Abstract
Prenatal exposure to metabolism-disrupting chemicals (MDCs) has been linked to birth weight, but the molecular mechanisms remain largely unknown. In this study, we investigated gene expressions and biological pathways underlying the associations between MDCs and birth weight, using microarray transcriptomics, in a Belgian birth cohort. Whole cord blood measurements of dichlorodiphenyldichloroethylene (p,p’-DDE), polychlorinated biphenyls 153 (PCB-153), perfluorooctanoic acid (PFOA), perfluorooctane sulfonic acid (PFOS), and transcriptome profiling were conducted in 192 mother–child pairs. A workflow including a transcriptome-wide association study, pathway enrichment analysis with a meet-in-the-middle approach, and mediation analysis was performed to characterize the biological pathways and intermediate gene expressions of the MDC–birth weight relationship. Among 26,170 transcriptomic features, we successfully annotated five overlapping metabolism-related gene expressions associated with both an MDC and birth weight, comprising BCAT2, IVD, SLC25a16, HAS3, and MBOAT2. We found 11 overlapping pathways, and they are mostly related to genetic information processing. We found no evidence of any significant mediating effect. In conclusion, this exploratory study provides insights into transcriptome perturbations that may be involved in MDC-induced altered birth weight.
1. Introduction
Metabolism-disrupting chemicals (MDCs) have been defined as natural or anthropogenic endocrine-disrupting chemicals (EDCs) that can promote metabolic changes and ultimately lead to obesity, type 2 diabetes and/or non-alcoholic fatty liver disease (NAFLD) [1]. In line with the Developmental Origins of Health and Disease (DOHaD) hypothesis [1], the prenatal period is a highly sensitive and vulnerable phase during which stressors, such as MDCs, can alter cell numbers and fate, gene expression, and protein levels that may lead to changes in tissue and organ function and contribute to increased susceptibility to a variety of non-communicable diseases later in life [2]. This may be the result of differences in toxicokinetics between children and adults and from time-dependent programming during early development [3].
Both high and low birth weight (HBW and LBW) are considered important predictors of later perturbed metabolic outcomes in children and adults [4,5,6]. While some observational studies have demonstrated associations between exposure to MDCs [including dichlorodiphenyldichloroethylene (p,p’-DDE), polychlorinated biphenyl-153 (PCB-153), perfluorooctanoic acid (PFOA), and perfluorooctane sulfonic acid (PFOS)] and birth weight [7,8,9,10], the molecular mechanisms of action remain poorly understood. The field of omics, based on high-throughput biochemical data, provides promising opportunities to advance and enhance our understanding of the impact of MDCs on child health, including by revealing changes in the gene expression using transcriptome profiling [11,12].
Assessing the effects of various chemical exposures on gene expression may help to uncover cellular mechanisms through which exposures influence the development of metabolic disorders in human populations. Several recent epidemiological studies using transcriptomics data have increased our understanding of how exposure to MDCs may perturb gene expression, and have identified regulatory pathways that may be affected by these exposures [13,14,15], as well as links between gene expression and birth weight [16,17,18,19]. However, to our knowledge, a study assessing the transcriptome in relation to both MDCs and birth weight in the same study population has not been performed.
Based on results from our previous birth cohort study [15], several MDCs (p,p’-DDE, PCB-153, PFOA, and PFOS) were suggested to play a role in transcriptional changes which are related to metabolic health outcomes. This led us to hypothesize that prenatal exposure to MDCs induces transcriptional modifications that, in turn, affect birth weight and have adverse effects on human health. Here, we aim to identify transcriptomic alterations in the cord blood of Belgian mother–child pairs that are associated with both prenatal MDC levels and birth weight in order to better understand the molecular effects and the underlying mechanisms.
2. Results
2.1. Population Characteristics
Demographic and exposure information for participants are shown in Table 1 and Table S1. The median gestational age was 40 weeks. Most children (98%) had a birth weight at or more than 2500 g, with a median of 3540 g. The median concentrations were 75.9 ng/g lipid, 28.7 ng/g lipid, 1600 ng/L, and 2700 ng/L for p,p’-DDE, PCB-153, PFOA, and PFOS, respectively (Table S1). The majority of the mothers had completed a high level of education (59%), had a normal pre-pregnancy body mass index (BMI) between 18.5 and 25 kg/m2 (71%), and did not smoke during pregnancy (85%). In addition, 38% of mothers were nulliparous and 57% were above 30 years of age at delivery.

Table 1.
Study population characteristics of 193 mother–child pairs, Flanders, Belgium.
2.2. Gene Expression Associated with MDCs and Birth Weight
Using a transcriptome-wide association study (TWAS) approach, we failed to select any features from models (1) or (2) with significance levels of false discovery rate (FDR) <0.05 or 0.20, and selected only a few features with a stringent p-value < 0.01 (Table 2). In order to avoid excluding weak but possibly relevant features, we used a relatively lenient p-value < 0.05 to select features for further analyses as an exploratory study. With p-value < 0.05, we found that 2110 out of 26,170 features were associated with one or more MDCs (777, 623, 333, and 624 for p,p’-DDE, PCB-153, PFOA, and PFOS, respectively; Table 2), and 775 features were associated with birth weight. A similar number of associated features were found in the sensitivity analyses of gestational age-unadjusted MDC–transcriptome associations (Table S2). In addition, as shown in the volcano plots (Figure S1a–e), the significance and directionality of gene expression obtained with and without adjustment for gestational age were consistent in the TWAS models for MDCs and features.

Table 2.
Number of features associated with MDCs and birth weight at different significance levels.
At p-value < 0.05, we found overlapping features associated with an MDC (p,p’-DDE, PCB-153, PFOA, or PFOS) and birth weight (12, 31, 17, and 40, respectively; Figure 1). These features were annotated to corresponding unique gene symbols, and according to the Human Protein Atlas and GeneCards [20,21], several were components of metabolism-related pathways, including branched-chain aminotransferase 2 (BCAT2) (amino acid metabolism; valine, leucine, and isoleucine degradation; valine, leucine, and isoleucine biosynthesis; and pantothenate and CoA biosynthesis), isovaleryl-CoA dehydrogenase (IVD) (valine, leucine, and isoleucine degradation), solute carrier family 25-A16 (SLC25A16) (pantothenate and CoA biosynthesis), Hyaluronan Synthase 3 (HAS3) (carbohydrate metabolism and glycosaminoglycan metabolism), and Membrane Bound O-Acyltransferase Domain Containing 2 (MBOAT2) (glycerophospholipid metabolism) (Table 3). However, with the mediation analysis, we did not observe any overlapping gene expression playing a significant mediating role, given the relatively large FDR values (Table 3). In addition, the individual associations of these five gene expressions with an MDC or birth weight is shown in Table S3.
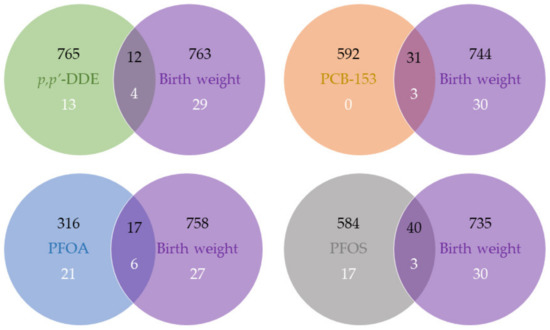
Figure 1.
Venn diagram of features and enriched pathways associated with MDCs and birth weight. Number in black refers to the number of features at p-value < 0.05, and number in white refers to the number of enriched pathways at FDR < 0.05.

Table 3.
The ACMEs of an MDC on birth weight via overlapping gene expression.
2.3. Pathways Associated with MDCs and Birth Weight
The MDC- or birth weight-associated pathways at FDR < 0.05 with at least five genes involved are represented in Table S4. There were 17, 3, 27, 20, and 33 pathways associated with p,p’-DDE, PCB-153, PFOA, PFOS, and birth weight, respectively; most of them were related to genetic information processing and organismal systems. Notably, one metabolic pathway [glycosaminoglycan biosynthesis] was linked to p,p’-DDE; three [metabolism of xenobiotics by cytochrome P450, drug metabolism, and type 1 diabetes mellitus (T1D)] were linked to PFOA, two [amino sugar and nucleotide sugar metabolism, type I diabetes mellitus] were linked to PFOS, and five [oxidative phosphorylation (OXPHOS), non-alcoholic fatty liver disease (NAFLD), cysteine and methionine metabolism, sulfur metabolism, and valine, leucine, and isoleucine degradation] were linked to birth weight.
At FDR < 0.05, we found that four, three, six, and three pathways associated with birth weight ovrlapped with p,p’-DDE, PCB-153, PFOA, and PFOS, respectively (Figure 1). They mostly belong to the “genetic information processing” category in the Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Database [22], and none of them were metabolism-related pathways (Table 4). The PC1 scores used to represent pathways in the mediation analysis explained 37–57% of the variance in the involved genes, and given the insignificant average causal mediation effects (ACMEs) with large FDR values, we did not observe any pathway that mediated both MDC and birth weight (Table 4).

Table 4.
The ACMEs of an MDC on birth weight via overlapping pathways.
3. Discussion
Transcriptome changes in early life may act in response to environmental exposures and subsequently lead to adverse health outcomes later in life; however, epidemiological studies are scarce. This is the first paper that evaluated the cord blood transcriptome with MDC exposures and birth weight. We examined differences in transcriptomics at the gene and pathway levels.
The five gene expressions that are metabolism-related and were found to be associated with both an MDC and birth weight are BCAT2, IVD, SLC25a16, HAS3, and MBOAT2. Birth weight may be altered by an MDC through one of these gene expressions, although we did not find a mediating effect to be significant. Branched-chain amino acids (BCAAs) are associated with the progression of obesity-related metabolic disorders [23]; additionally, BCAA catabolism is suggested to play a role in the pathogenesis of metabolic disturbances, and BCAT2 is an important enzyme that catalyzes the initial step of BCAA catabolism [24,25]. In a recent human study [26], BCAT2 variants were detected in Spanish infants suspected of having maple syrup urine disease—a rare metabolic disorder that some babies are born with. In line with our finding on higher BCAT2 expression with high birth weight, LBW pigs were found to express less BCAT2 mRNA in the longissimus dorsi muscle compared to normal birth weight pigs [27]. Similarly, higher BCAT2 mRNA was revealed in the blastocysts of diabetic rabbits compared to control blastocysts [28]. We observed an inverse association of IVD expression with birth weight. It has been demonstrated that the deficiency of the mitochondrial enzyme IVD may lead to isovaleric acidemia (IVA), an inherited metabolic disorder that may cause problems with the breakdown of the amino acid leucine [29]. Children with this condition may fail to gain weight and often experience developmental delays [30]. SLC25a16 has been considered as a carrier of Grave’s disease, which causes hypothyroidism [31]. On the other hand, hypothyroidism is thought to cause HBW [32,33], which may explain the association we found between SLC25a16 and higher birth weight, but this needs to be further explored. Our results on gene expression suggest new insights into birth weight changes indirectly caused by MDCs, and also provide some support, albeit weak signals, for the existing evidence from transcriptomics–birth weight research. However, it is also important to note that none of the features selected for further analyses from the TWAS models passed the FDR correction threshold; our gene expression results should therefore be viewed as exploratory and hypothesis-generating.
Metabolism-related pathways linked to both an MDC and birth weight were not observed in this study. However, some results on the metabolism-related pathways associated with an MDC or birth weight are noteworthy. For PFOA, we have observed positive associations with the metabolism of xenobiotics by cytochrome P450 and drug metabolism and inverse association with T1D. In a mouse study, PFOA was found to induce the cytochrome P450 enzyme by activating constitutive androstane receptor (CAR) nuclear receptors [34]. Another mouse study has shown that PFOA may induce drug metabolism, and then lead to liver injury [35]. For PFOS, we observed inverse associations with amino sugar and nucleotide sugar metabolism, and T1D. Consistently, PFOS-induced altered amino sugar and nucleotide sugar metabolism were found in a recent zebrafish study, as well as in Hispanic children [36,37]. In a large U.S. study, PFOA and PFOS were associated with a reduced risk of T1D in adults [38], but in a recent Finnish study, they both were associated with an increased risk of T1D in newborns [39]. For birth weight, lower birth weight was found to be associated with six metabolism-related pathways, comprising OXPHOS, NAFLD, cysteine and methionine metabolism, sulfur metabolism, valine, leucine and isoleucine degradation, and fatty acid biosynthesis. Consistent with our findings, LBW was shown to be associated with OXPHOS in the skeletal muscle and myotubes of Danish individuals [40,41]. A study investigating the relationship between birth weight and NAFLD, in 538 children, also showed an overrepresentation of LBW in those with NAFLD compared with the general U.S. population [42]. This inverse relationship between birth weight and NAFLD occurrence was also confirmed in a large French prospective cohort study of 55,034 adults [43]. Likewise, a recent systematic review and meta-analysis demonstrated that excess methionine and cysteine led to lower birth weight [44]. The effect of branched chain amino acids (valine, leucine and isoleucine) on birth weight was not yet clear, and most of the existing studies were animal studies [44]. In addition, there is growing evidence that there may be an association between high fatty acid levels and LBW [45,46,47,48].
The strengths of our study include the well-defined sampling frame and the use of omics techniques, which allow for the investigation of multiple genes and pathways simultaneously, in order to explore the impact of MDCs on the transcriptome perturbations and the subsequent impact on the birth weight. We also acknowledge several limitations of this study. First, the relatively small sample size (n = 193 mother–child pairs) of our study population was prone to modest statistical power in detecting associations. Also for this reason, we did not perform sex-specific analysis despite that EDCs have been shown to exert different adverse effects in males and females, both in laboratory animals and in humans [49]. Second, it should also be noted that the concentrations of p,p’-DDE, PCB-153, and PFOS in our study population were relatively low compared with the median exposure levels observed in other studies that found associations with birth weight [7,50,51], and they may not have been high enough to have a measurable effect, or the limited contrast in exposures may have limited statistical power to detect associations; PFOA levels were more comparable with levels in other studies. Third, the cross-sectional design of the study precluded establishing a temporal or causal relationship between MDC concentrations, transcriptome, and birth weight. Last, as with any other observational epidemiological study, there may be residual confounding bias due to uncontrolled unmeasured confounders, but we expected these to be minimal, as we carefully adjusted for a set of covariates that have been shown to be important with the help of directed acyclic graphs (DAGs).
In addition, the mechanisms are complex and sensitive windows, for exposure to MDCs may vary depending on the specific chemical. Alterations at the molecular level caused by MDCs may also differ according to the specific outcome being studied. Therefore, different exposure windows and outcomes should be assessed in further studies investigating the metabolism-disrupting effects of chemicals.
4. Materials and Methods
4.1. Study Population
We used data from the second cycle of the Flemish Environment and Health Study (FLEHS II, 2008–2009), whose design and recruitment have been previously described in detail [52]. In short, 255 mother–child pairs were recruited from Flanders, Belgium, using a two-stage sampling procedure, with provinces as the primary sampling unit and maternity units as the secondary sampling unit. Mothers who had lived for at least 10 years in Flanders and were able to fill in Dutch questionnaires were invited to participate. The number of participants in each province was proportional to the number of inhabitants. Among the mother–child pairs, 195 were randomly selected for transcriptome profiling. We restricted our analyses to the 193 term births (gestational age ≥37 weeks) in this study because preterm birth is a potential mediator of the effects of chemical exposures on birth weight [53].
4.2. Exposure Assessment
Several classes of environmental chemicals were measured in cord blood samples. Here, we have focused our analyses on MDC exposures that could be detected in at least 60% of the cord blood samples [54]: p,p’-DDE, PCB-153, PFOA, and PFOS (see Supplementary Material, Table S1 for detection rates, which ranged from 97 to 100% for these four chemicals). Samples were collected immediately after birth and stored at −80 °C until the measurements. MDC concentrations were measured using gas chromatography-electron capture negative ionization mass spectrometry (for p,p’-DDE and PCB-153) and high-performance liquid chromatography with tandem mass spectrometry detection (for PFOA and PFOS), as previously described [55,56]. All of the samples had quantifiable concentrations of p,p’-DDE, PFOA, and PFOS, while for PCB-153, 3% of the samples had values below the limit of quantification (LOQ, 300 ng/L). These values were then imputed using maximum likelihood estimation, assuming a censored log-normal distribution for values above the LOQ and conditional on the observed values for other biomarkers [54,57]. Lipid-standardized p,p’-DDE and PCB-153 concentrations were calculated based on estimated total lipids [total lipids = 50.49 + 1.32 × (cholesterol + triglycerides) (mg/dL)] and expressed as ng/g lipids for subsequent analyses [15]. All MDC concentrations were log2-transformed in order to reduce the potential influence of extreme values.
4.3. Transcriptome Profiling and Processing
As previously described [15,58], total RNA was extracted from the cord blood samples and stored at −80 °C. Amplified and labeled cRNA were then hybridized to 4 × 44 K Agilent Whole Human Genome Microarray (design 014850, one-color experimental setup with Cy3-labeling; Santa Clara, CA, USA), according to the manufacturer’s protocol. Preprocessing, quality assessment, and normalization of the microarray data were performed as described previously [15]. Briefly, the arrays were scanned with an Agilent scanner (G2565BA) and were subjected to primary quality control using the Agilent Feature Extraction Software (Version 10.7; Santa Clara, CA, USA). Furthermore, for each feature on the array, the quantile-normalized and log2-transformed signal intensity derived from Cy3 fluorescent dye was used for subsequent analyses. For replicated features on the array, the mean of signal intensities was calculated. After control and noise filtering by removing features with signal intensity below 3, 33,543 features retained. Thereafter, we used the R package Combat to eliminate possible batch effects related to different hybridization dates (28 dates from 14 September 2011 to 11 January 2012) [59,60]. Lastly, 26,170 (78.02%) features were annotated to a total of 17,880 unique gene symbols according to the Molecular Signatures Database (MSigDB) and were subjected to further statistical and functional analyses [61].
4.4. Outcome Assessment and Covariates
We considered birth weight (g) as our outcome of interest. DAGs were used to guide the selection of covariates (Figures S1b and S2a). The set of minimally sufficient covariates included sex of the child (girl, boy), smoking during pregnancy (smoking, non-smoking), parity (0, 1, ≥2), maternal education (low, medium, high), maternal age at delivery (<27, 27 < 30, 30 < 33, ≥33 years), pre-pregnancy BMI (<18.5, 18.5 < 25, 25 < 30, ≥30 kg/m2), and gestational age (weeks). Birth weight and child sex were collected from maternity medical records. Other covariate data was obtained from questionnaires. Missing data in covariates and exposures that were completely missing (1–3% and 1% of participants had one of more missing values, respectively) were singly imputed using the R package mice [62].
4.5. TWAS
TWASs were conducted in order to investigate the association of global transcriptomics with (1) MDCs and (2) birth weight. We used the following multivariable linear models to evaluate the effects of MDC exposures and potential predictors of birth weight, for each feature and MDC separately:
where i indexes the study subjects and Model (1) describes the association between a single transcriptomic feature and a single MDC, while Model (2) describes the association between birth weight and a single transcriptomic feature. Parameters β0 and γ0 are the model intercepts, while β1 and γ1 refer to the effect estimates (slopes) for a single MDC on a single transcriptomic feature, and for a single transcriptomic feature on birth weight, respectively. Parameters β2–8 and γ2–8 are coefficients corresponding to other covariates in the model, and ε1i and ε2i represent the residual errors, which are assumed to follow a normal distribution.
log2(feature intensityi) = β0 + β1 log2(MDCi) + β2 sexi + β3 smoking during pregnancyi + β4 parityi + β5 educationi + β6 age at deliveryi + β7 pre-pregnancy BMIi + β8 gestational agei + ε1i
birth weighti = γ0 + γ1 log2(feature intensityi) + γ2 sexi + γ3 smoking during pregnancyi + γ4 parityi + γ5 educationi + γ6 age at deliveryi + γ7 pre-pregnancy BMIi + γ8 gestational agei + ε2i
According to observed p-values for β1 and γ1, we estimated FDR using the method of Benjamini and Hochberg to correct for multiple testing and to select significant features [63].
4.6. Enrichment Pathway Analysis
In order to find pathways associated with MDC exposures and birth weight, we carried out Gene Set Enrichment Analyses (GSEA) using the WEB-based GEne SeT AnaLysis Toolkit (WebGestalt; Los Angeles, CA, USA) tool with pathway gene sets from the KEGG database [22,64,65]. First, we generated the respective ranked lists of all 26,170 features, sorted by their degree of differential expression (log2-fold change) in cord blood in relation to MDCs and birth weight, i.e., β1 and γ1 obtained from Models (1) and (2) [66,67]. Subsequently, the normalized enrichment scores were calculated, reflecting the degree to which pathways were enriched by ranked genes, where positive and negative values represent positive and inverse associations of pathways with MDCs or birth weight, respectively [68]. We restricted to pathways with at least five genes involved, and estimated the statistical significance using 1000 gene set permutations with FDR correction for multiple testing. Pathways with FDR < 0.05 were considered significant.
4.7. Mediation Analysis
Figure 2 outlines the workflow of the meet-in-the-middle approach used in this study [69]. The overlapping selected features and pathways observed in association with any of the four MDCs and birth weight were further explored by mediation analysis using the R package mediation [70] to explore potential biological mechanisms and mediating effects linking exposure and outcome. When assessing an overlapping feature as a mediator, we included it in the mediation model, and computed ACMEs (also known as indirect effects) using 1000 bootstrapped samples with FDR correction, and it was considered as a potential mediating feature if the FDR < 0.05. When examining an overlapping pathway as a mediator, we first performed a principal component analysis (PCA) on the genes belonging to that pathway, and then used the first principal component score (PC1) to represent that pathway in the mediation model [71,72]. ACMEs with FDR < 0.05 were generated to identify potential mediating pathways.
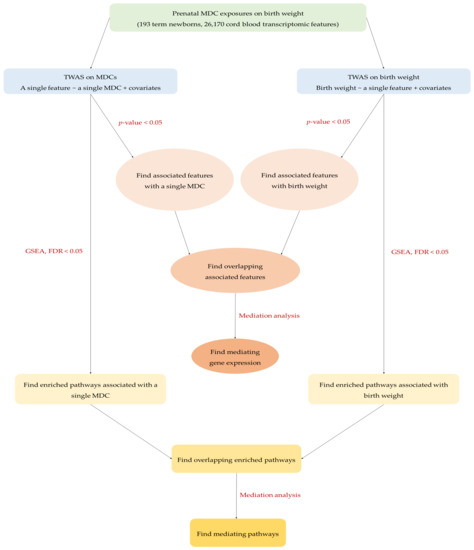
Figure 2.
The workflow of meet-in-the-middle approach in the present study.
4.8. Sensitivity Analysis
Recognizing that gestational age could be associated with MDC exposure and impact transcriptome levels [15], combined with several other studies also showing that the transcriptome was substantially influenced by gestational age [73,74], gestational age was included as a control variable in our primary regression model of MDCs and transcriptome (TWAS Model (1)). On the other hand, the causal direction of the association between gestational age and MDC is not entirely clear, and it is possible that gestational age mediates the outcome [75]. Therefore, in a sensitivity analysis, we assessed MDC and transcriptome associations without adjusting for gestational age in order to avoid adjustment for a potential mediator [53].
All statistical analyses were performed in R version 4.1.0 [76].
5. Conclusions
In summary, we integrated cord blood TWASs in order to identify gene expressions and pathways associated with MDCs and birth weight. Taken together, our study suggested five gene expressions associated with at least one MDC and birth weight. This provides insight into the etiology of higher and lower birth weight and possible later metabolic disorders, but again, this is an exploratory study with weak signals. In order to validate our results and further understand the potential link between MDC exposures and birth weight, and to elucidate the underlying mechanisms, studies with larger sample sizes and prospective study designs combined with advanced omics techniques are warranted.
Supplementary Materials
The supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms24087607/s1.
Author Contributions
Conceptualization, R.V. and S.R.; methodology, A.C., L.P., R.V., V.L. and S.R.; formal analysis, A.C.; data curation, A.C. and S.R.; writing—original draft preparation, A.C.; writing—review and editing, L.P., G.E., J.L., R.V., V.L. and S.R.; supervision, V.L. and S.R.; funding acquisition, J.L. and R.V. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the European Union’s Horizon 2020 Research and Innovation Action, under grant agreement GOLIATH no. 825489.
Institutional Review Board Statement
The study was approved by the ethical committee of the University of Antwerp (Reference UA A08 09).
Informed Consent Statement
Written informed consent was provided by all participating mothers.
Data Availability Statement
Data available on request due to privacy restrictions.
Acknowledgments
The authors would like to thank the Flemish Center of Expertise on Environment and Health, who carried out the FLEHS studies, and the Ministry of the Flemish Community, who commissioned, financed, and steered the studies. The authors also would like to thank all participants for their generous collaboration.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Heindel, J.J.; Blumberg, B.; Cave, M.; Machtinger, R.; Mantovani, A.; Mendez, M.A.; Nadal, A.; Palanza, P.; Panzica, G.; Sargis, R.; et al. Metabolism Disrupting Chemicals and Metabolic Disorders. Reprod. Toxicol. 2017, 68, 3–33. [Google Scholar] [CrossRef] [PubMed]
- Heindel, J.J.; vom Saal, F.S. Role of Nutrition and Environmental Endocrine Disrupting Chemicals during the Perinatal Period on the Aetiology of Obesity. Mol. Cell. Endocrinol. 2009, 304, 90–96. [Google Scholar] [CrossRef]
- Perlroth, N.H.; Branco, C.W.C. Current Knowledge of Environmental Exposure in Children during the Sensitive Developmental Periods. J. Pediatr. 2017, 93, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Binkin, N.J.; Yip, R.; Fleshood, L.; Trowbridge, F.L. Birth Weight and Childhood Growth. Pediatrics 1988, 82, 828–834. [Google Scholar] [CrossRef] [PubMed]
- Knop, M.R.; Geng, T.T.; Gorny, A.W.; Ding, R.; Li, C.; Ley, S.H.; Huang, T. Birth Weight and Risk of Type 2 Diabetes Mellitus, Cardiovascular Disease, and Hypertension in Adults: A Meta-Analysis of 7,646,267 Participants From 135 Studies. J. Am. Heart Assoc. 2018, 7, e008870. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Y.; Ma, J.; Wang, Y.; Li, W.; Katzmarzyk, P.T.; Chaput, J.-P.; Fogelholm, M.; Johnson, W.D.; Kuriyan, R.; Kurpad, A.; et al. Birth Weight and Childhood Obesity: A 12-Country Study. Int. J. Obes. Suppl. 2015, 5 (Suppl. S2), S74–S79. [Google Scholar] [CrossRef]
- Casas, M.; Nieuwenhuijsen, M.; Martínez, D.; Ballester, F.; Basagaña, X.; Basterrechea, M.; Chatzi, L.; Chevrier, C.; Eggesbø, M.; Fernandez, M.F.; et al. Prenatal Exposure to PCB-153, p,p’-DDE and Birth Outcomes in 9000 Mother-Child Pairs: Exposure-Response Relationship and Effect Modifiers. Environ. Int. 2015, 74, 23–31. [Google Scholar] [CrossRef]
- Govarts, E.; Nieuwenhuijsen, M.; Schoeters, G.; Ballester, F.; Bloemen, K.; de Boer, M.; Chevrier, C.; EggesbØ, M.; Guxens, M.; Krämer, U.; et al. Birth Weight and Prenatal Exposure to Polychlorinated Biphenyls (PCBs) and Dichlorodiphenyldichloroethylene (DDE): A Meta-Analysis within 12 European Birth Cohorts. Environ. Health Perspect. 2012, 120, 162–170. [Google Scholar] [CrossRef]
- Dzierlenga, M.W.; Crawford, L.; Longnecker, M.P. Birth Weight and Perfluorooctane Sulfonic Acid: A Random-Effects Meta-Regression Analysis. Environ. Epidemiol. 2020, 4, e095. [Google Scholar] [CrossRef]
- Steenland, K.; Barry, V.; Savitz, D. Serum Perfluorooctanoic Acid and Birthweight: An Updated Meta-Analysis with Bias Analysis. Epidemiology 2018, 29, 765–776. [Google Scholar] [CrossRef]
- Messerlian, C.; Martinez, R.M.; Hauser, R.; Baccarelli, A.A. ‘Omics’ and Endocrine Disrupting Chemicals—New Paths Forward. Nat. Rev. Endocrinol. 2017, 13, 740–748. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Hollinger, H.; Radke, E.G. ‘Omics in Environmental Epidemiological Studies of Chemical Exposures: A Systematic Evidence Map. Environ. Int. 2022, 164, 107243. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.; Yan, L.; Zhang, J.; Wang, Z.; Tian, M.; Shen, H. An Integrated Metabonomics and Transcriptomics Approach to Understanding Metabolic Pathway Disturbance Induced by Perfluorooctanoic Acid. J. Pharm. Biomed. Anal. 2013, 86, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Espín-Pérez, A.; Hebels, D.G.A.J.; Kiviranta, H.; Rantakokko, P.; Georgiadis, P.; Botsivali, M.; Bergdahl, I.A.; Palli, D.; Späth, F.; Johansson, A.; et al. Identification of Sex-Specific Transcriptome Responses to Polychlorinated Biphenyls (PCBs). Sci. Rep. 2019, 9, 746. [Google Scholar] [CrossRef] [PubMed]
- Remy, S.; Govarts, E.; Wens, B.; De Boever, P.; Den Hond, E.; Croes, K.; Sioen, I.; Baeyens, W.; van Larebeke, N.; Koppe, J.; et al. Metabolic Targets of Endocrine Disrupting Chemicals Assessed by Cord Blood Transcriptome Profiling. Reprod. Toxicol. 2016, 65, 307–320. [Google Scholar] [CrossRef]
- Alfano, R.; Chadeau-Hyam, M.; Ghantous, A.; Keski-Rahkonen, P.; Chatzi, L.; Perez, A.E.; Herceg, Z.; Kogevinas, M.; de Kok, T.M.; Nawrot, T.S.; et al. A Multi-Omic Analysis of Birthweight in Newborn Cord Blood Reveals New Underlying Mechanisms Related to Cholesterol Metabolism. Metabolism 2020, 110, 154292. [Google Scholar] [CrossRef]
- Cox, B.; Tsamou, M.; Vrijens, K.; Neven, K.Y.; Winckelmans, E.; De Kok, T.M.; Plusquin, M.; Nawrot, T.S. A Co-Expression Analysis of the Placental Transcriptome in Association with Maternal Pre-Pregnancy BMI and Newborn Birth Weight. Front. Genet. 2019, 10, 354. [Google Scholar] [CrossRef]
- Gillberg, L.; Perfilyev, A.; Brøns, C.; Thomasen, M.; Grunnet, L.G.; Volkov, P.; Rosqvist, F.; Iggman, D.; Dahlman, I.; Risérus, U.; et al. Adipose Tissue Transcriptomics and Epigenomics in Low Birthweight Men and Controls: Role of High-Fat Overfeeding. Diabetologia 2016, 59, 799–812. [Google Scholar] [CrossRef]
- Peng, S.; Deyssenroth, M.A.; Di Narzo, A.F.; Cheng, H.; Zhang, Z.; Lambertini, L.; Rusualepp, A.; Kovacic, J.C.; Bjorkegren, J.L.M.; Marsit, C.J.; et al. Genetic Regulation of the Placental Transcriptome Underlies Birth Weight and Risk of Childhood Obesity. PLOS Genet. 2018, 14, e1007799. [Google Scholar] [CrossRef]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Tissue-Based Map of the Human Proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Stein, T.I.; Nudel, R.; Lieder, I.; Mazor, Y.; et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr. Protoc. Bioinform. 2016, 54, 1.30.1–1.30.33. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Zhang, F.; Sun, D.; Wang, X.; Zhang, X.; Zhang, J.; Yan, F.; Huang, C.; Xie, H.; Lin, C.; et al. Branched-Chain Amino Acids Exacerbate Obesity-Related Hepatic Glucose and Lipid Metabolic Disorders via Attenuating Akt2 Signaling. Diabetes 2020, 69, 1164–1177. [Google Scholar] [CrossRef] [PubMed]
- Vanweert, F.; Schrauwen, P.; Phielix, E. Role of Branched-Chain Amino Acid Metabolism in the Pathogenesis of Obesity and Type 2 Diabetes-Related Metabolic Disturbances BCAA Metabolism in Type 2 Diabetes. Nutr. Diabetes 2022, 12, 35. [Google Scholar] [CrossRef]
- Lei, M.Z.; Li, X.X.; Zhang, Y.; Li, J.T.; Zhang, F.; Wang, Y.P.; Yin, M.; Qu, J.; Lei, Q.Y. Acetylation Promotes BCAT2 Degradation to Suppress BCAA Catabolism and Pancreatic Cancer Growth. Signal Transduct. Target. Ther. 2020, 5, 70. [Google Scholar] [CrossRef]
- Navarrete, R.; Leal, F.; Vega, A.I.; Morais-López, A.; Garcia-Silva, M.T.; Martín-Hernández, E.; Quijada-Fraile, P.; Bergua, A.; Vives, I.; García-Jiménez, I.; et al. Value of Genetic Analysis for Confirming Inborn Errors of Metabolism Detected through the Spanish Neonatal Screening Program. Eur. J. Hum. Genet. 2019, 27, 556–562. [Google Scholar] [CrossRef]
- Yonke, J.A.; Seymour, K.A.; El-Kadi, S.W. Leucine Supplementation Does Not Enhance Translation Initiation of Low Birth-Weight Neonatal Pigs Despite Greater Sestrin2 Expression in Skeletal Muscle. FASEB J. 2020, 34, 1. [Google Scholar] [CrossRef]
- Gürke, J.; Hirche, F.; Thieme, R.; Haucke, E.; Schindler, M.; Stangl, G.I.; Fischer, B.; Santos, A.N. Maternal Diabetes Leads to Adaptation in Embryonic Amino Acid Metabolism during Early Pregnancy. PLoS ONE 2015, 10, e0127465. [Google Scholar] [CrossRef]
- Vockley, J.; Ensenauer, R. Isovaleric Acidemia: New Aspects of Genetic and Phenotypic Heterogeneity. Am. J. Med. Genet. Part C Semin. Med. Genet. 2006, 142C, 95–103. [Google Scholar] [CrossRef]
- Grünert, S.C.; Wendel, U.; Lindner, M.; Leichsenring, M.; Schwab, K.O.; Vockley, J.; Lehnert, W.; Ensenauer, R. Clinical and Neurocognitive Outcome in Symptomatic Isovaleric Acidemia. Orphanet J. Rare Dis. 2012, 7, 9. [Google Scholar] [CrossRef]
- Gutiérrez-Aguilar, M.; Baines, C.P. Physiological and Pathological Roles of Mitochondrial SLC25 Carriers. Biochem. J. 2013, 454, 371–386. [Google Scholar] [CrossRef]
- Derakhshan, A.; Peeters, R.P.; Taylor, P.N.; Bliddal, S.; Carty, D.M.; Meems, M.; Vaidya, B.; Chen, L.; Knight, B.A.; Ghafoor, F.; et al. Association of Maternal Thyroid Function with Birth Weight: A Systematic Review and Individual-Participant Data Meta-Analysis. Lancet Diabetes Endocrinol. 2020, 8, 501–510. [Google Scholar] [CrossRef] [PubMed]
- Andersen, S.L.; Olsen, J.; Wu, C.S.; Laurberg, P. Low Birth Weight in Children Born to Mothers with Hyperthyroidism and High Birth Weight in Hypothyroidism, Whereas Preterm Birth Is Common in Both Conditions: A Danish National Hospital Register Study. Eur. Thyroid. J. 2013, 2, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Klaassen, C.D. Perfluorocarboxylic Acids Induce Cytochrome P450 Enzymes in Mouse Liver through Activation of PPAR-α and CAR Transcription Factors. Toxicol. Sci. 2008, 106, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Safe, S.H. Polychlorinated Biphenyls (PCBs): Environmental Impact, Biochemical and Toxic Responses, and Implications for Risk Assessment. Crit. Rev. Toxicol. 1994, 24, 87–149. [Google Scholar] [CrossRef]
- Lee, H.; Sung, E.J.; Seo, S.; Min, E.K.; Lee, J.Y.; Shim, I.; Kim, P.; Kim, T.Y.; Lee, S.; Kim, K.T. Integrated Multi-Omics Analysis Reveals the Underlying Molecular Mechanism for Developmental Neurotoxicity of Perfluorooctanesulfonic Acid in Zebrafish. Environ. Int. 2021, 157, 106802. [Google Scholar] [CrossRef]
- Alderete, T.L.; Jin, R.; Walker, D.I.; Valvi, D.; Chen, Z.; Jones, D.P.; Peng, C.; Gilliland, F.D.; Berhane, K.; Conti, D.V.; et al. Perfluoroalkyl Substances, Metabolomic Profiling, and Alterations in Glucose Homeostasis among Overweight and Obese Hispanic Children: A Proof-of-Concept Analysis. Environ. Int. 2019, 126, 445–453. [Google Scholar] [CrossRef]
- Conway, B.; Innes, K.E.; Long, D. Perfluoroalkyl Substances and Beta Cell Deficient Diabetes. J. Diabetes Complicat. 2016, 30, 993–998. [Google Scholar] [CrossRef]
- McGlinchey, A.; Sinioja, T.; Lamichhane, S.; Sen, P.; Bodin, J.; Siljander, H.; Dickens, A.M.; Geng, D.; Carlsson, C.; Duberg, D.; et al. Prenatal Exposure to Perfluoroalkyl Substances Modulates Neonatal Serum Phospholipids, Increasing Risk of Type 1 Diabetes. Environ. Int. 2020, 143, 105935. [Google Scholar] [CrossRef]
- Hansen, N.S.; Hjort, L.; Broholm, C.; Gillberg, L.; Schrölkamp, M.; Schultz, H.S.; Mortensen, B.; Jørgensen, S.W.; Friedrichsen, M.; Wojtaszewski, J.F.P.; et al. Metabolic and Transcriptional Changes in Cultured Muscle Stem Cells from Low Birth Weight Subjects. J. Clin. Endocrinol. Metab. 2016, 101, 2254–2264. [Google Scholar] [CrossRef]
- Brøns, C.; Jacobsen, S.; Nilsson, E.; Rönn, T.; Jensen, C.B.; Storgaard, H.; Poulsen, P.; Groop, L.; Ling, C.; Astrup, A.; et al. Deoxyribonucleic Acid Methylation and Gene Expression of PPARGC1A in Human Muscle Is Influenced by High-Fat Overfeeding in a Birth-Weight-Dependent Manner. J. Clin. Endocrinol. Metab. 2010, 95, 3048–3056. [Google Scholar] [CrossRef] [PubMed]
- Newton, K.P.; Feldman, H.S.; Chambers, C.D.; Wilson, L.; Behling, C.; Clark, J.M.; Molleston, J.P.; Chalasani, N.; Sanyal, A.J.; Fishbein, M.H.; et al. Low and High Birth Weights Are Risk Factors for Nonalcoholic Fatty Liver Disease in Children. J. Pediatr. 2017, 187, 141–146.e1. [Google Scholar] [CrossRef] [PubMed]
- Amadou, C.; Nabi, O.; Serfaty, L.; Lacombe, K.; Boursier, J.; Mathurin, P.; Ribet, C.; de Ledinghen, V.; Zins, M.; Charles, M.A. Association between Birth Weight, Preterm Birth, and Nonalcoholic Fatty Liver Disease in a Community-Based Cohort. Hepatology 2022, 76, 1438–1451. [Google Scholar] [CrossRef] [PubMed]
- Terstappen, F.; Tol, A.J.C.; Gremmels, H.; Wever, K.E.; Paauw, N.D.; Joles, J.A.; van der Beek, E.M.; Lely, A.T. Prenatal Amino Acid Supplementation to Improve Fetal Growth: A Systematic Review and Meta-Analysis. Nutrients 2020, 12, 2535. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Vilhjálmsdóttir, B.L.; Rohde, J.F.; Walker, K.C.; Runstedt, S.E.; Lauritzen, L.; Heitmann, B.L.; Specht, I.O. Systematic Literature Review and Meta-Analysis of the Relationship Between Polyunsaturated and Trans Fatty Acids During Pregnancy and Offspring Weight Development. Front. Nutr. 2021, 8, 625596. [Google Scholar] [CrossRef]
- Lee, E.; Kim, H.; Kim, H.; Ha, E.H.; Chang, N. Association of Maternal Omega-6 Fatty Acid Intake with Infant Birth Outcomes: Korean Mothers and Children’s Environmental Health (MOCEH). Nutr. J. 2018, 17, 47. [Google Scholar] [CrossRef]
- Meher, A.; Randhir, K.; Mehendale, S.; Wagh, G.; Joshi, S. Maternal Fatty Acids and Their Association with Birth Outcome: A Prospective Study. PLoS ONE 2016, 11, e0147359. [Google Scholar] [CrossRef]
- Van Eijsden, M.; Hornstra, G.; Van Der Wal, M.F.; Vrijkotte, T.G.M.; Bonsel, G.J. Maternal N-3, n-6, and Trans Fatty Acid Profile Early in Pregnancy and Term Birth Weight: A Prospective Cohort Study. Am. J. Clin. Nutr. 2008, 87, 887–895. [Google Scholar] [CrossRef]
- Van Larebeke, N.; Fucic, A. Chapter 5: Sex-Specific Actions of Endocrine Disruptors. In Challenges in Endocrine Disruptor Toxicology and Risk Assessment; Royal Society of Chemistry: London, UK, 2020; pp. 121–154. [Google Scholar] [CrossRef]
- Wolff, M.S.; Engel, S.; Berkowitz, G.; Teitelbaum, S.; Siskind, J.; Barr, D.B.; Wetmur, J. Prenatal Pesticide and PCB Exposures and Birth Outcomes. Pediatr. Res. 2007, 61, 243–250. [Google Scholar] [CrossRef]
- Wikström, S.; Lin, P.I.; Lindh, C.H.; Shu, H.; Bornehag, C.G. Maternal Serum Levels of Perfluoroalkyl Substances in Early Pregnancy and Offspring Birth Weight. Pediatr. Res. 2020, 87, 1093–1099. [Google Scholar] [CrossRef]
- Schoeters, G.; Den Hond, E.; Colles, A.; Loots, I.; Morrens, B.; Keune, H.; Bruckers, L.; Nawrot, T.; Sioen, I.; De Coster, S.; et al. Concept of the Flemish Human Biomonitoring Programme. Int. J. Hyg. Environ. Health 2012, 215, 102–108. [Google Scholar] [CrossRef] [PubMed]
- Wilcox, A.J.; Weinberg, C.R.; Basso, O. On the Pitfalls of Adjusting for Gestational Age at Birth. Am. J. Epidemiol. 2011, 174, 1062–1068. [Google Scholar] [CrossRef] [PubMed]
- Lubin, J.H.; Colt, J.S.; Camann, D.; Davis, S.; Cerhan, J.R.; Severson, R.K.; Bernstein, L.; Hartge, P. Epidemiologic Evaluation of Measurement Data in the Presence of Detection Limits. Environ. Health Perspect. 2004, 112, 1691–1696. [Google Scholar] [CrossRef] [PubMed]
- Midasch, O.; Drexler, H.; Hart, N.; Beckmann, M.W.; Angerer, J. Transplacental Exposure of Neonates to Perfluorooctanesulfonate and Perfluorooctanoate: A Pilot Study. Int. Arch. Occup. Environ. Health 2007, 80, 643–648. [Google Scholar] [CrossRef]
- Covaci, A.; Voorspoels, S. Optimization of the Determination of Polybrominated Diphenyl Ethers in Human Serum Using Solid-Phase Extraction and Gas Chromatography-Electron Capture Negative Ionization Mass Spectrometry. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2005, 827, 216–223. [Google Scholar] [CrossRef]
- Ottenbros, I.; Govarts, E.; Lebret, E.; Vermeulen, R.; Schoeters, G.; Vlaanderen, J. Network Analysis to Identify Communities Among Multiple Exposure Biomarkers Measured at Birth in Three Flemish General Population Samples. Front. Public Health 2021, 9, 590038. [Google Scholar] [CrossRef]
- Remy, S.; Govarts, E.; Bruckers, L.; Paulussen, M.; Wens, B.; Den Hond, E.; Nelen, V.; Baeyens, W.; Van Larebeke, N.; Loots, I.; et al. Expression of the SFLT1 Gene in Cord Blood Cells Is Associated to Maternal Arsenic Exposure and Decreased Birth Weight. PLoS ONE 2014, 9, e92677. [Google Scholar] [CrossRef]
- Espín-Pérez, A.; Portier, C.; Chadeau-Hyam, M.; van Veldhoven, K.; Kleinjans, J.C.S.; de Kok, T.M.C.M. Comparison of Statistical Methods and the Use of Quality Control Samples for Batch Effect Correction in Human Transcriptome Data. PLoS ONE 2018, 13, e0202947. [Google Scholar] [CrossRef]
- Johnson, W.E.; Li, C.; Rabinovic, A. Adjusting Batch Effects in Microarray Expression Data Using Empirical Bayes Methods. Biostatistics 2007, 8, 118–127. [Google Scholar] [CrossRef]
- Liberzon, A.; Subramanian, A.; Pinchback, R.; Thorvaldsdóttir, H.; Tamayo, P.; Mesirov, J.P. Molecular Signatures Database (MSigDB) 3.0. Bioinformatics 2011, 27, 1739–1740. [Google Scholar] [CrossRef]
- van Buuren, S.; Groothuis-Oudshoorn, K. Mice: Multivariate Imputation by Chained Equations in R. J. Stat. Softw. 2011, 45, 1–67. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Liao, Y.; Wang, J.; Jaehnig, E.J.; Shi, Z.; Zhang, B. WebGestalt 2019: Gene Set Analysis Toolkit with Revamped UIs and APIs. Nucleic Acids Res. 2019, 47, W199–W205. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene Set Enrichment Analysis: A Knowledge-Based Approach for Interpreting Genome-Wide Expression Profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Drizik, E.; Corbett, S.; Zheng, Y.; Vermeulen, R.; Dai, Y.; Hu, W.; Ren, D.; Duan, H.; Niu, Y.; Xu, J.; et al. Transcriptomic Changes in The Nasal Epithelium Associated with Diesel Engine Exhaust Exposure. Environ. Int. 2020, 137, 105506. [Google Scholar] [CrossRef] [PubMed]
- Winckelmans, E.; Vrijens, K.; Tsamou, M.; Janssen, B.G.; Saenen, N.D.; Roels, H.A.; Kleinjans, J.; Lefebvre, W.; Vanpoucke, C.; De Kok, T.M.; et al. Newborn Sex-Specific Transcriptome Signatures and Gestational Exposure to Fine Particles: Findings from the ENVIRONAGE Birth Cohort. Environ. Health 2017, 16, 52. [Google Scholar] [CrossRef]
- Reimand, J.; Isserlin, R.; Voisin, V.; Kucera, M.; Tannus-Lopes, C.; Rostamianfar, A.; Wadi, L.; Meyer, M.; Wong, J.; Xu, C.; et al. Pathway Enrichment Analysis and Visualization of Omics Data Using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nat. Protoc. 2019, 14, 482–517. [Google Scholar] [CrossRef]
- Chadeau-Hyam, M.; Athersuch, T.J.; Keun, H.C.; De Iorio, M.; Ebbels, T.M.D.; Jenab, M.; Sacerdote, C.; Bruce, S.J.; Holmes, E.; Vineis, P. Meeting-in-the-Middle Using Metabolic Profiling—A Strategy for the Identification of Intermediate Biomarkers in Cohort Studies. Biomarkers 2011, 16, 83–88. [Google Scholar] [CrossRef]
- Tingley, D.; Yamamoto, H.T.; Hirose, K.; Keele, L.; Princeton, K.I. Mediation: R Package for Causal Mediation Analysis. J. Stat. Softw. 2014, 59, v059. [Google Scholar] [CrossRef]
- Carmeli, C.; Kutalik, Z.; Mishra, P.P.; Porcu, E.; Delpierre, C.; Delaneau, O.; Kelly-Irving, M.; Bochud, M.; Dhayat, N.A.; Ponte, B.; et al. Gene Regulation Contributes to Explain the Impact of Early Life Socioeconomic Disadvantage on Adult Inflammatory Levels in Two Cohort Studies. Sci. Rep. 2021, 11, 3100. [Google Scholar] [CrossRef]
- Dagnino, S.; Bodinier, B.; Guida, F.; Smith-Byrne, K.; Petrovic, D.; Whitaker, M.D.; Nøst, T.H.; Agnoli, C.; Palli, D.; Sacerdote, C.; et al. Prospective Identification of Elevated Circulating CDCP1 in Patients Years before Onset of Lung Cancer. Cancer Res. 2021, 81, 3738–3748. [Google Scholar] [CrossRef] [PubMed]
- Stünkel, W.; Pan, H.; Chew, S.B.; Tng, E.; Tan, J.H.; Chen, L.; Joseph, R.; Cheong, C.Y.; Ong, M.L.; Lee, Y.S.; et al. Transcriptome Changes Affecting Hedgehog and Cytokine Signalling in the Umbilical Cord: Implications for Disease Risk. PLoS ONE 2012, 7, e39744. [Google Scholar] [CrossRef] [PubMed]
- Kleinbrink, E.L.; Gomez-Lopez, N.; Ju, D.; Done, B.; Goustin, A.S.; Tarca, A.L.; Romero, R.; Lipovich, L. Gestational Age Dependence of the Maternal Circulating Long Non-Coding RNA Transcriptome During Normal Pregnancy Highlights Antisense and Pseudogene Transcripts. Front. Genet. 2021, 12, 760849. [Google Scholar] [CrossRef] [PubMed]
- Street, M.E.; Bernasconi, S. Endocrine-Disrupting Chemicals in Human Fetal Growth. Int. J. Mol. Sci. 2020, 21, 1430. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: The R Project for Statistical Computing. Available online: https://www.r-project.org/index.html (accessed on 19 August 2021).
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).