Abstract
Chronic myeloid leukemia (CML) is a clonal myeloproliferative neoplasia characterized by the BCR::ABL1 fusion gene, which codifies the BCR-ABL protein with increased tyrosine kinase activity. Despite the clinical results for the outstanding tyrosine kinase inhibitors (TKIs), drug resistance is a problem in CML management. Genetic variants that alter redox homeostasis by changing antioxidant enzyme expression or activity may influence patient responses and could enhance patient stratification. We aimed to assess the association of SOD2, CAT GPX1, NRF2, and KEAP1 genetic variants with TKI response and disease prognosis. For this purpose, we genotyped the variants rs4880 (SOD2), rs1050450 (GPX1), rs1001179 (CAT), rs6721961, rs4893819, rs35652124, rs6706649, rs13001694 (NFE2L2), and rs113540846 (KEAP1) via PCR in 187 CML patients. Our results show that variants in genes related to oxidative stress influence the development and degree of TKI resistance (allele G and GG genotypes of GPX1 and CT genotype of NFE2L2 rs4893819), the appearance of mutations in the BCR::ABL1 gene (AG genotype of NFE2L2 rs13001694 and genetic profile GGCTTCCCGG of the NFE2L2/KEAP1 axis), disease evolution (AG genotype of SOD2 and CT genotype of NFE2L2 rs4893819), and overall survival (CC genotype of CAT and GG genotype of NFE2L2 rs13001694) of CML patients. Our study found that variants in oxidative stress-related genes impact treatment response and outcomes in CML.
1. Introduction
Chronic myeloid leukemia (CML) is a clonal myeloproliferative neoplasia characterized by the presence of the BCR::ABL1 fusion gene as a consequence of a reciprocal translocation between chromosomes 9 and 22 [1]. Currently, and based on the constitutive tyrosine kinase activity of BCR-ABL oncoprotein, the therapeutic protocols are centered on tyrosine kinase inhibitors (TKIs) such as imatinib, dasatinib, and nilotinib [2]. Despite the excellent clinical results of TKIs, the emergence of drug resistance has become a problem in managing CML [3]. Multiple molecular mechanisms contribute to TKI resistance, including BCR::ABL1 point mutations, altered drug transporter activity, DNA repair and genomic instability, activation of alternative signaling pathways, epigenetic dysregulation, and oxidative stress [4].
Oxidative stress (OS) is related to the development and progression of various pathologies, including cancer [5]. This stress state results from the imbalance between reactive oxygen species (ROS) production and the antioxidant defense levels, which neutralize the former molecules [6,7,8]. At the physiological levels, and due to their role as second messengers in intracellular signaling pathways, ROS control cell division, proliferation, and survival [9]. However, long-term ROS exposure induces damage in proteins, lipids, and DNA, contributing to neoplasia development, progression, and drug resistance [10]. In CML, as in other hematological malignancies, the increase in ROS has been described and associated with the carcinogenic process [11]. Particularly in CML, BCR-ABL oncoprotein activity leads to ROS generation, resulting in an oxidative stress environment prone to inducing DNA damage and genomic instability [12]. Multiple drugs, as in the case of TKIs, increase the levels of exogenous ROS, inducing cell death [5]. The nuclear factor erythroid 2-related factor 2 (NRF2)/kelch-like ECH-associated protein 1 (KEAP1) pathway is one of the main regulators of oxidative homeostasis. In the presence of oxidative stress, NRF2 dissociates from KEAP1 and translocates to the nucleus [13]. This transcription factor, codified by the NFE2L2 gene, induces the expression of genes that play a role in redox homeostasis, DNA repair, drug excretion, survival, and autophagy. The cytoprotective and antioxidant genes, such as GPX1, CAT, and SOD2, are some examples of NRF2-regulated genes that could be implicated in tumor progression, metastasis, and resistance to chemotherapy [14,15,16]. The genetic background of each patient can influence all these intricate signaling pathways associated with cellular redox homeostasis. We hypothesize that germinative genetic variants, such as single nucleotide variants (SNVs), which are associated with altered expression levels and enzymatic activity in OS key player genes, may influence CML development, prognosis, and treatment response.
In this context, and given the relevance of oxidative stress homeostasis in CML treatment and prognosis, we evaluated the possible association of NFE2L2, KEAP1, SOD2, CAT, and GPX1 genetic variants with TKI response (including the response rates, number of required lines of TKI treatment, and presence of BCR::ABL1 mutations) and disease prognosis (progression and overall survival).
2. Results
2.1. Characteristics of CML Patients
Our cohort of CML patients included 187 CML patients, composed of 110 (58.8%) males and 77 (41.2%) females with a median age of 54 years (range: 15–86). Most patients were diagnosed in the chronic phase (94.6%, n = 177) (Table 1). Patients were classified as resistant if they required two or more lines of TKI treatment. In this context, 138 (73.8%) patients were categorized as TKI responders, while 49 (26.2%) were classified as TKI-resistant. No statistically significant differences were found between these two sub-groups of patients according to demographic features and the clinical parameters not associated with resistance (Table 1). Nearly all CML patients received imatinib as a first-line TKI (95.2%, n = 178). In the resistant sub-group, all the patients were treated with imatinib up-front. Furthermore, 12 patients (6.4%) were exposed to three or more TKI lines during treatment, and 22 (21.2%) developed BCR::ABL1 mutations. In this cohort, only 8 patients progressed (4.3%) to advanced forms, with all of them belonging to the TKI-resistant group, and 27 patients (14.4%) died.

Table 1.
Characteristics of CML patients.
2.2. Genetic Variants Associated with TKI Response and BCR::ABL1 Mutational Status
The association of selected genetic variants with the response to TKI treatment was evaluated to infer the contribution of these SNVs in the response profile. We performed multiple analyses according to allele and genotype distribution and applied four genetic models described in the Materials and Methods section. To achieve this goal, we correlated the SNVs with TKI response (sensitive and resistant patient subgroups), the number of TKI lines of treatment needed, and the BCR::ABL1 mutational status.
The allele distribution in TKI-sensitive and TKI-resistant patients is represented in Table 2. Patients carrying allele G of GPX1 rs1050450 had a higher probability of developing TKI resistance (OR = 1.841, 95%CI = 1.108–3.059, p = 0.020). The same association was observed in allele G of KEAP1 rs113540846, with a probability of becoming resistant to TKI treatment that was 31 times higher (OR = 31.07, 95%CI = 1.886–511.9, p < 0.0001) (Table 2).

Table 2.
Allele distribution based on TKI response.
We observed significant associations between specific genotypes and the response profiles (Table 3). All genotypes among the study groups were in Hardy–Weinberg equilibrium (HWE). For GPX1 rs1050450, the patients homozygotic for allele G presented a risk of failing the first-line TKI treatment which was two times higher (MD: OR = 2.199, 95%CI = 1.120–4.316, p = 0.022) (Table 3). Furthermore, the resistant patients may require more than two lines of treatment, and this could be related to the SNVs observed in the study. We observed that patients heterozygotic to the NFE2L2 rs4893819 variant had 5.6 times higher risk of requiring three or more lines of treatment (highly resistant profile) during the disease’s course (MOD: OR = 5.600, 95%CI = 1.218–25.751, p = 0.027) (Table 3). Moreover, the presence of a BCR::ABL1 point mutation was the most common resistance mechanism evaluated in the case of lack of response. Over the multiple genetic variants evaluated, NFE2L2 rs13001694 was associated with the BCR::ABL1 mutational status. Patients with the genotype AG presented a probability of developing mutations in the fusion gene which was 8.5 times higher (MCD: OR = 8.571, 95%CI = 1.004–73.210, p = 0.050; MOD: OR = 12.000, 95%CI = 1.429–100.754, p = 0.022) (Table 3). Additionally, the impact of gender was also assessed, and no significant associations were identified.

Table 3.
The distribution of significant genotypes of the selected SNVs in CML patients according to TKI response profile.
To assess the impact of the multiple genetic variants on the response profile, we performed haplotype and genotypic profile (GP) analysis using Arlequin software. The haplotype analysis was performed for the NFE2L2 (rs6721961/rs4893819/rs35652124/rs6706649) genetic variants. The GP analysis was performed and grouped in three categories: (1) the global profile with seven SNVs (NFE2L2: rs6721961/rs4893819/rs35652124/rs6706649/SOD2: rs4880/GPX1: rs1050450/KEAP1: rs113540846); (2) the NFE2L2/KEAP1 axis (NFE2L2: rs6721961/rs4893819/rs35652124/rs6706649/KEAP1: rs113540846); and (3) antioxidant defenses (SOD2 rs4880/GPX1 rs1050450). For all the variants related to TKI response, we did not observe any association with NFE2L2 haplotypes. Regarding the GP analysis of the NFE2L2/KEAP1 axis, patients with GP GG CT TC CC GG presented an increased risk of BCR::ABL1 mutations (OR = 6.000, 95%CI = 1.730–20.810, p = 0.006).
2.3. Impacts of Studied SNVs on Progression and Overall Survival
The link between the selected genetic variants and prognosis was evaluated by the association with CML progression and evolution and overall survival. In terms of allele distribution, we did not observe any association with disease progression or survival. However, the genotypic analysis revealed an association between CAT rs1001179 and the CML patients’ overall survival. The CC genotype showed an increased risk of death (MD: OR = 5.100, 95%CI = 1.125–23.117, p = 0.035) while the CT genotype showed a protective effect (MCD: OR = 0.163, 95%CI = 0.027–0.969, p = 0.046) (Table 4).

Table 4.
Significant genotype distribution of CAT rs1001179 according to overall survival.
The impact of different SNVs on CML prognosis was also evaluated by estimating the rate of disease progression and overall survival. This analysis was performed using the Kaplan-Meier method, and patients were stratified according to their SNV genotypes. As observed in Figure 1, the NFE2L2 rs4893819 CT genotype (HR = 7.844, 95%CI = 1.82–29.93, p = 0.020) and the SOD2 rs4880 AG genotype (HR = 7.190, 95%CI = 1.62–31.96, p = 0.035) were significantly associated with higher progression rates in comparison with the homozygotic genotypes of NFE2L2 rs4893819 and SOD2 rs4880, respectively (Figure 1a,b). Moreover, CML patients carrying the GG genotype for NFE2L2 rs13001694 had a significantly lower overall survival time (HR = 11.86, 95%CI = 1.39–100.7, p = 0.023) compared with those that were allele A carriers (Figure 1c), with an average overall survival time of 11.7 ± 2.5 years compared with 24.3 ± 1.6 years for patients with the AA and AG genotypes.
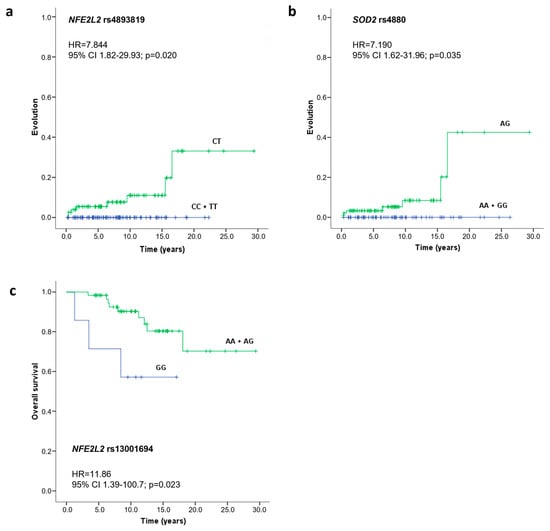
Figure 1.
Evolution and overall survival curves of CML patients according to NFE2L2 rs4893819 (a), SOD2 rs4880 (b), and NFE2L2 rs13001694 (c) genotypes. Time to evolution and survival analysis were performed using the Kaplan Meier method, differences in survival were tested with a log-rank test, and the hazard ratio (HR) with a 95% confidence interval (CI) was calculated using the Cox proportional hazard model.
Haplotype and genotypic profile (GP) analyses of the studied SNVs were performed as previously described, and the contributions to CML prognosis were assessed. According to the NFE2L2 haplotype analysis, we identified 11 haplotypes in our CML patient’s cohort. Three of them (GCC, GCTC, and TTTT) were identified only in patients who showed disease progression, while GCTC, GTCC, GTCT, GTTT, and TCTT were only observed in the opposite group. The haplotype GTTC was associated with six times higher risk of disease progression (OR = 6.160, 95%CI = 1.338–28.36, p = 0.036). Furthermore, the genotypic analysis identified an association between the GP of antioxidant defenses with CML progression. The GP AG GG antioxidant defenses (SOD2 rs4880/GPX1 rs1050450) conferred a higher risk of disease progression (OR = 4.615, 95%CI = 1.083–19.67, p = 0.047).
2.4. Influence of Studied SNVs on Gene Expression Levels and Protein Function
Genotype-tissue expression (GTEx) analysis in whole blood samples was possible for seven of the nine SNVs tested, with missing information for NFE2L2 rs6706649 and rs13001694. Over the SNVs evaluated, homozygous samples for the polymorphic allele showed significantly lower expression levels than those homozygous for the reference allele. CAT rs1001179 and KEAP1 rs113540846 were the exceptions, as SNVs resulted in higher gene expression levels according to GTEx analysis (Supplementary Figure S1).
For the SNVs that resulted in a missense variant, we used predictive tools to assess the impact on protein function and stability. Regarding protein function, SOD2 rs4880 and GPX1 rs1050450 were classified as tolerated effects according to Sort Intolerant from Tolerant (SIFT) and benign by Polymorphism Phenotyping v2 (PolyPhen-2) analysis. Concerning protein stability, the predicted stability change (ΔΔGStability) was −0.23 kcal/mol for SOD2 rs4880 and −0.56 kal/mol for GPX1 rs1050450, indicating a destabilizing effect from the variants on the protein structure.
3. Discussion
Oxidative stress is a key player in the development and progression of neoplasms but also has a relevant role in the treatment efficacy of these diseases [15]. The genetic variability of each patient influences tumor susceptibility, the progression and survival when neoplasms are already installed, as well as the response to oncological treatments [15,17,18,19,20]. Genetic variants that impact redox homeostasis, by altering the expression levels of antioxidant enzymes or redox regulator transcription factors or altering enzyme activities, may present a relevant role in patients’ responses, and they could contribute to better patient stratification. Based on this, we conducted a study focusing on genetic variants of genes essential for maintaining oxidative stress balance and that have been linked to decreased gene expression or lower protein activity.
Focusing on the NRF2 transcription factor (TF), the genetic variants on the NFE2L2 gene that codify this TF have been described as associated with the development and progression of various solid and hematological cancers [19,21]. However, the link between TKI response in CML patients and these variants has still not been explored. In our study, we observed an association of the NFE2L2 rs4893819 CT genotype with a higher TKI-resistant phenotype, since the patients carrying this genotype had a 5.6 times higher probability of needing three or more lines of treatment. In accordance with this, the same genotype was correlated with a higher risk of disease evolution. In the literature, the studies evaluating this SNV are scarce, and according to Nunes Dos Santos et al. (2019), this SNV did not present any association with another disease as alcoholic hepatitis [22]. Another NFE2L2 variant, rs13001694, has been associated with an increased risk of developing myelodysplastic neoplasia and breast, colon, and stomach cancer [23,24]. According to Gonçalves et al. (2017), individuals carrying allele G are more likely to develop myelodysplastic neoplasia when compared with AA carriers [19]. In the context of TKI response, we found that CML patients with AG genotypes for this variant have a higher risk of BCR::ABL1 mutations, which are highly associated with TKI resistance. On the other hand, the GG genotype gives CML patients treated with TKIs a shorter average life expectancy. Despite the other NFE2L2 variants studied (rs6721961, rs6706649, and rs35652124) being linked with neoplasia development and other pathologies [25,26,27,28], in our study, we did not find any correlation between them and CML patients’ responses and prognoses. More comprehensively, we also studied the contributions of the different selected SNVs on the NFE2L2 gene to CML patient’s characteristics via haplotype analysis. In our CML cohort, the NFE2L2 haplotype GTTC (allele G of SNV rs6721961, T of rs4893819, T of rs35652124, and C of rs6706649) was associated with six times higher risk of disease progression. A similar analysis was performed by Arisawa et al. (2008), and they found that the TC haplotype (NFE2L2 rs35652124 and rs6706649) was associated with higher levels of inflammation and a greater likelihood of developing gastric mucosal atrophy [29]. Corroborating our results, the GTC haplotype (NFE2L2 rs6721961, rs35652124, and rs6706649) has been described as having low promoter activity and consequently lower gene expression, which could explain the impact on disease evolution [30].
Germline and somatic variants in the KEAP1 gene result in reduced activity for the KEAP1 protein, leading to accumulation of the NRF2 transcription factor and, consequently, resistance to chemotherapy [31]. Despite these relevant roles, the SNVs in this gene have not been explored in the context of hematological neoplasms. Studies on the KEAP1 rs113540846 variant are not described in the literature. However, we found a strong correlation between allele G and an increased risk of developing resistance to TKIs, and GTEx analysis indicated that this allele correlated with lower gene expression levels. Further analysis is needed to explore the relevance and impact of KEAP1 SNVs in the CML scenario.
Genetic variants of antioxidant enzymes, namely in SOD2, GPX1, and CAT, were already associated with the development of various neoplasms and treatment efficacy [10,32]. For SOD2, allele G in the rs4880 variant has been associated with lower gene expression and lower stability of mRNA, which affects the entry of SOD2 into mitochondria. As a result, the antioxidant activity of SOD2 decreases by 30–40%, thus reducing the neutralizing capacity of superoxide anions [33,34]. This particular SNV was associated with the development of chronic kidney disease, breast cancer, lung cancer, and acute myeloid leukemia, among other pathologies [28,32]. According to Xu et al. (2015), the AG and GG genotypes have a negative impact on survival for Chinese gastric cancer patients who receive platinum- and fluorouracil-based adjuvant chemotherapy [35]. Along the same line, the G carrier genotypes of this variant were associated with higher stages of disease in papillary thyroid carcinoma [36]. Moreover, the SOD2 rs4880 AG genotype was associated with non-chemotherapy response in breast cancer patients [37]. However, the contribution to CML response and prognosis is poorly explored. In accordance, in our study, we observed a correlation of the AG genotype with disease progression, where AG patients progressed seven times faster compared with those with the AA and GG genotypes. A crucial antioxidant defense to detoxification of hydrogen peroxide is glutathione peroxidase 1, and genetic variants on GPX1 have been linked with neoplasia development [38,39,40]. In particular, the rs1050450 variant, which consists of a substitution amino acid change from proline to leucine, causes a decline in GPX1 activity due to a conformational change in the protein [41,42]. According to Kagita et al. (2021), the GA and GG genotypes of GPX1 rs1050450 were identified as being a risk factor of CML development [43]. Furthermore, in the same study, patients homozygous to allele A of the rs1050450 variant, which encodes an enzyme with reduced activity [38,43], were associated with a less profound molecular response and the development of BCR::ABL1 mutations. In contrast, in our CML cohort, the allele G and GG genotypes were associated with a worse response to TKIs, and these patients were more likely to become TKI-resistant. In accordance with our results, the GG genotype was associated with a higher risk of breast cancer and with higher detoxification activity due to higher GPX1 activity in these individuals [41,44].
Another relevant enzyme in the detoxification of hydrogen peroxide is catalase, which is encoded by the CAT gene. For this gene, several genetic variants have been studied, with CAT rs1001179 being the one known in more detail. This variant occurs in the promoter region and affects the association of transcription factors to the promoter, leading to changes in the transcription rate [45,46]. As a consequence, individuals carrying TT genotypes present lower CAT activity in comparison with CC genotype carriers [47,48]. Due to its important role in redox homeostasis, this variant has been identified as a risk factor for CML, hepatocellular carcinoma, breast cancer, and gastric cancer development [43,49,50,51]. However, the reports with this variant are not consensual, and some authors failed to identify this risk association, particularly for CML [52]. In our study, the presence of the CC genotype was associated with an increased probability of death, while the opposite effect was observed for the CT genotype. According to our results, the presence of allele T was associated with a higher probability of survival. In accordance with the findings previously described by Kagita et al. (2021) [43], we did not find any association between this CAT SNV and the BCR::ABL1 mutational status.
For a more comprehensive and integrated investigation of the impact of several SNVs at the same time, we performed a genotypic profile (GP) analysis to identify profiles that could contribute to a better prediction of CML TKI response and prognosis. The GP of the NFE2L2/KEAP1 axis, GG CT TC CC GG, was related to a higher risk of BCR::ABL1 mutations, a crucial event for TKI efficacy, while the antioxidant defense GP (SOD2 rs4880/GPX1 rs1050450) AG GG was associated with a high risk of disease progression. In previous work from our group in the same patient cohort, we identified genetic profiles in the ABC and SLC drug transporter genes related to TKI response and the degree of resistance [20]. However, a similar analysis with oxidative stress-related genes was not described in the CML field.
The studies of genetic variants and their link with neoplasia prognosis and treatment response have been commonly associated with conflicting results. These heterogeneous results can be explained by limitations in sample size, SNV ethnic differences, treatment, and response criteria [53]. Furthermore, epigenetic mechanisms, such as DNA methylation or post-translational modifications of histones, can also influence gene transcription, altering expression regardless of the germline genetic variant present [54]. In particular, we recognize that the sample size may constitute a limitation in our work, and further analysis could be performed for a more complete view of the selected SNVs’ contribution to CML treatment response and prognosis. In this line, future analyses could complement the performed functional studies with predictive tools, particularly to assess gene expression and protein activity in a patient’s cohort. Still, the identification of relevant SNV profiles can significantly impact therapeutic monitoring and improve TKI response, namely by improving the mutation screening in patients with higher risk of BCR::ABL1 mutations or switching to a different TKI in cases of patients at high risk of needing multiple lines of treatment. The inclusion of SNV analysis can increase patient quality of life and consequently reduce the burden on the healthcare system.
4. Materials and Methods
4.1. Study Population
In this retrospective study, we enrolled 187 CML patients recruited from three hospitals—Centro Hospitalar e Universitário de Coimbra (ULSC/CHUC), Hospital Distrital da Figueira da Foz (HDFF), and Instituto Português de Oncologia de Lisboa (IPO-Lisboa)—between September 2014 and August 2017. Patients were diagnosed based on the World Health Organization (WHO) classification, and treatment response criteria were defined according to European Leukemia Net (ELN) guidelines [2]. Blood samples were obtained during monitoring consultations conducted by the clinical team.
The characteristics of the CML patients included in this study are summarized in Table 1. The patient group was subdivided into responsive (n = 138) or resistant (n = 49) to TKIs, based on the therapeutic outcomes for prognostic analysis (Table 1). To avoid confounding effects, the patients intolerant to therapy were excluded. This study was conducted according to the Helsinki Declaration, and all participants provided written informed consent for participation before enrolment. All research procedures were approved by the Faculty of Medicine Ethics Committee (University of Coimbra, Portugal) (ref. CE-014/2014).
4.2. Gene and SNV Selection
The genes to be studied were selected based on their relationship with oxidative stress. According to the literature, the NFE2L2 and KEAP1 genes encode the NRF2 and KEAP1 proteins, respectively, and are crucial players in redox homeostasis. The CAT, GPX1, and SOD2 genes encode the antioxidant proteins catalase, glutathione peroxidase 1, and manganese superoxide dismutase, respectively. The genetic variants of these genes were selected based on the frequency of the minor allele (MAF), which had to be greater than 10% in the European population (ALFA allele frequency). After this screening, the variants selected were rs6721961, rs4893819, rs35652124, rs6706649, and rs13001694 from the NFE2L2 gene, rs113540846 from the KEAP1 gene, rs4880 from the SOD2 gene, rs1050450 from the GPX1 gene, and rs1001179 from the CAT gene. In the Supplementary Materials (Table S1), the characterization of the selected SNVs is detailed.
4.3. Genotyping
Blood samples were collected in EDTA tubes, and NZYol reagent (NZYtech, Lisbon, Portugal) was used to extract genomic DNA from the whole blood according to the manufacturer’s protocol. The DNA quality and quantity were determined using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, NC, USA). For each genotyping reaction, 100 ng of DNA was used, and SNVs were genotyped through tetra-primer ARMS-PCR, ASO-PCR, and RFLP-PCR assays. The primers for the different reaction were designed using BatchPrimer3, version 1.0 (USDA-ARS, Albany, CA, USA) (http://probes.pw.usda.gov/batchprimer3/ (accessed on 5 October 2020)) [55]. The primers and PCR reaction conditions are described in the Supplementary Materials (Table S2). The results were initially confirmed via direct sequencing. Samples discovered to contain the three potential genotypes were employed as positive controls in each genotype assay. Genotyping was repeated in approximately 10% of the total samples as a quality check to ensure accuracy.
4.4. Genetic Analysis
In this work, we started by analyzing the allele and genotype distribution. In this analysis, it was important to understand the concepts of minor (m) and major (M) alleles. In a biallelic SNV, the minor allele represents the less frequent allele in the population, while the major (M) allele is the most frequent allele and serves as a reference [56]. Different genetic models can be applied to genotypic analysis, including the codominant model (MCD), dominant model (MD), recessive model (MR), and overdominant model (MOD). Each of these models represents different assumptions about genetic effects. The codominant model (MCD), where each genotype was compared with the homozygous of a major allele (mm or mM vs. MM), assumes each genotype individually and determines the risk associated with it compared to the reference genotype. The dominant model, in which minor allele carriers are analyzed against the major allele homozygous (mm + mM vs. MM), assumes that having one or more copies of the major allele increases risk. When minor allele homozygotes are compared to major allele carriers (mm vs. MM + mM), this corresponds to the recessive model (MR), which assumes the need for two copies of a minor allele to alter the risk associated with that variant. Lastly, the overdominant model (MOD), where heterozygous individuals are compared to homozygous individuals (mM vs. MM + mm), explores the combination of both alleles to confer risk [57,58].
4.5. Statistical Analysis
The statistical analysis was conducted in collaboration with the Laboratory of Biostatistics and Medical Informatics at the Faculty of Medicine of the University of Coimbra (FMUC). The frequencies of alleles and genotypes were determined by direct counting. The frequencies of the genotypic profile (GP) and the Hardy–Weinberg equilibrium (HWE) in the studied groups were determined using Arlequin software version 3.5.1.2 (CMPG, University of Bern, Switzerland) [59]. The comparison of allele frequencies was conducted using Fisher’s exact test in GraphPad Prism, version 9.0 (GraphPad Software, San Diego, CA, USA). The associations between genotype, haplotype, GP, and clinical characteristics were analyzed using odds ratio (OR) and 95% confidence interval (CI) calculations. This was performed through unconditioned logistic regression in SPSS, version 29.0 (IBM, New York, NY, USA). We used the Kaplan–Meier method in SPSS to estimate the time to disease progression and overall survival of patients grouped based on their genotypes. We tested for differences using the log-rank statistic. The hazard ratio (HR) and its 95% confidence interval (CI) were calculated using the Cox proportional hazard model. All statistical analyses were two-sided, and we considered p < 0.05 statistically significant.
GTEx portal v10 (https://www.gtexportal.org/home/) was accessed on 10 April 2025 to evaluate the effect of the selected SNPs on gene expression in whole blood samples. The SIFT algorithm (https://sift.bii.a-star.edu.sg/) and PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/) were also accessed on 10 April 2025 to estimate the functional impact of the SNP and the associated amino acid substitution on protein function [60]. The effect of the selected SNPs on protein stability was evaluated using DynaMut online tools (https://biosig.lab.uq.edu.au/dynamut2/), accessed on 10 April 2025.
5. Conclusions
In conclusion, the results obtained show that variants in genes related to oxidative stress influence the clinical characteristics of CML patients, namely the development and degree of TKI resistance, mutations in the BCR::ABL1 gene, the evolution of the disease, and the overall survival of CML patients. Determining these SNVs and combining them with other known variants could improve the prognostic characterization of CML patients and enable a better understanding of inter-individual differences, thus allowing for a more personalized and informed therapeutic choice and contributing to precision medicine.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/ijms26125682/s1.
Author Contributions
R.A., A.C.G. and A.B.S.-R. designed the experiments. R.A. and A.C.G. drafted the manuscript. R.A., F.V. and J.J. performed the experiments. R.A., F.V., A.C.G. and B.O. performed the statistical analysis. A.P., P.F.-T. and A.M.A. recruited and provided the clinical information of the participants. G.M., M.C. and J.D. provided the DNA samples. A.C.G. and A.B.S.-R. revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by ACIMAGO (Project 18/12) and Liga Portuguesa Contra o Cancro—Núcleo regional do Centro. The Centre for Innovative Biomedicine and Biotechnology (CIBB) and the Institute for Clinical and Biomedical Research (iCBR) are supported by the Foundation for Science and Technology (FCT) of Portugal through Strategic Projects UIDB/04539/2020 (10.54499/UIDB/04539/2020) and UIDP/04539/2020 (10.54499/UIDP/04539/2020) and Associated Laboratory funding LA/P/0058/2020 (10.54499/LA/P/0058/2020).
Institutional Review Board Statement
This study was conducted according to the Helsinki Declaration, and all research procedures were approved by the Ethics Committee of the Faculty of Medicine (University of Coimbra, Portugal) (ref. CE-014/2014 and 31 March 2014).
Informed Consent Statement
All participants provided written informed consent for participation before enrolment.
Data Availability Statement
The original contributions presented in this study are included in the article or Supplementary Materials. Further inquiries can be directed to the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the study’s design; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Abbreviations
The following abbreviations are used in this manuscript:
| ASO-PCR | Allele-specific oligonucleotide polymerase chain reaction |
| CAT | Catalase |
| CI | Confidence interval |
| CML | Chronic myeloid leukemia |
| ELN | European Leukemia Net |
| GP | Genotypic profile |
| GPX1 | Glutathione peroxidase 1 |
| HR | Hazard ratio |
| HWE | Hardy–Weinberg equilibrium |
| KEAP1 | Kelch-like ECH-associated protein 1 |
| M | Major allele |
| m | Minor allele |
| MAF | Frequency of the minor allele |
| MCD | Codominant model |
| MD | Dominant model |
| MgCL2 | Magnesium chloride |
| MOD | Overdominant model |
| MR | Recessive model |
| NFE2L2 | Nuclear factor erythroid 2-related factor 2 gene |
| NRF2 | Nuclear factor erythroid 2-related factor 2 |
| OD | Odds ratio |
| OS | Oxidative stress |
| RFLP-PCR | Restriction fragment length polymorphism polymerase chain reaction |
| Ref | Reference allele or genotype |
| ROS | Reactive oxygen species |
| SNV | Single nucleotide variants |
| SOD2 | Superoxide dismutase [Mn], mitochondrial |
| Ta | Annealing temperature |
| TETRA-ARMS-PCR | Tetra-primer amplification refractory mutation system-polymerase chain reaction |
| TF | Transcription factor |
| TKI | Tyrosine kinase inhibitor |
| WHO | World Health Organization |
References
- Ali, M.A. Chronic Myeloid Leukemia in the Era of Tyrosine Kinase Inhibitors: An Evolving Paradigm of Molecularly Targeted Therapy. Mol. Diagn. Ther. 2016, 20, 315–333. [Google Scholar] [CrossRef] [PubMed]
- Baccarani, M.; Castagnetti, F.; Gugliotta, G.; Rosti, G. A review of the European LeukemiaNet recommendations for the management of CML. Ann. Hematol. 2015, 94 (Suppl. S2), S141–S147. [Google Scholar] [CrossRef] [PubMed]
- Hochhaus, A.; Saussele, S.; Rosti, G.; Mahon, F.X.; Janssen, J.; Hjorth-Hansen, H.; Richter, J.; Buske, C.; Committee, E.G. Chronic myeloid leukaemia: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2017, 28, iv41–iv51. [Google Scholar] [CrossRef]
- Alves, R.; Goncalves, A.C.; Rutella, S.; Almeida, A.M.; De Las Rivas, J.; Trougakos, I.P.; Sarmento Ribeiro, A.B. Resistance to Tyrosine Kinase Inhibitors in Chronic Myeloid Leukemia-From Molecular Mechanisms to Clinical Relevance. Cancers 2021, 13, 4820. [Google Scholar] [CrossRef]
- Allegra, A.; Mirabile, G.; Caserta, S.; Stagno, F.; Russo, S.; Pioggia, G.; Gangemi, S. Oxidative Stress and Chronic Myeloid Leukemia: A Balance between ROS-Mediated Pro- and Anti-Apoptotic Effects of Tyrosine Kinase Inhibitors. Antioxidants 2024, 13, 461. [Google Scholar] [CrossRef]
- Valko, M.; Leibfritz, D.; Moncol, J.; Cronin, M.T.; Mazur, M.; Telser, J. Free radicals and antioxidants in normal physiological functions and human disease. Int. J. Biochem. Cell Biol. 2007, 39, 44–84. [Google Scholar] [CrossRef] [PubMed]
- Hayes, J.D.; Dinkova-Kostova, A.T.; Tew, K.D. Oxidative Stress in Cancer. Cancer Cell 2020, 38, 167–197. [Google Scholar] [CrossRef]
- Birben, E.; Sahiner, U.M.; Sackesen, C.; Erzurum, S.; Kalayci, O. Oxidative stress and antioxidant defense. World Allergy Organ. J. 2012, 5, 9–19. [Google Scholar] [CrossRef]
- Parvez, S.; Long, M.J.C.; Poganik, J.R.; Aye, Y. Redox Signaling by Reactive Electrophiles and Oxidants. Chem. Rev. 2018, 118, 8798–8888. [Google Scholar] [CrossRef]
- Pascu, V.E.G.; AM, G.A. Involvement of Oxidative Stress in Resistance to Tyrosine-Kinase Inhibitors Therapy in Chronic Myeloid Leukemia. Curr. Health Sci. J. 2020, 46, 420–432. [Google Scholar]
- Hole, P.S.; Darley, R.L.; Tonks, A. Do reactive oxygen species play a role in myeloid leukemias? Blood 2011, 117, 5816–5826. [Google Scholar] [CrossRef] [PubMed]
- Slupianek, A.; Falinski, R.; Znojek, P.; Stoklosa, T.; Flis, S.; Doneddu, V.; Pytel, D.; Synowiec, E.; Blasiak, J.; Bellacosa, A.; et al. BCR-ABL1 kinase inhibits uracil DNA glycosylase UNG2 to enhance oxidative DNA damage and stimulate genomic instability. Leukemia 2013, 27, 629–634. [Google Scholar] [CrossRef] [PubMed]
- Tonelli, C.; Chio, I.I.C.; Tuveson, D.A. Transcriptional Regulation by Nrf2. Antioxid. Redox Signal. 2018, 29, 1727–1745. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q. Role of nrf2 in oxidative stress and toxicity. Annu. Rev. Pharmacol. Toxicol. 2013, 53, 401–426. [Google Scholar] [CrossRef]
- de la Vega, M.R.; Chapman, E.; Zhang, D.D. NRF2 and the Hallmarks of Cancer. Cancer Cell 2018, 34, 21–43. [Google Scholar] [CrossRef]
- Khodakarami, A.; Adibfar, S.; Karpisheh, V.; Abolhasani, S.; Jalali, P.; Mohammadi, H.; Gholizadeh Navashenaq, J.; Hojjat-Farsangi, M.; Jadidi-Niaragh, F. The molecular biology and therapeutic potential of Nrf2 in leukemia. Cancer Cell Int. 2022, 22, 241. [Google Scholar] [CrossRef]
- Scharfe, C.P.I.; Tremmel, R.; Schwab, M.; Kohlbacher, O.; Marks, D.S. Genetic variation in human drug-related genes. Genome Med. 2017, 9, 117. [Google Scholar] [CrossRef]
- Assaraf, Y.G.; Brozovic, A.; Goncalves, A.C.; Jurkovicova, D.; Line, A.; Machuqueiro, M.; Saponara, S.; Sarmento-Ribeiro, A.B.; Xavier, C.P.R.; Vasconcelos, M.H. The multi-factorial nature of clinical multidrug resistance in cancer. Drug Resist. Updates 2019, 46, 100645. [Google Scholar] [CrossRef]
- Goncalves, A.C.; Alves, R.; Baldeiras, I.; Cortesao, E.; Carda, J.P.; Branco, C.C.; Oliveiros, B.; Loureiro, L.; Pereira, A.; Nascimento Costa, J.M.; et al. Genetic variants involved in oxidative stress, base excision repair, DNA methylation, and folate metabolism pathways influence myeloid neoplasias susceptibility and prognosis. Mol. Carcinog. 2017, 56, 130–148. [Google Scholar] [CrossRef]
- Alves, R.; Goncalves, A.C.; Jorge, J.; Marques, G.; Ribeiro, A.B.; Tenreiro, R.; Coucelo, M.; Diamond, J.; Oliveiros, B.; Pereira, A.; et al. Genetic Variants of ABC and SLC Transporter Genes and Chronic Myeloid Leukaemia: Impact on Susceptibility and Prognosis. Int. J. Mol. Sci. 2022, 23, 9815. [Google Scholar] [CrossRef]
- Yang, W.; Liu, H.; Duan, B.; Xu, X.; Carmody, D.; Luo, S.; Walsh, K.M.; Abbruzzese, J.L.; Zhang, X.; Chen, X.; et al. Three novel genetic variants in NRF2 signaling pathway genes are associated with pancreatic cancer risk. Cancer Sci. 2019, 110, 2022–2032. [Google Scholar] [CrossRef] [PubMed]
- Nunes Dos Santos, K.; Florentino, R.M.; Franca, A.; Lima Filho, A.C.M.; Santos, M.L.D.; Missiaggia, D.; Fonseca, M.C.; Brasil Costa, I.; Vidigal, P.V.T.; Nathanson, M.H.; et al. Polymorphism in the Promoter Region of NFE2L2 Gene Is a Genetic Marker of Susceptibility to Cirrhosis Associated with Alcohol Abuse. Int. J. Mol. Sci. 2019, 20, 3589. [Google Scholar] [CrossRef]
- Sjoblom, T.; Jones, S.; Wood, L.D.; Parsons, D.W.; Lin, J.; Barber, T.D.; Mandelker, D.; Leary, R.J.; Ptak, J.; Silliman, N.; et al. The consensus coding sequences of human breast and colorectal cancers. Science 2006, 314, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Sporn, M.B.; Liby, K.T. NRF2 and cancer: The good, the bad and the importance of context. Nat. Rev. Cancer 2012, 12, 564–571. [Google Scholar] [CrossRef]
- Okano, Y.; Nezu, U.; Enokida, Y.; Lee, M.T.; Kinoshita, H.; Lezhava, A.; Hayashizaki, Y.; Morita, S.; Taguri, M.; Ichikawa, Y.; et al. SNP (-617C>A) in ARE-like loci of the NRF2 gene: A new biomarker for prognosis of lung adenocarcinoma in Japanese non-smoking women. PLoS ONE 2013, 8, e73794. [Google Scholar] [CrossRef] [PubMed]
- Hartikainen, J.M.; Tengstrom, M.; Kosma, V.M.; Kinnula, V.L.; Mannermaa, A.; Soini, Y. Genetic polymorphisms and protein expression of NRF2 and Sulfiredoxin predict survival outcomes in breast cancer. Cancer Res. 2012, 72, 5537–5546. [Google Scholar] [CrossRef]
- Marzec, J.M.; Christie, J.D.; Reddy, S.P.; Jedlicka, A.E.; Vuong, H.; Lanken, P.N.; Aplenc, R.; Yamamoto, T.; Yamamoto, M.; Cho, H.Y.; et al. Functional polymorphisms in the transcription factor NRF2 in humans increase the risk of acute lung injury. FASEB J. 2007, 21, 2237–2246. [Google Scholar] [CrossRef]
- Zazueta, C.; Jimenez-Uribe, A.P.; Pedraza-Chaverri, J.; Buelna-Chontal, M. Genetic Variations on Redox Control in Cardiometabolic Diseases: The Role of Nrf2. Antioxidants 2022, 11, 507. [Google Scholar] [CrossRef]
- Arisawa, T.; Tahara, T.; Shibata, T.; Nagasaka, M.; Nakamura, M.; Kamiya, Y.; Fujita, H.; Yoshioka, D.; Okubo, M.; Hirata, I.; et al. Nrf2 gene promoter polymorphism and gastric carcinogenesis. Hepatogastroenterology 2008, 55, 750–754. [Google Scholar]
- Matana, A.; Ziros, P.G.; Chartoumpekis, D.V.; Renaud, C.O.; Polasek, O.; Hayward, C.; Zemunik, T.; Sykiotis, G.P. Rare and common genetic variations in the Keap1/Nrf2 antioxidant response pathway impact thyroglobulin gene expression and circulating levels, respectively. Biochem. Pharmacol. 2020, 173, 113605. [Google Scholar] [CrossRef]
- Islam, S.S.; Qassem, K.; Islam, S.; Parag, R.R.; Rahman, M.Z.; Farhat, W.A.; Yeger, H.; Aboussekhra, A.; Karakas, B.; Noman, A.S.M. Genetic alterations of Keap1 confers chemotherapeutic resistance through functional activation of Nrf2 and Notch pathway in head and neck squamous cell carcinoma. Cell Death Dis. 2022, 13, 696. [Google Scholar] [CrossRef] [PubMed]
- Crawford, A.; Fassett, R.G.; Geraghty, D.P.; Kunde, D.A.; Ball, M.J.; Robertson, I.K.; Coombes, J.S. Relationships between single nucleotide polymorphisms of antioxidant enzymes and disease. Gene 2012, 501, 89–103. [Google Scholar] [CrossRef] [PubMed]
- Sutton, A.; Imbert, A.; Igoudjil, A.; Descatoire, V.; Cazanave, S.; Pessayre, D.; Degoul, F. The manganese superoxide dismutase Ala16Val dimorphism modulates both mitochondrial import and mRNA stability. Pharm. Genomics 2005, 15, 311–319. [Google Scholar] [CrossRef] [PubMed]
- Kinnula, V.L.; Crapo, J.D. Superoxide dismutases in malignant cells and human tumors. Free Radic. Biol. Med. 2004, 36, 718–744. [Google Scholar] [CrossRef]
- Xu, Z.; Chen, Y.; Gu, D.; Lee, N.P.; Sun, S.; Gong, W.; Tan, Y.; Luk, J.M.; Chen, J. SOD2 rs4880 CT/CC genotype predicts poor survival for Chinese gastric cancer patients received platinum and fluorouracil based adjuvant chemotherapy. Am. J. Transl. Res. 2015, 7, 401–410. [Google Scholar] [CrossRef]
- Salimi, S.; Harati-Sadegh, M.; Eskandari, M.; Heidari, Z. The effects of the genetic polymorphisms of antioxidant enzymes on susceptibility to papillary thyroid carcinoma. IUBMB Life 2020, 72, 1045–1053. [Google Scholar] [CrossRef]
- Gallegos-Arreola, M.P.; Ramirez-Patino, R.; Sanchez-Lopez, J.Y.; Zuniga-Gonzalez, G.M.; Figuera, L.E.; Delgado-Saucedo, J.I.; Gomez-Meda, B.C.; Rosales-Reynoso, M.A.; Puebla-Perez, A.M.; Lemus-Varela, M.L.; et al. SOD2 Gene Variants (rs4880 and rs5746136) and Their Association with Breast Cancer Risk. Curr. Issues Mol. Biol. 2022, 44, 5221–5233. [Google Scholar] [CrossRef]
- Kucukgergin, C.; Sanli, O.; Amasyali, A.S.; Tefik, T.; Seckin, S. Genetic variants of MnSOD and GPX1 and susceptibility to bladder cancer in a Turkish population. Med. Oncol. 2012, 29, 1928–1934. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, R.; Chen, N.; Yang, L.; Wang, Y.; Sun, Y.; Huang, L.; Zhu, M.; Ji, Y.; Li, W. Association between glutathione peroxidase-1 (GPX1) Rs1050450 polymorphisms and cancer risk. Int. J. Clin. Exp. Pathol. 2017, 10, 9527–9540. [Google Scholar]
- Ravn-Haren, G.; Olsen, A.; Tjonneland, A.; Dragsted, L.O.; Nexo, B.A.; Wallin, H.; Overvad, K.; Raaschou-Nielsen, O.; Vogel, U. Associations between GPX1 Pro198Leu polymorphism, erythrocyte GPX activity, alcohol consumption and breast cancer risk in a prospective cohort study. Carcinogenesis 2006, 27, 820–825. [Google Scholar] [CrossRef]
- Jablonska, E.; Gromadzinska, J.; Peplonska, B.; Fendler, W.; Reszka, E.; Krol, M.B.; Wieczorek, E.; Bukowska, A.; Gresner, P.; Galicki, M.; et al. Lipid peroxidation and glutathione peroxidase activity relationship in breast cancer depends on functional polymorphism of GPX1. BMC Cancer 2015, 15, 657. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wang, H.; Zhou, J.; Shao, Q. Glutathione Peroxidase GPX1 and Its Dichotomous Roles in Cancer. Cancers 2022, 14, 2560. [Google Scholar] [CrossRef]
- Kagita, S.; Digumarti, R.; Gundeti, S. Role of Antioxidant Gene Polymorphisms in Risk and Prognosis of Chronic Myeloid Leukemia. Asian Pac. J. Cancer Biol. 2021, 6, 27–36. [Google Scholar] [CrossRef]
- Hong, Z.; Tian, C.; Zhang, X. GPX1 gene Pro200Leu polymorphism, erythrocyte GPX activity, and cancer risk. Mol. Biol. Rep. 2013, 40, 1801–1812. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Liu, X.; Wang, M.; Wang, X.; Kang, H.; Lin, S.; Yang, P.; Dai, C.; Xu, P.; Li, S.; et al. Two common functional catalase gene polymorphisms (rs1001179 and rs794316) and cancer susceptibility: Evidence from 14,942 cancer cases and 43,285 controls. Oncotarget 2016, 7, 62954–62965. [Google Scholar] [CrossRef]
- Galasso, M.; Dalla Pozza, E.; Chignola, R.; Gambino, S.; Cavallini, C.; Quaglia, F.M.; Lovato, O.; Dando, I.; Malpeli, G.; Krampera, M.; et al. The rs1001179 SNP and CpG methylation regulate catalase expression in chronic lymphocytic leukemia. Cell. Mol. Life Sci. 2022, 79, 521. [Google Scholar] [CrossRef]
- Eras, N.; Turkoz, G.; Tombak, A.; Tiftik, N.; Yalin, S.; Berkoz, M.; Erden, S.; Akbas, E. An investigation of the relation between catalase C262T gene polymorphism and catalase enzyme activity in leukemia patients. Arch. Med. Sci. 2021, 17, 928–933. [Google Scholar] [CrossRef]
- Ahn, J.; Nowell, S.; McCann, S.E.; Yu, J.; Carter, L.; Lang, N.P.; Kadlubar, F.F.; Ratnasinghe, L.D.; Ambrosone, C.B. Associations between catalase phenotype and genotype: Modification by epidemiologic factors. Cancer Epidemiol. Biomark. Prev. 2006, 15, 1217–1222. [Google Scholar] [CrossRef]
- Liu, Y.; Xie, L.; Zhao, J.; Huang, X.; Song, L.; Luo, J.; Ma, L.; Li, S.; Qin, X. Association between catalase gene polymorphisms and risk of chronic hepatitis B, hepatitis B virus-related liver cirrhosis and hepatocellular carcinoma in Guangxi population: A case-control study. Medicine 2015, 94, e702. [Google Scholar] [CrossRef]
- Saadat, M.; Saadat, S. Genetic Polymorphism of CAT C-262 T and Susceptibility to Breast Cancer, a Case-Control Study and Meta-Analysis of the Literatures. Pathol. Oncol. Res. 2015, 21, 433–437. [Google Scholar] [CrossRef]
- Ebrahimpour, S.; Saadat, I. Association of CAT C-262T and SOD1 A251G single nucleotide polymorphisms susceptible to gastric cancer. Mol. Biol. Res. Commun. 2014, 3, 223–229. [Google Scholar] [PubMed]
- Banescu, C.; Trifa, A.P.; Voidazan, S.; Moldovan, V.G.; Macarie, I.; Benedek Lazar, E.; Dima, D.; Duicu, C.; Dobreanu, M. CAT, GPX1, MnSOD, GSTM1, GSTT1, and GSTP1 genetic polymorphisms in chronic myeloid leukemia: A case-control study. Oxid. Med. Cell Longev. 2014, 2014, 875861. [Google Scholar] [CrossRef]
- Ben Hassine, I.; Gharbi, H.; Soltani, I.; Ben Hadj Othman, H.; Farrah, A.; Amouri, H.; Teber, M.; Ghedira, H.; Ben Youssef, Y.; Safra, I.; et al. Molecular study of ABCB1 gene and its correlation with imatinib response in chronic myeloid leukemia. Cancer Chemother. Pharmacol. 2017, 80, 829–839. [Google Scholar] [CrossRef] [PubMed]
- Neul, C.; Schaeffeler, E.; Sparreboom, A.; Laufer, S.; Schwab, M.; Nies, A.T. Impact of Membrane Drug Transporters on Resistance to Small-Molecule Tyrosine Kinase Inhibitors. Trends Pharmacol. Sci. 2016, 37, 904–932. [Google Scholar] [CrossRef]
- You, F.M.; Huo, N.; Gu, Y.Q.; Luo, M.C.; Ma, Y.; Hane, D.; Lazo, G.R.; Dvorak, J.; Anderson, O.D. BatchPrimer3: A high throughput web application for PCR and sequencing primer design. BMC Bioinform. 2008, 9, 253. [Google Scholar] [CrossRef]
- Mansur, Y.A.; Rojano, E.; Ranea, J.A.G.; Perkins, J.R. Chapter 7—Analyzing the Effects of Genetic Variation in Noncoding Genomic Regions. In Precision Medicine; Deigner, H.-P., Kohl, M., Eds.; Academic Press: Cambridge, MA, USA, 2018; pp. 119–144. [Google Scholar]
- Lewis, C.M. Genetic association studies: Design, analysis and interpretation. Brief. Bioinform. 2002, 3, 146–153. [Google Scholar] [CrossRef]
- Setu, T.; Basak, T. An Introduction to Basic Statistical Models in Genetics. Open J. Stat. 2021, 11, 1017–1025. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Adzhubei, I.A.; Schmidt, S.; Peshkin, L.; Ramensky, V.E.; Gerasimova, A.; Bork, P.; Kondrashov, A.S.; Sunyaev, S.R. A method and server for predicting damaging missense mutations. Nat. Methods 2010, 7, 248–249. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).