Abstract
Breast cancer (BC) is one of the biggest health problems worldwide, characterized by intricate metabolic and biochemical complexities stemming from pronounced variations across dysregulated molecular pathways. If BC is not diagnosed early, complications may lead to death. Thus, the pursuit of novel therapeutic avenues persists, notably focusing on epigenetic pathways such as histone deacetylases (HDACs). The compound N-(2-hydroxyphenyl)-2-propylpentanamide (HO-AAVPA), a derivative of valproic acid (VPA), has emerged as a promising candidate warranting pre-clinical investigation. HO-AAVPA is an HDAC inhibitor with antiproliferative effects on BC, but its molecular mechanism has yet to be deciphered. Furthermore, in the present study, we determined the metabolomic effects of HO-AAVPA and VPA on cells of luminal breast cancer (MCF-7) and triple-negative breast cancer (MDA-MB-231) subtypes. The LC-MS untargeted metabolomic study allowed for the simultaneous measurement of multiple metabolites and pathways, identifying that both compounds affect glycerophospholipid and sphingolipid metabolism in the MCF-7 and MDA-MB-231 cell lines, suggesting that other biological targets were different from HDACs. In addition, there are different dysregulate metabolites, possibly due to the physicochemical differences between HO-AAVPA and VPA.
1. Introduction
Breast cancer (BC) has the highest incidence and is the leading cause of death due to cancer worldwide [1]. Early BC detection usually results in a favorable prognosis; however, if early detection fails, invasive BC treatment options are ineffective [1,2,3]. One of the biggest challenges for BC treatment is its heterogeneity, exemplified by BC subtypes that show a high diversity of genetic and epigenetic origins, which determine its classification and treatment and are directly related to the state of relevant receptors such as estrogen receptor (ER), progesterone receptor (PR), and human epithelial receptor 2 (HER2) [2]. Differences in the expression of these receptors have been used to indicate prognosis; the latter ranges from BC luminal A (LA) to triple-negative BC (TNBC), with highly contrasting prognoses: better prognosis and lower aggressiveness for LA and worse prognosis and high aggressiveness for TNBC [2].
The role of epigenetic modifications in carcinogenesis and cancer development has become more relevant due to environmental factors [4]. In particular, histone acetylation and deacetylation by histone acetyltransferases (HAT) and histone deacetylases (HDAC), respectively, have gained great importance due to their involvement in the regulation of transcription processes for genes implicated in carcinogenesis and cancer development [5,6]. In normal conditions, HDACs maintain histones positively charged, favoring their interactions with DNA and regulating the expression of genes involved in tumorigenesis. However, in cancer cells, HDAC overexpression disrupts gene regulation, causing the expression of genes associated with carcinogenesis and cancer development [5,7]. In this regard, the deregulation of HDAC has been identified as one of the main components of the origin of breast cancer, and thus, these proteins are considered high-value targets for its treatment [6,8,9,10,11]. Then, special attention has been given to HDAC inhibitors (HDACi) due to their capacity to restore altered epigenetic pathways, enabling the correct function of mechanisms that deal with cancer cells [7,12,13,14,15,16,17,18,19,20]. One of these compounds is valproic acid (VPA), an HDACi that acts on classes I and IIa HDACs [7,21]. In vitro studies have demonstrated that VPA can induce cell cycle arrest and apoptosis and affect cell migration in multiple breast cancer cell lines [22,23,24,25]. Similar to suberoylanilide hydroxamic acid (SAHA) and other HDACi, VPA also regulates cellular metabolism in cancer cells [26,27,28,29]. This metabolic regulation occurs independently of the transcriptional regulation of specific metabolism-related genes, a mechanism that also contributes to its anticancer activity [26].
Despite its therapeutic effects, the use of VPA is limited by its high concentrations, which can lead to toxicity. As a result, several VPA-derived molecules have been proposed; the most notable is compound N-(2-hydroxyphenyl)-2-propylpentanamide (HO-AAVPA), which is a VPA derivative [30,31,32,33,34]. HO-AAVPA exerts similar effects to VPA in different experimental contexts. For example, HO-AAVPA possesses an anticonvulsant effect comparable to VPA but with lower toxicity and teratogenic effects in an in vivo model [35]. Additionally, HO-AAVPA modifies HMGB1 acetylation, a nonhistone protein involved in DNA stability, repair, transcription, and recombination processes and in the generation of reactive oxygen species (ROS), activities which are also found in VPA with lower potency [36]. Although HO-AAVPA was designed as an HDACi against HDAC8 [37] based on docking affinity measurement, in vitro experiments have shown that HO-AAVPA has a higher affinity for HDAC1 [32]. In their original publication, Prestegui-Martlel et al. (2016) demonstrated that HO-AAVPA has antiproliferative activity against BC cells at lower concentrations compared to VPA [37]. Accumulating evidence supports the similar effects produced by HO-AAVPA and VPA, as well as the discrepancies between the molecular target for which it was designed and the findings obtained in vitro [35,36,37]. Furthermore, many pharmacological properties of HO-AAVPA remain to be elucidated, particularly its impact on intracellular metabolism of distinct subtypes of BC cells, such as LA and TNBC, two cell lines with varying levels of aggressiveness and distinct biological targets, including HDACs [9] and metabolomic profiles [38].
Due to the large magnitude and impact of epigenetic modifications regulated by HDACs, powerful tools are currently used to measure a large set of molecules in a reduced number of experiments. One example of such a tool is metabolomics, which uses experimental and bioinformatic methods to study metabolites. Unlike targeted metabolomics, the non-targeted approach allows for the simultaneous detection and quantification of thousands of small molecules, a global metabolic profile of the analyzed samples [39,40,41]. Through this strategy, it becomes possible to compare metabolic changes resulting from a particular treatment. This helps enhance comprehension of the biological processes and the metabolic pathways that are implicated. Although untargeted metabolomics is a valuable technique, it has some challenges. Signals do not represent metabolites directly, and they are considered “features” that have a specific m/z value, abundance, and retention time. Features are then processed to determine which of them are isotopes from adducts that belong to the same molecule, called “compound”. One reliable way to determine the identity of these compounds is by comparing the fragmentation spectra of specific “compounds” to those in metabolite libraries like the Human Metabolome Database (HMDB). If the two spectra match, the metabolite can be given a putative identity (metabolite identification confidence level 2) [42].
Under this approach, the present study aims to explore the metabolic changes induced by HO-AAVPA in two subtypes of breast cancer cells, contrasting with the effects produced by VPA. We hypothesize that HO-AAVPA impacts deregulated metabolic pathways in BC cells similar to VPA [43,44,45], but their physicochemical difference properties could yield different metabolomics profiles. The results of this work can increase the amount of knowledge about the effects elicited in cellular metabolites as well as contribute new evidence about the metabolic pathways that, when restored, induce the activation of the mechanisms involved in the elimination of BC cells from different subtypes, LA, represented by MCF-7 (less aggressive BC), and TNBC, represented by MDA-MB-231 (more aggressive BC). The following untargeted metabolomic study was carried out in two steps. Firstly, LC-MS was used to detect any features that were dysregulated by the treatments. Subsequently, LC-MS/MS was employed to gather fragmentation information to annotate the corresponding features as putative metabolites.
2. Results and Discussion
2.1. Inhibitory Concentration Assays
First, the effect on cell viability was determined for each compound in both cell lines (Figure 1). Then, the IC15 values were calculated from IC50 values (Table 1) and compared with those of previous reports. By observing the range of concentrations tested for both compounds (140 µM to 36 mM VPA and 9 µM to 2 mM HO-AAVPA), it is clear that HO-AAVPA has a higher potency, attributed to the 2-hydroxybenzamide fragment that functions as a zinc-binding group [35,36,37]. In their published study of HO-AAVPA, Prestegui-Martel et al. (2016) calculated IC50 values of 280 and 190 µM for MDA-MB-231 and MCF-7 cells, respectively [37]; according to our results, the calculated IC50 value was quite similar for MDA-MB-231 cells (291.8 µM) but not for MCF-7 cells (476.1 µM), although it was in the same order of magnitude. For VPA, a direct comparison was not possible because, in the aforementioned work, antiproliferative effects were not observed either in MDA-MB-231 or in MCF-7 cells at the maximum concentration tested; thus, authors determined that the effect was present at >450 and >350 µM for MDA-MB-231 and MCF-7 cells, respectively. However, Mawatari et al. (2015) found IC50 values for these cell lines on the same order of magnitude: 5.4 mM for MDA-MB-231 and 8.1 mM for MCF-7, compared with 7.29 and 7.11 mM in our study, respectively [25]. Other studies with different evaluation times have reported IC50 values ranging from 1.5 to 16 mM for MDA-MB-231 cells and from 0.7 to 8.1 mM for MCF-7 cells [24,46,47,48].
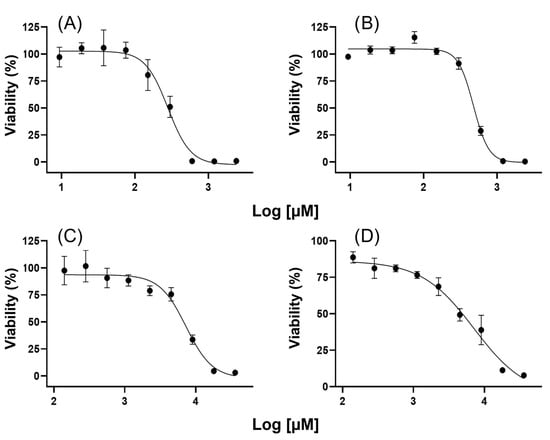
Figure 1.
The antiproliferative effects of different concentrations of HO-AAVPA and VPA on cell viability. (A) Effect of HO-AAVPA on MDA-MB-231 cells; (B) Effect of HO-AAVPA on MCF-7 cells; (C) Effect of VPA on MDA-MB-231 cells; (D) Effect of VPA on MCF-7 cells.

Table 1.
IC values of HO-AAVPA and VPA in triple-negative breast cancer and luminal breast cancer cells.
2.2. HO-AAVPA and VPA Effects on Breast Cancer Cells
Once IC15 values were obtained, the corresponding concentrations were applied to cell cultures to obtain the metabolomics samples. Through these experiments, 17 and 123 putative metabolites were found to be dysregulated by HO-AAVPA, and 12 and 20 were dysregulated by VPA in MDA-MB-231 (Table 2) and MCF-7 (Table 3) cells, respectively. For MDA-MB-231 cells, 10 putative metabolites were commonly affected by both compounds, although the trend (or direction) of dysregulation differed. Tables S1 and S2 summarize the putative identification values for the deregulated entities in the MDA-MB-231 and MCF7 cell lines, respectively.

Table 2.
Putative metabolites dysregulated in MDA-MB-231 cells.

Table 3.
Putative metabolites dysregulated in MCF-7 cells.
In MCF-7 cells, a higher number of shared putative metabolites were affected by both compounds (15 compounds), and, contrary to what we found in MDA-MB-231 cells, 6 were dysregulated in the same direction (downregulated). MCF-7-treated cells showed a tendency toward downregulation; however, the observed trend for MDA-MB-231-treated cells was the opposite: metabolites from HO-AAVPA-treated cells were mainly downregulated, while metabolites from VPA-treated cells were predominantly upregulated.
2.3. Effects of HO-AAVPA and VPA on the Metabolic Pathways of Breast Cancer Cells
Determination of the main affected pathways was performed through Pathway Analysis from Metaboanalyst, which considers both the number of altered elements in a pathway with respect to the total and the position they occupy in the pathway to calculate the impact score (Figure 2); however, this analysis does not consider the fold change direction [49]. As a result of the pathway impact analysis, some of the pathways were found to be unimpactful and are reported with a low score value (even zero in some cases), which means that a low number of members of the corresponding pathway (it might be only one member) was found in the dataset analyzed. Although MCF-7 cells treated with HO-AAVPA showed the highest number of dysregulated metabolites (Table 3), through pathway impact analysis, we were able to determine that the number of significantly dysregulated metabolic pathways was evenly distributed among treatments, ranging from two to three pathways. Even though the number of dysregulated metabolites varies, the number and identity of metabolic pathways impacted are similar. Among these pathways, lipid metabolism seems to be the main aspect affected, particularly glycerophospholipid and sphingolipid metabolism, although evidence of alteration of other pathways was also found, with lower impact. For TNBC cells, the sphingolipid metabolism impact score was lower than that for LA cells; for TNBC cells, glycerophospholipid metabolism was the most dysregulated. In contrast, sphingolipid metabolism was followed by glycerophospholipid metabolism as the most dysregulated pathway for LA cells. As stated by Zhao et al. (2013) and Nagarajan et al. (2021), modifications in lipid metabolism are one of the major components that enable cancer cells to adapt, which allows them to generate the necessary components for their development and tumorigenesis, such as increases in lipid uptake, lipogenesis, membrane synthesis, or signaling processes through phospholipids [50,51,52]. In another study, T. Burg et al. (2021) demonstrated that there is a relationship between epigenetic modifications and lipid metabolism defects in the spinal cord of a model of amyotrophic lateral sclerosis (ALS) in FUS mice according to transcriptomic and lipidomic analysis of treatment with a hydroxamic-derived selective HDAC6 inhibitor, ACY-738 [53]. The correlation of the previous studies with our results highlights the relevant role of the inhibition of HDACs in the metabolic modification of fatty acids in cancer cells, which, as is well known, are capable of reprogramming for their structural benefit and energy [54].
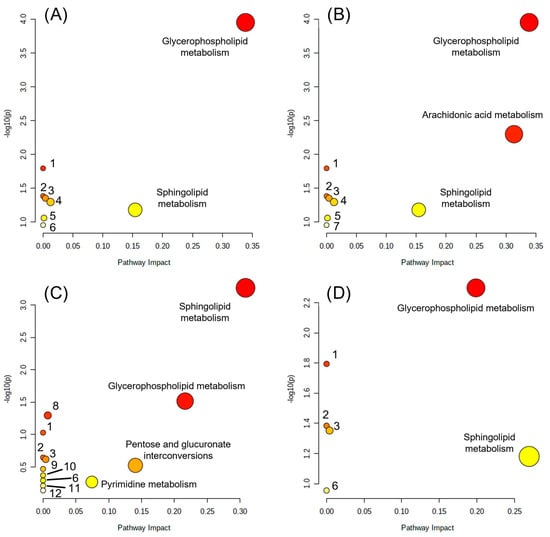
Figure 2.
Pathway impact analysis. (A) HO-AAVPA on MDA-MB-231; (B) VPA on MDA-MB-231; (C) HO-AAVPA on MCF-7; (D) VPA on MCF-7. Linoleic acid metabolism (1), alpha-linolenic acid metabolism (2), glycosylphosphatidylinositol (GPI)-anchor biosynthesis (3), glycerolipid metabolism (4), phosphatidylinositol signaling system (5), arachidonic acid metabolism (6), biosynthesis of unsaturated fatty acids (7), pantothenate and CoA biosynthesis (8), beta-alanine metabolism (9), alanine, aspartate, and glutamate metabolism (10), aminoacyl-tRNA biosynthesis (11), purine metabolism (12). The circle size correlates with the corresponding x-axis value, and the color (from yellow to red) correlates with the corresponding y-axis value.
As mentioned above, HO-AAVPA was designed considering the VPA and hydroxyarylamide pharmacophore groups to optimize the HDAC inhibitory effects; later, it was also found that HO-AAVPA possessed some other VPA effects but with better results, such as anticonvulsant activity, antiproliferative effects, promotion of translocation of HMGB1 protein, production of ROS imbalance, and modification of the acetylation state of proteins, which have led to the idea that both compounds share a number of similar mechanisms of action but HO-AAVPA has lower toxicological and teratogenic profile effects [32,35,36,37]. The evidence of similarity between HO-AAVPA and VPA is further reinforced by our metabolomics analysis, which obtained identical impact scores for HO-AAVPA and VPA with TNBC cells and quite similar scores for LA cells for glycerophospholipid and sphingolipid metabolism (Table 4). Thus, we generated three sets of results for each cell line: (1) the metabolic pathways that were affected by both compounds in a meaningful way (high impact score); (2) metabolic pathways that were differentially affected; and (3) metabolic pathways with no impact (low-impact score), but whose presence might give us some information when compared to other studies.

Table 4.
Pathway impact analysis of HO-AAVPA and VPA on TNB and LA cells.
Zhou et al. (2019) investigated the effect of VPA on the BC cell lines MDA-MB-231 and MCF-7 via a metabolomics approach. Among the multiple pathways on which VPA exerted its effect, they found sphingolipid metabolism within their MCF-7 dataset, although with a nonsignificant impact, contrary to what we found for the same pathway and both cell lines [44]. Interestingly, several pathways that were considered to have no impact (aminoacyl-tRNA biosynthesis, beta-alanine metabolism, pantothenate, and CoA biosynthesis, alanine, aspartate, glutamate, pyrimidine, and purine metabolisms) in our MCF-7 pathway analysis of HO-AAVPA-treated cells were found to be impactful in VPA-treated cells, which might indicate that HO-AAVPA and VPA indeed share mechanisms to exert their antiproliferative effect [44]. For MDA-MB-231, we found no similarities between our pathway analysis and the one performed by Zhou et al. (2019). In fact, these authors discovered that taurine and hypotaurine metabolism and beta-alanine metabolism are altered in both cell lines after VPA treatment. Additionally, alanine, aspartate, and glutamate metabolisms are mainly affected in MCF-7 cells and pyrimidine metabolism in MDA-MB-231 cells [44].
Estrada-Pérez et al. (2022) evaluated VPA in MCF-7 cells and reported the downregulation of members of important pathways, mainly the nonoxidative branch of the pentose phosphate pathway (PPP) and 2′deoxy-α-D-ribose-1-phosphate degradation, by administering the corresponding IC50 concentration of VPA [43,44]. Although none of these pathways were altered in our experiments, PPP was altered in the experiments performed by Zhou et al. (2019) in the same line using a concentration of 4 mM, halfway between our calculated IC15 and IC50 for that cell line (Table 1), so treating MCF-7 cells with VPA might first affect lipid metabolism, and as the VPA concentration increases, the pathways affected widen [44].
Gomes et al. (2020) also found that administering a moderate HDACi, resveratrol, modifies the lipid composition of MDA-MB-231 and MCF-7 cell lines [55]. Resveratrol decreases the biosynthesis of phosphatidylcholine (PC), phosphatidylinositol (PI), and lysophosphatidylcholine (LPC) in both cell lines, while phosphatidylethanolamine (PE) is also decreased in MDA-MB-231 cells and increased in MCF-7 cells [55]. Both HO-AAVPA and VPA also downregulated PC in both cell lines; PE was downregulated in all cases, except in MDA-MB-231 cells treated with VPA; LPC and lysophosphatidylethanolamine (LPE) were also downregulated, but such changes were detected only in MCF-7 cells treated with HO-AAVPA. Resveratrol is considered a pan inhibitor of HDAC [56,57], which has major activity on isoforms 1, 10, 4, and 9 of HDAC [56], all inhibited by VPA as well, except for HDAC10, so this might explain the similarities found between HDACi and VPA and HO-AAVPA [7].
The effect of VPA on different lipids has been reported in other contexts. Xu et al. (2019) evaluated the effect of VPA on lipids such as PE, PC, LPE, LPC, PI, sphingomyelin (SM), diacylglycerol (DG), and triacylglycerol (TG) and participants in sphingolipid metabolism, such as ceramides, from patients’ serum with abnormal liver function (ALF). Additionally, LPC, SM, and ceramides showed a significant decrease in concentration [58]. We found a higher number of dysregulated ceramides (upregulated) in MCF-7 cells than in MDA-MB-231 cells (downregulated) due to the effect of HO-AAVPA, while only one ceramide was found to be downregulated in MCF-7 VPA-treated cells. Xu et al. (2019) also evaluated the intracellular lipid content of L02 hepatic cells and observed an increase in lipid content due to VPA, which highlights the role of lipids in the effect of VPA and, in consequence, HO-AAVPA. They also noticed an increase in TG levels in L02 cells in a concentration-dependent manner, which we also observed as eight upregulated TGs in MCF-7 cells treated with HO-AAVPA.
Arachidonic acid (AA) was observed to have an upregulated tendency as a result of the treatment of MDA-MB-231 with VPA. The effect of VPA on AA has been studied before by Shimshoni et al. (2011) for bipolar disorder; in their work, they found that VPA inhibited the conversion of AA into acyl-CoA [58,59]. A similar event could occur in MDA-MB-231 cells exposed to VPA, provoking its accumulation seen as upregulation, a mechanism not found in HO-AAVPA-treated cells. Borin et al. (2017) focused on the AA pathway to collect information in their review of its implication on BC cell migration and invasion capacity; they noted that both invasion and metastasis decrease through the inhibition of the synthesis of 20-hydroxy-eicosatetraenoic acid (20-HETE) [60], so AA pathway dysregulation through VPA could be relevant to its antiproliferative effect in MDA-MB-231 cells treated with VPA but not in HO-AAVPA-treated cells.
As mentioned by Rosario et al. (2018) in their research on dysregulated pathways using transcripts from tumor and nontumor samples, the pentose glucuronate interconversion (PGI) pathway seems to be a commonly and significantly dysregulated pathway across multiple types of cancer, including BC [61], and this specific alteration may be how HO-AAVPA exerts its effect on MCF-7 cells. In the same study, the authors explored changes in the metabolism of BC cells versus normal cells and detected that glycerophospholipid and AA metabolism are dysregulated in BC cells, while sphingolipid and alpha-linolenic acid are not.
The differences observed between the studies mentioned above might be due to differences in the sample preparation methods, the compound concentration used, the exposure time to the compound, storage, data acquisition methods (including the acquisition platform), and other factors, as has been noted in other publications [62,63,64].
Although the scope of this work is limited to the putative annotation of metabolites, and definitive confirmation of metabolite identity requires direct comparison with reference compounds (metabolite identification confidence level 1, identity validation) [42], our experimental and analytical strategy used strict criteria to ensure high-quality annotations and conclusions, preventing false positives. This strategy provides us with a rigorous way to evaluate the possible identity of the features found. Results gathered through this study reveal interesting insights that required further confirmation by a targeted approach. Both the direction of dysregulated metabolites and the impact on pathways need to be addressed in further studies complemented with studies like proteomics.
3. Materials and Methods
3.1. Chemicals and Materials
Cell culture plastic material was purchased from TPP (Trasadingen, Switzerland), and fetal bovine serum (FBS) and trypsin-EDTA were acquired from Biowest (Riverside, MO, USA). LC-MS grade methanol was purchased from Honeywell Burdick and Jackson (Morristown, NJ, USA); 2-propanol and sodium hydroxide were purchased from Merck (Toluca, Mexico); chloroform, HEPES, urea, bicinchoninic acid, and sodium tartrate dihydrate were purchased from Sigma-Aldrich (Toluca, Mexico); sodium carbonate and sodium bicarbonate were purchased from Fermont (Mexico City, Mexico); and cupric sulfate pentahydrate was purchased from Golden Bell (Mexico City, Mexico). Ultrapure water was obtained from a Direct-Q 3 system (Millipore, Burlington, MA, USA). HO-AAVPA was synthesized as published by Prestegui-Martel et al. (2016) with a few modifications [37]. VPA, formic acid, ammonium acetate, and ammonium formate were acquired from Sigma-Aldrich (Toluca, México).
3.2. Cell Culture
MCF-7 and MDA-MB-231 cells were kindly donated by Dr. Gisela Ceballos Cancino (INMEGEN, Mexico City, Mexico). Both cell lines were grown in Dulbecco’s Modified Eagle’s Medium/High Modified (DMEM) without phenol red supplemented with 10% fetal bovine serum (FBS) at 37 °C and 5% of CO2 in a humidified atmosphere [43]. Cells were handled in a LabGard ES NU-540-400, Class II, Type A2 Laminar Flow (NUAIRE, Plymouth, MN, USA). The cells were detached using trypsin-EDTA upon reaching 75 ± 5% confluence, counted using a CytoSMART (Corning, Glendale, AZ, USA), and seeded in 60.1 cm2 tissue culture dishes until the required cell number was reached for the corresponding experiment.
3.3. Inhibitory Concentration Assays
Inhibitory concentration (IC) determinations were measured through MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assays in triplicate. Briefly, 1 × 104 cells in 100 µL of media were seeded in each well of a 96-well tissue culture plate and incubated for 24 h. Afterward, the culture media was replaced with media with one of nine concentrations evaluated, from 140.625 µM to 36 mM VPA and 9.375 µM to 2.4 mM HO-AAVPA, in 150 µL of media, and the cells were incubated for 72 h. HO-AAVPA was first dissolved in DMSO up to a final concentration of 0.1% in culture media; 0.1% DMSO in culture media was used as a diluent control. After the incubation period, the medium was replaced with 100 µL of MTT solution in PBS (0.5 mg/mL), incubated for 4 h, and then replaced with DMSO. Absorbance was registered at 550 nm in a Multiskan Sky with Cuvette and Touch Screen (Thermo Scientific, Waltham, MA, USA) spectrophotometer with 15 s of agitation. IC15 was calculated with GraphPad Prism 8 (version 8.01 for Windows) using the equation for log (inhibitor) versus response variable slope (four parameters).
3.4. Cell Treatment and Metabolite Extraction
For metabolomics, assays with seven independent replicates were conducted. For each replicate, four 147.8 cm2 tissue culture dishes with 7 × 106 viable cells in 13 mL culture media were prepared for the control, diluent control (DMSO 0.1%), VPA, and HO-AAVPA samples (IC15 treated to normalize effects for both compounds and cell lines) [44,63]. Before any treatment, the cells were allowed to grow for 24 h, and then the medium was replaced with the corresponding treatment and incubated for 48 h. VPA and HO-AAVPA cells were treated with 3.48 and 1.64 mM VPA and 175.6 and 313.8 µM HO-AAVPA for MDA-MB-231 and MCF-7, respectively. An additional extraction control was prepared in the same way as the diluent control, except that no cells were seeded in it.
Metabolite extraction was based on the Bligh–Dyer method for polar and nonpolar compounds, reported elsewhere [65], with modifications proposed by Agilent Technologies (Santa Clara, CA, USA) application note 5991-3528EN and a randomized extraction order. Briefly, cells were maintained in wet ice; the media was discarded, and then the cells were washed three times with 10 mL of 0.9% NaCl. Liquid nitrogen was applied to the monolayer to stop cell metabolism. Then, 2 mL of methanol was added, and the cells were scraped with a cell scraper. Cells were recovered in a 2 mL plastic vial and maintained in dry ice until all cells were harvested; then, the samples were transferred to −80 °C storage for further processing. Once all replicates were obtained, 50 µL of a solution of acetaminophen (1200 ppm) and carbamazepine (1200 ppm) was added to each sample, mixed, sonicated with a Vibra-Cell VC 130 Ultrasonic Processor (Sonics and Materials, Newtown, CT, USA) by applying pulses with a frequency of 40 kHz with an on and off cycle of 5 and 1 s, respectively, five times, and then the samples were split in two. Sequentially, 250 µL of chloroform, 350 of water, and 250 µL of chloroform were added and mixed for 10 s in a vortex after each addition. Separation of phases was achieved by centrifugation at 5000 rpm at 4 °C for 30 min. The aqueous and organic phases of samples that were previously split were merged in plastic vials and stored at −80 °C (including the protein disc). The organic phase and protein disc (for protein quantitation) were dried in an orbital incubator (INO-650 M, Prendo, Puebla, Mexico) at 30 °C, and the aqueous phase was dried in a Vacufuge plus (Eppendorf, Hamburg, Germany). Once dried, the samples were stored at −80 °C for further analysis.
3.5. Protein Quantification
Protein was quantitated by the bicinchoninic acid method [66]. Briefly, a nine-level calibration curve (0, 25, 125, 250, 500, 750, 1000, 1500, and 2000 µg/mL BSA) was prepared by serial dilution (diluent solution: 8 M urea and 20 mM HEPES, pH 8) [67]. Protein discs from MDA-MB-231 and MCF-7 cells were diluted in 1.0 and 1.6 mL of diluent solution, respectively, and mixed for 2 min with a vortexer. Fifty microliters of protein solution from MCF-7 cells were further diluted in 150 µL of diluent. Then, 25 µL of each level of calibration curve or sample (in duplicate) was added to a 96-well flat bottom plate (Corning) and mixed with 200 µL of working solution. The plates were incubated in an orbital incubator with constant agitation at 37 °C for 30 min in darkness. Absorbance values were registered at 562 nm with 10 s of agitation in a Multiskan Sky with Cuvette and Touch Screen (Thermo Scientific) spectrophotometer [68]. The working solution was prepared by mixing 50 parts of reagent A solution (0.1 g bicinchoninic acid, 2 g sodium carbonate, 0.16 g sodium tartrate dihydrate, 0.4 g sodium hydroxide and 0.95 g sodium bicarbonate in 100 mL of water) and 1 part of reagent B solution (0.4 g cupric sulfate pentahydrate in 10 mL of water). The protein concentration of each sample was obtained by substituting the corresponding absorbance values in the equation of the straight line.
3.6. LC-MS Data Acquisition
Data acquisition for each cell line was performed separately using an UHPLC 1290 Infinity II: 1290 Flexible Pump (G7104A) and 1290 vial sampler with integrated column compartment (G7129B), coupled with a Q-TOF (G6545A) with Dual AJS ESI as ionization source (G1959A), all from Agilent Technologies. A total of four acquisition conditions were used: HILIC-ESI(+)-MS; HILIC-ESI-(−)-MS (hydrophilic interaction liquid chromatography for polar metabolites); RPLC-ESI(+)-MS; and RPLC-ESI-(−)-MS (reversed-phase liquid chromatography for nonpolar metabolites). The sample injection order was as follows: the LC–MS system was first equilibrated by injecting 10 µL of the blank sample until no chromatographic variation was observed (blank injection); then, blank and blank + standards were injected twice. Afterward, an extraction blank was injected in triplicate, and later, to equilibrate the LC–MS system to biological samples, quality control samples (QC) were injected until no chromatographic variation was observed. QC samples were injected before and after every five biological sample injections, and the sequence order of the biological samples was randomly assigned [69,70].
Polar compounds were separated using a Poroshell 120 HILIC-Z (2.1 × 150 mm, 2.7 µm) column with a Poroshell 120 HILIC-Z (2.1 × 5 mm, 2.7 µm) guard column (Agilent Technologies, Santa Clara, CA, USA) through a nonlinear gradient for both positive and negative acquisition (application note: 5994-1492EN, Agilent Technologies, for positive and negative LC and MS acquisition). MDA-MB-231 cells were resuspended in 120 µL of ACN/MeOH/H2O 70:20:10 (v/v/v) (HILIC diluent), and MCF-7 cells were first diluted in 150 µL of HILIC diluent and mixed. Then, 12.5 µL of this mixture was further diluted with 47.5 µL of HILIC diluent to obtain similar signal intensities between MDA-MB-231 and MCF-7 metabolites. Positive acquisition used ammonium formate (AmF) 10 mM and formic acid (FA) 0.125% in water (solvent A) and AmF 10 mM and FA 0.125% in ACN/H2O 90:10 (v/v) (solvent B) as follows: 0–3 min of 98% B; 70% B at 11 min; 60% B at 14 min; and 10% B from 18 to 20 min with 4 min of post time. The column temperature was maintained at 25.0 ± 0.5 °C. Negative acquisition used ammonium acetate (AmAc) 10 mM in water (solvent A) and AmAc 10 mM in ACN/H2O 85:15 (v/v) (solvent B) as follows: 0–2 min of 92% B; 84% B from 5.5 to 8.5 min; 82% B from 9 to 14 min; 78% B at 17 min; 65% B at 21 min; 40%% B from 23 to 25 min; and 5% B from 27 to 29 min with 4 min of post time. The column temperature was maintained at 25.0 ± 0.5 °C and 40.0 ± 0.5 °C for positive and negative acquisition, respectively. The flow rate was 0.25 mL/min with 10 µL of volume injected and 15 s of needle washing with methanol for both cases.
The spectrometric conditions for polar compounds were as follows: drying gas temperature of 225 and 225 °C; drying gas flow of 8 and 13 L/min; sheath gas temperature of 225 and 350 °C; sheath gas flow of 10 and 12 L/min; nebulizer pressure of 40 and 35 psig; capillary voltage of 3000 and 3500 V; nozzle voltage of 0 and 0 V; fragmentor of 125 and 125 V; skimmer of 65 and 45 V; and octupole RFF of 450 and 750 V for positive and negative acquisition, respectively. The scan rate was 3 spectra/s and 50–1700 m/z for mass range; correction was performed with 121.05087300 and 922.00979800 m/z and 68.99575800 and 1033.98810900 m/z as mass references for positive and negative acquisition, respectively.
Nonpolar compounds were separated using a Zorbax Eclipse Plus C18 (2.1 × 150 mm, 1.8 µm) column with a Zorbax Eclipse Plus C18 (2.1 × 5 mm, 1.8 µm) guard column (Agilent Technologies) through a nonlinear gradient for both positive and negative acquisition [65]. MDA-MB-231 cells were resuspended in 120 µL of IPA/ACN 90:10 (v/v) (RPLC diluent); MCF-7 cells were first diluted in 150 µL of RPLC diluent and mixed. Then, 12.5 µL of this mixture was further diluted with 47.5 µL of RPLC diluent to obtain similar signal intensities between MDA-MB-231 and MCF-7 metabolites. Positive acquisition used AmF 10 mM and FA 0.1% in ACN/H2O 60:40 (v/v) (solvent A) and AmF 10 mM and FA 0.1% in IPA/ACN 90:10 (v/v) (solvent B) as follows: 32% B at 0 min; 40% B from 2 to 3 min; 45% B at 8 min; 50% B at 10 min; 60% B at 16 min; 70% B at 22 min; and 90% B from 28 to 36 min with a 3 min post time. Negative acquisition used 10 mM AmAc in 60:40 (v/v) ACN/H2O (solvent A) and 10 mM AmAc in 90:10 (v/v) IPA/ACN (solvent B) and the same gradient configuration as in positive acquisition. The column temperature was maintained at 60.0 ± 0.5 °C; the injection volume was 2 µL; the flow rate was 0.3 mL/min, and the column was washed with IPA/ACN 90:10 (v/v) for both acquisitions.
The spectrometric conditions for nonpolar compounds were as follows (application note: 5991-9280EN, Agilent Technologies): drying gas temperature of 200 °C; drying gas flow of 13 L/min; sheath gas temperature of 350 °C; sheath gas flow of 11 L/min; nebulizer pressure of 35 psig; capillary voltage of 3500 V; nozzle voltage of 1000 V; fragmentor of 175 V; skimmer of 65 V; voltage octupole RFF of 750 V; scan rate of 3 spectra/s and 50–1700 m/z for mass range for positive and negative acquisition, respectively; correction was performed with 121.05087300 and 922.00979800 m/z and 68.99575800 and 1033.98810900 m/z as mass references for positive and negative acquisition, respectively.
3.7. LC-MS Data Processing
Optimization of the feature extraction parameters for the Molecular Feature Extraction (MFE) algorithm, the retention time drift tolerance, ionic species included, and compound threshold were determined with MassHunter Workstation Software Qualitative Analysis (version B.07.00, build 7.7.7024.29, SP2, Agilent Technologies, Santa Clara, CA, USA) [71]. These parameters were optimized for each condition acquisition and for each cell line separately through inspection of the corresponding initial QC. Once the extraction parameters were optimized, all blank samples were first batch-analyzed separately to generate an exclusion feature list (a list with features present in the blank, except added standards and mass references, that need to be excluded from the analysis of samples) and then all QCs and biological samples were batch-analyzed, excluding features present in the exclusion list. Batch alignment and extraction were performed with MassHunter Profinder (version B.08.00, SP3, Agilent Technologies, Santa Clara, CA, USA) using the Batch Recursive Feature Extraction for small molecule/peptide algorithm using common organic molecules (no halogens) as an isotopic model, allowing for a maximum of two charges and only compounds with two or more ions to avoid false-positives. To further reduce false positives, only compounds present in four and six files (for the MFE and Find by Ion or FBIon algorithms, respectively) in at least one sample group (pseudo-replicates) and with an MFE score ≥ 70.0 were retained for further analysis and exported as a profinder archive (.pfa with median mass and retention time). For features present in blanks, only those present in one file and 100% of files in at least one sample group were retained in the MFE and FBIon algorithms, respectively. The results were inspected, particularly the correct integration of added standards. Before batch alignment, a time alignment was performed for all samples (blanks and QCs included) versus the initial QC to further correct retention time drift as follows: features with counts higher than 1000; maximum time shift of 0.5 min + 0.5%; and polynomial interpolation as a fitting model to reduce variation in the retention time.
Chemometric comparisons between all treatments for a given cell line were performed in Mass Profiler Professional (MPP) (version 14.9.1, Agilent Technologies, Santa Clara, CA, USA). First, normalization by internal standard (carbamazepine for HILIC-ESI(+) and acetaminophen for the rest of the conditions) and external scalar (protein concentration) were performed to correct for variation due to the extraction process and quantity of biological material, respectively. Then, principal component analysis (PCA) was performed on all samples to be grouped. Afterward, entities (features are called entities in MPP) were filtered by variability, only keeping those with standard deviation ≤ 1.0 for all conditions (treatments). The Shapiro–Wilk (p-value of 0.05) normality test was carried out, and two entity lists were created, one for entities with normal distribution and the other for entities with nonnormal distribution. Entities within the first list were analyzed with one-way ANOVA, Tukey’s HSD post hoc test, asymptotic computation of p-value, and multiple testing correction Benjamini Hochberg FDR. Entities within the second list were analyzed with the Kruskal–Wallis test, asymptotic computation of p-value, and multiple testing correction Benjamini Hochberg FDR. Pairwise contrast was performed by contrasting the treatments with their respective control, culture with medium for VPA, and vehicle control (DMSO) for HO-AAVPA. Only entities with a p-value ≤ 0.01 and a fold change value ≥ 2.0 were kept for both lists, excluding missing values for calculation. Entities were used to create an inclusion list, namely, a list of ions that are fragmented during LC-MS/MS data acquisition.
3.8. LC-MS/MS Data Acquisition and Metabolite Annotation
The first LC-MS/MS data acquisition was performed with the same chromatographic and spectrometric conditions previously described but with the following modifications: auto MS/MS mode with a list of preferred ions (features found to be relevant in the statistical analysis) by cell line; fragmentation under three different collision energies (10, 20 and 40 V); a number of charges = 1 (z = 1); isolation window width of ~4 amu; and retention time delta of 0.3 min. The scan rate for MS/MS was kept at 3 spectra/s in a range of 50–1700 m/z. This second data acquisition was performed by injecting 3 µL of the corresponding cell line quality control sample [43,71].
Scan spectra and extracted compound chromatograms from fragmented compounds were recovered through MassHunter Qualitative Analysis’s Find by Auto MS/MS algorithm, as well as product ion spectra by collision energy. Product ion spectra abundances were transformed from counts to percentage values and then searched against the HMDB 5.0 [72] using the LC-MS/MS Search tool (https://hmdb.ca/spectra/ms_ms/search, accessed from 1 April 2022 to 30 June 2022) as follows: low, medium, and high energy (10, 25 and 40 V, respectively); parent ion mass and mass/charge tolerance of 10 ppm; and enabling the inclusion of predicted spectra. The results with Fit, RFit, and Purity values ≥ 0.80 were kept, and the result with the highest value was assigned to annotate the corresponding compound as a putative metabolite. The annotation information for each compound was updated using the IDBrowser tool from MPP (version 14.9.1, Agilent Technologies, Santa Clara, CA, USA).
3.9. Effect on Metabolic Pathways
To determine the corresponding compound’s impact on metabolic pathways, we used the Metaboanalyst 5.0 platform with the Pathway Analysis module (https://dev.metaboanalyst.ca/MetaboAnalyst/upload/PathUploadView.xhtml, accessed on 1 July 2022 to 31 July 2022) [49]. HMDB accession numbers were used for the search (with the HMDB ID as the input type); scatter plots were used as the visualization method; the hypergeometric test was applied as the enrichment method; relative betweenness centrality was applied as topology analysis, and Homo sapiens (KEGG) was used as the pathway library [73,74,75,76,77].
4. Conclusions
In the present study, we tentatively identified the metabolic pathways modulated by two structurally similar HDACi, HO-AAVPA and VPA, in BC cells. The results highlight their main effect on metabolic pathways related to lipid metabolism in MCF-7 and MDA-MB-231 cell lines, supporting the hypothesis of similar mechanisms of action, but other deregulated metabolites found suggest the presence of different biological targets. However, a proteomic approach is required to support this claim. Furthermore, the observed differences in the metabolic pathways altered by each compound may be key to understanding how HO-AAVPA enhances VPA potency. Future studies are required to confirm the identity of the putative metabolites reported in this work. Despite these limitations, these findings provide a solid basis for understanding the mechanisms of metabolic regulation exerted by two HDACis and offer valuable insights for developing therapeutic strategies in breast cancer.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/ijms241914543/s1.
Author Contributions
A.R.E.-P. conducted the experiments, analyzed acquired data, and wrote this manuscript; M.C.R.-H. and J.C.-B. contributed to the experimental design, review, and editing J.B.G.-V. and H.L.M.-F. contributed to experimental design and helped in the discussion of results; C.F.-P. contributed to the writing—review and editing of this manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This project was supported by the Secretaría de Investigación y Posgrado (SIP) of the Instituto Politécnico Nacional (IPN), who provided grants such as BEIFI, COFAA-IPN: SIP-20212109, SIP-IPN multidisciplinario 20230834, and the Proyecto de innovación SIP-20221145. Additionally, support was received from CONACYT programs: Ciencia de Frontera CF-11312/2020, Ciencia básica y/o de frontera convocatoria paradigmas y controversias de la ciencia 319355-2022, and CONACYT 254600, 284243.
Institutional Review Board Statement
This study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Biosecurity Committee of Escuela Superior de Medicina (protocol code CBS-01/and date of approval: 11 December 2017).
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are openly available in Zenodo at https://doi.org/10.5281/zenodo.7991779 since 31 July 2023.
Acknowledgments
The authors thank the Instituto Politécnico Nacional (SIP), BEIFI, COFAA-IPN. Alan Rubén Estrada-Pérez acknowledges the support provided by the National Council of Science and Technology (CONACyT) for its support by granting a doctoral fellowship. Cynthia Fernández-Pomares acknowledges the support provided by CONACyT, under the program “Estancias Posdoctorales por México” 2022–2023.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Cheng, H.; Bai, Z.; Li, J. Breast Cancer Cell Line Classification and Its Relevance with Breast Tumor Subtyping. J. Cancer 2017, 8, 3131–3141. [Google Scholar] [CrossRef] [PubMed]
- Waks, A.G.; Winer, E.P. Breast Cancer Treatment. JAMA 2019, 321, 288–300. [Google Scholar] [CrossRef]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef]
- Glozak, M.A.; Seto, E. Histone Deacetylases and Cancer. Oncogene 2007, 26, 5420–5432. [Google Scholar] [CrossRef] [PubMed]
- Peserico, A.; Simone, C. Physical and Functional HAT/HDAC Interplay Regulates Protein Acetylation Balance. J. Biomed. Biotechnol. 2011, 2011, 371832. [Google Scholar] [CrossRef] [PubMed]
- Eckschlager, T.; Plch, J.; Stiborova, M.; Hrabeta, J. Histone Deacetylase Inhibitors as Anticancer Drugs. Int. J. Mol. Sci. 2017, 18, 1414. [Google Scholar] [CrossRef]
- Gil, J.; Ramírez-Torres, A.; Encarnación-Guevara, S. Lysine Acetylation and Cancer: A Proteomics Perspective. J. Proteom. 2017, 150, 297–309. [Google Scholar] [CrossRef]
- Park, S.; Jun, J.; Jeong, K.; Heo, H.; Sohn, J.; Lee, H.; Park, C.; Kang, J. Histone Deacetylases 1, 6 and 8 Are Critical for Invasion in Breast Cancer. Oncol. Rep. 2011, 25, 1677–1681. [Google Scholar] [CrossRef]
- Parbin, S.; Kar, S.; Shilpi, A.; Sengupta, D.; Deb, M.; Rath, S.K.; Patra, S.K. Histone Deacetylases. J. Histochem. Cytochem. 2013, 62, 11–33. [Google Scholar] [CrossRef]
- Aldana-Masangkay, G.I.; Sakamoto, K.M. The Role of HDAC6 in Cancer. J. Biomed. Biotechnol. 2011, 2011, 875824. [Google Scholar] [CrossRef]
- Hull, E.E.; Montgomery, M.R.; Leyva, K.J. HDAC Inhibitors as Epigenetic Regulators of the Immune System: Impacts on Cancer Therapy and Inflammatory Diseases. BioMed Res. Int. 2016, 2016, 8797206. [Google Scholar] [CrossRef]
- Conte, M.; Palma, R.D.; Altucci, L. HDAC Inhibitors as Epigenetic Regulators for Cancer Immunotherapy. Int. J. Biochem. Cell Biol. 2018, 98, 65–74. [Google Scholar] [CrossRef]
- Damaskos, C.; Garmpis, N.; Valsami, S.; Kontos, M.; Spartalis, E.; Kalampokas, T.; Kalampokas, E.; Athanasiou, A.; Moris, D.; Daskalopoulou, A.; et al. Histone Deacetylase Inhibitors: An Attractive Therapeutic Strategy Against Breast Cancer. Anticancer Res. 2017, 37, 35–46. [Google Scholar] [CrossRef]
- West, A.C.; Johnstone, R.W. New and Emerging HDAC Inhibitors for Cancer Treatment. J. Clin. Investig. 2014, 124, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Johnstone, R.W. Histone-Deacetylase Inhibitors: Novel Drugs for the Treatment of Cancer. Nat. Rev. Drug Discov. 2002, 1, 287–299. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Bishayee, A.; Pandey, A.K. Targeting Histone Deacetylases with Natural and Synthetic Agents: An Emerging Anticancer Strategy. Nutrients 2018, 10, 731. [Google Scholar] [CrossRef] [PubMed]
- Linares, A.; Dalenc, F.; Balaguer, P.; Boulle, N.; Cavailles, V. Manipulating Protein Acetylation in Breast Cancer: A Promising Approach in Combination with Hormonal Therapies? J. Biomed. Biotechnol. 2011, 2011, 856985. [Google Scholar] [CrossRef]
- Ali, S.R.; Humphreys, K.J.; McKinnon, R.A.; Michael, M.Z. Impact of Histone Deacetylase Inhibitors on MicroRNA Expression and Cancer Therapy: A Review. Drug Dev. Res. 2015, 76, 296–317. [Google Scholar] [CrossRef]
- Li, Y.; Seto, E. HDACs and HDAC Inhibitors in Cancer Development and Therapy. Cold Spring Harb. Perspect. Med. 2016, 6, a026831. [Google Scholar] [CrossRef]
- Sixto-López, Y.; Bello, M.; Correa-Basurto, J. Exploring the Inhibitory Activity of Valproic Acid against the HDAC Family Using an MMGBSA Approach. J. Comput. Aided Mol. Des. 2020, 34, 857–878. [Google Scholar] [CrossRef] [PubMed]
- Giordano, F.; Paolì, A.; Forastiero, M.; Marsico, S.; Amicis, F.D.; Marrelli, M.; Naimo, G.D.; Mauro, L.; Panno, M.L. Valproic Acid Inhibits Cell Growth in Both MCF-7 and MDA-MB231 Cells by Triggering Different Responses in a Cell Type-Specific Manner. J. Transl. Med. 2023, 21, 165. [Google Scholar] [CrossRef] [PubMed]
- Ozman, Z.; Iptec, B.O.; Sahin, E.; Eskiler, G.G.; Ozkan, A.D.; Kaleli, S. Regulation of Valproic Acid Induced EMT by AKT/GSK3β/β-Catenin Signaling Pathway in Triple Negative Breast Cancer. Mol. Biol. Rep. 2021, 48, 1335–1343. [Google Scholar] [CrossRef] [PubMed]
- Fortunati, N.; Bertino, S.; Costantino, L.; Bosco, O.; Vercellinatto, I.; Catalano, M.G.; Boccuzzi, G. Valproic Acid Is a Selective Antiproliferative Agent in Estrogen-Sensitive Breast Cancer Cells. Cancer Lett. 2008, 259, 156–164. [Google Scholar] [CrossRef]
- Mawatari, T.; Ninomiya, I.; Inokuchi, M.; Harada, S.; Hayashi, H.; Oyama, K.; Makino, I.; Nakagawara, H.; Miyashita, T.; Tajima, H.; et al. Valproic acid inhibits proliferation of HER2-expressing breast cancer cells by inducing cell cycle arrest and apoptosis through Hsp70 acetylation. Int. J. Oncol. 2015, 47, 2073–2081. [Google Scholar] [CrossRef]
- Wardell, S.E.; Ilkayeva, O.R.; Wieman, H.L.; Frigo, D.E.; Rathmell, J.C.; Newgard, C.B.; McDonnell, D.P. Glucose Metabolism as a Target of Histone Deacetylase Inhibitors. Mol. Endocrinol. 2009, 23, 388–401. [Google Scholar] [CrossRef]
- Fang, E.; Wang, J.; Hong, M.; Zheng, L.; Tong, Q. Valproic Acid Suppresses Warburg Effect and Tumor Progression in Neuroblastoma. Biochem. Biophys. Res. Commun. 2019, 508, 9–16. [Google Scholar] [CrossRef]
- Geng, H.-W.; Yin, F.-Y.; Zhang, Z.-F.; Gong, X.; Yang, Y. Butyrate Suppresses Glucose Metabolism of Colorectal Cancer Cells via GPR109a-AKT Signaling Pathway and Enhances Chemotherapy. Front. Mol. Biosci. 2021, 8, 634874. [Google Scholar] [CrossRef]
- Chittur, S.V.; Sangster-Guity, N.; McCormick, P.J. Histone Deacetylase Inhibitors: A New Mode for Inhibition of Cholesterol Metabolism. BMC Genom. 2008, 9, 507. [Google Scholar] [CrossRef]
- Marcos, X.; Sixto-López, Y.; Pérez-Casas, S.; Correa-Basurto, J. Computational Study of DMPC Liposomes Loaded with the N-(2-Hydroxyphenyl)-2-Propylpentanamide (HO-AAVPA) and Determination of Its Antiproliferative Activity in Vitro in NIH-3T3 Cells. J. Biomol. Struct. Dyn. 2021, 40, 11448–11459. [Google Scholar] [CrossRef]
- López-Bautista, M.C.; Avendaño-Alejo, M.; Castañeda, L.; Peralta-Ángeles, J.A.; Reyes-Esqueda, J.A. Study of Nonlinear Properties of N-(2-Hydroxyphenyl)-2-Propylpentanamide in Polymeric Solution. Optik 2019, 180, 724–732. [Google Scholar] [CrossRef]
- Sixto-López, Y.; Rosales-Hernández, M.C.; de Oca, A.C.-M.; Fragoso-Morales, L.G.; Mendieta-Wejebe, J.E.; Correa-Basurto, A.M.; Abarca-Rojano, E.; Correa-Basurto, J. N-(2′-Hydroxyphenyl)-2-Propylpentanamide (HO-AAVPA) Inhibits HDAC1 and Increases the Translocation of HMGB1 Levels in Human Cervical Cancer Cells. Int. J. Mol. Sci. 2020, 21, 5873. [Google Scholar] [CrossRef] [PubMed]
- Correa-Basurto, A.M.; Romero-Castro, A.; Correa-Basurto, J.; Hernández-Rodríguez, M.; Soriano-Ursúa, M.A.; García-Machorro, J.; Tolentino-López, L.E.; Rosales-Hernández, M.C.; Mendieta-Wejebe, J.E. Pharmacokinetics and Tissue Distribution of N-(2-Hydroxyphenyl)-2-Propylpentanamide in Wistar Rats and Its Binding Properties to Human Serum Albumin. J. Pharm. Biomed. Anal. 2019, 162, 130–139. [Google Scholar] [CrossRef] [PubMed]
- Mendieta-Wejebe, J.; Silva-Trujillo, A.; Bello, M.; Mendoza-Figueroa, H.L.; Galindo-Alvarez, N.; Albores, A.; Tamay-Cach, F.; Rosales-Hernández, M.; Romero-Castro, A.; Correa-Basurto, J. Exploring the Biotransformation of N-(2-hydroxyphenyl)-2-propylpentanamide (an Aryl Valproic Acid Derivative) by CYP2C11, Using in Silico Predictions and in Vitro Studies. J. Pharm. Pharmacol. 2020, 72, 938–955. [Google Scholar] [CrossRef] [PubMed]
- Cristóbal-Luna, J.; Correa-Basurto, J.; Mendoza-Figueroa, H.L.; Chamorro-Cevallos, G. Anti-Epileptic Activity, Toxicity and Teratogenicity in CD1 Mice of a Novel Valproic Acid Arylamide Derivative, N-(2-Hydroxyphenyl)-2-Propylpentanamide. Toxicol. Appl. Pharm. 2020, 399, 115033. [Google Scholar] [CrossRef] [PubMed]
- Contis-Montes de Oca, A.; Rodarte Valle, E.; Rosales-Hernández, M.C.; Abarca-Rojano, E.; Rojas-Hernández, S.; Fragoso-Vázquez, M.J.; Mendieta-Wejebe, J.E.; Correa-Basurto, A.M.; Vázquez-Moctezuma, I.; Correa-Basurto, J. N-(2′-Hydroxyphenyl)-2-propylpentanamide (OH-VPA), a histone deacetylase inhibitor, induces the release of nuclear HMGB1 and modifies ROS levels in HeLa cells. Oncotarget 2018, 9, 33368–33381. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Prestegui-Martel, B.; Bermúdez-Lugo, J.A.; Chávez-Blanco, A.; Dueñas-González, A.; García-Sánchez, J.R.; Pérez-González, O.A.; Padilla-Martínez, I.I.; Fragoso-Vázquez, M.J.; Mendieta-Wejebe, J.E.; Correa-Basurto, A.M.; et al. N-(2-Hydroxyphenyl)-2-Propylpentanamide, a Valproic Acid Aryl Derivative Designed in Silico with Improved Anti-Proliferative Activity in HeLa, Rhabdomyosarcoma and Breast Cancer Cells. J. Enzym. Inhib. Med. Chem. 2016, 31, 140–149. [Google Scholar] [CrossRef]
- Subramani, R.; Poudel, S.; Smith, K.D.; Estrada, A.; Lakshmanaswamy, R. Metabolomics of Breast Cancer: A Review. Metabolites 2022, 12, 643. [Google Scholar] [CrossRef]
- D’Alessandro, A.; Zolla, L. Proteomics and Metabolomics in Cancer Drug Development. Expert Rev. Proteom. 2013, 10, 473–488. [Google Scholar] [CrossRef]
- Dettmer, K.; Aronov, P.A.; Hammock, B.D. Mass Spectrometry-based Metabolomics. Mass Spectrom. Rev. 2007, 26, 51–78. [Google Scholar] [CrossRef]
- Wishart, D.S. Emerging Applications of Metabolomics in Drug Discovery and Precision Medicine. Nat. Rev. Drug Discov. 2016, 15, 473–484. [Google Scholar] [CrossRef] [PubMed]
- Schrimpe-Rutledge, A.C.; Codreanu, S.G.; Sherrod, S.D.; McLean, J.A. Untargeted Metabolomics Strategies—Challenges and Emerging Directions. J. Am. Soc. Mass Spectrom. 2016, 27, 1897–1905. [Google Scholar] [CrossRef]
- Estrada-Pérez, A.R.; Rosales-Hernández, M.C.; García-Vázquez, J.B.; Bakalara, N.; Fromager, B.; Correa-Basurto, J. Untargeted LC-MS/MS Metabolomics Study on the MCF-7 Cell Line in the Presence of Valproic Acid. Int. J. Mol. Sci. 2022, 23, 2645. [Google Scholar] [CrossRef]
- Zhou, X.; Li, Z.; Wang, X.; Jiang, G.; Shan, C.; Liu, S. Metabolomics Reveals the Effect of Valproic Acid on MCF-7 and MDA-MB-231 Cells. Xenobiotica 2019, 50, 252–260. [Google Scholar] [CrossRef] [PubMed]
- Granit, A.; Mishra, K.; Barasch, D.; Peretz-Yablonsky, T.; Eyal, S.; Kakhlon, O. Metabolomic Profiling of Triple Negative Breast Cancer Cells Suggests That Valproic Acid Can Enhance the Anticancer Effect of Cisplatin. Front. Cell Dev. Biol. 2022, 10, 1014798. [Google Scholar] [CrossRef] [PubMed]
- Wawruszak, A.; Luszczki, J.J.; Grabarska, A.; Gumbarewicz, E.; Dmoszynska-Graniczka, M.; Polberg, K.; Stepulak, A. Assessment of Interactions between Cisplatin and Two Histone Deacetylase Inhibitors in MCF7, T47D and MDA-MB-231 Human Breast Cancer Cell Lines—An Isobolographic Analysis. PLoS ONE 2015, 10, e0143013. [Google Scholar] [CrossRef]
- Hsu, K.-W.; Huang, C.-Y.; Tam, K.-W.; Lin, C.-Y.; Huang, L.-C.; Lin, C.-L.; Hsieh, W.-S.; Chi, W.-M.; Chang, Y.-J.; Wei, P.-L.; et al. The Application of Non-Invasive Apoptosis Detection Sensor (NIADS) on Histone Deacetylation Inhibitor (HDACi)-Induced Breast Cancer Cell Death. Int. J. Mol. Sci. 2018, 19, 452. [Google Scholar] [CrossRef]
- Cody, J.J.; Markert, J.M.; Hurst, D.R. Histone Deacetylase Inhibitors Improve the Replication of Oncolytic Herpes Simplex Virus in Breast Cancer Cells. PLoS ONE 2014, 9, e92919. [Google Scholar] [CrossRef]
- Xia, J.; Wishart, D.S. MetPA: A Web-Based Metabolomics Tool for Pathway Analysis and Visualization. Bioinformatics 2010, 26, 2342–2344. [Google Scholar] [CrossRef]
- Zhao, Y.; Butler, E.B.; Tan, M. Targeting Cellular Metabolism to Improve Cancer Therapeutics. Cell Death Dis. 2013, 4, e532. [Google Scholar] [CrossRef]
- Nagarajan, S.R.; Butler, L.M.; Hoy, A.J. The Diversity and Breadth of Cancer Cell Fatty Acid Metabolism. Cancer Metab. 2021, 9, 2. [Google Scholar] [CrossRef]
- Schiliro, C.; Firestein, B.L. Mechanisms of Metabolic Reprogramming in Cancer Cells Supporting Enhanced Growth and Proliferation. Cells 2021, 10, 1056. [Google Scholar] [CrossRef] [PubMed]
- Burg, T.; Rossaert, E.; Moisse, M.; Damme, P.V.; Bosch, L.V.D. Histone Deacetylase Inhibition Regulates Lipid Homeostasis in a Mouse Model of Amyotrophic Lateral Sclerosis. Int. J. Mol. Sci. 2021, 22, 11224. [Google Scholar] [CrossRef] [PubMed]
- Koundouros, N.; Poulogiannis, G. Reprogramming of Fatty Acid Metabolism in Cancer. Br. J. Cancer 2020, 122, 4–22. [Google Scholar] [CrossRef]
- Gomes, L.; Viana, L.; Silva, J.L.; Mermelstein, C.; Atella, G.; Fialho, E. Resveratrol Modifies Lipid Composition of Two Cancer Cell Lines. BioMed Res. Int. 2020, 2020, 5393041. [Google Scholar] [CrossRef] [PubMed]
- Urias, B.S.; Pavan, A.R.; Albuquerque, G.R.; Prokopczyk, I.M.; Alves, T.M.F.; de Melo, T.R.F.; Sartori, G.R.; da Silva, J.H.M.; Chin, C.M.; Santos, J.L.D. Optimization of Resveratrol Used as a Scaffold to Design Histone Deacetylase (HDAC-1 and HDAC-2) Inhibitors. Pharmaceuticals 2022, 15, 1260. [Google Scholar] [CrossRef] [PubMed]
- Venturelli, S.; Berger, A.; Böcker, A.; Busch, C.; Weiland, T.; Noor, S.; Leischner, C.; Schleicher, S.; Mayer, M.; Weiss, T.S.; et al. Resveratrol as a Pan-HDAC Inhibitor Alters the Acetylation Status of Jistone Proteins in Human-Derived Hepatoblastoma Cells. PLoS ONE 2013, 8, e73097. [Google Scholar] [CrossRef]
- Xu, S.; Chen, Y.; Ma, Y.; Liu, T.; Zhao, M.; Wang, Z.; Zhao, L. Lipidomic Profiling Reveals Disruption of Lipid Metabolism in Valproic Acid-Induced Hepatotoxicity. Front. Pharmacol. 2019, 10, 819. [Google Scholar] [CrossRef]
- Shimshoni, J.A.; Basselin, M.; Li, L.O.; Coleman, R.A.; Rapoport, S.I.; Modi, H.R. Valproate Uncompetitively Inhibits Arachidonic Acid Acylation by Rat Acyl-CoA Synthetase 4: Relevance to Valproate’s Efficacy against Bipolar Disorder. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2011, 1811, 163–169. [Google Scholar] [CrossRef]
- Borin, T.F.; Angara, K.; Rashid, M.H.; Achyut, B.R.; Arbab, A.S. Arachidonic Acid Metabolite as a Novel Therapeutic Target in Breast Cancer Metastasis. Int. J. Mol. Sci. 2017, 18, 2661. [Google Scholar] [CrossRef]
- Rosario, S.R.; Long, M.D.; Affronti, H.C.; Rowsam, A.M.; Eng, K.H.; Smiraglia, D.J. Pan-Cancer Analysis of Transcriptional Metabolic Dysregulation Using the Cancer Genome Atlas. Nat. Commun. 2018, 9, 5330. [Google Scholar] [CrossRef] [PubMed]
- Sostare, J.; Guida, R.D.; Kirwan, J.; Chalal, K.; Palmer, E.; Dunn, W.B.; Viant, M.R. Comparison of Modified Matyash Method to Conventional Solvent Systems for Polar Metabolite and Lipid Extractions. Anal. Chim. Acta 2018, 1037, 301–315. [Google Scholar] [CrossRef]
- Bi, H.; Krausz, K.W.; Manna, S.K.; Li, F.; Johnson, C.H.; Gonzalez, F.J. Optimization of Harvesting, Extraction, and Analytical Protocols for UPLC-ESI-MS-Based Metabolomic Analysis of Adherent Mammalian Cancer Cells. Anal. Bioanal. Chem. 2013, 405, 5279–5289. [Google Scholar] [CrossRef] [PubMed]
- Dudzik, D.; Barbas-Bernardos, C.; García, A.; Barbas, C. Quality Assurance Procedures for Mass Spectrometry Untargeted Metabolomics. a Review. J. Pharm. Biomed. Anal. 2018, 147, 149–173. [Google Scholar] [CrossRef] [PubMed]
- Ulmer, C.Z.; Yost, R.A.; Chen, J.; Mathews, C.E.; Garrett, T.J. Liquid Chromatography-Mass Spectrometry Metabolic and Lipidomic Sample Preparation Workflow for Suspension-Cultured Mammalian Cells Using Jurkat T Lymphocyte Cells. J. Proteom. Bioinform. 2015, 8, 126–132. [Google Scholar] [CrossRef]
- Walker, J.M. The Bicinchoninic Acid (BCA) Assay for Protein Quantitation. In The Protein Protocols Handbook; Humana Press Inc.: Totowa, NJ, USA, 1996; pp. 11–14. [Google Scholar] [CrossRef]
- Cui, Y.; Wang, X.; Xu, J.; Liu, X.; Wang, X.; Pang, J.; Song, Y.; Yu, M.; Song, W.; Luo, X.; et al. Proteomic Analysis of Taenia Solium Cyst Fluid by Shotgun LC-MS/MS. J. Parasitol. 2021, 107, 799–809. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Qiu, J.; Xu, Y.; Liao, G.; Jia, Q.; Pan, Y.; Wang, T.; Qian, Y. Integrated Non-Targeted Lipidomics and Metabolomics Analyses for Fluctuations of Neonicotinoids Imidacloprid and Acetamiprid on Neuro-2a Cells. Environ. Pollut. 2021, 284, 117327. [Google Scholar] [CrossRef]
- Broadhurst, D.; Goodacre, R.; Reinke, S.N.; Kuligowski, J.; Wilson, I.D.; Lewis, M.R.; Dunn, W.B. Guidelines and Considerations for the Use of System Suitability and Quality Control Samples in Mass Spectrometry Assays Applied in Untargeted Clinical Metabolomic Studies. Metabolomics 2018, 14, 72. [Google Scholar] [CrossRef]
- Dunn, W.B.; Wilson, I.D.; Nicholls, A.W.; Broadhurst, D. The Importance of Experimental Design and QC Samples in Large-Scale and MS-Driven Untargeted Metabolomic Studies of Humans. Bioanalysis 2012, 4, 2249–2264. [Google Scholar] [CrossRef]
- Estrada-Pérez, A.R.; Bakalara, N.; García-Vázquez, J.B.; Rosales-Hernández, M.C.; Fernández-Pomares, C.; Correa-Basurto, J. LC–MS Based Lipidomics Depict Phosphatidylethanolamine as Biomarkers of TNBC MDA-MB-231 over NTNBC MCF-7 Cells. Int. J. Mol. Sci. 2022, 23, 12074. [Google Scholar] [CrossRef]
- Wishart, D.S.; Guo, A.; Oler, E.; Wang, F.; Anjum, A.; Peters, H.; Dizon, R.; Sayeeda, Z.; Tian, S.; Lee, B.L.; et al. HMDB 5.0: The Human Metabolome Database for 2022. Nucleic Acids Res. 2021, 50, D622–D631. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Sinelnikov, I.V.; Han, B.; Wishart, D.S. MetaboAnalyst 3.0—Making Metabolomics More Meaningful. Nucleic Acids Res. 2015, 43, W251–W257. [Google Scholar] [CrossRef]
- Pang, Z.; Zhou, G.; Ewald, J.; Chang, L.; Hacariz, O.; Basu, N.; Xia, J. Using MetaboAnalyst 5.0 for LC–HRMS Spectra Processing, Multi-Omics Integration and Covariate Adjustment of Global Metabolomics Data. Nat. Protoc. 2022, 17, 1735–1761. [Google Scholar] [CrossRef]
- Chong, J.; Wishart, D.S.; Xia, J. Using MetaboAnalyst 4.0 for Comprehensive and Integrative Metabolomics Data Analysis. Curr. Protoc. Bioinform. 2019, 68, e86. [Google Scholar] [CrossRef]
- Xia, J.; Wishart, D.S. Web-Based Inference of Biological Patterns, Functions and Pathways from Metabolomic Data Using MetaboAnalyst. Nat. Protoc. 2011, 6, 743–760. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Chong, J.; Zhou, G.; de Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.-É.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the Gap between Raw Spectra and Functional Insights. Nucleic Acids Res. 2021, 49, gkab382. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).