Regulation by Different Types of Chaperones of Amyloid Transformation of Proteins Involved in the Development of Neurodegenerative Diseases
Abstract
1. Introduction
2. Types of Chaperones That May Be Associated with Pathological Transformation of Proteins
3. Effect of Artificial Chaperones on the Pathological Transformation of Amyloidogenic Proteins
4. Influence of Chaperones on Pathological Transformation of Amyloidogenic Proteins
4.1. Alpha-Synuclein
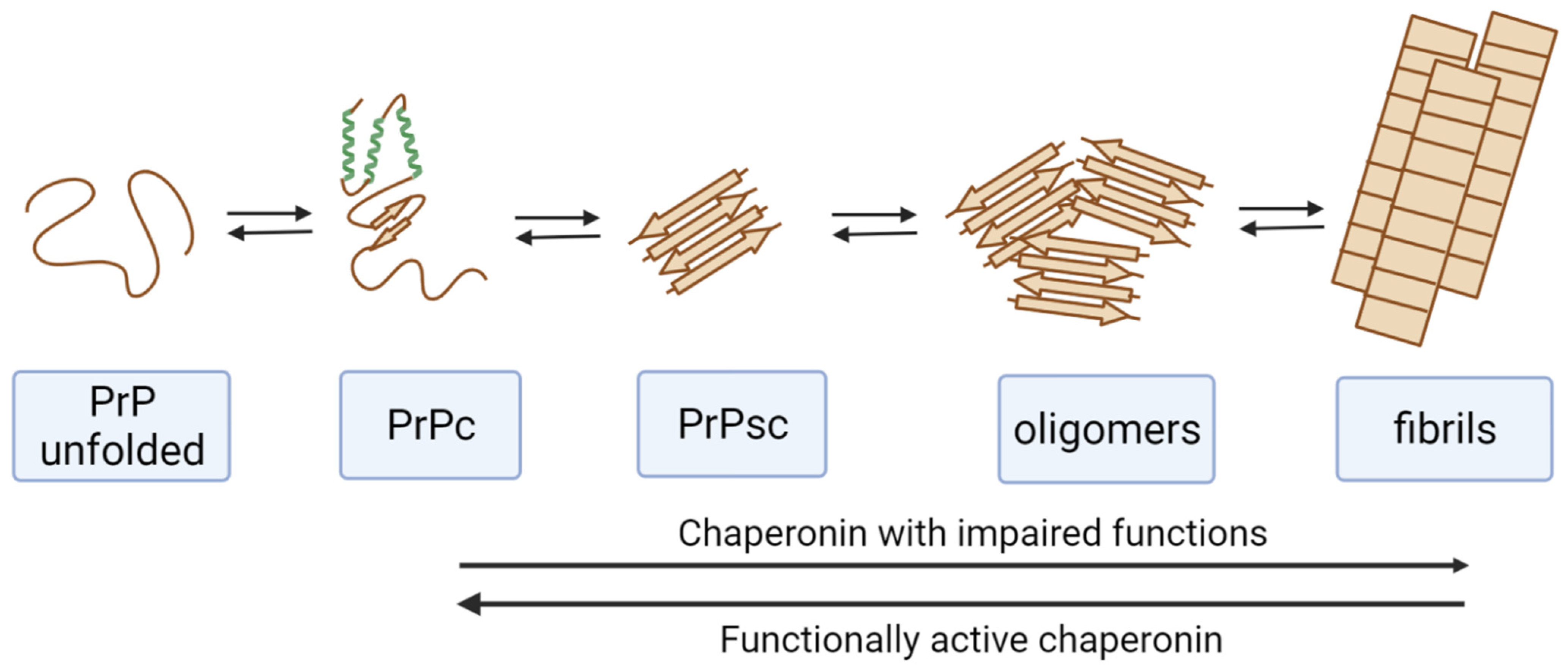
4.2. Prion Protein
5. Mechanisms of the Formation of Misfolded Proteins
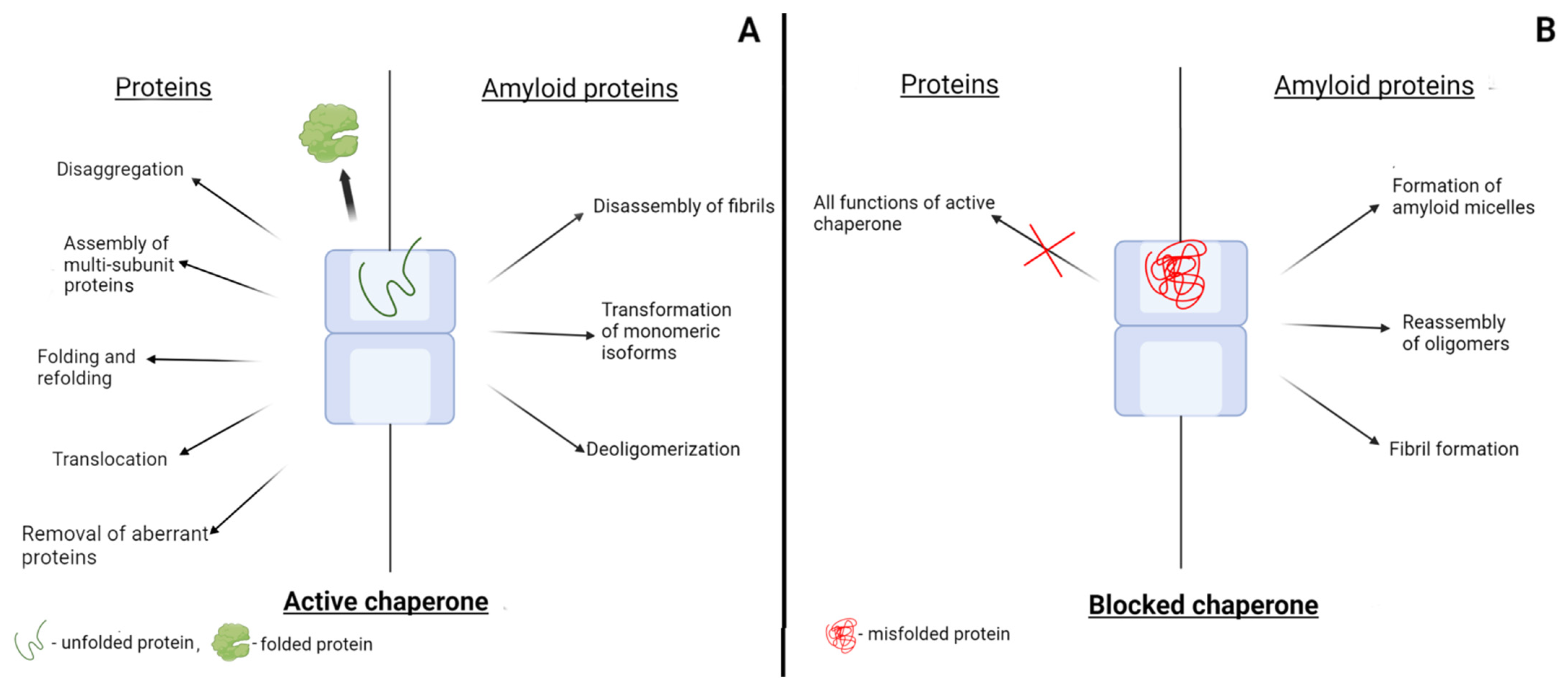
6. Blocking of Chaperones by Misfolded Proteins and Amyloidogenic Proteins
7. Post-Translational Modifications and Functioning of Chaperones
8. Participation of Bacterial Chaperones of Gut Microbiota in the Pathological Transformation of Amyloidogenic Proteins
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
Abbreviations
| BiP/Grp78 | the eukaryotic chaperone |
| ER | endoplasmic reticulum |
| FDHs | flavin-dependent halogenases |
| GAPDH | glyceraldehyde-3-phosphate dehydrogenase |
| GPI | glycophosphatidylinositol anchor |
| GroE | the bacterial chaperonin GroEL/GroES complex |
| GroEL | the bacterial chaperonin |
| GroES | co-chaperonin |
| HSPs | heat shock proteins |
| PrPSc | the scrapie isoform of the prion protein |
| PrPc | the native prion protein |
| RNS | reactive nitrogen species |
| ROS | reactive oxygen species |
| TRiC | the eukaryotic chaperonin (Hsp60) |
| UPR | unfolded protein response |
References
- Chiti, F.; Dobson, C.M. Protein Misfolding, Functional Amyloid, and Human Disease. Annu. Rev. Biochem. 2006, 75, 333–366. [Google Scholar] [CrossRef] [PubMed]
- Chiti, F.; Dobson, C.M. Protein Misfolding, Amyloid Formation, and Human Disease: A Summary of Progress over the Last Decade. Annu. Rev. Biochem. 2017, 86, 27–68. [Google Scholar] [CrossRef] [PubMed]
- Soto, C.; Estrada, L.; Castilla, J. Amyloids, prions and the inherent infectious nature of misfolded protein aggregates. Trends Biochem. Sci. 2006, 31, 150–155. [Google Scholar] [CrossRef]
- Toyama, B.H.; Weissman, J.S. Amyloid Structure: Conformational Diversity and Consequences. Annu. Rev. Biochem. 2011, 80, 557–585. [Google Scholar] [CrossRef]
- Ciechanover, A.; Kwon, Y.T. Protein Quality Control by Molecular Chaperones in Neurodegeneration. Front. Neurosci. 2017, 11, 185. [Google Scholar] [CrossRef]
- Guzhova, I.V.; Lazarev, V.F.; Kaznacheeva, A.V.; Ippolitova, M.V.; Muronetz, V.I.; Kinev, A.V.; Margulis, B.A. Novel mechanism of Hsp70 chaperone-mediated prevention of polyglutamine aggregates in a cellular model of huntington disease. Hum. Mol. Genet. 2011, 20, 3953–3963. [Google Scholar] [CrossRef]
- Tittelmeier, J.; Nachman, E.; Nussbaum-Krammer, C. Molecular Chaperones: A Double-Edged Sword in Neurodegenerative Diseases. Front. Aging Neurosci. 2020, 12, 581374. [Google Scholar] [CrossRef]
- Kampinga, H.H.; Craig, E.A. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 2010, 11, 579–592. [Google Scholar] [CrossRef]
- Bukau, B.; Weissman, J.; Horwich, A. Molecular Chaperones and Protein Quality Control. Cell 2006, 125, 443–451. [Google Scholar] [CrossRef]
- Wentink, A.; Nussbaum-Krammer, C.; Bukau, B. Modulation of Amyloid States by Molecular Chaperones. Cold Spring Harb. Perspect. Biol. 2019, 11, a033969. [Google Scholar] [CrossRef]
- Kampinga, H.H.; Bergink, S. Heat shock proteins as potential targets for protective strategies in neurodegeneration. Lancet Neurol. 2016, 15, 748–759. [Google Scholar] [CrossRef]
- Hartl, F.U.; Bracher, A.; Hayer-Hartl, M. Molecular chaperones in protein folding and proteostasis. Nature 2011, 475, 324–332. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.E.; Hipp, M.S.; Bracher, A.; Hayer-Hartl, M.; Ulrich Hartl, F. Molecular Chaperone Functions in Protein Folding and Proteostasis. Annu. Rev. Biochem. 2013, 82, 323–355. [Google Scholar] [CrossRef]
- Skjaerven, L.; Cuéllar, J.; Martinez, A.; Valpuesta, J. Dynamics, flexibility, and allostery in molecular chaperonins. FEBS Lett. 2015, 589 Pt A, 2522–2532. [Google Scholar] [CrossRef]
- Webster, J.M.; Darling, A.L.; Uversky, V.N.; Blair, L.J. Small Heat Shock Proteins, Big Impact on Protein Aggregation in Neurodegenerative Disease. Front. Pharmacol. 2019, 10, 1047. [Google Scholar] [CrossRef]
- Semenyuk, P.I.; Kurochkina, L.P.; Gusev, N.B.; Izumrudov, V.A.; Muronetz, V.I. Chaperone-like activity of synthetic polyanions can be higher than the activity of natural chaperones at elevated temperature. Biochem. Biophys. Res. Commun. 2017, 489, 200–205. [Google Scholar] [CrossRef] [PubMed]
- Muronetz, V.I.; Kazakov, S.V.; Dainiak, M.B.; Izumrudov, V.A.; Galaev, I.Y.; Mattiasson, B. Interaction of antibodies and antigens conjugated with synthetic polyanions: On the way of creating an artificial chaperone. Biochim. Biophys. Acta 2000, 1475, 141–150. [Google Scholar] [CrossRef]
- Dainiak, M.B.; Izumrudov, V.A.; Muronetz, V.I.; Galaev, I.Y.; Mattiasson, B. Conjugates of monoclonal antibodies with polyelectrolyte complexes—an attempt to make an artificial chaperone. Biochim. Biophys. Acta 1998, 1381, 279–285. [Google Scholar] [CrossRef]
- Dainiak, M.B.; Izumrudov, V.A.; Muronetz, V.I.; Galaev, I.Y.; Mattiasson, B. Reactivation of glyceraldehyde-3-phosphate dehydrogenase using conjugates of monoclonal antibodies with polyelectrolyte complexes. An attempt to make an artificial chaperone. J. Mol. Recognit. 1998, 11, 25–27. [Google Scholar] [CrossRef]
- Sorokina, S.A.; Stroylova, Y.Y.; Shifrina, Z.B.; Muronetz, V.I. Disruption of Amyloid Prion Protein Aggregates by Cationic Pyridylphenylene Dendrimers. Macromol. Biosci. 2016, 16, 266–275. [Google Scholar] [CrossRef]
- Semenyuk, P.I.; Moiseeva, E.V.; Stroylova, Y.Y.; Lotti, M.; Izumrudov, V.A.; Muronetz, V.I. Sulfated and sulfonated polymers are able to solubilize efficiently the protein aggregates of different nature. Arch. Biochem. Biophys. 2015, 567, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Sorokina, S.; Semenyuk, P.; Stroylova, Y.; Muronetz, V.; Shifrina, Z. Complexes between cationic pyridylphenylene dendrimers and ovine prion protein: Do hydrophobic interactions matter? RSC Adv. 2017, 7, 16565–16574. [Google Scholar] [CrossRef]
- Sorokina, S.A.; Stroilova, Y.Y.; Muronets, V.I.; Shifrina, Z.B. Role of the Structural Characteristics of Dendrimers in the Manifestation of the Antiamyloid Properties. Ineos Open 2020, 3, 140–145. [Google Scholar] [CrossRef]
- Klajnert, B.; Cortijo-Arellano, M.; Cladera, J.; Bryszewska, M. Influence of dendrimer’s structure on its activity against amyloid fibril formation. Biochem. Biophys. Res. Commun. 2006, 345, 21–28. [Google Scholar] [CrossRef]
- Semenyuk, P.; Kurochkina, L.; Barinova, K.; Muronetz, V. Alpha-Synuclein Amyloid Aggregation Is Inhibited by Sulfated Aromatic Polymers and Pyridinium Polycation. Polymers 2020, 12, 517. [Google Scholar] [CrossRef]
- Caughey, B.; Raymond, G.J. Sulfated polyanion inhibition of scrapie-associated PrP accumulation in cultured cells. J. Virol. 1993, 67, 643–650. [Google Scholar] [CrossRef]
- Van Horssen, J.; Wesseling, P.; Van Den Heuvel, L.P.; De Waal, R.M.; Verbeek, M.M. Heparan sulphate proteoglycans in Alzheimer’s disease and amyloid-related disorders. Lancet Neurol. 2003, 2, 482–492. [Google Scholar] [CrossRef]
- Iannuzzi, C.; Irace, G.; Sirangelo, I. The Effect of Glycosaminoglycans (GAGs) on Amyloid Aggregation and Toxicity. Molecules 2015, 20, 2510–2528. [Google Scholar] [CrossRef]
- Nishitsuji, K.; Uchimura, K. Sulfated glycosaminoglycans in protein aggregation diseases. Glycoconj. J. 2017, 34, 453–466. [Google Scholar] [CrossRef]
- Cohlberg, J.A.; Li, J.; Uversky, V.N.; Fink, A.L. Heparin and other glycosaminoglycans stimulate the formation of amyloid fibrils from alpha-synuclein in vitro. Biochemistry 2002, 41, 1502–1511. [Google Scholar] [CrossRef]
- McLaurin, J.; Franklin, T.; Zhang, X.; Deng, J.; Fraser, P.E. Interactions of Alzheimer amyloid-beta peptides with glycosaminoglycans effects on fibril nucleation and growth. Eur. J. Biochem. 1999, 266, 1101–1110. [Google Scholar] [CrossRef] [PubMed]
- Timmer, N.M.; Schirris, T.J.J.; Bruinsma, I.B.; Otte-Höller, I.; van Kuppevelt, T.H.; de Waal, R.M.W.; Verbeek, M.M. Aggregation and cytotoxic properties towards cultured cerebrovascular cells of Dutch-mutated Abeta40 (DAbeta(1-40)) are modulated by sulfate moieties of heparin. Neurosci Res. 2010, 66, 380–389. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, M.; Crowther, R.A.; Jakes, R.; Goedert, M. Alzheimer-like changes in microtubule-associated protein Tau induced by sulfated glycosaminoglycans. Inhibition of microtubule binding, stimulation of phosphorylation, and filament assembly depend on the degree of sulfation. J. Biol. Chem. 1997, 272, 33118–33124. [Google Scholar] [CrossRef] [PubMed]
- Townsend, D.; Hughes, E.; Hussain, R.; Siligardi, G.; Baldock, S.J.; Madine, J.; Middleton, D.A. Heparin and Methionine Oxidation Promote the Formation of Apolipoprotein A-I Amyloid Comprising α-Helical and β-Sheet Structures. Biochemistry 2017, 56, 1632–1644. [Google Scholar] [CrossRef] [PubMed]
- Enshaei, H.; Puiggalí-Jou, A.; Saperas, N.; Alemán, C. Conducting polymer nanoparticles for a voltage-controlled release of pharmacological chaperones. Soft Matter. 2021, 17, 3314–3321. [Google Scholar] [CrossRef] [PubMed]
- Semenyuk, P.; Muronetz, V. Protein Interaction with Charged Macromolecules: From Model Polymers to Unfolded Proteins and Post-Translational Modifications. Int. J. Mol. Sci. 2019, 20, 1252. [Google Scholar] [CrossRef] [PubMed]
- Ulmer, T.S.; Bax, A.; Cole, N.B.; Nussbaum, R.L. Structure and Dynamics of Micelle-bound Human α-Synuclein. J. Biol. Chem. 2005, 280, 9595–9603. [Google Scholar] [CrossRef]
- Goedert, M.; Spillantini, M.G.; Del Tredici, K.; Braak, H. 100 years of Lewy pathology. Nat. Rev. Neurol. 2013, 9, 13–24. [Google Scholar] [CrossRef]
- Roberts, H.L.; Brown, D.R. Seeking a Mechanism for the Toxicity of Oligomeric α-Synuclein. Biomolecules 2015, 5, 282–305. [Google Scholar] [CrossRef]
- Cox, D.; Selig, E.; Griffin, M.; Carver, J.; Ecroyd, H. Small Heat-shock Proteins Prevent α-Synuclein Aggregation via Transient Interactions and Their Efficacy Is Affected by the Rate of Aggregation. J. Biol. Chem. 2016, 291, 22618–22629. [Google Scholar] [CrossRef]
- Waudby, C.; Knowles, T.; Devlin, G.L.; Skepper, J.N.; Ecroyd, H.; Carver, J.; Welland, M.E.; Christodoulou, J.; Dobson, C.M.; Meehan, S. The Interaction of αB-Crystallin with Mature α-Synuclein Amyloid Fibrils Inhibits Their Elongation. Biophys. J. 2010, 98, 843–851. [Google Scholar] [CrossRef] [PubMed]
- Cox, D.; Whiten, D.R.; Brown, J.W.P.; Horrocks, M.H.; Gil, R.S.; Dobson, C.M.; Klenerman, D.; van Oijen, A.M.; Ecroyd, H. The small heat shock protein Hsp27 binds α-synuclein fibrils, preventing elongation and cytotoxicity. J. Biol. Chem. 2018, 293, 4486–4497. [Google Scholar] [CrossRef]
- Selig, E.E.; Zlatic, C.O.; Cox, D.; Mok, Y.-F.; Gooley, P.R.; Ecroyd, H.; Griffin, M.D.W. N- and C-terminal regions of αB-crystallin and Hsp27 mediate inhibition of amyloid nucleation, fibril binding, and fibril disaggregation. J. Biol. Chem. 2020, 295, 9838–9854. [Google Scholar] [CrossRef]
- Gaspar, R.; Garting, T.; Stradner, A. Eye lens crystallin proteins inhibit the autocatalytic amyloid amplification nature of mature α-synuclein fibrils. PLoS ONE 2020, 15, e0235198. [Google Scholar] [CrossRef] [PubMed]
- Luk, K.C.; Mills, I.P.; Trojanowski, J.Q.; Lee, V.M.-Y. Interactions between Hsp70 and the Hydrophobic Core of α-Synuclein Inhibit Fibril Assembly. Biochemistry 2008, 47, 12614–12625. [Google Scholar] [CrossRef] [PubMed]
- Pemberton, S.; Madiona, K.; Pieri, L.; Kabani, M.; Bousset, L.; Melki, R. Hsc70 Protein Interaction with Soluble and Fibrillar α-Synuclein. J. Biol. Chem. 2011, 286, 34690–34699. [Google Scholar] [CrossRef]
- Tao, J.; Berthet, A.; Citron, Y.R.; Tsiolaki, P.L.; Stanley, R.; Gestwicki, J.E.; Agard, D.A.; McConlogue, L. Hsp70 chaperone blocks α-synuclein oligomer formation via a novel engagement mechanism. J. Biol. Chem. 2021, 296, 100613. [Google Scholar] [CrossRef]
- Dedmon, M.M.; Christodoulou, J.; Wilson, M.; Dobson, C.M. Heat Shock Protein 70 Inhibits α-Synuclein Fibril Formation via Preferential Binding to Prefibrillar Species. J. Biol. Chem. 2005, 280, 14733–14740. [Google Scholar] [CrossRef]
- Roodveldt, C.; Bertoncini, C.W.; Andersson, A.; van der Goot, A.T.; Hsu, S.-T.; Fernández-Montesinos, R.; de Jong, J.; van Ham, T.J.; Nollen, E.A.; Pozo, D.; et al. Chaperone proteostasis in Parkinson’s disease: Stabilization of the Hsp70/alpha-synuclein complex by Hip. EMBO J. 2009, 28, 3758–3770. [Google Scholar] [CrossRef]
- Gao, X.; Carroni, M.; Nussbaum-Krammer, C.; Mogk, A.; Nillegoda, N.B.; Szlachcic, A.; Guilbride, D.L.; Saibil, H.R.; Mayer, M.P.; Bukau, B. Human Hsp70 disaggregase reverses Parkinson’s-linked α-synuclein amyloid fibrils. Mol. Cell 2015, 59, 781–793. [Google Scholar] [CrossRef]
- Yamamoto, H.; Fukui, N.; Adachi, M.; Saiki, E.; Yamasaki, A.; Matsumura, R.; Kuroyanagi, D.; Hongo, K.; Mizobata, T.; Kawata, Y. Human molecular chaperone Hsp60 and its apical domain suppress amyloid fibril formation of α-synuclein. Int. J. Mol. Sci. 2019, 21, 47. [Google Scholar] [CrossRef] [PubMed]
- Fukui, N.; Araki, K.; Hongo, K.; Mizobata, T.; Kawata, Y. Modulating the effects of the bacterial chaperonin GroEL on fibrillogenic polypeptides through modification of domain hinge architecture. J. Biol. Chem. 2016, 291, 25217–25226. [Google Scholar] [CrossRef] [PubMed]
- Ojha, B.; Fukui, N.; Hongo, K.; Mizobata, T.; Kawata, Y. Suppression of amyloid fibrils using the GroEL apical domain. Sci. Rep. 2016, 6, 31041. [Google Scholar] [CrossRef] [PubMed]
- Sot, B.; Rubio-Muñoz, A.; Leal-Quintero, A.; Martínez-Sabando, J.; Marcilla, M.; Roodveldt, C.; Valpuesta, J.M. The chaperonin CCT inhibits assembly of α-synuclein amyloid fibrils by a specific, conformation-dependent interaction. Sci Rep. 2017, 7, 40859. [Google Scholar] [CrossRef] [PubMed]
- Falsone, S.F.; Kungl, A.J.; Rek, A.; Cappai, R.; Zangger, K. The molecular chaperone Hsp90 modulates intermediate steps of amyloid assembly of the Parkinson-related protein alpha-synuclein. J. Biol. Chem. 2009, 284, 31190–31199. [Google Scholar] [CrossRef]
- Tittelmeier, J.; Sandhof, C.A.; Ries, H.M.; Druffel-Augustin, S.; Mogk, A.; Bukau, B.; Nussbaum-Krammer, C. The HSP110/HSP70 disaggregation system generates spreading-competent toxic α-synuclein species. EMBO J. 2020, 39, e103954. [Google Scholar] [CrossRef]
- Kiselev, G.G.; Naletova, I.N.; Sheval, E.V.; Stroylova, Y.Y.; Schmalhausen, E.V.; Haertlé, T.; Muronetz, V.I. Chaperonins induce an amyloid-like transformation of ovine prion protein: The fundamental difference in action between eukaryotic TRiC and bacterial GroEL. Biochim. Biophys. Acta 2011, 1814, 1730–1738. [Google Scholar] [CrossRef]
- Pan, K.M.; Baldwin, M.; Nguyen, J.; Gasset, M.; Serban, A.; Groth, D.; Mehlhorn, I.; Huang, Z.; Fletterick, R.J.; Cohen, F.E. Conversion of alpha-helices into beta-sheets features in the formation of the scrapie prion proteins. Proc. Natl. Acad. Sci. USA 1993, 90, 10962–10966. [Google Scholar] [CrossRef]
- Bosques, C.J.; Imperiali, B. The interplay of glycosylation and disulfide formation influences fibrillization in a prion protein fragment. Proc. Natl. Acad. Sci. USA 2003, 100, 7593–7598. [Google Scholar] [CrossRef]
- Stahl, N.; Borchelt, D.R.; Hsiao, K.; Prusiner, S.B. Scrapie prion protein contains a phosphatidylinositol glycolipid. Cell 1987, 51, 229–240. [Google Scholar] [CrossRef]
- Haraguchi, T.; Fisher, S.; Olofsson, S.; Endo, T.; Groth, D.; Tarentino, A.; Borchelt, D.R.; Teplow, D.; Hood, L.; Burlingame, A.; et al. Asparagine-linked glycosylation of the scrapie and cellular prion proteins. Arch. Biochem. Biophys. 1989, 274, 1–13. [Google Scholar] [CrossRef]
- Ford, M.J.; Burton, L.J.; Morris, R.J.; Hall, S.M. Selective expression of prion protein in peripheral tissues of the adult mouse. Neuroscience 2002, 113, 177–192. [Google Scholar] [CrossRef]
- Mironov, A.; Latawiec, D.; Wille, H.; Bouzamondo-Bernstein, E.; Legname, G.; Williamson, R.A.; Burton, D.; DeArmond, S.J.; Prusiner, S.B.; Peters, P.J. Cytosolic prion protein in neurons. J. Neurosci. 2003, 23, 7183–7193. [Google Scholar] [CrossRef] [PubMed]
- Roucou, X.; Guo, Q.; Zhang, Y.; Goodyer, C.G.; LeBlanc, A.C. Cytosolic prion protein is not toxic and protects against Bax-mediated cell death in human primary neurons. J. Biol. Chem. 2003, 278, 40877–40881. [Google Scholar] [CrossRef] [PubMed]
- Braakman, I.; Hebert, D.N. Protein folding in the endoplasmic reticulum. Cold Spring Harb. Perspect. Biol. 2013, 5, a013201. [Google Scholar] [CrossRef]
- Hebert, D.N.; Garman, S.C.; Molinari, M. The glycan code of the endoplasmic reticulum: Asparagine-linked carbohydrates as protein maturation and quality-control tags. Trends Cell Biol. 2005, 15, 364–370. [Google Scholar] [CrossRef]
- Ron, D.; Walter, P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 2007, 8, 519–529. [Google Scholar] [CrossRef]
- Hetz, C.; Papa, F.R. The unfolded protein response and cell fate control. Mol. Cell 2018, 69, 169–181. [Google Scholar] [CrossRef]
- Rodriguez, D.; Rojas-Rivera, D.; Hetz, C. Integrating stress signals at the endoplasmic reticulum: The BCL-2 protein family rheostat. Biochim. Biophys. Acta 2011, 1813, 564–574. [Google Scholar] [CrossRef]
- Krance, S.H.; Luke, R.; Shenouda, M.; Israwi, A.R.; Colpitts, S.J.; Darwish, L.; Strauss, M.; Watts, J.C. Cellular models for discovering prion disease therapeutics: Progress and challenges. J. Neurochem. 2020, 153, 150–172. [Google Scholar] [CrossRef]
- Brandner, S.; Jaunmuktane, Z. Prion disease: Experimental models and reality. Acta Neuropathol. 2017, 133, 197–222. [Google Scholar] [CrossRef] [PubMed]
- Hetz, C.A.; Soto, C. Stressing out the ER: A role of the unfolded protein response in prion-related disorders. Curr. Mol. Med. 2006, 6, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Hetz, C.; Russelakis-Carneiro, M.; Maundrell, K.; Castilla, J.; Soto, C. Caspase-12 and endoplasmic reticulum stress mediate neurotoxicity of pathological prion protein. EMBO J. 2003, 22, 5435–5445. [Google Scholar] [CrossRef] [PubMed]
- Kopp, M.C.; Larburu, N.; Durairaj, V.; Adams, C.J.; Ali, M.M.U. UPR proteins IRE1 and PERK switch BiP from chaperone to ER stress sensor. Nat. Struct. Mol. Biol. 2019, 26, 1053–1062. [Google Scholar] [CrossRef]
- Karlin, S.; Brocchieri, L. Heat shock protein 70 family: Multiple sequence comparisons, function, and evolution. J. Mol. Evol. 1998, 47, 565–577. [Google Scholar] [CrossRef]
- Bakunts, A.; Orsi, A.; Vitale, M.; Cattaneo, A.; Lari, F.; Tadè, L.; Sitia, R.; Raimondi, A.; Bachi, A.; van Alken, E. Ratiometric sensing of BiP-client versus BiP levels by the unfolded protein response determines its signaling amplitude. Elife 2017, 6, e27518. [Google Scholar] [CrossRef]
- Jin, T.; Gu, Y.; Zanusso, G.; Sy, M.; Kumar, A.; Cohen, M.; Gambetti, P.; Singh, N. The chaperone protein BiP binds to a mutant prion protein and mediates its degradation by the proteasome. J. Biol. Chem. 2000, 275, 38699–38704. [Google Scholar] [CrossRef]
- Peters, S.L.; Déry, M.-A.; LeBlanc, A.C. Familial prion protein mutants inhibit Hrd1-mediated retrotranslocation of misfolded proteins by depleting misfolded protein sensor BiP. Hum. Mol. Genet. 2016, 25, 976–988. [Google Scholar] [CrossRef]
- Torres, M.; Castillo, K.; Armisén, R.; Stutzin, A.; Soto, C.; Hetz, C. Prion protein misfolding affects calcium homeostasis and sensitizes cells to endoplasmic reticulum stress. PLoS ONE 2010, 5, e15658. [Google Scholar] [CrossRef]
- Hetz, C.; Russelakis-Carneiro, M.; Wälchli, S.; Carboni, S.; Vial-Knecht, E.; Maundrell, K.; Castilla, J.; Soto, C. The disulfide isomerase Grp58 is a protective factor against prion neurotoxicity. J. Neurosci. 2005, 25, 2793–2802. [Google Scholar] [CrossRef]
- Laszlo, L.; Lowe, J.; Self, T.; Kenward, N.; Landon, M.; McBride, T.; Farquhar, C.; McConnell, I.; Brown, J.; Hope, J.; et al. Lysosomes as key organelles in the pathogenesis of prion encephalopathies. J. Pathol. 1992, 166, 333–341. [Google Scholar] [CrossRef]
- Zhu, T.; Chen, J.-L.; Wang, Q.; Shao, W.; Qi, B. Modulation of mitochondrial dynamics in neurodegenerative diseases: An insight into prion diseases. Front. Aging Neurosci. 2018, 10, 336. [Google Scholar] [CrossRef] [PubMed]
- Brown, C.R.; Martin, R.L.; Hansen, W.J.; Beckmann, R.P.; Welch, W.J. The constitutive and stress inducible forms of hsp 70 exhibit functional similarities and interact with one another in an ATP-dependent fashion. J. Cell Biol. 1993, 120, 1101–1112. [Google Scholar] [CrossRef]
- Kenward, N.; Hope, J.; Landon, M.; Mayer, R.J. Expression of polyubiquitin and heat-shock protein 70 genes increases in the later stages of disease progression in scrapie-infected mouse brain. J. Neurochem. 1994, 62, 1870–1877. [Google Scholar] [CrossRef]
- Mamchur, A.A.; Moiseenko, A.V.; Panina, I.S.; Yaroshevich, I.A.; Kudryavtseva, S.S.; Pichkur, E.B.; Sokolova, O.S.; Muronetz, V.I.; Stanishneva-Konovalova, T.B. Structural and computational study of the GroEL-prion protein complex. Biomedicines 2021, 9, 1649. [Google Scholar] [CrossRef]
- Kudryavtseva, S.S.; Stroylova, Y.Y.; Zanyatkin, I.A.; Haertle, T.; Muronetz, V.I. Inhibition of chaperonin GroEL by a monomer of ovine prion protein and its oligomeric forms. Biochemistry 2016, 81, 1213–1220. [Google Scholar] [CrossRef]
- Kudryavtseva, S.S.; Stroylova, Y.Y.; Kurochkina, L.P.; Muronetz, V.I. The chaperonin TRiC is blocked by native and glycated prion protein. Arch. Biochem. Biophys. 2020, 683, 108319. [Google Scholar] [CrossRef]
- Kushnirov, V.V.; Dergalev, A.A.; Alexandrov, A.I. Amyloid fragmentation and disaggregation in yeast and animals. Biomolecules 2021, 11, 1884. [Google Scholar] [CrossRef]
- Paushkin, S.V.; Kushnirov, V.V.; Smirnov, V.N.; Ter-Avanesyan, M.D. Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J. 1996, 15, 3127–3134. [Google Scholar] [CrossRef]
- Chernova, T.A.; Chernoff, Y.O.; Wilkinson, K.D. Prion-based memory of heat stress in yeast. Prion 2017, 11, 151–161. [Google Scholar] [CrossRef]
- Chernoff, Y.O.; Uptain, S.M.; Lindquist, S.L. Analysis of prion factors in yeast. Methods Enzymol. 2002, 351, 499–538. [Google Scholar]
- Rikhvanov, E.G.; Romanova, N.V.; Chernoff, Y.O. Chaperone effects on prion and nonprion aggregates. Prion 2007, 1, 217–222. [Google Scholar] [CrossRef]
- Rubel, A.A.; Ryzhova, T.A.; Antonets, K.S.; Chernoff, Y.O.; Galkin, A. Identification of PrP sequences essential for the interaction between the PrP polymers and Aβ peptide in a yeast-based assay. Prion 2013, 7, 469–476. [Google Scholar] [CrossRef]
- Levinthal, C. Are there pathways for protein folding? J. Chim. Phys. 1968, 65, 44–45. [Google Scholar] [CrossRef]
- Ptitsyn, O.B.; Bychkova, V.E.; Uversky, V.N. Kinetic and equilibrium folding intermediates. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1995, 348, 35–41. [Google Scholar]
- Garbuzynskiy, S.O.; Ivankov, D.N.; Bogatyreva, N.S.; Finkelstein, A.V. Golden triangle for folding rates of globular proteins. Proc. Natl. Acad. Sci. USA 2013, 110, 147–150. [Google Scholar] [CrossRef]
- Finkelstein, A.V. 50+ years of protein folding. Biochemistry 2018, 83 (Suppl. 1), S3–S18. [Google Scholar] [CrossRef]
- Ivankov, D.N.; Finkelstein, A.V. Solution of Levinthal’s paradox and a physical theory of protein folding times. Biomolecules 2020, 10, 250. [Google Scholar] [CrossRef]
- Chah, S.; Kumar, C.V.; Hammond, M.R.; Zare, R.N. Denaturation and renaturation of self-assembled yeast iso-1-cytochrome c on Au. Anal. Chem. 2004, 76, 2112–2117. [Google Scholar] [CrossRef]
- Nájera, H.; Costas, M.; Fernández-Velasco, D.A. Thermodynamic characterization of yeast triosephosphate isomerase refolding: Insights into the interplay between function and stability as reasons for the oligomeric nature of the enzyme. Biochem. J. 2003, 370 Pt 3, 785–792. [Google Scholar] [CrossRef]
- Ibarra-Molero, B.; Naganathan, A.N.; Sanchez-Ruiz, J.M.; Muñoz, V. Modern analysis of protein folding by differential scanning calorimetry. Methods Enzymol. 2016, 567, 281–318. [Google Scholar]
- Blumlein, A.; McManus, J.J. Reversible and non-reversible thermal denaturation of lysozyme with varying pH at low ionic strength. Biochim. Biophys. Acta 2013, 1834, 2064–2070. [Google Scholar] [CrossRef]
- Kang, T.; Hong, S.; Kim, H.J.; Moon, J.; Oh, S.; Paik, S.R.; Yi, J. Characterization of surface-confined alpha-synuclein by surface plasmon resonance measurements. Langmuir 2006, 22, 13–17. [Google Scholar] [CrossRef]
- Guidry, J.J.; Moczygemba, C.K.; Steede, N.K.; Landry, S.J.; Wittung-Stafshede, P. Reversible denaturation of oligomeric human chaperonin 10: Denatured state depends on chemical denaturant. Protein Sci. 2000, 9, 2109–2117. [Google Scholar] [CrossRef]
- Tang, C.; Lew, S.; He, D. Using a second-order differential model to fit data without baselines in protein isothermal chemical denaturation. Protein Sci. 2016, 25, 898–904. [Google Scholar] [CrossRef]
- Biswas, H.; Chattopadhyaya, R. Thermal, chemical and pH induced unfolding of turmeric root lectin: Modes of denaturation. PLoS ONE 2014, 9, e103579. [Google Scholar] [CrossRef]
- Lapidus, L.J. Protein unfolding mechanisms and their effects on folding experiments. F1000Research 2017, 6, 1723. [Google Scholar] [CrossRef]
- Sorokina, I.; Mushegian, A. Modeling protein folding in vivo. Biol. Direct. 2018, 13, 13. [Google Scholar] [CrossRef]
- Braselmann, E.; Chaney, J.L.; Clark, P.L. Folding the proteome. Trends Biochem. Sci. 2013, 38, 337–344. [Google Scholar] [CrossRef]
- Dee, D.R.; Yada, R.Y. The prosegment catalyzes pepsin folding to a kinetically trapped native state. Biochemistry 2010, 49, 365–371. [Google Scholar] [CrossRef]
- Lawton, J.M.; Doonan, S. Thermal inactivation and chaperonin-mediated renaturation of mitochondrial aspartate aminotransferase. Biochem. J. 1998, 334 Pt 1, 219–224. [Google Scholar] [CrossRef]
- Ahern, T.J.; Klibanov, A.M. Analysis of processes causing thermal inactivation of enzymes. Methods Biochem. Anal. 1988, 33, 91–127. [Google Scholar]
- Singh, A.; Upadhyay, V.; Upadhyay, A.K.; Singh, S.M.; Panda, A.K. Protein recovery from inclusion bodies of Escherichia coli using mild solubilization process. Microb. Cell Factories 2015, 14, 41. [Google Scholar] [CrossRef]
- Ventura, S.; Villaverde, A. Protein quality in bacterial inclusion bodies. Trends Biotechnol. 2006, 24, 179–185. [Google Scholar] [CrossRef]
- Villaverde, A.; Carrió, M.M. Protein aggregation in recombinant bacteria: Biological role of inclusion bodies. Biotechnol. Lett. 2003, 25, 1385–1395. [Google Scholar] [CrossRef]
- Wang, L.; Maji, S.K.; Sawaya, M.R.; Eisenberg, D.; Riek, R. Bacterial inclusion bodies contain amyloid-like structure. PLoS Biol. 2008, 6, e195. [Google Scholar] [CrossRef]
- Ramón, A.; Señorale-Pose, M.; Marín, M. Inclusion bodies: Not that bad…. Front. Microbiol. 2014, 5, 56. [Google Scholar]
- Zimmerman, S.B.; Trach, S.O. Estimation of macromolecule concentrations and excluded volume effects for the cytoplasm of Escherichia coli. J. Mol. Biol. 1991, 222, 599–620. [Google Scholar] [CrossRef]
- Kuznetsova, I.M.; Turoverov, K.K.; Uversky, V.N. What macromolecular crowding can do to a protein. Int. J. Mol. Sci. 2014, 15, 23090–23140. [Google Scholar] [CrossRef]
- Minton, A.P. The influence of macromolecular crowding and macromolecular confinement on biochemical reactions in physiological media. J. Biol. Chem. 2001, 276, 10577–10580. [Google Scholar] [CrossRef]
- Smith, M.J.; Prochownik, E.V. Inhibition of c-jun causes reversible proliferative arrest and withdrawal from the cell cycle. Blood 1992, 79, 2107–2115. [Google Scholar] [CrossRef] [PubMed]
- Chebotareva, N.A.; Roman, S.G.; Borzova, V.A.; Eronina, T.B.; Mikhaylova, V.V.; Kurganov, B.I. Chaperone-like activity of HSPB5: The effects of quaternary structure dynamics and crowding. Int. J. Mol. Sci. 2020, 21, 4940. [Google Scholar] [CrossRef] [PubMed]
- Sorokina, I.; Mushegian, A.R.; Koonin, E.V. Is protein folding a thermodynamically unfavorable, active, energy-dependent process? Int. J. Mol. Sci. 2022, 23, 521. [Google Scholar] [CrossRef] [PubMed]
- Sorokina, I.; Mushegian, A. Rotational restriction of nascent peptides as an essential element of co-translational protein folding: Possible molecular players and structural consequences. Biol. Direct. 2017, 12, 14. [Google Scholar] [CrossRef] [PubMed]
- Sorokina, I.; Mushegian, A. The role of the backbone torsion in protein folding. Biol. Direct. 2016, 11, 64. [Google Scholar] [CrossRef]
- Sahakyan, H.; Nazaryan, K.; Mushegian, A.; Sorokina, I. Energy-dependent protein folding: Modeling how a protein folding machine may work. F1000Research 2021, 10, 3. [Google Scholar] [CrossRef]
- Broom, A.; Jacobi, Z.; Trainor, K.; Meiering, E.M. Computational tools help improve protein stability but with a solubility tradeoff. J. Biol. Chem. 2017, 292, 14349–14361. [Google Scholar] [CrossRef]
- Siddiqui, K.S. Defying the activity-stability trade-off in enzymes: Taking advantage of entropy to enhance activity and thermostability. Crit. Rev. Biotechnol. 2017, 37, 309–322. [Google Scholar] [CrossRef]
- Klaips, C.L.; Jayaraj, G.G.; Hartl, F.U. Pathways of cellular proteostasis in aging and disease. J. Cell Biol. 2018, 217, 51–63. [Google Scholar] [CrossRef]
- Kurtishi, A.; Rosen, B.; Patil, K.S.; Alves, G.W.; Møller, S.G. Cellular proteostasis in neurodegeneration. Mol. Neurobiol. 2019, 56, 3676–3689. [Google Scholar] [CrossRef]
- Saibil, H.R. Chaperone machines in action. Curr. Opin. Struct. Biol. 2008, 18, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Polyakova, O.V.; Roitel, O.; Asryants, R.A.; Poliakov, A.A.; Branlant, G.; Muronetz, V.I. Misfolded forms of glyceraldehyde-3-phosphate dehydrogenase interact with GroEL and inhibit chaperonin-assisted folding of the wild-type enzyme. Protein Sci. 2005, 14, 921–928. [Google Scholar] [CrossRef] [PubMed]
- Naletova, I.N.; Muronetz, V.I.; Schmalhausen, E.V. Unfolded, oxidized, and thermoinactivated forms of glyceraldehyde-3-phosphate dehydrogenase interact with the chaperonin GroEL in different ways. Biochim. Biophys. Acta 2006, 1764, 831–838. [Google Scholar] [CrossRef] [PubMed]
- Barinova, K.; Khomyakova, E.; Semenyuk, P.; Schmalhausen, E.; Muronetz, V. Binding of alpha-synuclein to partially oxidized glyceraldehyde-3-phosphate dehydrogenase induces subsequent inactivation of the enzyme. Arch. Biochem. Biophys. 2018, 642, 10–22. [Google Scholar] [CrossRef]
- Burmann, B.M.; Gerez, J.A.; Matečko-Burmann, I.; Campioni, S.; Kumari, P.; Ghosh, D.; Mazur, A.; Aspholm, E.E.; Šulskis, D.; Wawrzyniuk, M.; et al. Regulation of α-synuclein by chaperones in mammalian cells. Nature 2020, 577, 127–132. [Google Scholar] [CrossRef]
- Franco, A.; Cuéllar, J.; Fernández-Higuero, J.Á.; de la Arada, I.; Orozco, N.; Valpuesta, J.M.; Prado, A.; Muga, A. Truncation-driven lateral association of α-synuclein hinders amyloid clearance by the Hsp70-based disaggregase. Int. J. Mol. Sci. 2021, 22, 12983. [Google Scholar] [CrossRef]
- Nakamura, T.; Oh, C.-K.; Zhang, X.; Lipton, S.A. Protein S-nitrosylation and oxidation contribute to protein misfolding in neurodegeneration. Free Radic. Biol. Med. 2021, 172, 562–577. [Google Scholar] [CrossRef]
- Giasson, B.I.; Duda, J.E.; Murray, I.V.; Chen, Q.; Souza, J.M.; Hurtig, H.I.; Ischiropoulos, H.; Trojanowski, J.Q.; Lee, V.M. Oxidative damage linked to neurodegeneration by selective alpha-synuclein nitration in synucleinopathy lesions. Science 2000, 290, 985–989. [Google Scholar] [CrossRef]
- Hayashi, J.; Ton, J.; Negi, S.; Stephens, D.E.K.M.; Pountney, D.L.; Preiss, T.; Carver, J.A. The effect of oxidized dopamine on the structure and molecular chaperone function of the small heat-shock proteins, αB-crystallin and Hsp27. Int. J. Mol. Sci. 2021, 22, 3700. [Google Scholar] [CrossRef]
- Franco, M.C.; Ye, Y.; Refakis, C.A.; Feldman, J.L.; Stokes, A.L.; Basso, M.; Fernández de Mera, R.M.M.; Sparrow, N.A.; Calingasan, N.Y.; Kiaei, M.; et al. Nitration of Hsp90 induces cell death. Proc. Natl. Acad. Sci. USA 2013, 110, E1102–E1111. [Google Scholar] [CrossRef]
- Zhao, S.; Song, T.-Y.; Wang, Z.-Y.; Gao, J.; Cao, J.-W.; Hu, L.-L.; Huang, Z.-R.; Xie, L.-P.; Ji, Y. S-nitrosylation of Hsp90 promotes cardiac hypertrophy in mice through GSK3β signaling. Acta Pharmacol. Sin. 2021, 21, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Ruiz, A.; Villanueva, L.; de Orduña, C.G.; López-Ferrer, D.; Higueras, M.A.; Tarín, C.; Rodríguez-Crespo, I.; Vázquez, J.; Lamas, S. S-nitrosylation of Hsp90 promotes the inhibition of its ATPase and endothelial nitric oxide synthase regulatory activities. Proc. Natl. Acad. Sci. USA 2005, 102, 8525–8530. [Google Scholar] [CrossRef] [PubMed]
- Kröncke, K.D.; Haase, H.; Beyersmann, D.; Kolb-Bachofen, V.; Hayer-Hartl, M.K. Nitric oxide inhibits the cochaperone activity of the RING finger-like protein DnaJ. Nitric Oxide Biol. Chem. 2001, 5, 289–295. [Google Scholar] [CrossRef] [PubMed]
- Jakobsson, M.E.; Moen, A.; Bousset, L.; Egge-Jacobsen, W.; Kernstock, S.; Melki, R.; Falnes, P.Ø. Identification and characterization of a novel human methyltransferase modulating Hsp70 protein function through lysine methylation. J. Biol. Chem. 2013, 288, 27752–27763. [Google Scholar] [CrossRef]
- D’Argenio, V.; Sarnataro, D. Microbiome influence in the pathogenesis of prion and Alzheimer’s diseases. Int. J. Mol. Sci. 2019, 20, 4704. [Google Scholar] [CrossRef]
- Sun, M.; Ma, K.; Wen, J.; Wang, G.; Zhang, C.; Li, Q.; Bao, X.; Wang, H. A review of the brain-gut-microbiome axis and the potential role of microbiota in Alzheimer’s disease. J. Alzheimer’s Dis. 2020, 73, 849–865. [Google Scholar] [CrossRef]
- Kim, M.-S.; Kim, Y.; Choi, H.; Kim, W.; Park, S.; Lee, D.; Kim, D.K.; Kim, H.J.; Choi, H.; Hyun, D.-W.; et al. Transfer of a healthy microbiota reduces amyloid and tau pathology in an Alzheimer’s disease animal model. Gut 2020, 69, 283–294. [Google Scholar] [CrossRef]
- Pistollato, F.; Sumalla Cano, S.; Elio, I.; Masias Vergara, M.; Giampieri, F.; Battino, M. Role of gut microbiota and nutrients in amyloid formation and pathogenesis of Alzheimer disease. Nutr. Rev. 2016, 74, 624–634. [Google Scholar] [CrossRef]
- Nishiwaki, H.; Ito, M.; Ishida, T.; Hamaguchi, T.; Maeda, T.; Kashihara, K.; Tsuboi, Y.; Ueyama, J.; Shimamura, T.; Mori, H.; et al. Meta-analysis of gut dysbiosis in Parkinson’s disease. Mov. Disord. 2020, 35, 1626–1635. [Google Scholar] [CrossRef]
- Nishiwaki, H.; Hamaguchi, T.; Ito, M.; Ishida, T.; Maeda, T.; Kashihara, K.; Tsuboi, Y.; Ueyama, J.; Shimamura, T.; Mori, H.; et al. Short-chain fatty acid-producing gut microbiota is decreased in Parkinson’s disease but not in rapid-eye-movement sleep behavior disorder. Msystems 2020, 5, e00797-20. [Google Scholar] [CrossRef]
- Holmqvist, S.; Chutna, O.; Bousset, L.; Aldrin-Kirk, P.; Li, W.; Björklund, T.; Wang, Z.-Y.; Roybon, L.; Melki, R.; Li, J.-Y. Direct evidence of Parkinson pathology spread from the gastrointestinal tract to the brain in rats. Acta Neuropathol. 2014, 128, 805–820. [Google Scholar] [CrossRef]
- Uemura, N.; Yagi, H.; Uemura, M.T.; Hatanaka, Y.; Yamakado, H.; Takahashi, R. Inoculation of α-synuclein preformed fibrils into the mouse gastrointestinal tract induces Lewy body-like aggregates in the brainstem via the vagus nerve. Mol. Neurodegener. 2018, 13, 21. [Google Scholar] [CrossRef] [PubMed]
- Chandra, R.; Hiniker, A.; Kuo, Y.-M.; Nussbaum, R.L.; Liddle, R.A. α-Synuclein in gut endocrine cells and its implications for Parkinson’s disease. JCI Insight 2017, 2, e92295. [Google Scholar] [CrossRef] [PubMed]
- Liddle, R.A. Parkinson’s disease from the gut. Brain Res. 2018, 1693 Pt B, 201–206. [Google Scholar] [CrossRef]
| Types of Chaperones | Molecular Mass of One Subunit, kDa | Number of Subunits | ATPase Activity | Inhibition of Aggregation or Fibrillation | Stimulation of Fibrillation | Fibril Disaggregation |
|---|---|---|---|---|---|---|
| Small heat shock proteins | 12–43 | 9–50 | - | A-Syn (40–44) | ||
| Hsp70 | 66–78 | 1–2 | + | A-Syn (45–48) PrP (77) | A-Syn (50, 56) | |
| Hsp90 | 81–99 | 2 | + | A-Syn (55) | ||
| Hsp100–110 | 100–199 | 6 | + | A-Syn (50, 56) Sup35p (89) | ||
| GroEL (Hsp60) | 55–64 | 14 | + | A-Syn (51–53) | PrP (57) | |
| GroEL + GroES | + | PrP (86) | PrP (86) | |||
| TRiC (Hsp60) | 55–64 | 16 | + | A-Syn (54) | PrP (87) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muronetz, V.I.; Kudryavtseva, S.S.; Leisi, E.V.; Kurochkina, L.P.; Barinova, K.V.; Schmalhausen, E.V. Regulation by Different Types of Chaperones of Amyloid Transformation of Proteins Involved in the Development of Neurodegenerative Diseases. Int. J. Mol. Sci. 2022, 23, 2747. https://doi.org/10.3390/ijms23052747
Muronetz VI, Kudryavtseva SS, Leisi EV, Kurochkina LP, Barinova KV, Schmalhausen EV. Regulation by Different Types of Chaperones of Amyloid Transformation of Proteins Involved in the Development of Neurodegenerative Diseases. International Journal of Molecular Sciences. 2022; 23(5):2747. https://doi.org/10.3390/ijms23052747
Chicago/Turabian StyleMuronetz, Vladimir I., Sofia S. Kudryavtseva, Evgeniia V. Leisi, Lidia P. Kurochkina, Kseniya V. Barinova, and Elena V. Schmalhausen. 2022. "Regulation by Different Types of Chaperones of Amyloid Transformation of Proteins Involved in the Development of Neurodegenerative Diseases" International Journal of Molecular Sciences 23, no. 5: 2747. https://doi.org/10.3390/ijms23052747
APA StyleMuronetz, V. I., Kudryavtseva, S. S., Leisi, E. V., Kurochkina, L. P., Barinova, K. V., & Schmalhausen, E. V. (2022). Regulation by Different Types of Chaperones of Amyloid Transformation of Proteins Involved in the Development of Neurodegenerative Diseases. International Journal of Molecular Sciences, 23(5), 2747. https://doi.org/10.3390/ijms23052747





