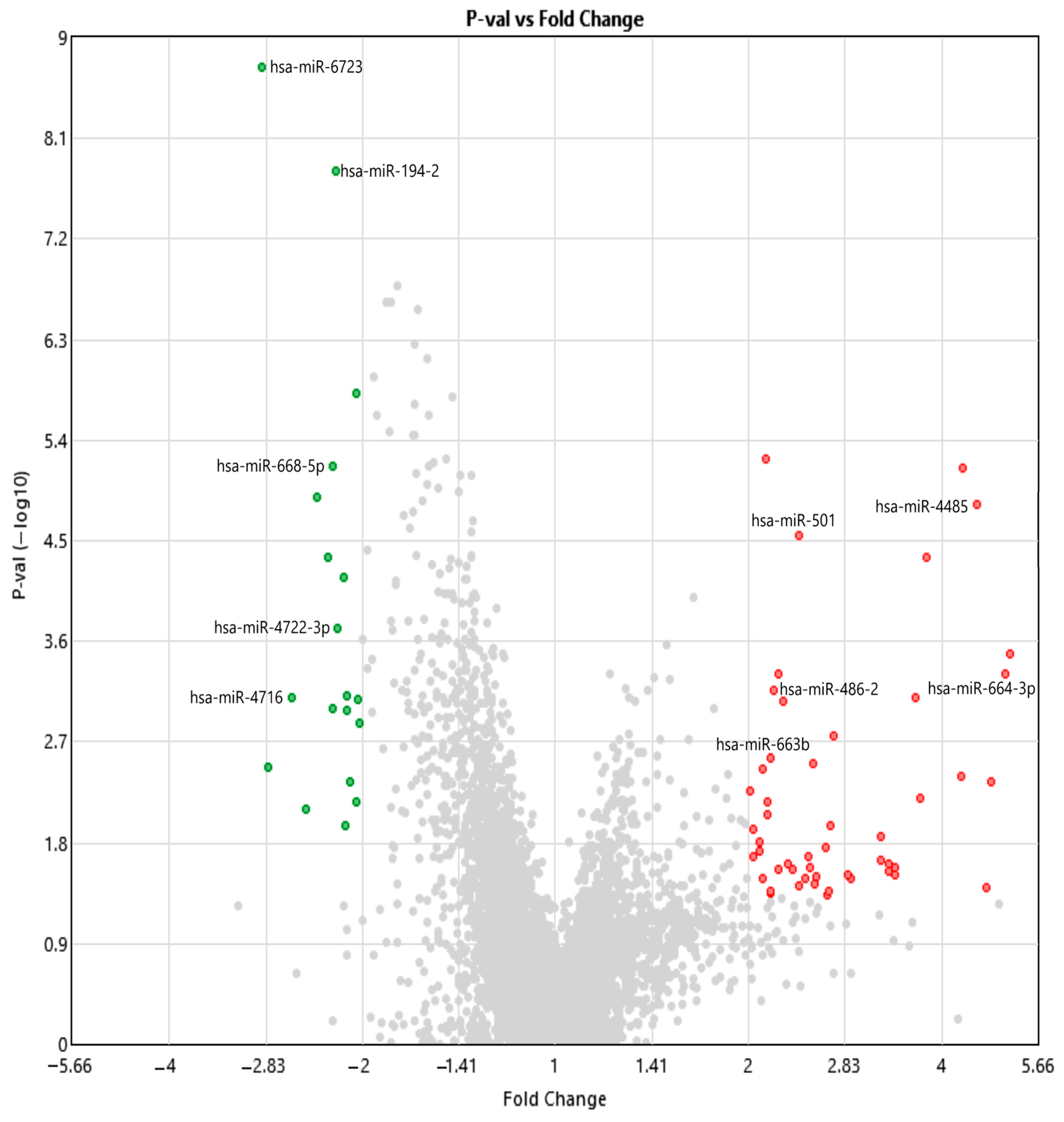
2.2. Analysis of Differentially Expressed miRNAs and Small RNAs Using Transcriptome Analysis Console
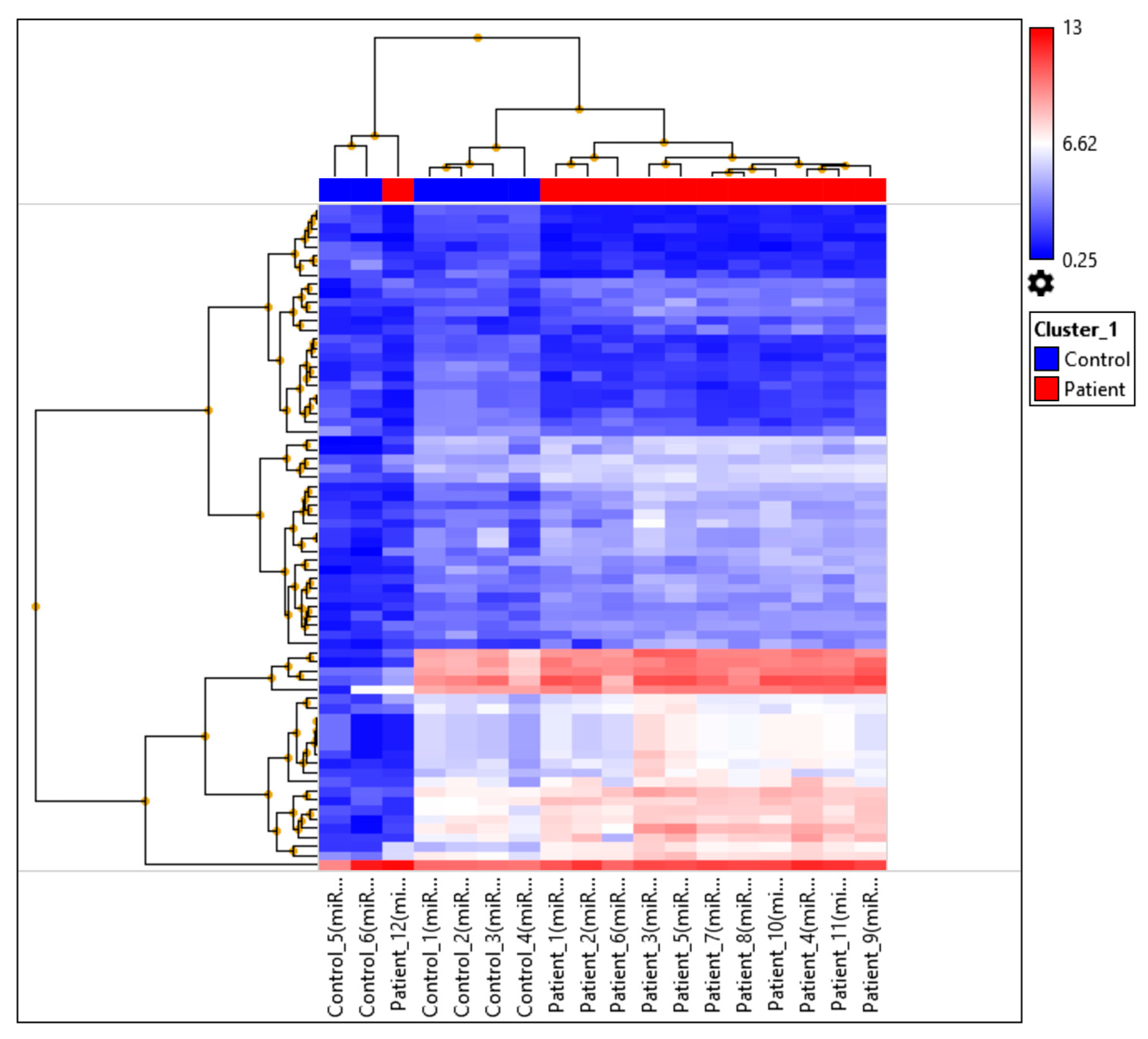
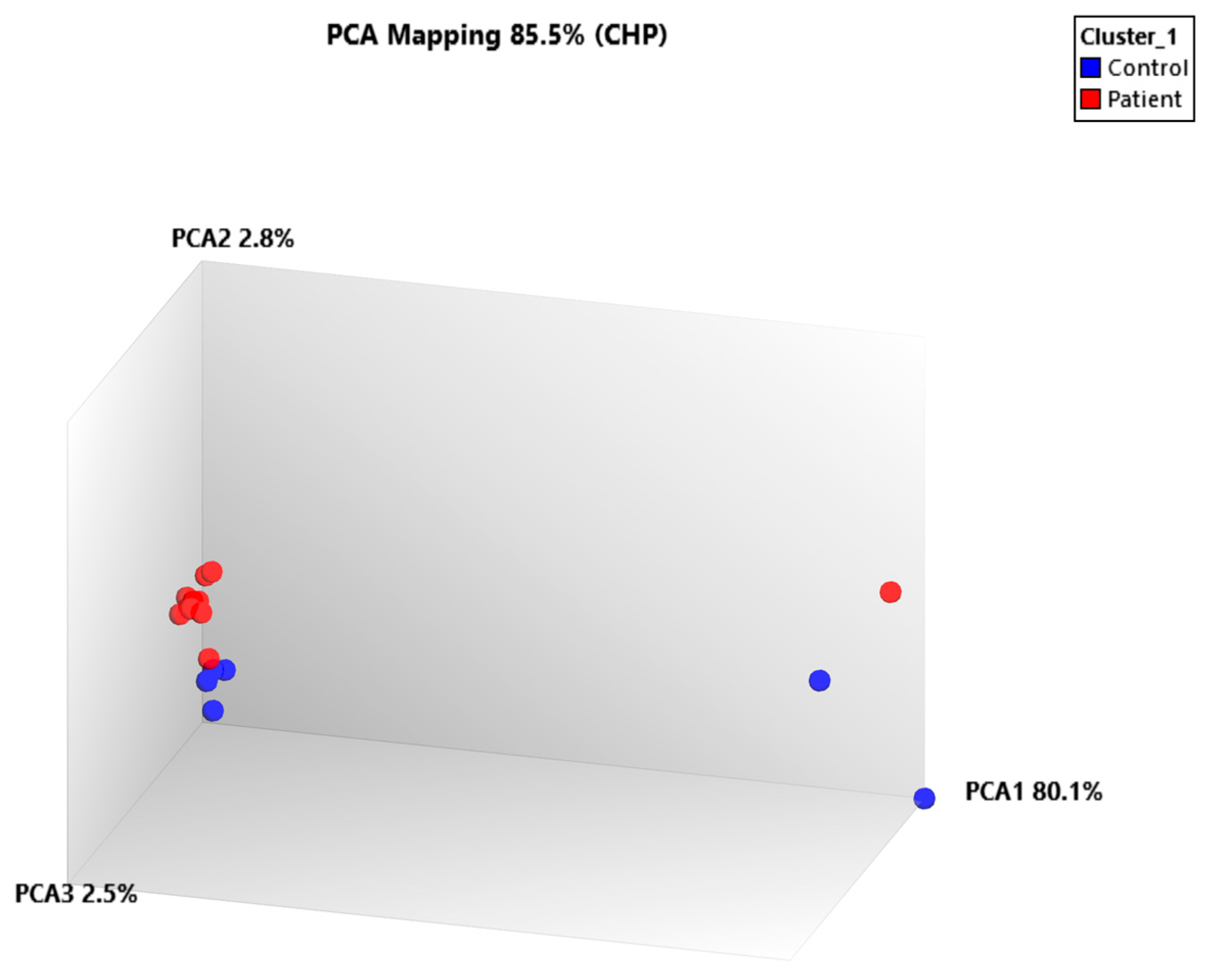
2.3. miRNA Profiling Results Based on Hierarchical Clustering and Principal Component Analysis (PCA) Mapping
2.4. Pathway Enrichment and Biomarker Candidate Analysis
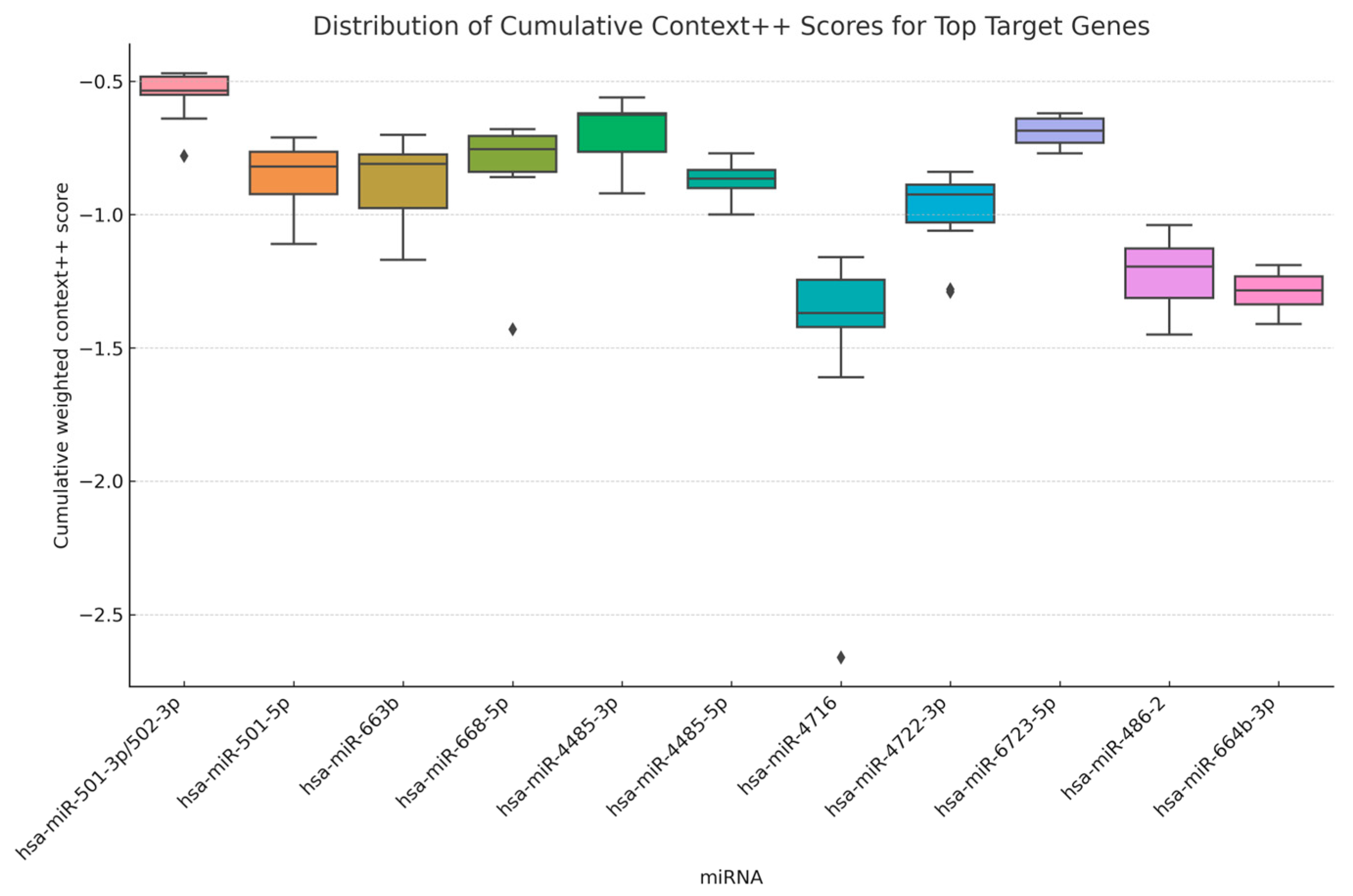
2.5. Distribution of Predicted miRNA–Target Binding Strengths
2.6. Gene Ontology (GO) and Pathway Enrichment Results
2.7. Druggability Analysis of miRNA Target Genes
3. Discussion
3.1. Overview of Key Findings and Relevance to NIHL
3.2. Differential Expression and Molecular Impact of Key miRNAs
3.3. Biological Pathways and GO Enrichment: Cell Cycle and Apoptosis
3.4. Diagnostic and Prognostic Utility of C-miRNAs
3.5. Functional Network Complexity Beyond miRNAs
3.6. Study Limitations and Future Directions
4. Materials and Methods
4.1. Clinical Specimens
4.2. Sample Processing and Quality Control
4.3. Total RNA Isolation from Blood Samples
4.4. MicroRNA Expression Array
4.5. Statistical Analysis
4.6. miRNA–Target Prediction and Score Distribution Analysis
4.7. Gene Ontology and Functional Enrichment Analysis
4.8. In Silico miRNA–mRNA Interaction and Drug Repositioning Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nelson, D.I.; Nelson, R.Y.; Concha-Barrientos, M.; Fingerhut, M. The global burden of occupational noise-induced hearing loss. Am. J. Ind. Med. 2005, 48, 446–458. [Google Scholar] [CrossRef]
- Wong, A.C.; Ryan, A.F. Mechanisms of sensorineural cell damage, death and survival in the cochlea. Front. Aging Neurosci. 2015, 7, 58. [Google Scholar] [CrossRef] [PubMed]
- Kobel, M.; Le Prell, C.G.; Liu, J.; Hawks, J.W.; Bao, J. Noise-induced cochlear synaptopathy: Past findings and future studies. Hear. Res. 2017, 349, 148–154. [Google Scholar] [CrossRef]
- Masterson, E.A.; Bushnell, P.T.; Themann, C.L.; Morata, T.C. Hearing Impairment Among Noise-Exposed Workers—United States, 2003–2012. MMWR Morb. Mortal. Wkly. Rep. 2016, 65, 389–394. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.H.; Su, S.B.; Chen, K.T. An overview of occupational noise-induced hearing loss among workers: Epidemiology, pathogenesis, and preventive measures. Environ. Health Prev. Med. 2020, 25, 65. [Google Scholar] [CrossRef]
- Le, T.N.; Straatman, L.V.; Lea, J.; Westerberg, B. Current insights in noise-induced hearing loss: A literature review of the underlying mechanism, pathophysiology, asymmetry, and management options. J. Otolaryngol. Head Neck Surg. 2017, 46, 41. [Google Scholar] [CrossRef] [PubMed]
- Mao, H.; Chen, Y. Noise-Induced Hearing Loss: Updates on Molecular Targets and Potential Interventions. Neural Plast. 2021, 2021, 4784385. [Google Scholar] [CrossRef]
- Le Prell, C.G.; Hackett, T.A.; Ramachandran, R. Noise-Induced Hearing Loss and its Prevention: Current Issues in Mammalian Hearing. Curr. Opin. Physiol. 2020, 18, 32–36. [Google Scholar] [CrossRef]
- Li, G.; Yang, H.; Zhang, P.; Guo, Y.; Yuan, L.; Xu, S.; Yuan, Y.; Xiong, H.; Yin, H. Insights into the molecular underlying mechanisms and therapeutic potential of endoplasmic reticulum stress in sensorineural hearing loss. Front. Mol. Neurosci. 2024, 17, 1443401. [Google Scholar] [CrossRef]
- Henderson, D.; Bielefeld, E.C.; Harris, K.C.; Hu, B.H. The role of oxidative stress in noise-induced hearing loss. Ear Hear. 2006, 27, 1–19. [Google Scholar] [CrossRef]
- Yamashita, D.; Jiang, H.Y.; Schacht, J.; Miller, J.M. Delayed production of free radicals following noise exposure. Brain Res. 2004, 1019, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Fujioka, M.; Okano, H.; Ogawa, K. Inflammatory and immune responses in the cochlea: Potential therapeutic targets for sensorineural hearing loss. Front. Pharmacol. 2014, 5, 287. [Google Scholar] [CrossRef] [PubMed]
- Matsui, J.I.; Ogilvie, J.M.; Warchol, M.E. Inhibition of caspases prevents ototoxic and ongoing hair cell death. J. Neurosci. 2002, 22, 1218–1227. [Google Scholar] [CrossRef]
- Cheng, A.G.; Cunningham, L.L.; Rubel, E.W. Mechanisms of hair cell death and protection. Curr. Opin. Otolaryngol. Head Neck Surg. 2005, 13, 343–348. [Google Scholar] [CrossRef]
- Kujawa, S.G.; Liberman, M.C. Adding insult to injury: Cochlear nerve degeneration after “temporary” noise-induced hearing loss. J. Neurosci. 2009, 29, 14077–14085. [Google Scholar] [CrossRef]
- Wang, J.; Puel, J.L. Toward Cochlear Therapies. Physiol. Rev. 2018, 98, 2477–2522. [Google Scholar] [CrossRef]
- Mencia, A.; Modamio-Hoybjor, S.; Redshaw, N.; Morin, M.; Mayo-Merino, F.; Olavarrieta, L.; Aguirre, L.A.; del Castillo, I.; Steel, K.P.; Dalmay, T.; et al. Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss. Nat. Genet. 2009, 41, 609–613. [Google Scholar] [CrossRef] [PubMed]
- Friedman, L.M.; Dror, A.A.; Mor, E.; Tenne, T.; Toren, G.; Satoh, T.; Biesemeier, D.J.; Shomron, N.; Fekete, D.M.; Hornstein, E.; et al. MicroRNAs are essential for development and function of inner ear hair cells in vertebrates. Proc. Natl. Acad. Sci. USA 2009, 106, 7915–7920. [Google Scholar] [CrossRef]
- Solda, G.; Robusto, M.; Primignani, P.; Castorina, P.; Benzoni, E.; Cesarani, A.; Ambrosetti, U.; Asselta, R.; Duga, S. A novel mutation within the MIR96 gene causes non-syndromic inherited hearing loss in an Italian family by altering pre-miRNA processing. Hum. Mol. Genet. 2012, 21, 577–585. [Google Scholar] [CrossRef]
- Mittal, R.; Liu, G.; Polineni, S.P.; Bencie, N.; Yan, D.; Liu, X.Z. Role of microRNAs in inner ear development and hearing loss. Gene 2019, 686, 49–55. [Google Scholar] [CrossRef]
- Prasad, K.N.; Bondy, S.C. MicroRNAs in Hearing Disorders: Their Regulation by Oxidative Stress, Inflammation and Antioxidants. Front. Cell. Neurosci. 2017, 11, 276. [Google Scholar] [CrossRef] [PubMed]
- Xiong, H.; Pang, J.; Yang, H.; Dai, M.; Liu, Y.; Ou, Y.; Huang, Q.; Chen, S.; Zhang, Z.; Xu, Y.; et al. Activation of miR-34a/SIRT1/p53 signaling contributes to cochlear hair cell apoptosis: Implications for age-related hearing loss. Neurobiol. Aging 2015, 36, 1692–1701. [Google Scholar] [CrossRef] [PubMed]
- Ha, S.M.; Hwang, K.R.; Park, I.H.; Park, S.; Choi, J.S.; Park, D.J.; Park, J.E.; Lee, S.H.; Lee, H.Y.; Seo, Y.J. Circulating microRNAs as potentially new diagnostic biomarkers of idiopathic sudden sensorineural hearing loss. Acta Otolaryngol. 2020, 140, 1013–1020. [Google Scholar] [CrossRef]
- Weston, M.D.; Pierce, M.L.; Jensen-Smith, H.C.; Fritzsch, B.; Rocha-Sanchez, S.; Beisel, K.W.; Soukup, G.A. MicroRNA-183 family expression in hair cell development and requirement of microRNAs for hair cell maintenance and survival. Dev. Dyn. 2011, 240, 808–819. [Google Scholar] [CrossRef]
- Gifford, R.; JAAA Special Issue on Hearing Therapeutics and Protective Therapies. The Future of Hearing Therapeutics and Protective Therapies and the Potentially Profound Impact on Patients and Providers. J. Am. Acad. Audiol. 2021, 32, 625–626. [Google Scholar] [CrossRef]
- Park, S.; Han, S.H.; Kim, B.G.; Suh, M.W.; Lee, J.H.; Oh, S.H.; Park, M.K. Changes in microRNA Expression in the Cochlear Nucleus and Inferior Colliculus after Acute Noise-Induced Hearing Loss. Int. J. Mol. Sci. 2020, 21, 8792. [Google Scholar] [CrossRef]
- Lavoro, A.; Gattuso, G.; Grillo, C.; Spandidos, D.; Salmeri, M.; Lombardo, C.; Candido, S.; Falzone, L. Role of microRNAs as novel diagnostic biomarkers and potential therapeutic targets for hearing disorders. Int. J. Epigenetics 2022, 2, 1–12. [Google Scholar] [CrossRef]
- Wang, Z.; Dai, R.; Ahmed, S.A. MicroRNA-183/96/182 cluster in immunity and autoimmunity. Front. Immunol. 2023, 14, 1134634. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Ji, C.; Guo, S.; Su, X.; Zhao, X.; Zhang, S.; Liu, G.; Qiu, X.; Zhang, Q.; Guo, H.; et al. The miR-486-5p plays a causative role in prostate cancer through negative regulation of multiple tumor suppressor pathways. Oncotarget 2017, 8, 72835–72846. [Google Scholar] [CrossRef]
- Cortada, M.; Levano, S.; Bodmer, D. mTOR Signaling in the Inner Ear as Potential Target to Treat Hearing Loss. Int. J. Mol. Sci. 2021, 22, 6368. [Google Scholar] [CrossRef]
- Montecucco, A.; Zanetta, F.; Biamonti, G. Molecular mechanisms of etoposide. EXCLI J. 2015, 14, 95–108. [Google Scholar]
- Huang, H.Y.; Lin, Y.C.; Cui, S.; Huang, Y.; Tang, Y.; Xu, J.; Bao, J.; Li, Y.; Wen, J.; Zuo, H.; et al. miRTarBase update 2022: An informative resource for experimentally validated miRNA-target interactions. Nucleic Acids Res. 2022, 50, D222–D230. [Google Scholar] [CrossRef] [PubMed]
- McGeary, S.E.; Lin, K.S.; Shi, C.Y.; Pham, T.M.; Bisaria, N.; Kelley, G.M.; Bartel, D.P. The biochemical basis of microRNA targeting efficacy. Science 2019, 366, 6472. [Google Scholar] [CrossRef] [PubMed]
- Natarajan, N.; Batts, S.; Stankovic, K.M. Noise-Induced Hearing Loss. J. Clin. Med. 2023, 12, 2347. [Google Scholar] [CrossRef] [PubMed]
- Kurabi, A.; Keithley, E.M.; Housley, G.D.; Ryan, A.F.; Wong, A.C. Cellular mechanisms of noise-induced hearing loss. Hear. Res. 2017, 349, 129–137. [Google Scholar] [CrossRef]
- Fan, B.; Lu, F.; Du, W.J.; Chen, J.; An, X.G.; Wang, R.F.; Li, W.; Song, Y.L.; Zha, D.J.; Chen, F.Q. PTEN inhibitor bisperoxovanadium protects against noise-induced hearing loss. Neural Regen. Res. 2023, 18, 1601–1606. [Google Scholar]
- Petitpre, C.; Faure, L.; Uhl, P.; Fontanet, P.; Filova, I.; Pavlinkova, G.; Adameyko, I.; Hadjab, S.; Lallemend, F. Single-cell RNA-sequencing analysis of the developing mouse inner ear identifies molecular logic of auditory neuron diversification. Nat. Commun. 2022, 13, 3878. [Google Scholar] [CrossRef]
- Matern, M.S.; Milon, B.; Lipford, E.L.; McMurray, M.; Ogawa, Y.; Tkaczuk, A.; Song, Y.; Elkon, R.; Hertzano, R. GFI1 functions to repress neuronal gene expression in the developing inner ear hair cells. Development 2020, 147, dev186015. [Google Scholar] [CrossRef]
- Sadler, E.; Ryals, M.M.; May, L.A.; Martin, D.; Welsh, N.; Boger, E.T.; Morell, R.J.; Hertzano, R.; Cunningham, L.L. Cell-Specific Transcriptional Responses to Heat Shock in the Mouse Utricle Epithelium. Front. Cell. Neurosci. 2020, 14, 123. [Google Scholar] [CrossRef]
- Altschuler, R.A.; Wys, N.; Prieskorn, D.; Martin, C.; DeRemer, S.; Bledsoe, S.; Miller, J.M. Treatment with Piribedil and Memantine Reduces Noise-Induced Loss of Inner Hair Cell Synaptic Ribbons. Sci. Rep. 2016, 6, 30821. [Google Scholar] [CrossRef]
- Cortada, M.; Levano, S.; Hall, M.N.; Bodmer, D. mTORC2 regulates auditory hair cell structure and function. iScience 2023, 26, 107687. [Google Scholar] [CrossRef]
- Altschuler, R.A.; Kabara, L.; Martin, C.; Kanicki, A.; Stewart, C.E.; Kohrman, D.C.; Dolan, D.F. Rapamycin Added to Diet in Late Mid-Life Delays Age-Related Hearing Loss in UMHET4 Mice. Front. Cell. Neurosci. 2021, 15, 658972. [Google Scholar] [CrossRef]
- Wu, W.; Chen, P.; Yang, J.; Liu, Y. A Low Dose of Rapamycin Promotes Hair Cell Differentiation by Enriching SOX2(+) Progenitors in the Neonatal Mouse Inner Ear Organoids. J. Assoc. Res. Otolaryngol. 2024, 25, 149–165. [Google Scholar] [CrossRef] [PubMed]
- Smith-Roe, S.L.; Nakamura, J.; Holley, D.; Chastain, P.D.2nd; Rosson, G.B.; Simpson, D.A.; Ridpath, J.R.; Kaufman, D.G.; Kaufmann, W.K.; Bultman, S.J. SWI/SNF complexes are required for full activation of the DNA-damage response. Oncotarget 2015, 6, 732–745. [Google Scholar] [CrossRef]
- Altschuler, R.A.; Halsey, K.; Kanicki, A.; Martin, C.; Prieskorn, D.; DeRemer, S.; Dolan, D.F. Small Arms Fire-like noise: Effects on Hearing Loss, Gap Detection and the Influence of Preventive Treatment. Neuroscience 2019, 407, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Shi, J.; Du, J.; Chen, K.; Dong, C.; Jiang, D.; Jiang, H. MicroRNA-194 Regulates the Development and Differentiation of Sensory Patches and Statoacoustic Ganglion of Inner Ear by Fgf4. Med. Sci. Monit. 2018, 24, 1712–1723. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Zhang, X.; Cao, H.; Jiang, D.; Wang, X.; Zhou, W.; Chen, K.; Zhou, J.; Jiang, H.; Ba, L. MiR-194 is involved in morphogenesis of spiral ganglion neurons in inner ear by rearranging actin cytoskeleton via targeting RhoB. Int. J. Dev. Neurosci. 2017, 63, 16–26. [Google Scholar] [CrossRef]
- Weston, M.D.; Pierce, M.L.; Rocha-Sanchez, S.; Beisel, K.W.; Soukup, G.A. MicroRNA gene expression in the mouse inner ear. Brain Res. 2006, 1111, 95–104. [Google Scholar] [CrossRef]
- Fritzsch, B.; Pan, N.; Jahan, I.; Duncan, J.S.; Kopecky, B.J.; Elliott, K.L.; Kersigo, J.; Yang, T. Evolution and development of the tetrapod auditory system: An organ of Corti-centric perspective. Evol. Dev. 2013, 15, 63–79. [Google Scholar] [CrossRef]
- Forouzanfar, F.; Asgharzade, S. MicroRNAs in Noise-Induced Hearing Loss and their Regulation by Oxidative Stress and Inflammation. Curr. Drug Targets 2020, 21, 1216–1224. [Google Scholar] [CrossRef]
- Ruiz, M.; Gonzalez, S.; Bonnet, C.; Deng, S.X. Extracellular miR-6723-5p could serve as a biomarker of limbal epithelial stem/progenitor cell population. Biomark. Res. 2022, 10, 36. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Wu, L.; Ma, M.; Yang, L.; Qin, C. MicroRNA-668-3p regulates oxidative stress and cell damage induced by Abeta1-42 by targeting the OXR1/p53-p21 axis. Ann. Transl. Med. 2022, 10, 928. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Xu, Y.; Yu, M. MicroRNA-4722-5p and microRNA-615-3p serve as potential biomarkers for Alzheimer’s disease. Exp. Ther. Med. 2022, 23, 241. [Google Scholar] [CrossRef]
- Nandwa, J.O.; Mehmood, A.; Mahjabeen, I.; Raheem, K.Y.; Hamadou, M.; Raimi, M.; Kayani, M.A. miR-4716-3p and the target AKT2 Gene/rs2304186 SNP are associated with blood cancer pathogenesis in Pakistani population. Noncoding RNA Res. 2024, 9, 695–703. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Zhu, Y.; Liu, Y.; Liu, X.; Ding, Y.; Li, D.; Zhang, X.; Liu, Y. Tailored apoptotic vesicles promote bone regeneration by releasing the osteoinductive brake. Int. J. Oral Sci. 2024, 16, 31. [Google Scholar] [CrossRef]
- Hu, Z.; Zhao, J.; Hu, T.; Luo, Y.; Zhu, J.; Li, Z. miR-501-3p mediates the activity-dependent regulation of the expression of AMPA receptor subunit GluA1. J. Cell Biol. 2015, 208, 949–959. [Google Scholar] [CrossRef]
- Zhao, L.Y.; Xin, G.J.; Tang, Y.Y.; Li, X.F.; Li, Y.Z.; Tang, N.; Ma, Y.H. miR-664b-3p inhibits colon cell carcinoma via negatively regulating Budding uninhibited by benzimidazole 3. Bioengineered 2022, 13, 4857–4868. [Google Scholar] [CrossRef]
- Lee, B.P.; Buric, I.; George-Pandeth, A.; Flurkey, K.; Harrison, D.E.; Yuan, R.; Peters, L.L.; Kuchel, G.A.; Melzer, D.; Harries, L.W. MicroRNAs miR-203-3p, miR-664-3p and miR-708-5p are associated with median strain lifespan in mice. Sci. Rep. 2017, 7, 44620. [Google Scholar] [CrossRef]
- Xiao, F.; Chen, W.; Yu, C.; Zhao, G. MicroRNA-663b enhances migration and invasion by targeting adenomatous polyposis coli 2 in colorectal carcinoma cells. Oncol. Lett. 2020, 19, 3701–3710. [Google Scholar] [CrossRef]
- You, X.; Sun, W.; Wang, Y.; Liu, X.; Wang, A.; Liu, L.; Han, S.; Sun, Y.; Zhang, J.; Guo, L.; et al. Cervical cancer-derived exosomal miR-663b promotes angiogenesis by inhibiting vinculin expression in vascular endothelial cells. Cancer Cell Int. 2021, 21, 684. [Google Scholar] [CrossRef]
- Liu, L.; Chen, Y.; Qi, J.; Zhang, Y.; He, Y.; Ni, W.; Li, W.; Zhang, S.; Sun, S.; Taketo, M.M.; et al. Wnt activation protects against neomycin-induced hair cell damage in the mouse cochlea. Cell Death Dis. 2016, 7, e2136. [Google Scholar] [CrossRef] [PubMed]
- Shashoua, V.E.; Adams, D.S.; Volodina, N.V.; Li, H. New synthetic peptides can enhance gene expression of key antioxidant defense enzymes in vitro and in vivo. Brain Res. 2004, 1024, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Sohmer, H. Pathophysiological mechanisms of hearing loss. J. Basic Clin. Physiol. Pharmacol. 1997, 8, 113–125. [Google Scholar] [CrossRef]
- Li, H.; Wan, H.Q.; Zhao, H.J.; Luan, S.X.; Zhang, C.G. Identification of candidate genes and miRNAs associated with neuropathic pain induced by spared nerve injury. Int. J. Mol. Med. 2019, 44, 1205–1218. [Google Scholar] [CrossRef]
- Watkins, N.J.; Bohnsack, M.T. The box C/D and H/ACA snoRNPs: Key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip. Rev. RNA 2012, 3, 397–414. [Google Scholar] [CrossRef]
- Piekna-Przybylska, D.; Decatur, W.A.; Fournier, M.J. The 3D rRNA modification maps database: With interactive tools for ribosome analysis. Nucleic Acids Res. 2008, 36, D178–D183. [Google Scholar] [CrossRef]
- Bratkovic, T.; Bozic, J.; Rogelj, B. Functional diversity of small nucleolar RNAs. Nucleic Acids Res. 2020, 48, 1627–1651. [Google Scholar] [CrossRef]
- Shen, L.P.; Zhang, W.C.; Deng, J.R.; Qi, Z.H.; Lin, Z.W.; Wang, Z.D. Advances in the mechanism of small nucleolar RNA and its role in DNA damage response. Mil. Med. Res. 2024, 11, 53. [Google Scholar] [CrossRef] [PubMed]
- Kiss, T. Small nucleolar RNAs: An abundant group of noncoding RNAs with diverse cellular functions. Cell 2002, 109, 145–148. [Google Scholar] [CrossRef]
- Langhendries, J.L.; Nicolas, E.; Doumont, G.; Goldman, S.; Lafontaine, D.L. The human box C/D snoRNAs U3 and U8 are required for pre-rRNA processing and tumorigenesis. Oncotarget 2016, 7, 59519–59534. [Google Scholar] [CrossRef]
- Henras, A.K.; Plisson-Chastang, C.; O’Donohue, M.F.; Chakraborty, A.; Gleizes, P.E. An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscip. Rev. RNA 2015, 6, 225–242. [Google Scholar] [CrossRef] [PubMed]
| Hearing Loss Group | 0.5 kHz | 1 kHz | 2 kHz | 4 kHz | 6 kHz | 8 kHz |
|---|---|---|---|---|---|---|
| Ear | M (SD) | M (SD) | M (SD) | M (SD) | M (SD) | M (SD) |
| Right Ear | 28.75 | 29.58 * | 39.17 * | 57.08 ¶ | 52.50 ¶ | 58.33 ¶ |
| 17.07 | 17.90 | 21.30 | 17.25 | 21.69 | 21.99 | |
| Left Ear | 33.75 * | 33.33 | 44.17 * | 53.92 ¶ | 53.75 ¶ | 56.67 ¶ |
| 30.83 | 31.65 | 30.88 | 19.27 | 24.23 | 27.74 | |
| Control Group | 16.67 | 13.33 | 14.17 | 12.50 | 13.33 | 11.67 |
| Right Ear | 2.58 | 2.58 | 3.76 | 2.74 | 2.58 | 2.58 |
| Left Ear | 13.33 | 11.67 | 13.33 | 11.67 | 12.50 | 11.67 |
| 2.58 | 2.58 | 2.58 | 2.58 | 2.74 | 2.58 |
| ID | Control Avg (log2) | Patient Avg (log2) | Fold Change | p-Val | FDR p-Val | Transcript ID (Array Design) |
|---|---|---|---|---|---|---|
| 20537465 | 2.58 | 1.06 | −2.86 | 1.84 × 10−9 | 1.22 × 10−5 | hsa-mir-6723 |
| 20534809 | 2.23 | 1.09 | −2.21 | 1.55 × 10−8 | 5.13 × 10−5 | hsa-mir-194-2 |
| 20536918 | 2.48 | 1.44 | −2.05 | 1.49 × 10−6 | 0.001 | hsa-mir-1343 |
| 20538277 | 2.14 | 3.23 | 2.13 | 5.80 × 10−6 | 0.002 | U96b |
| 20504554 | 2.49 | 1.34 | −2.22 | 6.79 × 10−6 | 0.002 | hsa-miR-668-5p |
| 20538276 | 4.94 | 7.05 | 4.32 | 6.98 × 10−6 | 0.002 | U96a |
| 20533389 | 2.89 | 1.65 | −2.35 | 1.27 × 10−5 | 0.0028 | ENSG00000238581 |
| 20536757 | 2.39 | 4.57 | 4.54 | 1.47 × 10−5 | 0.0031 | hsa-mir-4485 |
| 20535205 | 2.83 | 4.09 | 2.39 | 2.80 × 10−5 | 0.0046 | hsa-mir-501 |
| 20532687 | 2.29 | 1.11 | −2.27 | 4.40 × 10−5 | 0.0063 | ACA50 |
| 20538104 | 2.32 | 4.24 | 3.79 | 4.49 × 10−5 | 0.0063 | SNORA38B |
| 20537539 | 2.49 | 1.39 | −2.14 | 6.60 × 10−5 | 0.0081 | hsa-mir-6796 |
| 20519556 | 2.01 | 0.88 | −2.19 | 0.0002 | 0.0162 | hsa-miR-4722-3p |
| 20534225 | 2.44 | 4.79 | 5.1 | 0.0003 | 0.021 | HBII-166 |
| 20521811 | 1.61 | 3.94 | 5.03 | 0.0005 | 0.026 | hsa-miR-664b-3p |
| 20534600 | 1.92 | 3.08 | 2.23 | 0.0005 | 0.0261 | hsa-mir-142 |
| 20537879 | 4.02 | 5.15 | 2.19 | 0.0007 | 0.0305 | hsa-mir-486-2 |
| 20532682 | 2.44 | 1.36 | −2.12 | 0.0008 | 0.0318 | ACA47 |
| 20532647 | 1.97 | 3.83 | 3.63 | 0.0008 | 0.032 | ACA3-2 |
| 20536948 | 3.98 | 2.62 | −2.58 | 0.0008 | 0.032 | hsa-mir-4716 |
| 20533559 | 2.34 | 1.32 | −2.03 | 0.0008 | 0.0327 | ENSG00000238852 |
| 20532706 | 2.35 | 3.53 | 2.26 | 0.0009 | 0.0331 | ACA62 |
| 20538129 | 2.79 | 1.63 | −2.22 | 0.001 | 0.0352 | U108 |
| 20538128 | 2.82 | 1.73 | −2.12 | 0.001 | 0.0357 | U108 |
| 20505964 | 1.94 | 0.93 | −2.02 | 0.0013 | 0.042 | hsa-miR-924 |
| 20538224 | 4.7 | 6.14 | 2.71 | 0.0018 | 0.0508 | U68 |
| 20506797 | 1.34 | 2.45 | 2.16 | 0.0027 | 0.0627 | hsa-miR-663b |
| 20532669 | 2.96 | 4.29 | 2.51 | 0.0031 | 0.0667 | ACA40 |
| 20534329 | 3.28 | 1.79 | −2.8 | 0.0033 | 0.0706 | HBII-85-6 |
| 20538223 | 6.33 | 7.4 | 2.11 | 0.0035 | 0.0727 | U68 |
| 20532651 | 2.34 | 4.44 | 4.29 | 0.0039 | 0.0755 | ACA32 |
| 20536823 | 3.29 | 2.22 | −2.09 | 0.0045 | 0.0792 | hsa-mir-4539 |
| 20532671 | 2.41 | 4.67 | 4.79 | 0.0045 | 0.0798 | ACA41 |
| 20532621 | 2.06 | 3.07 | 2.01 | 0.0055 | 0.0905 | ACA13 |
| 20538125 | 3.7 | 5.59 | 3.7 | 0.0063 | 0.0982 | U105 |
| 20538208 | 2.74 | 1.71 | −2.05 | 0.0069 | 0.1032 | U58A |
| 20538203 | 4.54 | 5.64 | 2.14 | 0.0069 | 0.1032 | U54 |
| 20525457 | 3.09 | 1.8 | −2.45 | 0.0078 | 0.1093 | hsa-miR-6748-5p |
| 20532648 | 2.8 | 3.9 | 2.14 | 0.0087 | 0.1142 | ACA3-2 |
| 20538109 | 8.51 | 9.94 | 2.68 | 0.0109 | 0.1248 | SNORD119 |
| 20532670 | 2.43 | 1.34 | −2.12 | 0.0111 | 0.1255 | ACA41 |
| 20500128 | 10.29 | 11.31 | 2.03 | 0.0121 | 0.1331 | hsa-miR-16-5p |
| 20538175 | 4.82 | 6.51 | 3.21 | 0.014 | 0.1434 | U3 |
| 20534374 | 2.74 | 3.79 | 2.08 | 0.0152 | 0.1499 | hsa-mir-28 |
| 20518444 | 2.87 | 4.27 | 2.64 | 0.0171 | 0.1567 | hsa-miR-3150b-3p |
| 20500151 | 8.97 | 10.03 | 2.08 | 0.0184 | 0.1635 | hsa-miR-25-3p |
| 20534249 | 4.45 | 5.47 | 2.03 | 0.0211 | 0.1751 | HBII-436 |
| 20534244 | 2.93 | 4.24 | 2.47 | 0.0211 | 0.1751 | HBII-336 |
| 20534235 | 4.68 | 6.36 | 3.2 | 0.0223 | 0.1801 | HBII-251 |
| 20534242 | 2.69 | 4.42 | 3.3 | 0.0242 | 0.1885 | HBII-296B |
| 20532652 | 2.05 | 3.25 | 2.3 | 0.0245 | 0.1898 | ACA32 |
| 20538325 | 4.72 | 6.48 | 3.39 | 0.0262 | 0.1968 | U3-2B |
| 20538326 | 4.72 | 6.48 | 3.39 | 0.0262 | 0.1968 | U3-2 |
| 20538327 | 4.72 | 6.48 | 3.39 | 0.0262 | 0.1968 | U3-3 |
| 20538328 | 4.72 | 6.48 | 3.39 | 0.0262 | 0.1968 | U3-4 |
| 20538173 | 6.98 | 8.29 | 2.49 | 0.0266 | 0.1982 | U38B |
| 20538165 | 8.5 | 9.72 | 2.34 | 0.0267 | 0.1988 | U34 |
| 20538171 | 6.98 | 8.14 | 2.23 | 0.0272 | 0.2009 | U37 |
| 20500488 | 6.33 | 8.05 | 3.3 | 0.0284 | 0.2053 | hsa-miR-223-3p |
| 20534241 | 2.65 | 4.4 | 3.38 | 0.0299 | 0.2092 | HBII-296B |
| 20538199 | 9.44 | 10.95 | 2.85 | 0.0306 | 0.2102 | U50 |
| 20538304 | 6.43 | 7.77 | 2.55 | 0.032 | 0.2157 | snR39B |
| 20525436 | 2.54 | 3.61 | 2.1 | 0.0327 | 0.2183 | hsa-miR-6737-5p |
| 20506004 | 3.52 | 5.06 | 2.89 | 0.0329 | 0.2186 | hsa-miR-935 |
| 20504364 | 5.04 | 6.33 | 2.45 | 0.0333 | 0.2193 | hsa-miR-619-5p |
| 20500442 | 1.25 | 2.59 | 2.53 | 0.0374 | 0.2336 | hsa-miR-34a-5p |
| 20538209 | 8.57 | 9.83 | 2.39 | 0.038 | 0.2356 | U58B |
| 20500769 | 4.81 | 7.04 | 4.7 | 0.0392 | 0.2387 | hsa-miR-126-3p |
| 20532632 | 4.77 | 6.18 | 2.67 | 0.0426 | 0.2471 | ACA20 |
| 20538282 | 6.49 | 7.61 | 2.16 | 0.0426 | 0.2471 | Z17B |
| 20518834 | 6.85 | 7.96 | 2.16 | 0.0435 | 0.2502 | hsa-miR-4454 |
| 20504584 | 2.63 | 4.04 | 2.66 | 0.0469 | 0.2607 | hsa-miR-378d |
| Rank | miRNA ID | PC2 Loading Score * |
|---|---|---|
| 1 | hsa-miR-486-2 | 0.1028 |
| 2 | hsa-miR-4722-3p | −0.0983 |
| 3 | hsa-miR-194-2 | −0.0914 |
| 4 | hsa-miR-664b-3p | 0.0885 |
| 5 | hsa-miR-501 | 0.0867 |
| 6 | hsa-miR-663b | 0.0791 |
| 7 | hsa-miR-4716 | −0.0756 |
| 8 | hsa-miR-6723 | −0.0719 |
| 9 | hsa-miR-668-5p | −0.0684 |
| 10 | hsa-miR-4485 | 0.0651 |
| miRNA ID | Fold Change | p-Value | FDR | Regulation |
|---|---|---|---|---|
| hsa-miR-6723 | −2.86 | 1.84 × 10−9 | 1.22 × 10−5 | ↓ Down |
| hsa-miR-194-2 | −2.21 | 1.55 × 10−8 | 5.13 × 10−5 | ↓ Down |
| hsa-miR-668-5p | −2.22 | 6.79 × 10−6 | 0.002 | ↓ Down |
| hsa-miR-4722-3p | −2.19 | 0.0002 | 0.0162 | ↓ Down |
| hsa-miR-4716 | −2.58 | 0.0008 | 0.032 | ↓ Down |
| hsa-miR-4485 | 4.54 | 1.47 × 10−5 | 0.0031 | ↑ Up |
| hsa-miR-501 | 2.39 | 2.80 × 10−5 | 0.0046 | ↑ Up |
| hsa-miR-664b-3p | 5.03 | 0.0005 | 0.026 | ↑ Up |
| hsa-miR-486-2 | 2.19 | 0.0007 | 0.0305 | ↑ Up |
| hsa-miR-663b | 2.16 | 0.0027 | 0.0627 | ↑ Up |
| miRNA | Target Gene | Gene Name | Cumulative Weighted Context++ Score |
|---|---|---|---|
| hsa-miR-501-3p/502-3p | RPRD1B | regulation of nuclear pre-mRNA domain containing 1B | −0.78 |
| hsa-miR-501-3p/502-3p | MYNN | myoneurin | −0.64 |
| hsa-miR-501-3p/502-3p | ADAMTS3 | ADAM metallopeptidase with thrombospondin type 1 motif, 3 | −0.55 |
| hsa-miR-501-3p/502-3p | ARPP21 | cAMP-regulated phosphoprotein, 21kDa | −0.55 |
| hsa-miR-501-3p/502-3p | SAMD12 | sterile alpha motif domain containing 12 | −0.54 |
| hsa-miR-501-3p/502-3p | B4GALT5 | UDP-Gal:betaGlcNAc beta 1,4-galactosyltransferase, polypeptide 5 | −0.53 |
| hsa-miR-501-3p/502-3p | SEC63 | SEC63 homolog (S. cerevisiae) | −0.52 |
| hsa-miR-501-3p/502-3p | CLIC4 | chloride intracellular channel 4 | −0.47 |
| hsa-miR-501-3p/502-3p | JDP2 | Jun dimerization protein 2 | −0.47 |
| hsa-miR-501-3p/502-3p | COL10A1 | collagen, type X, alpha 1 | −0.47 |
| hsa-miR-501-5p | XKR4 | XK, Kell blood group complex subunit-related family, member 4 | −1.11 |
| hsa-miR-501-5p | KLHDC1 | kelch domain containing 1 | −0.99 |
| hsa-miR-501-5p | PEX12 | peroxisomal biogenesis factor 12 | −0.94 |
| hsa-miR-501-5p | NLRP11 | NLR family, pyrin domain containing 11 | −0.87 |
| hsa-miR-501-5p | SPRR2A | small proline-rich protein 2A | −0.83 |
| hsa-miR-501-5p | SPRR2F | small proline-rich protein 2F | −0.81 |
| hsa-miR-501-5p | HTN3 | histatin 3 | −0.78 |
| hsa-miR-501-5p | SPRR2E | small proline-rich protein 2E | −0.76 |
| hsa-miR-501-5p | CLCA4 | chloride channel accessory 4 | −0.74 |
| hsa-miR-501-5p | LPAR1 | lysophosphatidic acid receptor 1 | −0.71 |
| hsa-miR-663b | BSPRY | B-box and SPRY domain containing | −1.17 |
| hsa-miR-663b | GPRIN2 | G protein regulated inducer of neurite outgrowth 2 | −1.05 |
| hsa-miR-663b | CSF2 | colony-stimulating factor 2 (granulocyte–macrophage) | −1 |
| hsa-miR-663b | C1orf158 | chromosome 1 open reading frame 158 | −0.9 |
| hsa-miR-663b | LL22NC03-75H12.2 | novel protein; uncharacterized protein | −0.81 |
| hsa-miR-663b | OSBPL5 | oxysterol binding protein-like 5 | −0.81 |
| hsa-miR-663b | RIBC2 | RIB43A domain with coiled-coils 2 | −0.79 |
| hsa-miR-663b | PYCR1 | pyrroline-5-carboxylate reductase 1 | −0.77 |
| hsa-miR-663b | CDKN2A | cyclin-dependent kinase inhibitor 2A | −0.71 |
| hsa-miR-663b | SHARPIN | SHANK-associated RH domain interactor | −0.7 |
| hsa-miR-668-5p | IER3 | immediate early response 3 | −1.43 |
| hsa-miR-668-5p | SEPT5 | septin 5 | −0.86 |
| hsa-miR-668-5p | NNAT | neuronatin | −0.84 |
| hsa-miR-668-5p | MED10 | mediator complex subunit 10 | −0.84 |
| hsa-miR-668-5p | TSSK6 | testis-specific serine kinase 6 | −0.78 |
| hsa-miR-668-5p | TXNL4B | thioredoxin-like 4B | −0.73 |
| hsa-miR-668-5p | C1orf216 | chromosome 1 open reading frame 216 | −0.72 |
| hsa-miR-668-5p | NSUN5 | NOP2/Sun domain family, member 5 | −0.7 |
| hsa-miR-668-5p | LDOC1L | leucine zipper, down-regulated in cancer 1-like | −0.69 |
| hsa-miR-668-5p | GBGT1 | globoside alpha-1,3-N-acetylgalactosaminyltransferase 1 | −0.68 |
| hsa-miR-4485-3p | PARK7 | parkinson protein 7 | −0.92 |
| hsa-miR-4485-3p | GALNT14 | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) | −0.79 |
| hsa-miR-4485-3p | AC012215.1 | uncharacterized protein | −0.77 |
| hsa-miR-4485-3p | PPAPDC3 | phosphatidic acid phosphatase type 2 domain containing 3 | −0.75 |
| hsa-miR-4485-3p | C14orf37 | chromosome 14 open reading frame 37 | −0.63 |
| hsa-miR-4485-3p | RNF41 | ring finger protein 41 | −0.62 |
| hsa-miR-4485-3p | SYP | synaptophysin | −0.62 |
| hsa-miR-4485-3p | GPBAR1 | G protein-coupled bile acid receptor 1 | −0.62 |
| hsa-miR-4485-3p | STARD6 | StAR-related lipid transfer (START) domain containing 6 | −0.6 |
| hsa-miR-4485-3p | AZI1 | 5-azacytidine induced 1 | −0.56 |
| hsa-miR-4485-5p | AC005003.1 | CDNA FLJ20464 fis, clone KAT06158; HCG1777549; uncharacterized protein | −1 |
| hsa-miR-4485-5p | CCDC142 | coiled-coil domain containing 142 | −0.97 |
| hsa-miR-4485-5p | RNF165 | ring finger protein 165 | −0.9 |
| hsa-miR-4485-5p | DUSP13 | dual specificity phosphatase 13 | −0,9 |
| hsa-miR-4485-5p | ZNF667 | zinc finger protein 667 | −0.89 |
| hsa-miR-4485-5p | FICD | FIC domain containing | −0.84 |
| hsa-miR-4485-5p | TNFSF14 | tumor necrosis factor (ligand) superfamily, member 14 | −0.84 |
| hsa-miR-4485-5p | RRP36 | ribosomal RNA processing 36 homolog (S. cerevisiae) | −0.83 |
| hsa-miR-4485-5p | KLHL35 | kelch-like family member 35 | −0.78 |
| hsa-miR-4485-5p | RP11-94B19.4 | uncharacterized protein | −0.77 |
| hsa-miR-4716 | FN3K | fructosamine 3 kinase | −2.66 |
| hsa-miR-4716 | KCNC3 | potassium voltage-gated channel, Shaw-related subfamily, member 3 | −1.61 |
| hsa-miR-4716 | PDX1 | pancreatic and duodenal homeobox 1 | −1.43 |
| hsa-miR-4716 | MSI1 | musashi RNA-binding protein 1 | −1.4 |
| hsa-miR-4716 | FAM222B | family with sequence similarity 222, member B | −1.38 |
| hsa-miR-4716 | MT-ND4L | mitochondrially encoded NADH dehydrogenase 4L | −1.36 |
| hsa-miR-4716 | SRF | serum response factor (c-fos serum response element-binding transcription factor) | −1.29 |
| hsa-miR-4716 | PPP2R2D | protein phosphatase 2, regulatory subunit B, delta | −1.23 |
| hsa-miR-4716 | PDCD6 | programmed cell death 6 | −1.17 |
| hsa-miR-4716 | C6orf223 | chromosome 6 open reading frame 223 | −1.16 |
| hsa-miR-4722-3p | ITPKB | inositol-trisphosphate 3-kinase B | −1.29 |
| hsa-miR-4722-3p | TLCD2 | TLC domain containing 2 | −1.28 |
| hsa-miR-4722-3p | FAM83F | family with sequence similarity 83, member F | −1.06 |
| hsa-miR-4722-3p | TRAF1 | TNF receptor-associated factor 1 | −0.94 |
| hsa-miR-4722-3p | GGT6 | gamma-glutamyltransferase 6 | −0.93 |
| hsa-miR-4722-3p | CLCN2 | chloride channel, voltage-sensitive 2 | −0.92 |
| hsa-miR-4722-3p | SYT12 | synaptotagmin XII | −0.91 |
| hsa-miR-4722-3p | NFAM1 | NFAT-activating protein with ITAM motif 1 | −0.88 |
| hsa-miR-4722-3p | PRR15L | proline rich 15-like | −0.87 |
| hsa-miR-4722-3p | SRL | sarcalumenin | −0.84 |
| hsa-miR-6723-5p | GNGT1 | guanine nucleotide-binding protein (G protein), gamma transducing activity polypeptide 1 | −0.77 |
| hsa-miR-6723-5p | CCDC90B | coiled-coil domain containing 90B | −0.75 |
| hsa-miR-6723-5p | RNF186 | ring finger protein 186 | −0.74 |
| hsa-miR-6723-5p | PSG2 | pregnancy specific beta-1-glycoprotein 2 | −0.7 |
| hsa-miR-6723-5p | CHIC1 | cysteine-rich hydrophobic domain 1 | −0.69 |
| hsa-miR-6723-5p | IL17A | interleukin 17A | −0.68 |
| hsa-miR-6723-5p | PSG11 | pregnancy specific beta-1-glycoprotein 11 | −0.64 |
| hsa-miR-6723-5p | POC1B | POC1 centriolar protein B | −0.64 |
| hsa-miR-6723-5p | TMEM114 | transmembrane protein 114 | −0.63 |
| hsa-miR-6723-5p | PSG3 | pregnancy specific beta-1-glycoprotein 3 | −0.62 |
| hsa-miR-486-2 | PTEN | phosphatase and tensin homolog | −1.45 |
| hsa-miR-486-2 | FOXO1 | Forkhead box O1 | −1.36 |
| hsa-miR-486-2 | IGF1 | insulin-like growth factor 1 | −1.32 |
| hsa-miR-486-2 | CDK2 | cyclin-dependent kinase 2 | −1.29 |
| hsa-miR-486-2 | MMP9 | matrix metallopeptidase 9 | −1.22 |
| hsa-miR-486-2 | BCL2 | B-cell lymphoma 2 | −1.17 |
| hsa-miR-486-2 | AKT1 | AKT serine/threonine kinase 1 | −1.15 |
| hsa-miR-486-2 | CCND1 | cyclin D1 | −1.12 |
| hsa-miR-486-2 | SMAD4 | SMAD family member 4 | −1.08 |
| hsa-miR-486-2 | VEGFA | vascular endothelial growth factor A | −1.04 |
| hsa-miR-664b-3p | TTK | dual specificity protein kinase | −1.41 |
| hsa-miR-664b-3p | TRIP13 | thyroid hormone receptor interactor | −1.39 |
| hsa-miR-664b-3p | MCM10 | DNA replication licensing factor | −1.34 |
| hsa-miR-664b-3p | NUSAP1 | nucleolar spindle associated protein | −1.33 |
| hsa-miR-664b-3p | CCNA2 | cyclin A2 | −1.3 |
| hsa-miR-664b-3p | BIRC5 | survivin | −1.27 |
| hsa-miR-664b-3p | CDC25C | cell division cycle 25C | −1.24 |
| hsa-miR-664b-3p | TOP2A | DNA topoisomerase II alpha | −1.23 |
| hsa-miR-664b-3p | CDCA5 | cell division cycle associated 5 | −1.21 |
| hsa-miR-664b-3p | CENPF | centromere protein F | −1.19 |
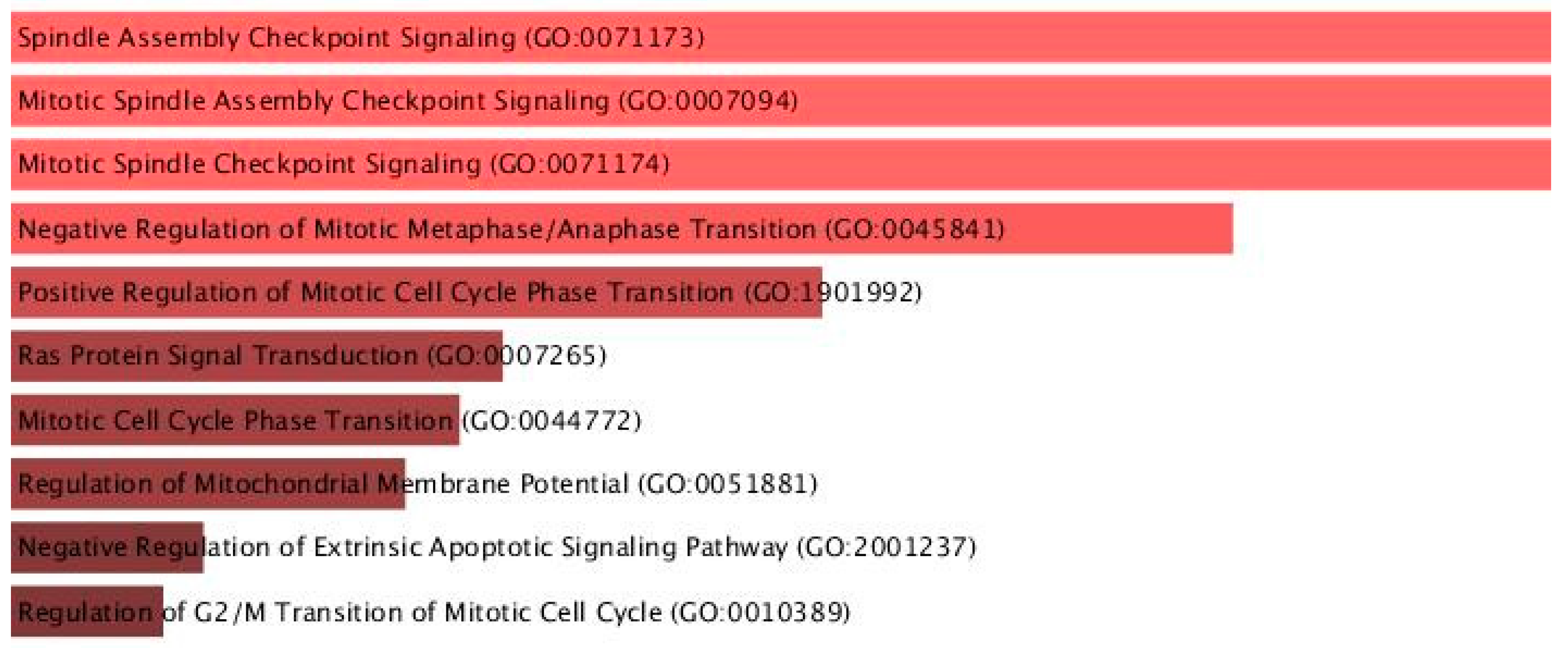
| Name | p-Value | Adjusted p-Value | Odds Ratio | Combined Score |
|---|---|---|---|---|
| Spindle Assembly Checkpoint Signaling (GO:0071173) | 0.00001379 | 0.005249 | 32.60 | 364.80 |
| Mitotic Spindle Assembly Checkpoint Signaling (GO:0007094) | 0.00001379 | 0.005249 | 32.60 | 364.80 |
| Mitotic Spindle Checkpoint Signaling (GO:0071174) | 0.00001379 | 0.005249 | 32.60 | 364.80 |
| Negative Regulation of Mitotic Metaphase/Anaphase Transition (GO:0045841) | 0.00001850 | 0.005282 | 29.98 | 326.76 |
| Positive Regulation of Mitotic Cell Cycle Phase Transition (GO:1901992) | 0.00002705 | 0.005677 | 16.01 | 168.35 |
| Ras Protein Signal Transduction (GO:0007265) | 0.00003634 | 0.005677 | 14.99 | 153.20 |
| Mitotic Cell Cycle Phase Transition (GO:0044772) | 0.00003782 | 0.005677 | 10.67 | 108.61 |
| Regulation of Mitochondrial Membrane Potential (GO:0051881) | 0.00003977 | 0.005677 | 24.17 | 244.94 |
| Negative Regulation of Extrinsic Apoptotic Signaling Pathway (GO:2001237) | 0.00004793 | 0.005679 | 14.09 | 140.12 |
| Regulation of G2/M Transition of Mitotic Cell Cycle (GO:0010389) | 0.00004974 | 0.005679 | 22.71 | 225.00 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Öztan, G.; İşsever, H.; Kurt, Ö.K.; Canbaz, S.; Oğuz, F.; İşsever, T.; Öztürk, Ö. Determination of Differential miRNA Expression Profile in People with Noise-Induced Hearing Loss. Int. J. Mol. Sci. 2025, 26, 6623. https://doi.org/10.3390/ijms26146623
Öztan G, İşsever H, Kurt ÖK, Canbaz S, Oğuz F, İşsever T, Öztürk Ö. Determination of Differential miRNA Expression Profile in People with Noise-Induced Hearing Loss. International Journal of Molecular Sciences. 2025; 26(14):6623. https://doi.org/10.3390/ijms26146623
Chicago/Turabian StyleÖztan, Gözde, Halim İşsever, Özlem Kar Kurt, Sevgi Canbaz, Fatma Oğuz, Tuğçe İşsever, and Özmen Öztürk. 2025. "Determination of Differential miRNA Expression Profile in People with Noise-Induced Hearing Loss" International Journal of Molecular Sciences 26, no. 14: 6623. https://doi.org/10.3390/ijms26146623
APA StyleÖztan, G., İşsever, H., Kurt, Ö. K., Canbaz, S., Oğuz, F., İşsever, T., & Öztürk, Ö. (2025). Determination of Differential miRNA Expression Profile in People with Noise-Induced Hearing Loss. International Journal of Molecular Sciences, 26(14), 6623. https://doi.org/10.3390/ijms26146623






