Activation of Hypoxia-Inducible Factor-1α Signaling Pathway Has the Protective Effect of Intervertebral Disc Degeneration
Abstract
1. Introduction
2. Results
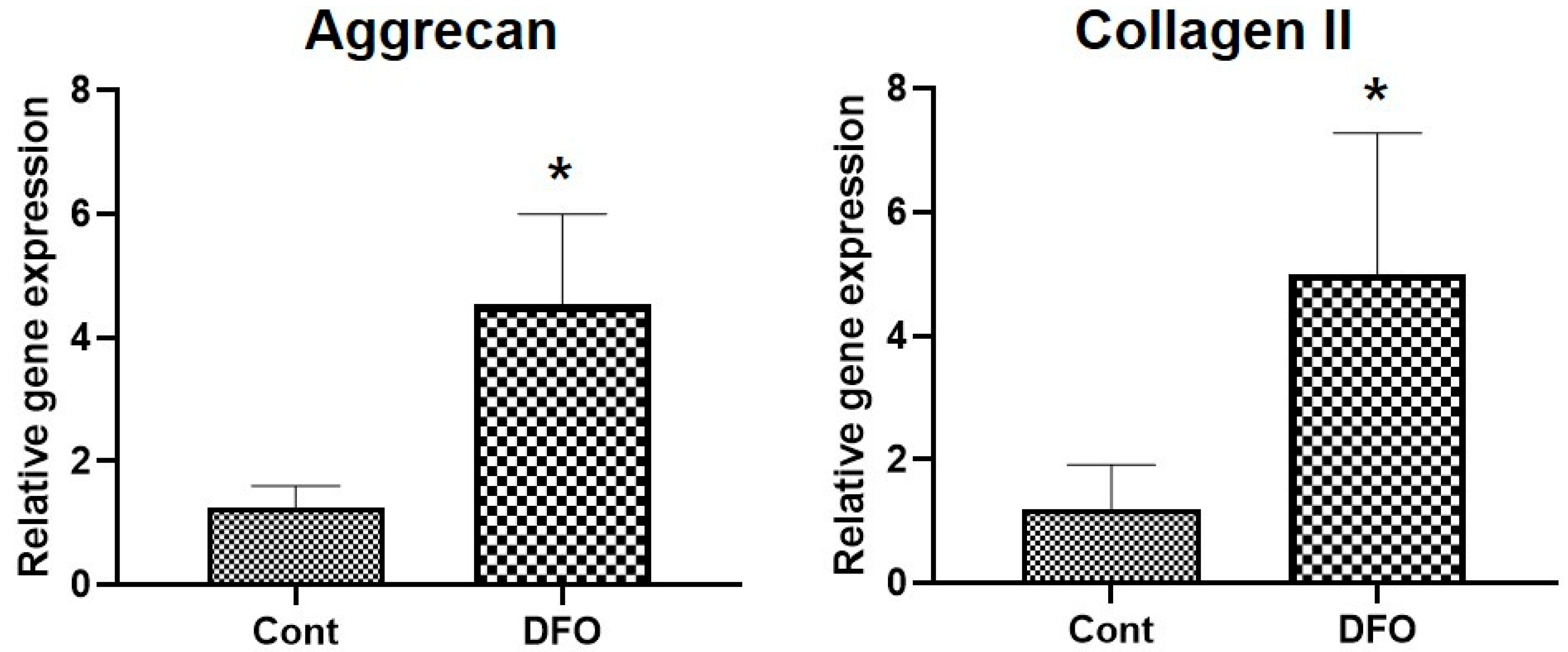
2.1. Hypoxia Increases Aggrecan and Collagen II mRNA Expression in Nucleus Pulposus (NP) Cells
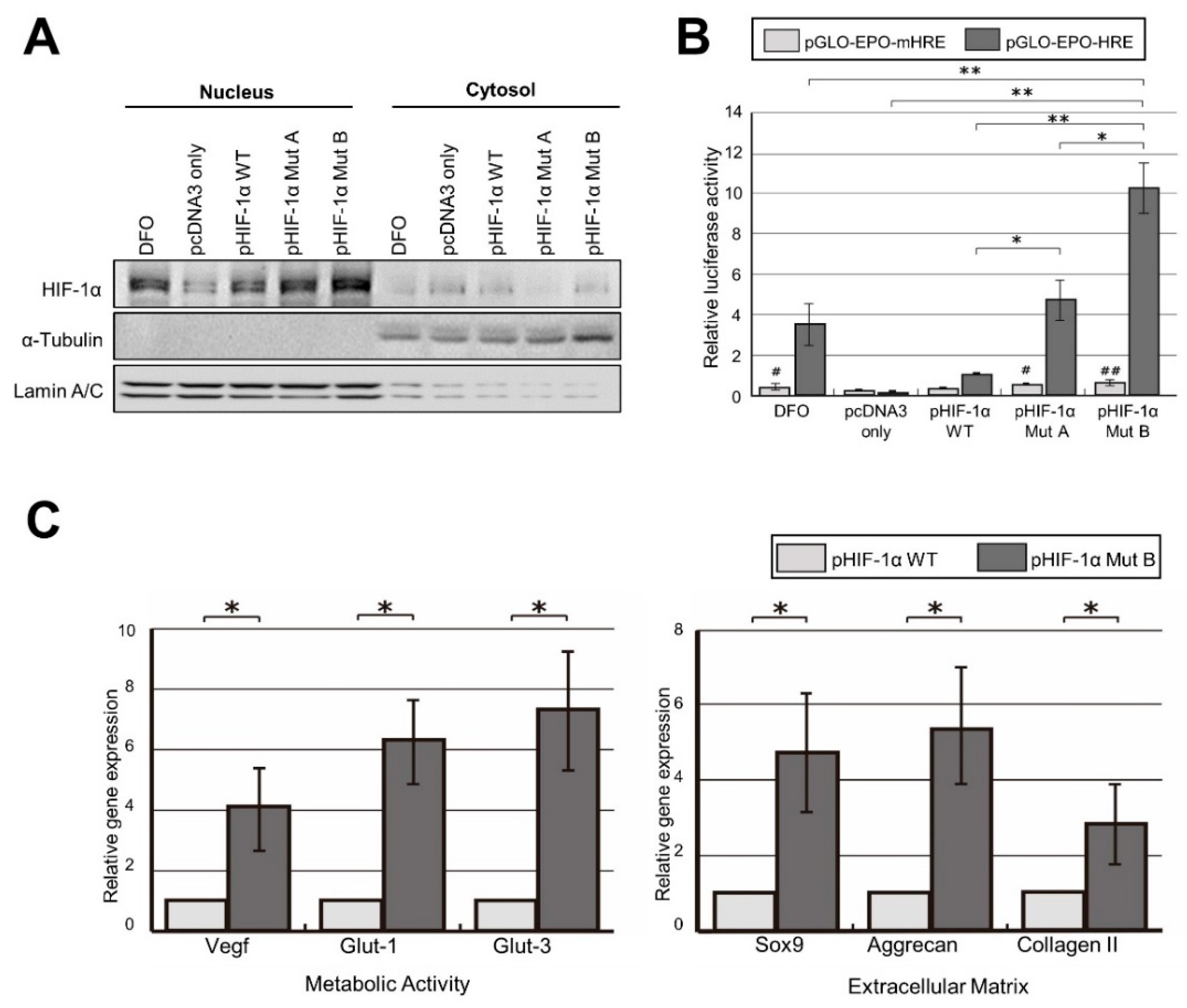
2.2. Robust Transcriptional Activity of pHIF-1α Mut
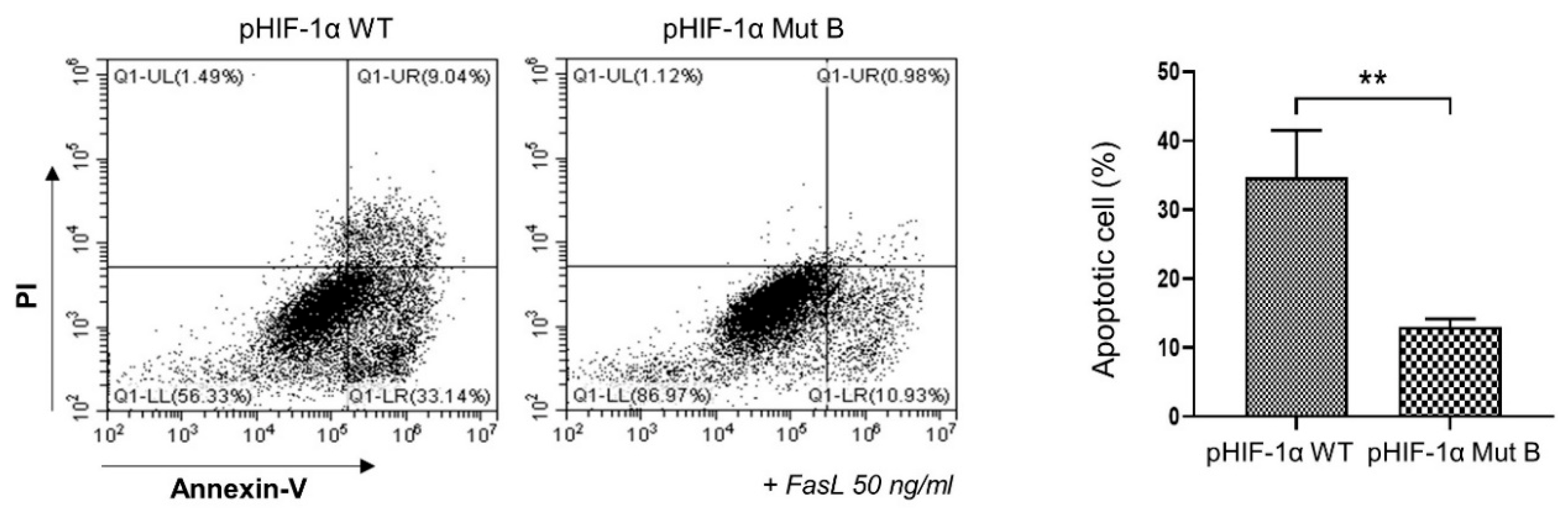
2.3. CA HIF-1α Decreased the Apoptosis of NP Cells
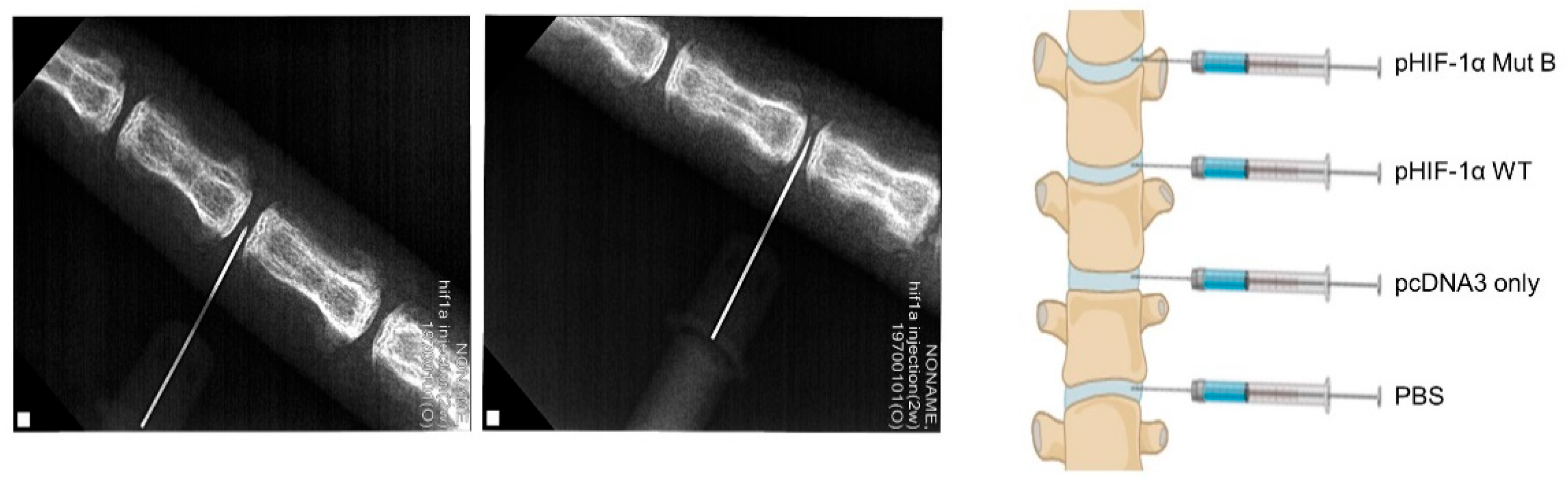
2.4. Radiographic and MRI Assessment, and Histological Evaluation of CA HIF-1α
3. Discussion
4. Materials and Methods
4.1. Construction of Mutant HIF-1α
4.2. Validation of HIF-1α Mut by Western Blot Analysis
4.3. Validation of pHIF-1α Mut by Luciferase Reporter Assay
4.4. Patients and Tissue Samples
4.5. Human NP Cell Isolation and Cultures
4.6. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
4.7. Preparation of Subcellular Fractions
4.8. Western Blotting
4.9. Apoptosis Assay
4.10. Preparation of Animals
4.11. Surgical Procedure
4.12. Radiographic and MRI Evaluation
4.13. Histological Analysis
4.14. Immunohistochemical Staining
4.15. Statistical Analyses
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Richardson, S.M.; Mobasheri, A.; Freemont, A.J.; Hoyland, J.A. Intervertebral disc biology, degeneration and novel tissue engineering and regenerative medicine therapies. Histol. Histopathol. 2007, 22, 1033–1041. [Google Scholar] [CrossRef] [PubMed]
- Peterson, C.K.; Bolton, J.E.; Wood, A.R. A cross-sectional study correlating lumbar spine degeneration with disability and pain. Spine 2000, 25, 218–223. [Google Scholar] [CrossRef] [PubMed]
- Stokes, I.A.; Iatridis, J.C. Mechanical conditions that accelerate intervertebral disc degeneration: Overload versus immobilization. Spine 2004, 29, 2724–2732. [Google Scholar] [CrossRef] [PubMed]
- Battié, M.C.; Videman, T. Lumbar disc degeneration: Epidemiology and genetics. J. Bone Jt. Surg. Am. 2006, 88 (Suppl. 2), 3–9. [Google Scholar] [CrossRef]
- Urban, J.P.; Smith, S.; Fairbank, J.C. Nutrition of the intervertebral disc. Spine 2004, 29, 2700–2709. [Google Scholar] [CrossRef]
- Zhao, C.Q.; Wang, L.M.; Jiang, L.S.; Dai, L.Y. The cell biology of intervertebral disc aging and degeneration. Ageing Res. Rev. 2007, 6, 247–261. [Google Scholar] [CrossRef]
- Maroudas, A.; Stockwell, R.A.; Nachemson, A.; Urban, J. Factors involved in the nutrition of the human lumbar intervertebral disc: Cellularity and diffusion of glucose in vitro. J. Anat. 1975, 120, 113–130. [Google Scholar]
- Ohshima, H.; Urban, J.P. The effect of lactate and pH on proteoglycan and protein synthesis rates in the intervertebral disc. Spine 1992, 17, 1079–1082. [Google Scholar] [CrossRef]
- Repanti, M.; Korovessis, P.G.; Stamatakis, M.V.; Spastris, P.; Kosti, P. Evolution of disc degeneration in lumbar spine: A comparative histological study between herniated and postmortem retrieved disc specimens. J. Spinal Disord. 1998, 11, 41–45. [Google Scholar] [CrossRef]
- Bibby, S.R.; Fairbank, J.C.; Urban, M.R.; Urban, J.P. Cell viability in scoliotic discs in relation to disc deformity and nutrient levels. Spine 2002, 27, 2220–2228, discussion 2227–2228. [Google Scholar] [CrossRef]
- Rajpurohit, R.; Risbud, M.V.; Ducheyne, P.; Vresilovic, E.J.; Shapiro, I.M. Phenotypic characteristics of the nucleus pulposus: Expression of hypoxia inducing factor-1, glucose transporter-1 and MMP-2. Cell Tissue Res. 2002, 308, 401–407. [Google Scholar] [CrossRef]
- Seagroves, T.N.; Ryan, H.E.; Lu, H.; Wouters, B.G.; Knapp, M.; Thibault, P.; Laderoute, K.; Johnson, R.S. Transcription factor HIF-1 is a necessary mediator of the pasteur effect in mammalian cells. Mol. Cell. Biol. 2001, 21, 3436–3444. [Google Scholar] [CrossRef]
- Epstein, A.C.; Gleadle, J.M.; McNeill, L.A.; Hewitson, K.S.; O’Rourke, J.; Mole, D.R.; Mukherji, M.; Metzen, E.; Wilson, M.I.; Dhanda, A.; et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 2001, 107, 43–54. [Google Scholar] [CrossRef]
- Maxwell, P.H.; Wiesener, M.S.; Chang, G.W.; Clifford, S.C.; Vaux, E.C.; Cockman, M.E.; Wykoff, C.C.; Pugh, C.W.; Maher, E.R.; Ratcliffe, P.J. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature 1999, 399, 271–275. [Google Scholar] [CrossRef]
- Mahon, P.C.; Hirota, K.; Semenza, G.L. FIH-1: A novel protein that interacts with HIF-1alpha and VHL to mediate repression of HIF-1 transcriptional activity. Genes Dev. 2001, 15, 2675–2686. [Google Scholar] [CrossRef]
- Kim, J.W.; Jeon, N.; Shin, D.E.; Lee, S.Y.; Kim, M.; Han, D.H.; Shin, J.Y.; Lee, S. Regeneration in Spinal Disease: Therapeutic Role of Hypoxia-Inducible Factor-1 Alpha in Regeneration of Degenerative Intervertebral Disc. Int. J. Mol. Sci. 2021, 22, 5281. [Google Scholar] [CrossRef]
- Lando, D.; Peet, D.J.; Gorman, J.J.; Whelan, D.A.; Whitelaw, M.L.; Bruick, R.K. FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev. 2002, 16, 1466–1471. [Google Scholar] [CrossRef]
- Chen, S.; Fang, X.Q.; Wang, Q.; Wang, S.W.; Hu, Z.J.; Zhou, Z.J.; Xu, W.B.; Wang, J.Y.; Qin, A.; Fan, S.W. PHD/HIF-1 upregulates CA12 to protect against degenerative disc disease: A human sample, in vitro and ex vivo study. Lab. Investig. 2016, 96, 561–569. [Google Scholar] [CrossRef][Green Version]
- Fujita, N.; Hirose, Y.; Tran, C.M.; Chiba, K.; Miyamoto, T.; Toyama, Y.; Shapiro, I.M.; Risbud, M.V. HIF-1-PHD2 axis controls expression of syndecan 4 in nucleus pulposus cells. FASEB J. 2014, 28, 2455–2465. [Google Scholar] [CrossRef]
- Zhang, H.; La Marca, F.; Hollister, S.J.; Goldstein, S.A.; Lin, C.Y. Developing consistently reproducible intervertebral disc degeneration at rat caudal spine by using needle puncture. J. Neurosurg. Spine 2009, 10, 522–530. [Google Scholar] [CrossRef]
- Zou, F.; Jiang, J.; Lu, F.; Ma, X.; Xia, X.; Wang, L.; Wang, H. Efficacy of intradiscal hepatocyte growth factor injection for the treatment of intervertebral disc degeneration. Mol. Med. Rep. 2013, 8, 118–122. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Li, J.; Teng, J.; Liu, Y.; Zhang, D.; Liu, L.; Zhang, W. microRNA-155-3p attenuates intervertebral disc degeneration via inhibition of KDM3A and HIF1α. Inflamm. Res. 2021, 70, 297–308. [Google Scholar] [CrossRef] [PubMed]
- Guillemin, K.; Krasnow, M.A. The hypoxic response: Huffing and HIFing. Cell 1997, 89, 9–12. [Google Scholar] [CrossRef]
- Jiang, B.H.; Zheng, J.Z.; Leung, S.W.; Roe, R.; Semenza, G.L. Transactivation and inhibitory domains of hypoxia-inducible factor 1alpha. Modulation of transcriptional activity by oxygen tension. J. Biol. Chem. 1997, 272, 19253–19260. [Google Scholar] [CrossRef]
- Hu, C.J.; Sataur, A.; Wang, L.; Chen, H.; Simon, M.C. The N-terminal transactivation domain confers target gene specificity of hypoxia-inducible factors HIF-1alpha and HIF-2alpha. Mol. Biol. Cell 2007, 18, 4528–4542. [Google Scholar] [CrossRef]
- Luo, Y.; Jiang, C.; Belanger, A.J.; Akita, G.Y.; Wadsworth, S.C.; Gregory, R.J.; Vincent, K.A. A constitutively active hypoxia-inducible factor-1alpha/VP16 hybrid factor activates expression of the human B-type natriuretic peptide gene. Mol. Pharmacol. 2006, 69, 1953–1962. [Google Scholar] [CrossRef]
- Patel, T.H.; Kimura, H.; Weiss, C.R.; Semenza, G.L.; Hofmann, L.V. Constitutively active HIF-1alpha improves perfusion and arterial remodeling in an endovascular model of limb ischemia. Cardiovasc. Res. 2005, 68, 144–154. [Google Scholar] [CrossRef]
- Roberts, S.; Caterson, B.; Menage, J.; Evans, E.H.; Jaffray, D.C.; Eisenstein, S.M. Matrix metalloproteinases and aggrecanase: Their role in disorders of the human intervertebral disc. Spine 2000, 25, 3005–3013. [Google Scholar] [CrossRef]
- Boos, N.; Nerlich, A.G.; Wiest, I.; von der Mark, K.; Aebi, M. Immunolocalization of type X collagen in human lumbar intervertebral discs during ageing and degeneration. Histochem. Cell Biol. 1997, 108, 471–480. [Google Scholar] [CrossRef]
- Inkinen, R.I.; Lammi, M.J.; Lehmonen, S.; Puustjärvi, K.; Kääpä, E.; Tammi, M.I. Relative increase of biglycan and decorin and altered chondroitin sulfate epitopes in the degenerating human intervertebral disc. J. Rheumatol. 1998, 25, 506–514. [Google Scholar]
- Le Maitre, C.L.; Freemont, A.J.; Hoyland, J.A. Localization of degradative enzymes and their inhibitors in the degenerate human intervertebral disc. J. Pathol. 2004, 204, 47–54. [Google Scholar] [CrossRef]
- Sive, J.I.; Baird, P.; Jeziorsk, M.; Watkins, A.; Hoyland, J.A.; Freemont, A.J. Expression of chondrocyte markers by cells of normal and degenerate intervertebral discs. Mol. Pathol. 2002, 55, 91–97. [Google Scholar] [CrossRef]
- Antoniou, J.; Steffen, T.; Nelson, F.; Winterbottom, N.; Hollander, A.P.; Poole, R.A.; Aebi, M.; Alini, M. The human lumbar intervertebral disc: Evidence for changes in the biosynthesis and denaturation of the extracellular matrix with growth, maturation, ageing, and degeneration. J. Clin. Investig. 1996, 98, 996–1003. [Google Scholar] [CrossRef]
- Bell, D.M.; Leung, K.K.; Wheatley, S.C.; Ng, L.J.; Zhou, S.; Ling, K.W.; Sham, M.H.; Koopman, P.; Tam, P.P.; Cheah, K.S. SOX9 directly regulates the type-II collagen gene. Nat. Genet. 1997, 16, 174–178. [Google Scholar] [CrossRef]
- Huang, W.; Zhou, X.; Lefebvre, V.; de Crombrugghe, B. Phosphorylation of SOX9 by cyclic AMP-dependent protein kinase A enhances SOX9’s ability to transactivate a Col2a1 chondrocyte-specific enhancer. Mol. Cell. Biol. 2000, 20, 4149–4158. [Google Scholar] [CrossRef]
- Wegner, M. From head to toes: The multiple facets of Sox proteins. Nucleic Acids Res. 1999, 27, 1409–1420. [Google Scholar] [CrossRef]
- Park, J.B.; Chang, H.; Kim, K.W. Expression of Fas ligand and apoptosis of disc cells in herniated lumbar disc tissue. Spine 2001, 26, 618–621. [Google Scholar] [CrossRef]
- Richardson, S.M.; Walker, R.V.; Parker, S.; Rhodes, N.P.; Hunt, J.A.; Freemont, A.J.; Hoyland, J.A. Intervertebral disc cell-mediated mesenchymal stem cell differentiation. Stem Cells 2006, 24, 707–716. [Google Scholar] [CrossRef]
- Nerlich, A.G.; Boos, N.; Wiest, I.; Aebi, M. Immunolocalization of major interstitial collagen types in human lumbar intervertebral discs of various ages. Virchows Arch. 1998, 432, 67–76. [Google Scholar] [CrossRef]
- Singleton, R.S.; Trudgian, D.C.; Fischer, R.; Kessler, B.M.; Ratcliffe, P.J.; Cockman, M.E. Quantitative mass spectrometry reveals dynamics of factor-inhibiting hypoxia-inducible factor-catalyzed hydroxylation. J. Biol. Chem. 2011, 286, 33784–33794. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, D.; Wang, X.; Yao, X.; Ye, C.; Zhang, S.; Wang, H.; Chang, C.; Xia, H.; Wang, Y.C.; et al. Hypoxia-inducible miR-182 enhances HIF1α signaling via targeting PHD2 and FIH1 in prostate cancer. Sci. Rep. 2015, 5, 12495. [Google Scholar] [CrossRef] [PubMed]
- Pfirrmann, C.W.; Metzdorf, A.; Zanetti, M.; Hodler, J.; Boos, N. Magnetic resonance classification of lumbar intervertebral disc degeneration. Spine 2001, 26, 1873–1878. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, A.H.; Hwang, D.; Ryan, D.A.; Freeman, A.K.; Kim, H. Degenerative anular changes induced by puncture are associated with insufficiency of disc biomechanical function. Spine 2009, 34, 998–1005. [Google Scholar] [CrossRef] [PubMed]
- Issy, A.C.; Castania, V.; Castania, M.; Salmon, C.E.; Nogueira-Barbosa, M.H.; Bel, E.D.; Defino, H.L. Experimental model of intervertebral disc degeneration by needle puncture in Wistar rats. Braz. J. Med. Biol. Res. 2013, 46, 235–244. [Google Scholar] [CrossRef]
- Nakayama, E.; Matsumoto, T.; Kazama, T.; Kano, K.; Tokuhashi, Y. Transplantation of dedifferentiation fat cells promotes intervertebral disc regeneration in a rat intervertebral disc degeneration model. Biochem. Biophys. Res. Commun. 2017, 493, 1004–1009. [Google Scholar] [CrossRef]
- Sakai, D.; Mochida, J.; Iwashina, T.; Hiyama, A.; Omi, H.; Imai, M.; Nakai, T.; Ando, K.; Hotta, T. Regenerative effects of transplanting mesenchymal stem cells embedded in atelocollagen to the degenerated intervertebral disc. Biomaterials 2006, 27, 335–345. [Google Scholar] [CrossRef]
- Masuda, K.; Aota, Y.; Muehleman, C.; Imai, Y.; Okuma, M.; Thonar, E.J.; Andersson, G.B.; An, H.S. A novel rabbit model of mild, reproducible disc degeneration by an anulus needle puncture: Correlation between the degree of disc injury and radiological and histological appearances of disc degeneration. Spine 2005, 30, 5–14. [Google Scholar] [CrossRef]

| Gene Name | Forward (5′–3′) | Reverse (5′–3′) |
|---|---|---|
| GAPDH | TCCCTGAGCTGAACGGGAAG | GGAGGAGTGGGTGTCGCTGT |
| VEGF | CCATGAACTTTCTGCTGTCTT | TCGATCGTTCTGTATCAGTCT |
| Glut-1 | CTTTGTGGCCTTCTTTGAAGT | CCACACAGTTGCTCCACAT |
| Glut-3 | AGGCTCGATGCTGTTCATCT | ACCGGCTTCCTCATTACCTT |
| SOX9 | GCGGAGGAAGTCGGTGAAGA | CCCTCTCGCTTCAGGTCAGC |
| Aggrecan | CACTGTTACCGCCACTTCCC | ACCAGCGGAAGTCCCCTTCG |
| Type II collagen | CTATCTGGACGAAGCAGCTGGCA | ATGGGTGCAATGTCAATGATGG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.-W.; An, H.-J.; Yeo, H.; Jeong, Y.; Lee, H.; Lee, J.; Nam, K.; Lee, J.; Shin, D.-E.; Lee, S. Activation of Hypoxia-Inducible Factor-1α Signaling Pathway Has the Protective Effect of Intervertebral Disc Degeneration. Int. J. Mol. Sci. 2021, 22, 11355. https://doi.org/10.3390/ijms222111355
Kim J-W, An H-J, Yeo H, Jeong Y, Lee H, Lee J, Nam K, Lee J, Shin D-E, Lee S. Activation of Hypoxia-Inducible Factor-1α Signaling Pathway Has the Protective Effect of Intervertebral Disc Degeneration. International Journal of Molecular Sciences. 2021; 22(21):11355. https://doi.org/10.3390/ijms222111355
Chicago/Turabian StyleKim, Jin-Woo, Hyun-Ju An, HyunJeong Yeo, Yunhui Jeong, HyeonHae Lee, Jusung Lee, Kisik Nam, Jongheon Lee, Dong-Eun Shin, and Soonchul Lee. 2021. "Activation of Hypoxia-Inducible Factor-1α Signaling Pathway Has the Protective Effect of Intervertebral Disc Degeneration" International Journal of Molecular Sciences 22, no. 21: 11355. https://doi.org/10.3390/ijms222111355
APA StyleKim, J.-W., An, H.-J., Yeo, H., Jeong, Y., Lee, H., Lee, J., Nam, K., Lee, J., Shin, D.-E., & Lee, S. (2021). Activation of Hypoxia-Inducible Factor-1α Signaling Pathway Has the Protective Effect of Intervertebral Disc Degeneration. International Journal of Molecular Sciences, 22(21), 11355. https://doi.org/10.3390/ijms222111355







