Abstract
Dual-specificity tyrosine phosphorylation-regulated kinases (DYRK1A, 1B, 2-4) and cdc2-like kinases (CLK1-4) belong to the CMGC group of serine/threonine kinases. These protein kinases are involved in multiple cellular functions, including intracellular signaling, mRNA splicing, chromatin transcription, DNA damage repair, cell survival, cell cycle control, differentiation, homocysteine/methionine/folate regulation, body temperature regulation, endocytosis, neuronal development, synaptic plasticity, etc. Abnormal expression and/or activity of some of these kinases, DYRK1A in particular, is seen in many human nervous system diseases, such as cognitive deficits associated with Down syndrome, Alzheimer’s disease and related diseases, tauopathies, dementia, Pick’s disease, Parkinson’s disease and other neurodegenerative diseases, Phelan-McDermid syndrome, autism, and CDKL5 deficiency disorder. DYRKs and CLKs are also involved in diabetes, abnormal folate/methionine metabolism, osteoarthritis, several solid cancers (glioblastoma, breast, and pancreatic cancers) and leukemias (acute lymphoblastic leukemia, acute megakaryoblastic leukemia), viral infections (influenza, HIV-1, HCMV, HCV, CMV, HPV), as well as infections caused by unicellular parasites (Leishmania, Trypanosoma, Plasmodium). This variety of pathological implications calls for (1) a better understanding of the regulations and substrates of DYRKs and CLKs and (2) the development of potent and selective inhibitors of these kinases and their evaluation as therapeutic drugs. This article briefly reviews the current knowledge about DYRK/CLK kinases and their implications in human disease.
1. Introduction
1.1. Protein Phosphorylation, Protein Kinases, Kinase Inhibitors, and Human Disease
Protein phosphorylation is probably one of the most important and most studied mechanism used by cells to regulate their proteins in terms of enzymatic activity, functions, localization, half-life, interactions with other proteins or other ligands, etc. It is also a key mechanism for signal transduction between cells and within cells. Protein phosphorylation occupies a central place in the scientific literature with 337,916 references (as of 1 June 2021). Protein phosphorylation on serine, threonine, and tyrosine residues is carried out by protein kinases, a family of enzymes known as the human kinome, comprising at least 538 members [1,2] divided into tyrosine kinases and serine/threonine kinases (some of the latter are so-called dual specificity, as they also phosphorylate tyrosine residues), histidine kinases, and pseudo-kinases (protein kinases: 573,472 references (as of 1 June 2021), i.e., one article published every 7 min for the last five years). Quite uniquely, four different Nobel Prizes in medicine or physiology have been awarded to this field (1989, 1992, 2000, 2001) (Figure 1).
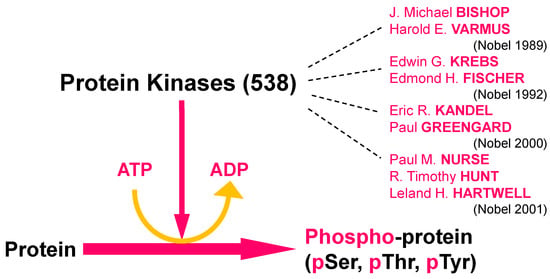
Figure 1.
Four Nobel Prizes in Physiology or Medicine awarded in the field of protein phosphorylation and protein kinases. Protein kinases catalyze the transfer of the γ-phosphate of ATP to the hydroxyl substituents of serine, threonine, or tyrosine residues in proteins, thereby altering the physiological properties of their protein substrates. The human kinome comprises 538 protein kinases. Michael Bishop and Harold E. Varmus received the Nobel Prize 1989 “for their discovery of the cellular origin of retroviral oncogenes” (src, the first described oncogene, which encodes a tyrosine kinase). Edmond H. Fischer and Edwin G. Krebs received the Nobel Prize 1992 “for their discoveries concerning reversible protein phosphorylation as a biological regulatory mechanism” (they are the true discoverers of protein kinases). The Nobel Prize 2000 was awarded jointly to Arvid Carlsson, Paul Greengard, and Eric R. Kandel “for their discoveries concerning signal transduction in the nervous system” (Paul Greengard investigated the mechanism of signal transduction of neurotransmitters in the central nervous system and demonstrated the key importance of phosphorylation by kinases such as CDK5, PKA, CK1, and CK2 and Eric Kandel the importance of PKA in memory in Aplysia). The Nobel Prize 2001 was awarded jointly to Leland H. Hartwell, Tim Hunt, and Paul M. Nurse “for their discoveries of key regulators of the cell cycle” (using yeast or sea urchin embryos, they discovered how the cell division cycle is regulated by CDKs). For more information on each of these awardees, see: https://www.nobelprize.org/prizes/medicine/ (accessed on 1 June 2021).
Since protein phosphorylation is involved in essentially all physiological events, abnormal phosphorylation is implicated in many human diseases. Abnormally expressed or abnormally active kinases represent the most frequent situation. Consequently, inhibiting disease-relevant kinases or normalizing their activities constitutes a rational approach to tackle numerous diseases. This is why protein kinases have become, in a few decades after their initial discovery [3], the first therapeutic targets—before G-protein-coupled receptors–in the pharmaceutical industry’s search for novel drug candidates (reviews: [2,4,5,6,7,8]). As of early February 2021, 62 kinase inhibitors have reached the market, mostly for the treatment of various cancer indications [9,10,11].
1.2. DYRKs and CLKs: Structure, Activation, Interactors, and Substrates
Among serine/threonine kinases, DYRKs and CLKs (Figure 2, Figure 3 and Figure 4) belong to a family of 62 kinases known as the CMGC group, which also includes mitogen-activated protein kinases (MAPKs), cyclin-dependent kinases (CDKs), and the glycogen synthase kinases 3 (GSK3) family. DYRKs and CLKs are two highly related and conserved kinase families (Table 1), usually sensitive to the same pharmacological inhibitors. The DYRK family comprises 5 members: DYRK1A and DYRK1B (class 1 DYRKs) and DYRK2, 3, and 4 (class 2 DYRKs) (reviews: [12,13,14]). The CLK family comprises 4 members: CLK1, 2, 3, and 4 (review: [15]).
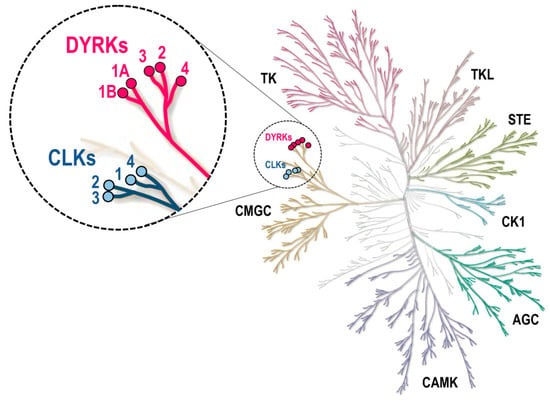
Figure 2.
DYRKs and CLKs within the human kinome phylogenetic tree. DYRK and CLK family members are highlighted with pink and blue circles, respectively. Kinome tree: courtesy of Cell Signaling Technology, Inc. (Danvers, MA, USA, www.cellsignal.com, accessed on 1 June 2021). AGC, cAMP-dependent protein kinase (PKA), cGMP-dependent protein kinase (PKG), and protein kinase C (PKC) families; CAMK, Ca2+/calmodulin-dependent kinases; CK1, casein kinases 1; CMGC, cyclin-dependent kinases (CDKs), mitogen-activated protein kinases (MAPK), glycogen synthase kinases (GSK3), dual-specificity, tyrosine phosphorylation-regulated kinases (DYRKs) and Cdc2-like kinases (CLKs); STE, homologs of yeast STE20 (MAP4K), STE11 (MAP3K), and STE7 (MAP2K) kinases; TK, tyrosine kinases; TKL, tyrosine kinase-like kinases.
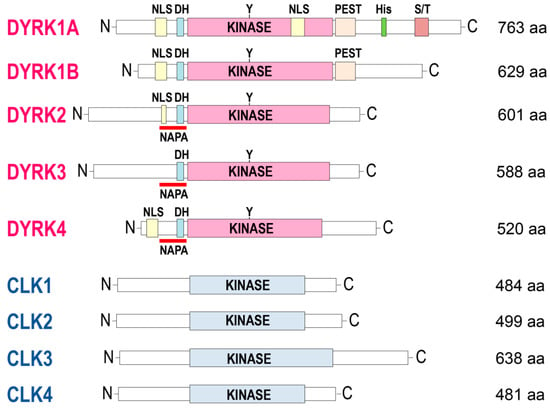
Figure 3.
Comparison of DYRKs and CLKs overall structures. Schematic representation of the canonical protein sequences of human CLKs and DYRKs (extracted from uniprot.org). NB: a NLS in DYRK4 is only found in isoform 4, the canonical sequence being isoform 5. aa, amino acids; C, C-terminal; DH, DYRK homology box; His, His domain (13 consecutive histidine residues region); KINASE, kinase domain; N, N-terminal; NAPA, N-terminal autophosphorylation accessory domain; NLS, nuclear localization signals domain; PEST, proline (P), glutamic acid (E), serine (S), and threonine (T)-rich domain; S/T, serine, and threonine-enriched domain; Y, Tyrosine residue autophosphorylated by DYRKs within the activation loop.

Table 1.
Sequence comparison of human CLK and DYRK family members. Numbers indicate percentage sequence identity and similarity among the nine kinase domains. Sequences were obtained from UniProtKB, and % of similarity and identity were calculated using BlastP (https://blast-ncbi-nlm-nih-gov.cov) (accessed on 1 June 2021).
Alignment of DYRKs and CLKs sequences shows the classical central kinase catalytic domain flanked by N-terminal and C-terminal extensions (Figure 3 and Figure 4). The N-terminal domain of all DYRKs displays a conserved DYRK homology box (DH) [16] that contributes to autophosphorylation of a conserved tyrosine in the kinase domain (Tyr321 in DYRK1A) during maturation of the kinase [17,18]. Autophosphorylation on the tyrosine residue is preceded by hydroxylation of a proline residue by the PHD1 prolyl hydroxylase, an absolute requirement for catalytic activation of the kinase [19]. The N-terminal domain of all DYRKs except DYRK3 contains a nuclear localization signal domain (NLS) [20]. DYRK2, DYRK3, and DYRK4 contain a conserved N-terminal autophosphorylation accessory (NAPA) domain essential for autophosphorylation of the activation loop tyrosine [21]. The C-terminal domain of DYRK1A and DYRK1B displays a region enriched in proline, glutamic acid, serine, and threonine known as a PEST sequence, which favors rapid degradation [22]. A region containing 13 consecutive histidine residues is present in the C-terminal region of DYRK1A but not in other DYRKs or CLKs. A comprehensive analysis of the human proteome revealed that only 86 proteins display such a histidine repeat stretch (5 or more histidines) [23]. The presence of a homopolymeric histidine repeat in nuclear proteins appears to be involved in the targeting/localization of these proteins to the nuclear speckles compartment. Many of these polyhistidine sequence-bearing proteins are expressed in the nervous system [23]. The unique polyhistidine sequence provides a natural His-tag which allows the purification/enrichment of DYRK1A using immobilized metal-affinity chromatography (IMAC) (nickel, cobalt) [24,25] [Sévère et al., unpublished]. DYRKs and CLKs have been highly conserved throughout evolution, and orthologs are found in yeast [26,27], plants [28,29,30,31,32], unicellular algae [33,34], and unicellular parasites such as Trypanosoma [35,36,37], Leishmania [38,39,40], and Plasmodium [41,42,43,44,45,46].
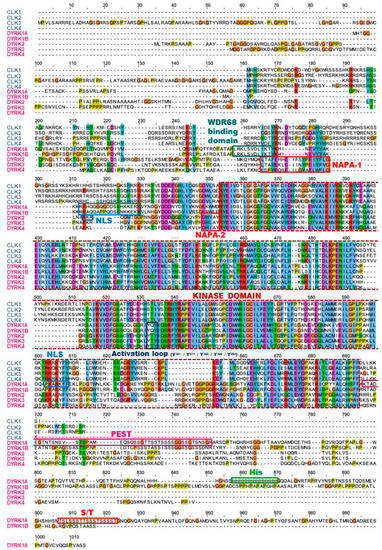
Figure 4.
Sequence alignment of human DYRKs and CLKs. Multiple sequence alignment of the canonical sequences of DYRK and CLK members was performed using Clustal Omega [41] (https://www.ebi.ac.uk) (accessed on 12 April 2021) and edited using Jalview [42]. Each residue in the alignment is assigned a colour if the amino acid profile of the alignment at that position meets some minimum criteria specific for the residue type (Clustal X Colour Scheme, http://www.jalview.org/help/html/colourSchemes/clustal.html) (accessed on 12 April 2021). Distinct sequences are indicated: Activation loop and tyrosine residue that is autophosphorylated (Yn); DH, DYRK homology box; His domain, 13 consecutive histidine residues region; kinase domain; NAPA, N-terminal autophosphorylation accessory domain; NLS, nuclear localization signal domain (NB: a NLS sequence is only found in isoform 4 of DYRK4, not in the canonical sequence); PEST, proline (P), glutamic acid (E), serine (S), and threonine (T)-rich domain; S/T, serine, and threonine-enriched domain; WDR68 binding domain.
Crystal structures of various DYRKs and CLKs, alone or in complex with inhibitors, have been solved (Table 2). These structures have allowed a detailed understanding of the mechanism of activation of DYRKs by autophosporylation on the tyrosine residue as well as an understanding of the binding mode of numerous inhibitors, providing very useful information for the structure-guided synthesis of improved pharmacological inhibitors.

Table 2.
Crystal structures of DYRKs and CLKs alone or in complex with inhibitors.
Table 2.
Crystal structures of DYRKs and CLKs alone or in complex with inhibitors.
| Kinase | Ligand | PDB | Reference |
|---|---|---|---|
| DYRK1A | DJM2005 | 2VX3, 2WO6 | [18] |
| Leucettine L41 | 4AZE | [43] | |
| Harmine | 3ANR | [44] | |
| INDY | 3ANQ | [44] | |
| Compounds 3 and 23 | 4MQ1, 4MQ2 | [45] | |
| LDN-211898 | 5AIK | Elkins, unpublished | |
| PKC412 | 4NCT | [46] | |
| Inhibitor 5t, 5s | 4YLL, 4YLK | [47] | |
| Compound 32, 14 | 6A1G, 6A1F | [48] | |
| XMD7-112, JWD-065 | 6EJ4, 6EIV | [49] | |
| [b]-annulated chloro-substituted indole 13 | 4YLJ | [50] | |
| KuFal319 | 6T6A | [50] | |
| AnnH75 | 4YU2 | [51] | |
| compound 2-2 (harmine derivative) | 6UWY | [52] | |
| GNF2133 | 6UIP | [53] | |
| DJM2005 (DB07608) | 2WO6 | [18] | |
| DYRK2 | - | 3K2L | [18] |
| Leucettine L41 | 4AZF | [43] | |
| Indirubin 6i | 3KVW | [54] | |
| EHT 5372, EHT 1610 | 5LXC, 5LXD | [55] | |
| DYRK3 | Harmine | 5Y86 | [56] |
| CLK1 | - | 6TW2 | [57] |
| compounds 8g, 16 | 6FT8, 6FT9 | [58] | |
| debromohymenialdisine | 1Z57 | [59] | |
| KH-CB19 | 2VAG | [60,61] | |
| Pyrido [3, 4-G] quinazolines 13, 14 | 5J1V, 5J1W | [62] | |
| Compound 25 | 5X8I | [63] | |
| CX-4945 | 6KHD | [64] | |
| CX-4945 | 6FYO | [65] | |
| Compounds 9m, 10i | 6Q8P, 6Q8K | [66] | |
| 5-iodotubercidin | 6G33 | [67] | |
| furanopyrimidines VN412, VN316, VN345 | 6I5H, 6I5L, 6I5K | [68] | |
| ETH1610 (Cpd 17) | 6YTI | [69] | |
| KH-CARB13 (Cpd 3) | 6YTG | [69] | |
| Tg003 (Cpd 2) | 6YTE | [69] | |
| GW807982X (Cpd 8) | 6ZLN | [69] | |
| imidazopyridazine (Cpd 1) | 6YTA | [69] | |
| CAF052 | 7AK3 | [70] | |
| TbCLK1 | AB1 | 6Q2A | [40] |
| CLK2 | 1RO, NR9 | 3NR9 | Knapp, unpublished |
| CX-4945 | 6KHE | [64] | |
| CX-4945 | 6FYL | [65] | |
| CLK3 | - | 2EU9, 2EXE | [59] |
| KH-CB19 | 2WU7 | [60] | |
| K00546 | 2WU6 | [60] | |
| Leucettine L41 | 3RAW | [71] | |
| CX-4945 | 6KHF | [64] | |
| CX-4945 | 6FYP | [65] | |
| KH-CARB13 (Cpd 3) | 6YU1 | [69] | |
| Tg003 | 6YTW | [70] | |
| compound 8a | 6FT7 | [58] | |
| CLK4 | CX-4945 | 6FYV | [65] |
The nuclear interactome of DYRK1A is highly enriched in DNA damage repair factors (RNF169), transcriptional elongation factors, and E3 ubiquitin ligases [72,73,74]. The interactome of all CMGC kinases, including DYRKs and CLKs, has been extensively studied [75]. Other large-scale interactome studies provide information on proteins binding to DYRKs and CLKs [76,77]. A detailed description of the DYRKs and CLKs interactomes is beyond the scope of this review. However, we would like to mention WDR68, also known as DCAF7 (DDB1-associated and CUL4-associated factor 7) or HAN11 (Human homolog of the Petunia hybrida an11 gene), a scaffolding protein of the WD40-repeat protein family [78] that binds class 1 DYRKs and HIPK2 (Homeodomain-interacting protein kinase 2). The interaction between WDR68 and DYRK1A/DYRK1B has been extensively studied [79,80,81]: it involves a conserved 12 amino acid sequence located in the N-terminal domain of DYRK1A/1B. This interaction mediates binding to other proteins, such as the adenovirus E1A oncoprotein [81] and RNA polymerase II [82], thereby probably favoring substrate recruitment for DYRK1A/1B and HIPK2. WDR68 is essential for craniofacial development, a process involving DYRK1A [83,84]. DYRK1A regulates the interaction between WDR68 and Huntington-associated protein 1 (Hap1), which may contribute to postnatal growth retardation in Down syndrome (DS) [85]. Expression of WDR68 regulates the level of expression of DYRK1A and DYRK1B [86].
DYRK and CLK kinases phosphorylate many substrates involved in signaling pathways, mRNA splicing, chromatin transcription, DNA damage repair, cell survival, cell cycle control, differentiation, homocysteine/methionine/folate regulation, endocytosis, neuronal development and functions, synaptic plasticity, etc. Reviewing substrates and cellular functions of all DYRKs and CLKs is beyond the scope of this brief review, although phosphorylation of substrates and their cellular and physiological consequences underlie normal functioning and pathological conditions.
2. DYRKs and Human Disease
There is growing evidence for the involvement of various DYRKs in human disease. We will briefly review these accumulating data (Table 3 and Figure 5A).
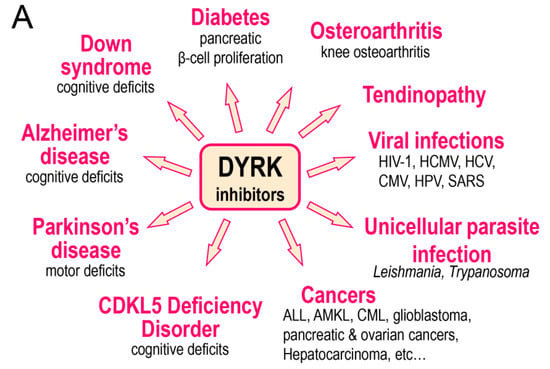
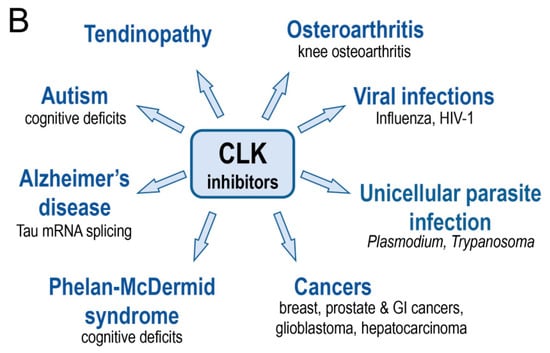
Figure 5.
DYRK and CLK inhibitors and their potential use. (A) DYRK inhibitors (in particular inhibitors of DYRK1A) have been investigated in the indicated diseases. (B) CLK inhibitors (in particular inhibitors of CLK1) have been investigated in the indicated diseases.
2.1. DYRK1A and Down Syndrome (DS)
The gene encoding DYRK1A is located on chromosome 21, within the Down syndrome critical region (DSCR), the triploidy of which is responsible for most DS-associated deficiencies (reviews: [13,14]) (Table 3 for more details). There is considerable genetical and pharmacological evidence showing that the mere 1.5-fold overexpression of DYRK1A is responsible for most cognitive deficits observed in DS patients (reviews: [14,87,88,89,90,91,92]). Genetical normalization of DYRK1A levels or pharmacological inhibition of its catalytic activity restores cognitive functions (Table 3 for specific references). The development of pharmacological inhibitors of DYRK1A is a major avenue for the treatment of cognitive deficits associated with DS (and Alzheimer’s disease) (reviews: [88,89,93]).
2.2. DYRK1A and Alzheimer’s Disease (AD)
There is mounting evidence for a role of DYRK1A in the onset of AD (reviews: [14,88,94,95]) (Table 3 for more details). DYRK1A phosphorylates key substrates involved in AD and dementia: Tau, septin 4, amyloid precursor protein (APP), presenilin 1, neprilysin, Munc18-1, α-synuclein, RCAN1, and β-tubulin. By modulating alternative splicing of Tau exon 10, DYRK1A favors the production of the 3R-Tau splice isoform (characteristic for DS/AD/tauopathy) over the 4R-Tau isoform [96,97,98]. Inhibition of DYRK1A and possibly of other DYRKs and CLKs promotes autophagy, which could counterbalance the autophagy deficit seen in AD.
2.3. DYRK1A and Parkinson’s Disease (PD)
Genome-wide association studies (GWAS) have revealed that DYRK1A is a risk factor for PD [99]. DYRK1A phosphorylates key factors for PD such as parkin, septin 4, and α-synuclein. Upregulation of micro-RNAs specific for PD targets DYRK1A expression [100]. There is further evidence that DYRK1A expression is increased in PD and in Pick’s disease [101].
2.4. DYRK1A and Mental Retardation Disease 7 (MRD7)
Haploinsufficiency of the DYRK1A gene, due to various truncation mutations, microdeletions, or missense variants resulting in reduced DYRK1A, is responsible for MRD7, an autism spectrum disorder displaying microcephaly, intellectual disability, speech impairment, and distinct facies (reviews: [91,102,103,104]).
2.5. DYRK1A and Viral Infections
DYRK1A and DYRK1B are utilized during human cytomegalovirus (HCMV) placental replication. Inhibition of DYRKs prevent replication of various viruses, including hepatitis C virus (HCV), human cytomegalovirus (HCMV), human immunodeficiency virus type 1 (HIV-1), and herpes simplex virus 1 (HSV-1) (Table 3 for more details).
2.6. DYRK1A and Diabetes
There is a growing body of evidence showing that DYRK1A/1B inhibitors induce the proliferation of insulin-producing pancreatic β-cells, making DYRK1A/1B kinases attractive therapeutic targets for β cell regeneration for both type 1 and type 2 diabetes [105,106] (Table 3 for more details).
2.7. DYRK1A and Cancers and Leukemias
There is abundant literature linking DYRK1A with solid cancers and leukemias (reviews: [107,108,109]). The most prominent examples are pancreatic cancer, brain tumor, acute megakaryoblastic leukemia (AMKL) [110], and acute lymphoblastic leukemia (ALL) [111] (Table 3 for more details). DYRK1A regulates DNA damage response [72,74]. In some situations, DYRK1A appears to function as a tumor-suppressor protein [112,113,114].
2.8. Other DYRKs and Human Disease
DYRK1B is involved in the replication of various viruses including HCV, Chikungunya virus, Dengue virus, SARS coronavirus, HCMV, and human papillomavirus (HPV). Like with DYRK1A, DYRK1B inhibition leads to the proliferation of pancreatic, insulin-producing β-cells. DYRK1B is involved in neuroinflammation [115]. Targeting DYRK1B provides a new rationale for treatment of various solid cancers such as liposarcoma or breast cancers (reviews: [116,117]) as well as in chronic myeloid leukemia (CML).
DYRK2, in association with GSK-3β, regulates neuronal morphogenesis [118]. DYRK2 is involved in various ways in cancer development (reviews: [119,120]).
DYRK3 promotes hepatocellular carcinoma [121] and glioblastoma [122]. DYRK3 is required for influenza virus replication [123]. DYRK3 couples stress granule condensation/dissolution to mechanistic target of rapamycin complex 1 (mTORC1) signaling [124]. DYRK3 regulates phase transition of membraneless organelles in mitosis [125]. DYRK3 and DYRK4 are involved in the regulation of cytoskeletal organization and process outgrowth in neurons.
DYRK1A decreases axon growth, DYRK3 and DYRK4 increase dendritic branching, and DYRK2 decreases both axon and dendrite growth and branching [126].

Table 3.
DYRKs and human disease. Evidence for causality and beneficial effects of pharmacological treatment by DYRKs inhibitors.
Table 3.
DYRKs and human disease. Evidence for causality and beneficial effects of pharmacological treatment by DYRKs inhibitors.
| Kinase Target | Disease | References |
|---|---|---|
| DYRK1A | Down syndrome (DS) | [127,128,129,130,131,132,133,134,135,136,137,138,139,140,141,142,143,144,145,146,147] |
| DYRK1A | Alzheimer’s disease (AD) and other Taupathies | [96,98,128,129,131,148,149,150,151,152,153,154,155,156,157,158,159,160,161,162,163] |
| DYRK1A | Parkinson’s disease | [99,100,101,131,164,165,166,167,168] |
| DYRK1A | Pick’s disease | [101] |
| DYRK1A | CDKL5 Deficiency Disorder | [169] |
| DYRK1A | Diabetes | [52,53,105,106,170,171,172,173,174,175,176,177,178,179] |
| DYRK1A | Regulation of folate and methionine metabolism | [180] |
| DYRK1A | Cancers (review) | [109] |
| DYRK1A | Glioblastoma | [181] |
| DYRK1A | Head and neck squamous cell carcinoma | [182] |
| DYRK1A | Pancreatic ductal adenocarcinoma | [183,184,185] |
| DYRK1A | Hepatocellular carcinoma | [186] |
| DYRK1A | Ovarian cancer | [187,188] |
| DYRK1A | Acute megakaryoblastic leukemia (AMKL) | [110,189] |
| DYRK1A | Acute lymphoblastic leukemia (ALL) | [111,190,191] |
| DYRK1A | Psoriasis | [192] |
| DYRK1A | Knee osteoarthritis | [193,194] |
| DYRK1A | Tendinopathy | [195] |
| DYRK1A | Human immunodeficiency virus type 1 (HIV-1) | [196,197,198] |
| DYRK1A DYRK1B | Human cytomegalovirus (HCMV) | [199] |
| DYRK1B | Hepatitis C virus (HCV), Chikungunya virus, Dengue virus, and severe acute respiratory syndrome (SARS) coronavirus Cytomegalovirus (CMV) Human papillomavirus (HPV) | [199,200,201] |
| DYRK1B | Diabetes | [105] |
| DYRK1B | Neuroinflammation | [115] |
| DYRK1B | Oral squamous cell carcinoma Liposarcoma Breast cancer Hedgehog/GLI-dependent cancer | [117,202,203,204,205] |
| DYRK2 | Cancers (reviews) | [119,120,206,207] |
| DYRK2 | Triple-negative breast cancer (TNBC) and multiple myeloma (MM) | [208,209] |
| DYRK2 | Lung adenocarcinoma | [210] |
| DYRK2 | Chronic myeloid leukemia (CML) | [211,212] |
| DYRK2 | Gliblastoma | [213] |
| DYRK2 | Colorectal cancer (tumor suppressor) | [214] |
| DYRK2 | Liver cancer (predictive marker) | [215] |
| DYRK2 | Trypanosoma cruzi | [216] |
| DYRK3 | Hepatocellular carcinoma | [121] |
| DYRK3 | Glioblastoma | [122] |
| DYRK3 | Influenza virus replication | [123] |
| DYRK3 | Anemia | [217] |
| DYRK3 | Osteoarthritis | [218] |
| DYRK4 | Breast cancer | [219] |
| DYRKs | Glioblastoma | [220] |
| DYRKs | Herpes virus, rhesus macaque cytomegalovirus, varicella-zoster virus, and herpes simplex virus (HSV-1) | [221] |
| LmDYRK1 | Leishmaniasis | [39] |
| TbDYRK | Trypanosoma brucei (sleeping sickness) | [35,36,37] |
| DYRKs/CLKs | Glioblastoma | [220] |
3. CLKs and Human Disease
The data supporting the involvement of various CLKs in human disease is briefly described below and in Table 4 and Figure 5B.
CLKs play essential functions in alternative splicing. CLKs act as a body-temperature sensors, which globally control alternative splicing and gene expression. The activity of CLKs is indeed highly responsive to physiological temperature changes, which is conferred by structural rearrangements within the kinase activation segment [57].
CLK1 triggers periodic alternative splicing during the cell division cycle [222]. CLK1 regulates influenza A virus mRNA splicing, and its inhibition prevents viral replication. CLK1 and CLK2 also regulate HIV-1 gene expression. CLK1 is an autophagy inducer. CLK1 inhibition may prevent chemoresistance in glioma, and CLK1 inhibition by TG693 allows the skipping of mutated exon 31 of the dystrophin gene in Duchenne Muscular Dystrophy. CLK1 autoregulates itself through exon skipping and intron retention [223].
Inhibition of CLK2 has been proposed as a way to improve neuronal functions and combat intellectual disability and autism in Phelan–McDermid syndrome (PMDS) [65]. Alternative splicing of Tau exon 10 is regulated by CLK2 and other CLKs, leading to changes in the 3R/4R isoform ratio and neurodegeneration in sporadic AD [224,225]. Dual inhibitions of CLK2 and DYRK1A by Lorecivivint (SM04690) and by its analogue SM04755 are potential disease-modifying approaches for knee osteoarthritis [193,194] and for tendinopathy, respectively [195]. CLK2 inhibition compromises MYC-driven breast tumors, triple-negative breast cancer, and glioblastoma. Inhibition of CLK2, CLK3, and/or CLK4 blocks HIV-1 production.
CLK3 contributes to hepatocellular carcinoma [226], prostate cancer [227], and cholangiocarcinoma [228]. CLK3 is abundantly expressed in testis and in spermatozoa.

Table 4.
CLKs and human disease. Evidence for causality and beneficial effects of pharmacological treatment by CLK inhibitors.
Table 4.
CLKs and human disease. Evidence for causality and beneficial effects of pharmacological treatment by CLK inhibitors.
| Kinase Target | Disease | References |
|---|---|---|
| CLK1 | Glioblastoma Small-cell lung cancer | [229] [230] |
| CLK1 | Duchenne muscular dystrophy | [231] |
| CLK1 | Influenza A West Nile and Chikungunya viruses | [232,233,234,235,236] [61] |
| CLK1/CLK2 | Triple-negative breast cancer | [237] |
| CLK2 | HIV-1 | [238] |
| CLK2 | Autism Phelan-McDermid syndrome (PMDS) | [239] [65] |
| CLK2 | Knee osteoarthritis Tendinopathy | [193,194] [195] |
| CLK2 | Breast cancer Triple-negative breast cancer Glioblastoma | [240,241] [242,243] [244,245] |
| CLK2 | Alzheimer’s disease (alternative splicing of Tau exon 10) | [224,225] |
| CLK3 | Hepatocellular carcinoma Prostate cancer Cholangiocarcinoma | [226] [227] [228] |
| CLKs | Body temperature | [57] |
| CLKs | Prostate cancer Gastrointestinal cancer Colorectal, ovarian cancers | [227] [246] [247] |
| PfCLKs | Plasmodium falciparum (malaria) | [248,249,250,251,252,253] |
| Tb CLK1/2 | Trypanosoma brucei (sleeping sickness) | [38,40] |
4. Therapeutic Potential of DYRK and CLK Inhibitors
Abnormal activities in DYRKs and CLKs have motivated numerous groups to search for, optimize, and characterize pharmacological inhibitors of these kinases for their use in various indications (reviews: [88,89,93]) (Figure 5). There is particular interest in the development of DYRKs/CLKs inhibitors as potential drug candidates to address cognitive deficits in DS and AD as well as to increase the pancreatic β-cell mass in both type 1 and type 2 diabetes (review: [106]) or to inhibit several cancers and leukemias by inhibiting cell proliferation. A few representative inhibitors are shown in Figure 6. Most DYRK1A inhibitors also inhibit, to various extent, DYRK1B, 2, 3, and 4 as well as the closely related CLK1, 2, 3, and 4 [93]. Apart from FINDY, which inhibits DYRK1A by interfering with its folding process [254], all reported inhibitors appear to act by competing with ATP in its binding to the catalytic site of the kinases (as demonstrated by enzymological studies as well as by co-crystallization with their kinase targets (Table 2)). Several DYRK1A inhibitors have been reported in recent years (reviews: [88,93,95]) which, like Leucettines and Leucettine L41 in particular, correct cognition deficits in DS and AD animal models [127,128,148].
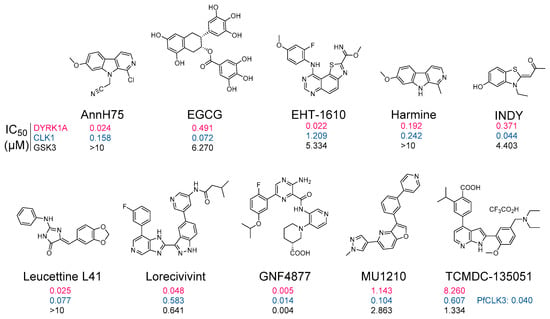
Figure 6.
DYRK and CLK inhibitors. A few representative pharmacological inhibitors: AnnH75 [51], EGCG [255], EHT-1610 [55], Harmine [256], INDY [44], Leucettine L41 [43,257], Lorecivivint [258,259], GNF4877 [170,179], MU1210 [68], and TCMDC-135051 [251,252]. Numbers under each structure indicates IC50 values (expressed in µM) towards DYRK1A, CLK1, and GSK3β (33PanQinase™ assay, Reaction Biology Corp.).
5. Conclusions
The limited studies that have been carried out so far with DYRKs and CLKs have opened up new avenues in our understanding of their regulation and functions. Yet, a great deal of work remains to be done to fully understand the cellular and physiological functions of each member of the DYRK and CLK families. Tissular and cellular distribution, polymorphism and mutations, regulation of expression levels, and post-translational modifications are just a few of the parameters that need to be investigated in detail. Conditional knock-out/knock-in and overexpression models will also contribute to the understanding of the unique roles of each of these kinases and their eventual redundancy. Very precious tools—antibodies, affinity reagents, pharmacological inhibitors, kinase inactive mutants, transgenic animals—have been developed, yet DYRK1A has been mostly studied, and other DYRKs and CLKs will require the development of specific tools.
The currently available data demonstrate major implications of several protein kinases of the DYRK and CLK families in several human diseases. The first inhibitors are reaching regulatory preclinical studies and early clinical studies. The next few years will certainly see the validation of specific DYRKs and CLKs inhibitors for specific clinical indications. It is still a bit early to speculate which one these will be. Clearly though, cognition in DS and AD, diabetes, cancers, and osteoarthritis are the most advanced examples of potential applications, but viral and unicellular parasite infections will certainly gain momentum as potential therapeutic indications for DYRKs/CLKs inhibitors. Higher potency and higher selectivity will also emerge in the near future. We can clearly anticipate that, as fundamental knowledge will accumulate on these protein kinases, more applied pharmaceutical work will result in well characterized, selective, and potent inhibitors leading to significant clinical improvements for patients.
Author Contributions
M.F.L. and L.M. wrote the article and composed the figures. Both authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by grants from the Foundation Jérôme Lejeune and the Agence Nationale pour la Recherche (ANR) (DYRK-DOWN). This project has received funding from the European Union’s Horizon 2020 research and innovation program under grant agreement No 848077. This reflects only the author’s view and the European Commission is not responsible for any use that may be made of the information it contains.
Conflicts of Interest
L. Meijer is a founder of Perha Pharmaceuticals and co-inventor in the Leucettine and Leucettinib patents.
Abbreviations
| AD | Alzheimer’s disease |
| AGC | PKA, PKG, and PKC family |
| ALL | acute lymphoblastic leukemia |
| AMKL | acute megakaryoblastic leukemia |
| APP | amyloid precursor protein |
| CAMK | Ca2+/calmodulin-dependent kinases |
| CDKs | cyclin-dependent kinases |
| CSNK1/CK1 | casein kinases 1 |
| CK2 | casein kinase 2 |
| CLKs | cdc2-like kinases |
| CML | chronic myeloid leukemia |
| DCAF7 | DDB1-associated and CUL4-associated factor 7 |
| DH | DYRK homology box |
| DS | Down syndrome |
| DYRKs | dual specificity, tyrosine phosphorylation regulated kinases |
| GSK3 | glycogen synthase kinase 3 |
| GWAS | genome-wide association studies |
| HAN11 | human homolog of the Petunia hybrida an11 gene |
| Hap1 | Huntington-associated protein 1 |
| HCMV | human cytomegalovirus |
| HCV | hepatitis C virus |
| HIV-1 | human immunodeficiency virus type 1 |
| HIPK2 | Homeodomain-interacting protein kinase 2 |
| HPV | human papillomavirus |
| HSV-1 | herpes simplex virus 1 |
| IMAC | immobilized metal-affinity chromatography |
| MAPKs | mitogen-activated protein kinases |
| MRD7 | mental retardation disease 7 |
| NAPA | N-terminal autophosphorylation accessory domain |
| NLS | nuclear localization signals domain |
| PD | Parkinson’s disease |
| PEST | region enriched in proline (P), glutamic acid (E), serine (S), and threonine (T) residues |
| PKA | cAMP-dependent protein kinase |
| PKC | protein kinase C |
| PKG | cGMP-dependent protein kinase |
| PMDS | Phelan-McDermid syndrome |
| STE | homologs of yeast STE7, STE11, and STE20 kinases |
| TK | tyrosine kinases |
| TKL | tyrosine kinase-like kinases |
| WDR68 | WD40-repeat protein 68 |
References
- Zhang, H.; Cao, X.; Tang, M.; Zhong, G.; Si, Y.; Li, H.; Zhu, F.; Liao, Q.; Li, L.; Zhao, J.; et al. A Subcellular Map of the Human Kinome. eLife 2021, 10. [Google Scholar] [CrossRef]
- Wilson, L.J.; Linley, A.; Hammond, D.E.; Hood, F.E.; Coulson, J.M.; MacEwan, D.J.; Ross, S.J.; Slupsky, J.R.; Smith, P.D.; Eyers, P.A.; et al. New Perspectives, Opportunities, and Challenges in Exploring the Human Protein Kinome. Cancer Res. 2018, 78, 15–29. [Google Scholar] [CrossRef]
- Fischer, E.H.; Krebs, E.G. Conversion of Phosphorylase b to Phosphorylase a in Muscle Extracts. J. Biol. Chem. 1955, 216, 121–132. [Google Scholar] [CrossRef]
- Roskoski, R. A Historical Overview of Protein Kinases and Their Targeted Small Molecule Inhibitors. Pharmacol. Res. 2015, 100, 1–23. [Google Scholar] [CrossRef]
- Ferguson, F.M.; Gray, N.S. Kinase Inhibitors: The Road Ahead. Nat. Rev. Drug Discov. 2018, 17, 353–377. [Google Scholar] [CrossRef]
- Wu, P.; Nielsen, T.E.; Clausen, M.H. Small-Molecule Kinase Inhibitors: An Analysis of FDA-Approved Drugs. Drug Discov. Today 2016, 21, 5–10. [Google Scholar] [CrossRef]
- Klaeger, S.; Heinzlmeir, S.; Wilhelm, M.; Polzer, H.; Vick, B.; Koenig, P.-A.; Reinecke, M.; Ruprecht, B.; Petzoldt, S.; Meng, C.; et al. The Target Landscape of Clinical Kinase Drugs. Science 2017, 358. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Nielsen, T.E.; Clausen, M.H. FDA-Approved Small-Molecule Kinase Inhibitors. Trends Pharmacol. Sci. 2015, 36, 422–439. [Google Scholar] [CrossRef]
- Roskoski, R. Properties of FDA-Approved Small Molecule Protein Kinase Inhibitors. Pharmacol. Res. 2019, 144, 19–50. [Google Scholar] [CrossRef]
- Roskoski, R. Properties of FDA-Approved Small Molecule Protein Kinase Inhibitors: A 2020 Update. Pharmacol. Res. 2020, 152, 104609. [Google Scholar] [CrossRef]
- Roskoski, R. Properties of FDA-Approved Small Molecule Protein Kinase Inhibitors: A 2021 Update. Pharmacol. Res. 2021, 165, 105463. [Google Scholar] [CrossRef]
- Aranda, S.; Laguna, A.; de la Luna, S. DYRK Family of Protein Kinases: Evolutionary Relationships, Biochemical Properties, and Functional Roles. FASEB J. 2011, 25, 449–462. [Google Scholar] [CrossRef]
- Becker, W.; Sippl, W. Activation, Regulation, and Inhibition of DYRK1A. FEBS J. 2011, 278, 246–256. [Google Scholar] [CrossRef]
- Arbones, M.L.; Thomazeau, A.; Nakano-Kobayashi, A.; Hagiwara, M.; Delabar, J.M. DYRK1A and Cognition: A Lifelong Relationship. Pharmacol. Ther. 2019, 194, 199–221. [Google Scholar] [CrossRef]
- Martín Moyano, P.; Němec, V.; Paruch, K. Cdc-Like Kinases (CLKs): Biology, Chemical Probes, and Therapeutic Potential. Int. J. Mol. Sci. 2020, 21, 7549. [Google Scholar] [CrossRef]
- Widowati, E.W.; Bamberg-Lemper, S.; Becker, W. Mutational Analysis of Two Residues in the DYRK Homology Box of the Protein Kinase DYRK1A. BMC Res. Notes 2018, 11, 297. [Google Scholar] [CrossRef]
- Himpel, S.; Panzer, P.; Eirmbter, K.; Czajkowska, H.; Sayed, M.; Packman, L.C.; Blundell, T.; Kentrup, H.; Grötzinger, J.; Joost, H.G.; et al. Identification of the Autophosphorylation Sites and Characterization of Their Effects in the Protein Kinase DYRK1A. Biochem. J. 2001, 359, 497–505. [Google Scholar] [CrossRef] [PubMed]
- Soundararajan, M.; Roos, A.K.; Savitsky, P.; Filippakopoulos, P.; Kettenbach, A.N.; Olsen, J.V.; Gerber, S.A.; Eswaran, J.; Knapp, S.; Elkins, J.M. Structures of Down Syndrome Kinases, DYRKs, Reveal Mechanisms of Kinase Activation and Substrate Recognition. Structure 2013, 21, 986–996. [Google Scholar] [CrossRef]
- Lee, S.B.; Ko, A.; Oh, Y.T.; Shi, P.; D’Angelo, F.; Frangaj, B.; Koller, A.; Chen, E.I.; Cardozo, T.; Iavarone, A.; et al. Proline Hydroxylation Primes Protein Kinases for Autophosphorylation and Activation. Mol. Cell 2020, 79, 376–389.e8. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.-C.; Hsia, K.-C. More than a Zip Code: Global Modulation of Cellular Function by Nuclear Localization Signals. FEBS J. 2020. [Google Scholar] [CrossRef] [PubMed]
- Kinstrie, R.; Luebbering, N.; Miranda-Saavedra, D.; Sibbet, G.; Han, J.; Lochhead, P.A.; Cleghon, V. Characterization of a Domain That Transiently Converts Class 2 DYRKs into Intramolecular Tyrosine Kinases. Sci. Signal 2010, 3, ra16. [Google Scholar] [CrossRef] [PubMed]
- Rechsteiner, M.; Rogers, S.W. PEST Sequences and Regulation by Proteolysis. Trends Biochem. Sci. 1996, 21, 267–271. [Google Scholar] [CrossRef]
- Salichs, E.; Ledda, A.; Mularoni, L.; Albà, M.M.; de la Luna, S. Genome-Wide Analysis of Histidine Repeats Reveals Their Role in the Localization of Human Proteins to the Nuclear Speckles Compartment. PLoS Genet. 2009, 5, e1000397. [Google Scholar] [CrossRef]
- Bornhorst, J.A.; Falke, J.J. Purification of Proteins Using Polyhistidine Affinity Tags. Methods Enzymol. 2000, 326, 245–254. [Google Scholar] [CrossRef]
- Raducanu, V.-S.; Isaioglou, I.; Raducanu, D.-V.; Merzaban, J.S.; Hamdan, S.M. Simplified Detection of Polyhistidine-Tagged Proteins in Gels and Membranes Using a UV-Excitable Dye and a Multiple Chelator Head Pair. J. Biol. Chem. 2020, 295, 12214–12223. [Google Scholar] [CrossRef]
- Kettenbach, A.N.; Deng, L.; Wu, Y.; Baldissard, S.; Adamo, M.E.; Gerber, S.A.; Moseley, J.B. Quantitative Phosphoproteomics Reveals Pathways for Coordination of Cell Growth and Division by the Conserved Fission Yeast Kinase Pom1. Mol. Cell Proteom. 2015, 14, 1275–1287. [Google Scholar] [CrossRef]
- Bhattacharjee, R.; Mangione, M.C.; Wos, M.; Chen, J.-S.; Snider, C.E.; Roberts-Galbraith, R.H.; McDonald, N.A.; Presti, L.L.; Martin, S.G.; Gould, K.L. DYRK Kinase Pom1 Drives F-BAR Protein Cdc15 from the Membrane to Promote Medial Division. Mol. Biol. Cell 2020, 31, 917–929. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Ntui, V.O.; Zhang, N.; Xiong, L. Arabidopsis Yak1 Protein (AtYak1) Is a Dual Specificity Protein Kinase. FEBS Lett. 2015, 589, 3321–3327. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.-Y.; Wu, Y.-C.; Pu, H.-Y.; Wang, Y.; Jang, G.-J.; Wu, S.-H. Plant Dual-Specificity Tyrosine Phosphorylation-Regulated Kinase Optimizes Light-Regulated Growth and Development in Arabidopsis. Plant Cell Environ. 2017, 40, 1735–1747. [Google Scholar] [CrossRef]
- Iwabuchi, K.; Ohnishi, H.; Tamura, K.; Fukao, Y.; Furuya, T.; Hattori, K.; Tsukaya, H.; Hara-Nishimura, I. ANGUSTIFOLIA Regulates Actin Filament Alignment for Nuclear Positioning in Leaves. Plant Physiol. 2019, 179, 233–247. [Google Scholar] [CrossRef]
- Barrada, A.; Djendli, M.; Desnos, T.; Mercier, R.; Robaglia, C.; Montané, M.-H.; Menand, B. A TOR-YAK1 Signaling Axis Controls Cell Cycle, Meristem Activity and Plant Growth in Arabidopsis. Development 2019, 146. [Google Scholar] [CrossRef]
- Forzani, C.; Duarte, G.T.; Van Leene, J.; Clément, G.; Huguet, S.; Paysant-Le-Roux, C.; Mercier, R.; De Jaeger, G.; Leprince, A.-S.; Meyer, C. Mutations of the AtYAK1 Kinase Suppress TOR Deficiency in Arabidopsis. Cell Rep. 2019, 27, 3696–3708.e5. [Google Scholar] [CrossRef]
- Colina, F.; Carbó, M.; Meijón, M.; Cañal, M.J.; Valledor, L. Low UV-C Stress Modulates Chlamydomonas Reinhardtii Biomass Composition and Oxidative Stress Response through Proteomic and Metabolomic Changes Involving Novel Signalers and Effectors. Biotechnol. Biofuels 2020, 13, 110. [Google Scholar] [CrossRef] [PubMed]
- Schulz-Raffelt, M.; Chochois, V.; Auroy, P.; Cuiné, S.; Billon, E.; Dauvillée, D.; Li-Beisson, Y.; Peltier, G. Hyper-Accumulation of Starch and Oil in a Chlamydomonas Mutant Affected in a Plant-Specific DYRK Kinase. Biotechnol. Biofuels 2016, 9, 55. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Miranda-Saavedra, D.; Luebbering, N.; Singh, A.; Sibbet, G.; Ferguson, M.A.J.; Cleghon, V. Deep Evolutionary Conservation of an Intramolecular Protein Kinase Activation Mechanism. PLoS ONE 2012, 7, e29702. [Google Scholar] [CrossRef]
- De Hiller, N.J.; Silva, N.A.A.E.; Faria, R.X.; Souza, A.L.A.; Resende, J.A.L.C.; Borges Farias, A.; Correia Romeiro, N.; de Luna Martins, D. Synthesis and Evaluation of the Anticancer and Trypanocidal Activities of Boronic Tyrphostins. ChemMedChem 2018, 13, 1395–1404. [Google Scholar] [CrossRef]
- Cayla, M.; McDonald, L.; MacGregor, P.; Matthews, K. An Atypical DYRK Kinase Connects Quorum-Sensing with Posttranscriptional Gene Regulation in Trypanosoma Brucei. eLife 2020, 9. [Google Scholar] [CrossRef] [PubMed]
- Ishii, M.; Akiyoshi, B. Characterization of Unconventional Kinetochore Kinases KKT10 and KKT19 in Trypanosoma Brucei. J. Cell Sci. 2020, 133. [Google Scholar] [CrossRef] [PubMed]
- Rocha, V.P.C.; Dacher, M.; Young, S.A.; Kolokousi, F.; Efstathiou, A.; Späth, G.F.; Soares, M.B.P.; Smirlis, D. Leishmania Dual-Specificity Tyrosine-Regulated Kinase 1 (DYRK1) Is Required for Sustaining Leishmania Stationary Phase Phenotype. Mol. Microbiol. 2020, 113, 983–1002. [Google Scholar] [CrossRef]
- Saldivia, M.; Fang, E.; Ma, X.; Myburgh, E.; Carnielli, J.B.T.; Bower-Lepts, C.; Brown, E.; Ritchie, R.; Lakshminarayana, S.B.; Chen, Y.-L.; et al. Targeting the Trypanosome Kinetochore with CLK1 Protein Kinase Inhibitors. Nat. Microbiol. 2020, 5, 1207–1216. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, Scalable Generation of High-Quality Protein Multiple Sequence Alignments Using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—A Multiple Sequence Alignment Editor and Analysis Workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef]
- Tahtouh, T.; Elkins, J.M.; Filippakopoulos, P.; Soundararajan, M.; Burgy, G.; Durieu, E.; Cochet, C.; Schmid, R.S.; Lo, D.C.; Delhommel, F.; et al. Selectivity, Cocrystal Structures, and Neuroprotective Properties of Leucettines, a Family of Protein Kinase Inhibitors Derived from the Marine Sponge Alkaloid Leucettamine B. J. Med. Chem. 2012, 55, 9312–9330. [Google Scholar] [CrossRef]
- Ogawa, Y.; Nonaka, Y.; Goto, T.; Ohnishi, E.; Hiramatsu, T.; Kii, I.; Yoshida, M.; Ikura, T.; Onogi, H.; Shibuya, H.; et al. Development of a Novel Selective Inhibitor of the Down Syndrome-Related Kinase Dyrk1A. Nat. Commun. 2010, 1, 1–9. [Google Scholar] [CrossRef]
- Anderson, K.; Chen, Y.; Chen, Z.; Dominique, R.; Glenn, K.; He, Y.; Janson, C.; Luk, K.-C.; Lukacs, C.; Polonskaia, A.; et al. Pyrido[2,3-d]Pyrimidines: Discovery and Preliminary SAR of a Novel Series of DYRK1B and DYRK1A Inhibitors. Bioorg. Med. Chem. Lett. 2013, 23, 6610–6615. [Google Scholar] [CrossRef] [PubMed]
- Alexeeva, M.; Åberg, E.; Engh, R.A.; Rothweiler, U. The Structure of a Dual-Specificity Tyrosine Phosphorylation-Regulated Kinase 1A–PKC412 Complex Reveals Disulfide-Bridge Formation with the Anomalous Catalytic Loop HRD(HCD) Cysteine. Acta Crystallogr. D Biol. Crystallogr. 2015, 71, 1207–1215. [Google Scholar] [CrossRef] [PubMed]
- Falke, H.; Chaikuad, A.; Becker, A.; Loaëc, N.; Lozach, O.; Abu Jhaisha, S.; Becker, W.; Jones, P.G.; Preu, L.; Baumann, K.; et al. 10-Iodo-11H-Indolo[3,2-c]Quinoline-6-Carboxylic Acids Are Selective Inhibitors of DYRK1A. J. Med. Chem. 2015, 58, 3131–3143. [Google Scholar] [CrossRef]
- Fukuda, T.; Uchida, K.; Nakayama, H.; Ano, Y. Short-Term Administration of Iso-α-Acids Increases Transthyretin Transcription in the Hippocampus. Biochem. Biophys. Res. Commun. 2018, 507, 471–475. [Google Scholar] [CrossRef]
- Czarna, A.; Wang, J.; Zelencova, D.; Liu, Y.; Deng, X.; Choi, H.G.; Zhang, T.; Zhou, W.; Chang, J.W.; Kildalsen, H.; et al. Novel Scaffolds for Dual Specificity Tyrosine-Phosphorylation-Regulated Kinase (DYRK1A) Inhibitors. J. Med. Chem. 2018, 61, 7560–7572. [Google Scholar] [CrossRef] [PubMed]
- Lechner, C.; Flaßhoff, M.; Falke, H.; Preu, L.; Loaëc, N.; Meijer, L.; Knapp, S.; Chaikuad, A.; Kunick, C. [B]-Annulated Halogen-Substituted Indoles as Potential DYRK1A Inhibitors. Molecules 2019, 24, 4090. [Google Scholar] [CrossRef]
- Wurzlbauer, A.; Rüben, K.; Gürdal, E.; Chaikuad, A.; Knapp, S.; Sippl, W.; Becker, W.; Bracher, F. How to Separate Kinase Inhibition from Undesired Monoamine Oxidase a Inhibition-The Development of the DYRK1A Inhibitor AnnH75 from the Alkaloid Harmine. Molecules 2020, 25, 5962. [Google Scholar] [CrossRef] [PubMed]
- Kumar, K.; Wang, P.; Wilson, J.; Zlatanic, V.; Berrouet, C.; Khamrui, S.; Secor, C.; Swartz, E.A.; Lazarus, M.; Sanchez, R.; et al. Synthesis and Biological Validation of a Harmine-Based, Central Nervous System (CNS)-Avoidant, Selective, Human β-Cell Regenerative Dual-Specificity Tyrosine Phosphorylation-Regulated Kinase A (DYRK1A) Inhibitor. J. Med. Chem. 2020, 63, 2986–3003. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.A.; Jin, Q.; Zou, Y.; Ding, Q.; Yan, S.; Wang, Z.; Hao, X.; Nguyen, B.; Zhang, X.; Pan, J.; et al. Selective DYRK1A Inhibitor for the Treatment of Type 1 Diabetes: Discovery of 6-Azaindole Derivative GNF2133. J. Med. Chem. 2020, 63, 2958–2973. [Google Scholar] [CrossRef]
- Myrianthopoulos, V.; Kritsanida, M.; Gaboriaud-Kolar, N.; Magiatis, P.; Ferandin, Y.; Durieu, E.; Lozach, O.; Cappel, D.; Soundararajan, M.; Filippakopoulos, P.; et al. Novel Inverse Binding Mode of Indirubin Derivatives Yields Improved Selectivity for DYRK Kinases. ACS Med. Chem. Lett. 2012, 4, 22–26. [Google Scholar] [CrossRef]
- Chaikuad, A.; Diharce, J.; Schröder, M.; Foucourt, A.; Leblond, B.; Casagrande, A.-S.; Désiré, L.; Bonnet, P.; Knapp, S.; Besson, T. An Unusual Binding Model of the Methyl 9-Anilinothiazolo[5,4-f] Quinazoline-2-Carbimidates (EHT 1610 and EHT 5372) Confers High Selectivity for Dual-Specificity Tyrosine Phosphorylation-Regulated Kinases. J. Med. Chem. 2016, 59, 10315–10321. [Google Scholar] [CrossRef]
- Kim, K.; Cha, J.S.; Cho, Y.-S.; Kim, H.; Chang, N.; Kim, H.-J.; Cho, H.-S. Crystal Structure of Human Dual-Specificity Tyrosine-Regulated Kinase 3 Reveals New Structural Features and Insights into Its Auto-Phosphorylation. J. Mol. Biol. 2018, 430, 1521–1530. [Google Scholar] [CrossRef]
- Haltenhof, T.; Kotte, A.; De Bortoli, F.; Schiefer, S.; Meinke, S.; Emmerichs, A.-K.; Petermann, K.K.; Timmermann, B.; Imhof, P.; Franz, A.; et al. A Conserved Kinase-Based Body-Temperature Sensor Globally Controls Alternative Splicing and Gene Expression. Mol. Cell 2020, 78, 57–69.e4. [Google Scholar] [CrossRef] [PubMed]
- Walter, A.; Chaikuad, A.; Helmer, R.; Loaëc, N.; Preu, L.; Ott, I.; Knapp, S.; Meijer, L.; Kunick, C. Molecular Structures of Cdc2-like Kinases in Complex with a New Inhibitor Chemotype. PLoS ONE 2018, 13, e0196761. [Google Scholar] [CrossRef]
- Bullock, A.N.; Das, S.; Debreczeni, J.É.; Rellos, P.; Fedorov, O.; Niesen, F.H.; Guo, K.; Papagrigoriou, E.; Amos, A.L.; Cho, S.; et al. Kinase Domain Insertions Define Distinct Roles of CLK Kinases in SR Protein Phosphorylation. Structure 2009, 17, 352–362. [Google Scholar] [CrossRef] [PubMed]
- Fedorov, O.; Huber, K.; Eisenreich, A.; Filippakopoulos, P.; King, O.; Bullock, A.N.; Szklarczyk, D.; Jensen, L.J.; Fabbro, D.; Trappe, J.; et al. Specific CLK Inhibitors from a Novel Chemotype for Regulation of Alternative Splicing. Chem. Biol. 2011, 18, 67–76. [Google Scholar] [CrossRef]
- Dekel, N.; Eisenberg-Domovich, Y.; Karlas, A.; Meyer, T.F.; Bracher, F.; Lebendiker, M.; Danieli, T.; Livnah, O. Expression, Purification and Crystallization of CLK1 Kinase—A Potential Target for Antiviral Therapy. Protein Expr. Purif. 2020, 176, 105742. [Google Scholar] [CrossRef]
- Esvan, Y.J.; Zeinyeh, W.; Boibessot, T.; Nauton, L.; Théry, V.; Knapp, S.; Chaikuad, A.; Loaëc, N.; Meijer, L.; Anizon, F.; et al. Discovery of Pyrido[3,4-g]Quinazoline Derivatives as CMGC Family Protein Kinase Inhibitors: Design, Synthesis, Inhibitory Potency and X-Ray Co-Crystal Structure. Eur. J. Med. Chem. 2016, 118, 170–177. [Google Scholar] [CrossRef]
- Sun, Q.-Z.; Lin, G.-F.; Li, L.-L.; Jin, X.-T.; Huang, L.-Y.; Zhang, G.; Yang, W.; Chen, K.; Xiang, R.; Chen, C.; et al. Discovery of Potent and Selective Inhibitors of Cdc2-Like Kinase 1 (CLK1) as a New Class of Autophagy Inducers. J. Med. Chem. 2017, 60, 6337–6352. [Google Scholar] [CrossRef]
- Lee, J.Y.; Yun, J.-S.; Kim, W.-K.; Chun, H.-S.; Jin, H.; Cho, S.; Chang, J.H. Structural Basis for the Selective Inhibition of Cdc2-Like Kinases by CX-4945. Biomed. Res. Int. 2019. [Google Scholar] [CrossRef]
- Kallen, J.; Bergsdorf, C.; Arnaud, B.; Bernhard, M.; Brichet, M.; Cobos-Correa, A.; Elhajouji, A.; Freuler, F.; Galimberti, I.; Guibourdenche, C.; et al. X-Ray Structures and Feasibility Assessment of CLK2 Inhibitors for Phelan-McDermid Syndrome. ChemMedChem 2018, 13, 1997–2007. [Google Scholar] [CrossRef] [PubMed]
- Tazarki, H.; Zeinyeh, W.; Esvan, Y.J.; Knapp, S.; Chatterjee, D.; Schröder, M.; Joerger, A.C.; Khiari, J.; Josselin, B.; Baratte, B.; et al. New Pyrido[3,4-g]Quinazoline Derivatives as CLK1 and DYRK1A Inhibitors: Synthesis, Biological Evaluation and Binding Mode Analysis. Eur. J. Med. Chem. 2019, 166, 304–317. [Google Scholar] [CrossRef]
- Heroven, C.; Georgi, V.; Ganotra, G.K.; Brennan, P.; Wolfreys, F.; Wade, R.C.; Fernández-Montalván, A.E.; Chaikuad, A.; Knapp, S. Halogen-Aromatic π Interactions Modulate Inhibitor Residence Times. Angew. Chem. Int. Ed. Engl. 2018, 57, 7220–7224. [Google Scholar] [CrossRef] [PubMed]
- Němec, V.; Hylsová, M.; Maier, L.; Flegel, J.; Sievers, S.; Ziegler, S.; Schröder, M.; Berger, B.-T.; Chaikuad, A.; Valčíková, B.; et al. Furo[3,2-b]Pyridine: A Privileged Scaffold for Highly Selective Kinase Inhibitors and Effective Modulators of the Hedgehog Pathway. Angew. Chem. Int. Ed. Engl. 2019, 58, 1062–1066. [Google Scholar] [CrossRef]
- Schröder, M.; Bullock, A.N.; Fedorov, O.; Bracher, F.; Chaikuad, A.; Knapp, S. DFG-1 Residue Controls Inhibitor Binding Mode and Affinity, Providing a Basis for Rational Design of Kinase Inhibitor Selectivity. J. Med. Chem. 2020, 63, 10224–10234. [Google Scholar] [CrossRef] [PubMed]
- Schröder, M.; Filippakopoulos, P.; Schwalm, M.P.; Ferrer, C.A.; Drewry, D.H.; Knapp, S.; Chaikuad, A. Crystal Structure and Inhibitor Identifications Reveal Targeting Opportunity for the Atypical MAPK Kinase ERK3. Int. J. Mol. Sci. 2020, 21, 7953. [Google Scholar] [CrossRef]
- Debdab, M.; Carreaux, F.; Renault, S.; Soundararajan, M.; Fedorov, O.; Filippakopoulos, P.; Lozach, O.; Babault, L.; Tahtouh, T.; Baratte, B.; et al. Leucettines, a Class of Potent Inhibitors of Cdc2-Like Kinases and Dual Specificity, Tyrosine Phosphorylation Regulated Kinases Derived from the Marine Sponge Leucettamine B: Modulation of Alternative Pre-RNA Splicing. J. Med. Chem. 2011, 54, 4172–4186. [Google Scholar] [CrossRef]
- Guard, S.E.; Poss, Z.C.; Ebmeier, C.C.; Pagratis, M.; Simpson, H.; Taatjes, D.J.; Old, W.M. The Nuclear Interactome of DYRK1A Reveals a Functional Role in DNA Damage Repair. Sci. Rep. 2019, 9, 6539. [Google Scholar] [CrossRef]
- Guard, S.E.; Ebmeier, C.C.; Old, W.M. Label-Free Immunoprecipitation Mass Spectrometry Workflow for Large-Scale Nuclear Interactome Profiling. J. Vis. Exp. 2019. [Google Scholar] [CrossRef]
- Roewenstrunk, J.; Di Vona, C.; Chen, J.; Borras, E.; Dong, C.; Arató, K.; Sabidó, E.; Huen, M.S.Y.; de la Luna, S. A Comprehensive Proteomics-Based Interaction Screen That Links DYRK1A to RNF169 and to the DNA Damage Response. Sci. Rep. 2019, 9, 6014. [Google Scholar] [CrossRef] [PubMed]
- Varjosalo, M.; Keskitalo, S.; Van Drogen, A.; Nurkkala, H.; Vichalkovski, A.; Aebersold, R.; Gstaiger, M. The Protein Interaction Landscape of the Human CMGC Kinase Group. Cell Rep. 2013, 3, 1306–1320. [Google Scholar] [CrossRef]
- Huttlin, E.L.; Ting, L.; Bruckner, R.J.; Gebreab, F.; Gygi, M.P.; Szpyt, J.; Tam, S.; Zarraga, G.; Colby, G.; Baltier, K.; et al. The BioPlex Network: A Systematic Exploration of the Human Interactome. Cell 2015, 162, 425–440. [Google Scholar] [CrossRef] [PubMed]
- Hein, M.Y.; Hubner, N.C.; Poser, I.; Cox, J.; Nagaraj, N.; Toyoda, Y.; Gak, I.A.; Weisswange, I.; Mansfeld, J.; Buchholz, F.; et al. A Human Interactome in Three Quantitative Dimensions Organized by Stoichiometries and Abundances. Cell 2015, 163, 712–723. [Google Scholar] [CrossRef]
- Song, R.; Wang, Z.-D.; Schapira, M. Disease Association and Druggability of WD40 Repeat Proteins. J. Proteome Res. 2017, 16, 3766–3773. [Google Scholar] [CrossRef]
- Miyata, Y.; Nishida, E. DYRK1A Binds to an Evolutionarily Conserved WD40-Repeat Protein WDR68 and Induces Its Nuclear Translocation. Biochim. Biophys. Acta BBA Mol. Cell Res. 2011, 1813, 1728–1739. [Google Scholar] [CrossRef] [PubMed]
- Miyata, Y.; Shibata, T.; Aoshima, M.; Tsubata, T.; Nishida, E. The Molecular Chaperone TRiC/CCT Binds to the Trp-Asp 40 (WD40) Repeat Protein WDR68 and Promotes Its Folding, Protein Kinase DYRK1A Binding, and Nuclear Accumulation. J. Biol. Chem. 2014, 289, 33320–33332. [Google Scholar] [CrossRef] [PubMed]
- Glenewinkel, F.; Cohen, M.J.; King, C.R.; Kaspar, S.; Bamberg-Lemper, S.; Mymryk, J.S.; Becker, W. The Adaptor Protein DCAF7 Mediates the Interaction of the Adenovirus E1A Oncoprotein with the Protein Kinases DYRK1A and HIPK2. Sci. Rep. 2016, 6, 28241. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Cattoglio, C.; Xue, Y.; Zhou, Q. A Complex between DYRK1A and DCAF7 Phosphorylates the C-Terminal Domain of RNA Polymerase II to Promote Myogenesis. Nucleic Acids Res. 2019, 47, 4462–4475. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Doan, D.; Roman Petersen, Y.; Alvarado, E.; Alvarado, G.; Bhandari, A.; Mohanty, A.; Mohanty, S.; Nissen, R.M. Wdr68 Requires Nuclear Access for Craniofacial Development. PLoS ONE 2013, 8, e54363. [Google Scholar] [CrossRef]
- Alvarado, E.; Yousefelahiyeh, M.; Alvarado, G.; Shang, R.; Whitman, T.; Martinez, A.; Yu, Y.; Pham, A.; Bhandari, A.; Wang, B.; et al. Wdr68 Mediates Dorsal and Ventral Patterning Events for Craniofacial Development. PLoS ONE 2016, 11, e0166984. [Google Scholar] [CrossRef] [PubMed]
- Xiang, J.; Yang, S.; Xin, N.; Gaertig, M.A.; Reeves, R.H.; Li, S.; Li, X.-J. DYRK1A Regulates Hap1–Dcaf7/WDR68 Binding with Implication for Delayed Growth in Down Syndrome. Proc. Natl. Acad. Sci. USA 2017, 114, E1224–E1233. [Google Scholar] [CrossRef]
- Yousefelahiyeh, M.; Xu, J.; Alvarado, E.; Yu, Y.; Salven, D.; Nissen, R.M. DCAF7/WDR68 Is Required for Normal Levels of DYRK1A and DYRK1B. PLoS ONE 2018, 13, e0207779. [Google Scholar] [CrossRef]
- Rueda, N.; Flórez, J.; Dierssen, M.; Martínez-Cué, C. Translational Validity and Implications of Pharmacotherapies in Preclinical Models of Down Syndrome. Prog. Brain Res. 2020, 251, 245–268. [Google Scholar] [CrossRef]
- Jarhad, D.B.; Mashelkar, K.K.; Kim, H.-R.; Noh, M.; Jeong, L.S. Dual-Specificity Tyrosine Phosphorylation-Regulated Kinase 1A (DYRK1A) Inhibitors as Potential Therapeutics. J. Med. Chem. 2018, 61, 9791–9810. [Google Scholar] [CrossRef]
- Feki, A.; Hibaoui, Y. DYRK1A Protein, A Promising Therapeutic Target to Improve Cognitive Deficits in Down Syndrome. Brain Sci. 2018, 8, 187. [Google Scholar] [CrossRef] [PubMed]
- Kay, L.J.; Smulders-Srinivasan, T.K.; Soundararajan, M. Understanding the Multifaceted Role of Human Down Syndrome Kinase DYRK1A. Adv. Protein Chem. Struct. Biol. 2016, 105, 127–171. [Google Scholar] [CrossRef] [PubMed]
- Duchon, A.; Herault, Y. DYRK1A, a Dosage-Sensitive Gene Involved in Neurodevelopmental Disorders, Is a Target for Drug Development in Down Syndrome. Front. Behav. Neurosci. 2016, 10. [Google Scholar] [CrossRef]
- Becker, W.; Soppa, U.; Tejedor, F.J. DYRK1A: A Potential Drug Target for Multiple Down Syndrome Neuropathologies. CNS Neurol. Disord. Drug Targets 2014, 13, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.L.; Fruit, C.; Hérault, Y.; Meijer, L.; Besson, T. Dual-Specificity Tyrosine Phosphorylation-Regulated Kinase 1A (DYRK1A) Inhibitors: A Survey of Recent Patent Literature. Expert Opin. Ther. Patents 2017, 27, 1183–1199. [Google Scholar] [CrossRef] [PubMed]
- Stotani, S.; Giordanetto, F.; Medda, F. DYRK1A Inhibition as Potential Treatment for Alzheimer’s Disease. Future Med. Chem. 2016, 8, 681–696. [Google Scholar] [CrossRef]
- Pathak, A.; Rohilla, A.; Gupta, T.; Akhtar, M.J.; Haider, M.R.; Sharma, K.; Haider, K.; Yar, M.S. DYRK1A Kinase Inhibition with Emphasis on Neurodegeneration: A Comprehensive Evolution Story-Cum-Perspective. Eur. J. Med. Chem. 2018, 158, 559–592. [Google Scholar] [CrossRef] [PubMed]
- Jin, N.; Yin, X.; Gu, J.; Zhang, X.; Shi, J.; Qian, W.; Ji, Y.; Cao, M.; Gu, X.; Ding, F.; et al. Truncation and Activation of Dual Specificity Tyrosine Phosphorylation-Regulated Kinase 1A by Calpain I: A molecular mechanism linked to tau pathology in alzheimer disease. J. Biol. Chem. 2015, 290, 15219–15237. [Google Scholar] [CrossRef]
- Yin, X.; Jin, N.; Gu, J.; Shi, J.; Zhou, J.; Gong, C.-X.; Iqbal, K.; Grundke-Iqbal, I.; Liu, F. Dual-Specificity Tyrosine Phosphorylation-Regulated Kinase 1A (Dyrk1A) Modulates Serine/Arginine-Rich Protein 55 (SRp55)-Promoted Tau Exon 10 Inclusion. J. Biol. Chem. 2012, 287, 30497–30506. [Google Scholar] [CrossRef]
- Yin, X.; Jin, N.; Shi, J.; Zhang, Y.; Wu, Y.; Gong, C.-X.; Iqbal, K.; Liu, F. Dyrk1A Overexpression Leads to Increase of 3R-Tau Expression and Cognitive Deficits in Ts65Dn Down Syndrome Mice. Sci. Rep. 2017, 7, 619. [Google Scholar] [CrossRef]
- Nalls, M.A.; Blauwendraat, C.; Vallerga, C.L.; Heilbron, K.; Bandres-Ciga, S.; Chang, D.; Tan, M.; Kia, D.A.; Noyce, A.J.; Xue, A.; et al. Identification of Novel Risk Loci, Causal Insights, and Heritable Risk for Parkinson’s Disease: A Meta-Analysis of Genome-Wide Association Studies. Lancet Neurol. 2019, 18, 1091–1102. [Google Scholar] [CrossRef]
- Chiu, C.-C.; Yeh, T.-H.; Chen, R.-S.; Chen, H.-C.; Huang, Y.-Z.; Weng, Y.-H.; Cheng, Y.-C.; Liu, Y.-C.; Cheng, A.-J.; Lu, Y.-C.; et al. Upregulated Expression of MicroRNA-204-5p Leads to the Death of Dopaminergic Cells by Targeting DYRK1A-Mediated Apoptotic Signaling Cascade. Front. Cell. Neurosci. 2019, 13. [Google Scholar] [CrossRef]
- Ferrer, I.; Barrachina, M.; Puig, B.; Martínez de Lagrán, M.; Martí, E.; Avila, J.; Dierssen, M. Constitutive Dyrk1A Is Abnormally Expressed in Alzheimer Disease, Down Syndrome, Pick Disease, and Related Transgenic Models. Neurobiol. Dis. 2005, 20, 392–400. [Google Scholar] [CrossRef] [PubMed]
- Ji, J.; Lee, H.; Argiropoulos, B.; Dorrani, N.; Mann, J.; Martinez-Agosto, J.A.; Gomez-Ospina, N.; Gallant, N.; Bernstein, J.A.; Hudgins, L.; et al. DYRK1A Haploinsufficiency Causes a New Recognizable Syndrome with Microcephaly, Intellectual Disability, Speech Impairment, and Distinct Facies. Eur. J. Hum. Genet. 2015, 23, 1473–1481. [Google Scholar] [CrossRef] [PubMed]
- Evers, J.M.G.; Laskowski, R.A.; Bertolli, M.; Clayton-Smith, J.; Deshpande, C.; Eason, J.; Elmslie, F.; Flinter, F.; Gardiner, C.; Hurst, J.A.; et al. Structural Analysis of Pathogenic Mutations in the DYRK1A Gene in Patients with Developmental Disorders. Hum. Mol. Genet. 2017, 26, 519–526. [Google Scholar] [CrossRef]
- Widowati, E.W.; Ernst, S.; Hausmann, R.; Müller-Newen, G.; Becker, W. Functional Characterization of DYRK1A Missense Variants Associated with a Syndromic Form of Intellectual Deficiency and Autism. Biol. Open 2018, 7. [Google Scholar] [CrossRef]
- Ackeifi, C.; Swartz, E.; Kumar, K.; Liu, H.; Chalada, S.; Karakose, E.; Scott, D.K.; Garcia-Ocaña, A.; Sanchez, R.; DeVita, R.J.; et al. Pharmacologic and Genetic Approaches Define Human Pancreatic β Cell Mitogenic Targets of DYRK1A Inhibitors. JCI Insight 2020, 5. [Google Scholar] [CrossRef] [PubMed]
- Kumar, K.; Suebsuwong, C.; Wang, P.; Garcia-Ocana, A.; Stewart, A.F.; DeVita, R.J. DYRK1A Inhibitors as Potential Therapeutics for β-Cell Regeneration for Diabetes. J. Med. Chem. 2021. [Google Scholar] [CrossRef]
- Abbassi, R.; Johns, T.G.; Kassiou, M.; Munoz, L. DYRK1A in Neurodegeneration and Cancer: Molecular Basis and Clinical Implications. Pharmacol. Ther. 2015, 151, 87–98. [Google Scholar] [CrossRef]
- Boni, J.; Rubio-Perez, C.; López-Bigas, N.; Fillat, C.; de la Luna, S. The DYRK Family of Kinases in Cancer: Molecular Functions and Therapeutic Opportunities. Cancers 2020, 12, 2106. [Google Scholar] [CrossRef]
- Laham, A.J.; Saber-Ayad, M.; El-Awady, R. DYRK1A: A down Syndrome-Related Dual Protein Kinase with a Versatile Role in Tumorigenesis. Cell Mol. Life Sci. 2021, 78, 603–619. [Google Scholar] [CrossRef]
- Malinge, S.; Bliss-Moreau, M.; Kirsammer, G.; Diebold, L.; Chlon, T.; Gurbuxani, S.; Crispino, J.D. Increased Dosage of the Chromosome 21 Ortholog Dyrk1a Promotes Megakaryoblastic Leukemia in a Murine Model of Down Syndrome. J. Clin. Investig. 2012, 122, 948–962. [Google Scholar] [CrossRef]
- Bhansali, R.S.; Rammohan, M.; Lee, P.; Laurent, A.P.; Wen, Q.; Suraneni, P.; Yip, B.H.; Tsai, Y.-C.; Jenni, S.; Bornhauser, B.; et al. DYRK1A Regulates B Cell Acute Lymphoblastic Leukemia through Phosphorylation of FOXO1 and STAT3. J. Clin. Investig. 2021, 131. [Google Scholar] [CrossRef]
- Lee, S.B.; Frattini, V.; Bansal, M.; Castano, A.M.; Sherman, D.; Hutchinson, K.; Bruce, J.N.; Califano, A.; Liu, G.; Cardozo, T.; et al. An ID2-Dependent Mechanism for VHL Inactivation in Cancer. Nature 2016, 529, 172–177. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Martínez, P.; Zahonero, C.; Sánchez-Gómez, P. DYRK1A: The Double-Edged Kinase as a Protagonist in Cell Growth and Tumorigenesis. Mol. Cell Oncol. 2015, 2. [Google Scholar] [CrossRef]
- Birger, Y.; Izraeli, S. DYRK1A in Down Syndrome: An Oncogene or Tumor Suppressor? J. Clin. Investig. 2012, 122, 807–810. [Google Scholar] [CrossRef] [PubMed][Green Version]
- He, M.; Gu, J.; Zhu, J.; Wang, X.; Wang, C.; Duan, C.; Ni, Y.; Lu, X.; Li, J. Up-Regulation of Dyrk1b Promote Astrocyte Activation Following Lipopolysaccharide-Induced Neuroinflammation. Neuropeptides 2018, 69, 76–83. [Google Scholar] [CrossRef] [PubMed]
- Becker, W. A Wake-up Call to Quiescent Cancer Cells—Potential Use of DYRK1B Inhibitors in Cancer Therapy. FEBS J. 2018, 285, 1203–1211. [Google Scholar] [CrossRef] [PubMed]
- Kokkorakis, N.; Gaitanou, M. Minibrain-Related Kinase/Dual-Specificity Tyrosine-Regulated Kinase 1B Implication in Stem/Cancer Stem Cells Biology. World J. Stem Cells 2020, 12, 1553–1575. [Google Scholar] [CrossRef]
- Woo, Y.; Kim, S.J.; Suh, B.K.; Kwak, Y.; Jung, H.-J.; Nhung, T.T.M.; Mun, D.J.; Hong, J.-H.; Noh, S.-J.; Kim, S.; et al. Sequential Phosphorylation of NDEL1 by the DYRK2-GSK3β Complex Is Critical for Neuronal Morphogenesis. eLife 2019, 8. [Google Scholar] [CrossRef]
- Yoshida, S.; Yoshida, K. Multiple Functions of DYRK2 in Cancer and Tissue Development. FEBS Lett. 2019, 593, 2953–2965. [Google Scholar] [CrossRef]
- Correa-Sáez, A.; Jiménez-Izquierdo, R.; Garrido-Rodríguez, M.; Morrugares, R.; Muñoz, E.; Calzado, M.A. Updating Dual-Specificity Tyrosine-Phosphorylation-Regulated Kinase 2 (DYRK2): Molecular Basis, Functions and Role in Diseases. Cell. Mol. Life Sci. 2020, 77, 4747–4763. [Google Scholar] [CrossRef]
- Ma, F.; Zhu, Y.; Liu, X.; Zhou, Q.; Hong, X.; Qu, C.; Feng, X.; Zhang, Y.; Ding, Q.; Zhao, J.; et al. Dual-Specificity Tyrosine Phosphorylation-Regulated Kinase 3 Loss Activates Purine Metabolism and Promotes Hepatocellular Carcinoma Progression. Hepatology 2019, 70, 1785–1803. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Lee, S.; Kang, H.; Shin, E.; Kim, H.Y.; Youn, H.; Youn, B. Dual Specificity Kinase DYRK3 Promotes Aggressiveness of Glioblastoma by Altering Mitochondrial Morphology and Function. Int. J. Mol. Sci. 2021, 22, 2982. [Google Scholar] [CrossRef] [PubMed]
- Bakre, A.; Andersen, L.E.; Meliopoulos, V.; Coleman, K.; Yan, X.; Brooks, P.; Crabtree, J.; Tompkins, S.M.; Tripp, R.A. Identification of Host Kinase Genes Required for Influenza Virus Replication and the Regulatory Role of MicroRNAs. PLoS ONE 2013, 8, e66796. [Google Scholar] [CrossRef]
- Wippich, F.; Bodenmiller, B.; Trajkovska, M.G.; Wanka, S.; Aebersold, R.; Pelkmans, L. Dual Specificity Kinase DYRK3 Couples Stress Granule Condensation/Dissolution to MTORC1 Signaling. Cell 2013, 152, 791–805. [Google Scholar] [CrossRef]
- Rai, A.K.; Chen, J.-X.; Selbach, M.; Pelkmans, L. Kinase-Controlled Phase Transition of Membraneless Organelles in Mitosis. Nature 2018, 559, 211–216. [Google Scholar] [CrossRef]
- Slepak, T.I.; Salay, L.D.; Lemmon, V.P.; Bixby, J.L. Dyrk Kinases Regulate Phosphorylation of Doublecortin, Cytoskeletal Organization, and Neuronal Morphology. Cytoskeleton 2012, 69, 514–527. [Google Scholar] [CrossRef]
- Nguyen, T.L.; Duchon, A.; Manousopoulou, A.; Loaëc, N.; Villiers, B.; Pani, G.; Karatas, M.; Mechling, A.E.; Harsan, L.-A.; Limanton, E.; et al. Correction of Cognitive Deficits in Mouse Models of Down Syndrome by a Pharmacological Inhibitor of DYRK1A. Dis. Models Mech. 2018, 11. [Google Scholar] [CrossRef] [PubMed]
- Souchet, B.; Audrain, M.; Billard, J.M.; Dairou, J.; Fol, R.; Orefice, N.S.; Tada, S.; Gu, Y.; Dufayet-Chaffaud, G.; Limanton, E.; et al. Inhibition of DYRK1A Proteolysis Modifies Its Kinase Specificity and Rescues Alzheimer Phenotype in APP/PS1 Mice. Acta Neuropathol. Commun. 2019, 7. [Google Scholar] [CrossRef]
- Sharma, A.; Chunduri, A.; Gopu, A.; Shatrowsky, C.; Crusio, W.E.; Delprato, A. Common Genetic Signatures of Alzheimer’s Disease in Down Syndrome. F1000Research 2020, 9, 1299. [Google Scholar] [CrossRef]
- Duchon, A.; Del Mar Muñiz Moreno, M.; Lorenzo, S.M.; de Souza, M.P.S.; Chevalier, C.; Nalesso, V.; Meziane, H.; de Sousa, P.L.; Noblet, V.; Armspach, J.-P.; et al. Multi-Influential Genetic Interactions Alter Behaviour and Cognition through Six Main Biological Cascades in Down Syndrome Mouse Models. Hum. Mol. Genet. 2021. [Google Scholar] [CrossRef]
- Kargbo, R.B. Selective DYRK1A Inhibitor for the Treatment of Neurodegenerative Diseases: Alzheimer, Parkinson, Huntington, and Down Syndrome. ACS Med. Chem. Lett. 2020, 11, 1795–1796. [Google Scholar] [CrossRef]
- Goodlett, C.R.; Stringer, M.; LaCombe, J.; Patel, R.; Wallace, J.M.; Roper, R.J. Evaluation of the Therapeutic Potential of Epigallocatechin-3-Gallate (EGCG) via Oral Gavage in Young Adult Down Syndrome Mice. Sci. Rep. 2020, 10, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Moroy, G.; Paul, J.-L.; Rebillat, A.-S.; Dierssen, M.; de la Torre, R.; Cieuta-Walti, C.; Dairou, J.; Janel, N. Molecular Rescue of Dyrk1A Overexpression Alterations in Mice with Fontup® Dietary Supplement: Role of Green Tea Catechins. Int. J. Mol. Sci. 2020, 21, 1404. [Google Scholar] [CrossRef] [PubMed]
- Chang, P.; Bush, D.; Schorge, S.; Good, M.; Canonica, T.; Shing, N.; Noy, S.; Wiseman, F.K.; Burgess, N.; Tybulewicz, V.L.J.; et al. Altered Hippocampal-Prefrontal Neural Dynamics in Mouse Models of Down Syndrome. Cell Rep. 2020, 30, 1152–1163.e4. [Google Scholar] [CrossRef] [PubMed]
- Sachse, S.M.; Lievens, S.; Ribeiro, L.F.; Dascenco, D.; Masschaele, D.; Horré, K.; Misbaer, A.; Vanderroost, N.; De Smet, A.S.; Salta, E.; et al. Nuclear Import of the DSCAM-Cytoplasmic Domain Drives Signaling Capable of Inhibiting Synapse Formation. EMBO J. 2019, 38. [Google Scholar] [CrossRef]
- Neumann, F.; Gourdain, S.; Albac, C.; Dekker, A.D.; Bui, L.C.; Dairou, J.; Schmitz-Afonso, I.; Hue, N.; Rodrigues-Lima, F.; Delabar, J.M.; et al. DYRK1A Inhibition and Cognitive Rescue in a Down Syndrome Mouse Model Are Induced by New Fluoro-DANDY Derivatives. Sci. Rep. 2018, 8, 2859. [Google Scholar] [CrossRef] [PubMed]
- García-Cerro, S.; Vidal, V.; Lantigua, S.; Berciano, M.T.; Lafarga, M.; Ramos-Cabrer, P.; Padro, D.; Rueda, N.; Martínez-Cué, C. Cerebellar Alterations in a Model of Down Syndrome: The Role of the Dyrk1A Gene. Neurobiol. Dis. 2018, 110, 206–217. [Google Scholar] [CrossRef] [PubMed]
- Stringer, M.; Goodlett, C.R.; Roper, R.J. Targeting Trisomic Treatments: Optimizing Dyrk1a Inhibition to Improve Down Syndrome Deficits. Mol. Genet. Genom. Med. 2017, 5, 451–465. [Google Scholar] [CrossRef]
- McElyea, S.D.; Starbuck, J.M.; Tumbleson-Brink, D.M.; Harrington, E.; Blazek, J.D.; Ghoneima, A.; Kula, K.; Roper, R.J. Influence of Prenatal EGCG Treatment and Dyrk1a Dosage Reduction on Craniofacial Features Associated with Down Syndrome. Hum. Mol. Genet. 2016, 25, 4856–4869. [Google Scholar] [CrossRef]
- Kim, H.; Lee, K.-S.; Kim, A.-K.; Choi, M.; Choi, K.; Kang, M.; Chi, S.-W.; Lee, M.-S.; Lee, J.-S.; Lee, S.-Y.; et al. A Chemical with Proven Clinical Safety Rescues Down-Syndrome-Related Phenotypes in through DYRK1A Inhibition. Dis. Models Mech. 2016, 9, 839–848. [Google Scholar] [CrossRef] [PubMed]
- Blazek, J.D.; Abeysekera, I.; Li, J.; Roper, R.J. Rescue of the Abnormal Skeletal Phenotype in Ts65Dn Down Syndrome Mice Using Genetic and Therapeutic Modulation of Trisomic Dyrk1a. Hum. Mol. Genet. 2015, 24, 5687–5696. [Google Scholar] [CrossRef]
- García-Cerro, S.; Martínez, P.; Vidal, V.; Corrales, A.; Flórez, J.; Vidal, R.; Rueda, N.; Arbonés, M.L.; Martínez-Cué, C. Overexpression of Dyrk1A Is Implicated in Several Cognitive, Electrophysiological and Neuromorphological Alterations Found in a Mouse Model of Down Syndrome. PLoS ONE 2014, 9, e106572. [Google Scholar] [CrossRef] [PubMed]
- De la Torre, R.; De Sola, S.; Pons, M.; Duchon, A.; de Lagran, M.M.; Farré, M.; Fitó, M.; Benejam, B.; Langohr, K.; Rodriguez, J.; et al. Epigallocatechin-3-Gallate, a DYRK1A Inhibitor, Rescues Cognitive Deficits in Down Syndrome Mouse Models and in Humans. Mol. Nutr. Food Res. 2014, 58, 278–288. [Google Scholar] [CrossRef] [PubMed]
- Altafaj, X.; Martín, E.D.; Ortiz-Abalia, J.; Valderrama, A.; Lao-Peregrín, C.; Dierssen, M.; Fillat, C. Normalization of Dyrk1A Expression by AAV2/1-ShDyrk1A Attenuates Hippocampal-Dependent Defects in the Ts65Dn Mouse Model of Down Syndrome. Neurobiol. Dis. 2013, 52, 117–127. [Google Scholar] [CrossRef]
- Park, J.; Oh, Y.; Chung, K.C. Two Key Genes Closely Implicated with the Neuropathological Characteristics in Down Syndrome: DYRK1A and RCAN1. BMB Rep. 2009, 42, 6–15. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Abalia, J.; Sahún, I.; Altafaj, X.; Andreu, N.; Estivill, X.; Dierssen, M.; Fillat, C. Targeting Dyrk1A with AAVshRNA Attenuates Motor Alterations in TgDyrk1A, a Mouse Model of Down Syndrome. Am. J. Hum. Genet. 2008, 83, 479–488. [Google Scholar] [CrossRef]
- Shi, J.; Zhang, T.; Zhou, C.; Chohan, M.O.; Gu, X.; Wegiel, J.; Zhou, J.; Hwang, Y.-W.; Iqbal, K.; Grundke-Iqbal, I.; et al. Increased Dosage of Dyrk1A Alters Alternative Splicing Factor (ASF)-Regulated Alternative Splicing of Tau in Down Syndrome. J. Biol. Chem. 2008, 283, 28660–28669. [Google Scholar] [CrossRef] [PubMed]
- Naert, G.; Ferré, V.; Meunier, J.; Keller, E.; Malmström, S.; Givalois, L.; Carreaux, F.; Bazureau, J.-P.; Maurice, T. Leucettine L41, a DYRK1A-Preferential DYRKs/CLKs Inhibitor, Prevents Memory Impairments and Neurotoxicity Induced by Oligomeric Aβ25-35 Peptide Administration in Mice. Eur. Neuropsychopharmacol. 2015, 25, 2170–2182. [Google Scholar] [CrossRef]
- Lee, Y.H.; Im, E.; Hyun, M.; Park, J.; Chung, K.C. Protein Phosphatase PPM1B Inhibits DYRK1A Kinase through Dephosphorylation of PS258 and Reduces Toxic Tau Aggregation. J. Biol. Chem. 2020. [Google Scholar] [CrossRef]
- Lee, H.; Woo, H.; Lee, H.-E.; Jeon, H.; Ryu, K.-Y.; Nam, J.H.; Jeon, S.G.; Park, H.; Lee, J.-S.; Han, K.-M.; et al. The Novel DYRK1A Inhibitor KVN93 Regulates Cognitive Function, Amyloid-Beta Pathology, and Neuroinflammation. Free Radic. Biol. Med. 2020, 160, 575–595. [Google Scholar] [CrossRef]
- Delabar, J.M.; Ortner, M.; Simon, S.; Wijkhuisen, A.; Feraudet-Tarisse, C.; Pegon, J.; Vidal, E.; Hirschberg, Y.; Dubois, B.; Potier, M.-C. Altered Age-Linked Regulation of Plasma DYRK1A in Elderly Cognitive Complainers (INSIGHT-PreAD Study) with High Brain Amyloid Load. Alzheimers Dement. 2020, 6, e12046. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, L.; Xie, F.; Wang, X.; Hou, Y.; Wang, X.; Liu, J. Overexpression of MiR-26a-5p Suppresses Tau Phosphorylation and Aβ Accumulation in Alzheimer’s Disease Mice by Targeting DYRK1A. Curr. Neurovasc. Res. 2020. [Google Scholar] [CrossRef]
- Velazquez, R.; Meechoovet, B.; Ow, A.; Foley, C.; Shaw, A.; Smith, B.; Oddo, S.; Hulme, C.; Dunckley, T. Chronic Dyrk1 Inhibition Delays the Onset of AD-Like Pathology in 3xTg-AD Mice. Mol. Neurobiol. 2019, 56, 8364–8375. [Google Scholar] [CrossRef] [PubMed]
- Branca, C.; Shaw, D.M.; Belfiore, R.; Gokhale, V.; Shaw, A.Y.; Foley, C.; Smith, B.; Hulme, C.; Dunckley, T.; Meechoovet, B.; et al. Dyrk1 Inhibition Improves Alzheimer’s Disease-like Pathology. Aging Cell 2017, 16, 1146–1154. [Google Scholar] [CrossRef]
- García-Cerro, S.; Rueda, N.; Vidal, V.; Lantigua, S.; Martínez-Cué, C. Normalizing the Gene Dosage of Dyrk1A in a Mouse Model of Down Syndrome Rescues Several Alzheimer’s Disease Phenotypes. Neurobiol. Dis. 2017, 106, 76–88. [Google Scholar] [CrossRef] [PubMed]
- Kawakubo, T.; Mori, R.; Shirotani, K.; Iwata, N.; Asai, M. Neprilysin Is Suppressed by Dual-Specificity Tyrosine-Phosphorylation Regulated Kinase 1A (DYRK1A) in Down-Syndrome-Derived Fibroblasts. Biol. Pharm. Bull. 2017, 40, 327–333. [Google Scholar] [CrossRef]
- Janel, N.; Alexopoulos, P.; Badel, A.; Lamari, F.; Camproux, A.C.; Lagarde, J.; Simon, S.; Feraudet-Tarisse, C.; Lamourette, P.; Arbones, M.; et al. Combined Assessment of DYRK1A, BDNF and Homocysteine Levels as Diagnostic Marker for Alzheimer’s Disease. Transl. Psychiatry 2017, 7, e1154. [Google Scholar] [CrossRef]
- Coutadeur, S.; Benyamine, H.; Delalonde, L.; de Oliveira, C.; Leblond, B.; Foucourt, A.; Besson, T.; Casagrande, A.-S.; Taverne, T.; Girard, A.; et al. A Novel DYRK1A (Dual Specificity Tyrosine Phosphorylation-Regulated Kinase 1A) Inhibitor for the Treatment of Alzheimer’s Disease: Effect on Tau and Amyloid Pathologies in Vitro. J. Neurochem. 2015, 133, 440–451. [Google Scholar] [CrossRef] [PubMed]
- Fant, X.; Durieu, E.; Chicanne, G.; Payrastre, B.; Sbrissa, D.; Shisheva, A.; Limanton, E.; Carreaux, F.; Bazureau, J.-P.; Meijer, L. Cdc-Like/Dual-Specificity Tyrosine Phosphorylation–Regulated Kinases Inhibitor Leucettine L41 Induces MTOR-Dependent Autophagy: Implication for Alzheimer’s Disease. Mol. Pharmacol. 2014, 85, 441–450. [Google Scholar] [CrossRef]
- Ryu, Y.S.; Park, S.Y.; Jung, M.-S.; Yoon, S.-H.; Kwen, M.-Y.; Lee, S.-Y.; Choi, S.-H.; Radnaabazar, C.; Kim, M.-K.; Kim, H.; et al. Dyrk1A-Mediated Phosphorylation of Presenilin 1: A Functional Link between Down Syndrome and Alzheimer’s Disease. J. Neurochem. 2010, 115, 574–584. [Google Scholar] [CrossRef]
- Ryoo, S.-R.; Cho, H.-J.; Lee, H.-W.; Jeong, H.K.; Radnaabazar, C.; Kim, Y.-S.; Kim, M.-J.; Son, M.-Y.; Seo, H.; Chung, S.-H.; et al. Dual-Specificity Tyrosine(Y)-Phosphorylation Regulated Kinase 1A-Mediated Phosphorylation of Amyloid Precursor Protein: Evidence for a Functional Link between Down Syndrome and Alzheimer’s Disease. J. Neurochem. 2008, 104, 1333–1344. [Google Scholar] [CrossRef] [PubMed]
- Ryoo, S.-R.; Jeong, H.K.; Radnaabazar, C.; Yoo, J.-J.; Cho, H.-J.; Lee, H.-W.; Kim, I.-S.; Cheon, Y.-H.; Ahn, Y.S.; Chung, S.-H.; et al. DYRK1A-Mediated Hyperphosphorylation of Tau. A Functional Link between Down Syndrome and Alzheimer Disease. J. Biol. Chem. 2007, 282, 34850–34857. [Google Scholar] [CrossRef] [PubMed]
- Kimura, R.; Kamino, K.; Yamamoto, M.; Nuripa, A.; Kida, T.; Kazui, H.; Hashimoto, R.; Tanaka, T.; Kudo, T.; Yamagata, H.; et al. The DYRK1A Gene, Encoded in Chromosome 21 Down Syndrome Critical Region, Bridges between β-Amyloid Production and Tau Phosphorylation in Alzheimer Disease. Hum. Mol. Genet. 2007, 16, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Fang, L.; Tang, B.-S.; Fan, K.; Wan, C.-M.; Yan, X.-X.; Guo, J.-F. Alzheimer’s Disease Susceptibility Genes Modify the Risk of Parkinson Disease and Parkinson’s Disease-Associated Cognitive Impairment. Neurosci. Lett. 2018, 677, 55–59. [Google Scholar] [CrossRef] [PubMed]
- Cen, L.; Xiao, Y.; Wei, L.; Mo, M.; Chen, X.; Li, S.; Yang, X.; Huang, Q.; Qu, S.; Pei, Z.; et al. Association of DYRK1A Polymorphisms with Sporadic Parkinson’s Disease in Chinese Han Population. Neurosci. Lett. 2016, 632, 39–43. [Google Scholar] [CrossRef]
- Im, E.; Chung, K.C. Dyrk1A Phosphorylates Parkin at Ser-131 and Negatively Regulates Its Ubiquitin E3 Ligase Activity. J. Neurochem. 2015, 134, 756–768. [Google Scholar] [CrossRef]
- Jones, E.L.; Aarsland, D.; Londos, E.; Ballard, C. A Pilot Study Examining Associations between DYRK1A and α-Synuclein Dementias. Neurodegener. Dis. 2012, 10, 229–231. [Google Scholar] [CrossRef]
- Sitz, J.H.; Baumgärtel, K.; Hämmerle, B.; Papadopoulos, C.; Hekerman, P.; Tejedor, F.J.; Becker, W.; Lutz, B. The Down Syndrome Candidate Dual-Specificity Tyrosine Phosphorylation-Regulated Kinase 1A Phosphorylates the Neurodegeneration-Related Septin 4. Neuroscience 2008, 157, 596–605. [Google Scholar] [CrossRef]
- Trovò, L.; Fuchs, C.; De Rosa, R.; Barbiero, I.; Tramarin, M.; Ciani, E.; Rusconi, L.; Kilstrup-Nielsen, C. The Green Tea Polyphenol Epigallocatechin-3-Gallate (EGCG) Restores CDKL5-Dependent Synaptic Defects in Vitro and in Vivo. Neurobiol. Dis. 2020, 104791. [Google Scholar] [CrossRef]
- Liu, Y.A.; Jin, Q.; Ding, Q.; Hao, X.; Mo, T.; Yan, S.; Zou, Y.; Huang, Z.; Zhang, X.; Gao, W.; et al. A Dual Inhibitor of DYRK1A and GSK3β for Β-Cell Proliferation: Aminopyrazine Derivative GNF4877. ChemMedChem 2020, 15, 1562–1570. [Google Scholar] [CrossRef]
- Kumar, K.; Wang, P.; Swartz, E.A.; Khamrui, S.; Secor, C.; Lazarus, M.B.; Sanchez, R.; Stewart, A.F.; DeVita, R.J. Structure-Activity Relationships and Biological Evaluation of 7-Substituted Harmine Analogs for Human β-Cell Proliferation. Molecules 2020, 25, 1983. [Google Scholar] [CrossRef]
- Hohmeier, H.E.; Zhang, L.; Taylor, B.; Stephens, S.; Lu, D.; McNamara, P.; Laffitte, B.; Newgard, C.B. Identification of a Small Molecule That Stimulates Human β-Cell Proliferation and Insulin Secretion, and Protects against Cytotoxic Stress in Rat Insulinoma Cells. PLoS ONE 2020, 15, e0224344. [Google Scholar] [CrossRef] [PubMed]
- Brial, F.; Alzaid, F.; Sonomura, K.; Kamatani, Y.; Meneyrol, K.; Le Lay, A.; Péan, N.; Hedjazi, L.; Sato, T.-A.; Venteclef, N.; et al. The Natural Metabolite 4-Cresol Improves Glucose Homeostasis and Enhances β-Cell Function. Cell Rep. 2020, 30, 2306–2320.e5. [Google Scholar] [CrossRef]
- Scavuzzo, M.A.; Borowiak, M. Two Drugs Converged in a Pancreatic β Cell. Sci. Transl. Med. 2020, 12. [Google Scholar] [CrossRef] [PubMed]
- Ackeifi, C.; Wang, P.; Karakose, E.; Manning Fox, J.E.; González, B.J.; Liu, H.; Wilson, J.; Swartz, E.; Berrouet, C.; Li, Y.; et al. GLP-1 Receptor Agonists Synergize with DYRK1A Inhibitors to Potentiate Functional Human β Cell Regeneration. Sci. Transl. Med. 2020, 12. [Google Scholar] [CrossRef]
- Lu, M.; Ma, L.; Shan, P.; Liu, A.; Yu, X.; Jiang, W.; Wang, X.; Zhao, X.; Ye, X.; Wang, T. DYRK1A Aggravates β Cell Dysfunction and Apoptosis by Promoting the Phosphorylation and Degradation of IRS2. Exp. Gerontol. 2019, 125, 110659. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Karakose, E.; Liu, H.; Swartz, E.; Ackeifi, C.; Zlatanic, V.; Wilson, J.; González, B.J.; Bender, A.; Takane, K.K.; et al. Combined Inhibition of DYRK1A, SMAD, and Trithorax Pathways Synergizes to Induce Robust Replication in Adult Human Beta Cells. Cell Metab. 2019, 29, 638–652.e5. [Google Scholar] [CrossRef]
- Wang, P.; Alvarez-Perez, J.-C.; Felsenfeld, D.P.; Liu, H.; Sivendran, S.; Bender, A.; Kumar, A.; Sanchez, R.; Scott, D.K.; Garcia-Ocaña, A.; et al. A High-Throughput Chemical Screen Reveals That Harmine-Mediated Inhibition of DYRK1A Increases Human Pancreatic Beta Cell Replication. Nat. Med. 2015, 21, 383–388. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; Taylor, B.; Jin, Q.; Nguyen-Tran, V.; Meeusen, S.; Zhang, Y.-Q.; Kamireddy, A.; Swafford, A.; Powers, A.F.; Walker, J.; et al. Inhibition of DYRK1A and GSK3B Induces Human β-Cell Proliferation. Nat. Commun. 2015, 6, 8372. [Google Scholar] [CrossRef]
- Zheng, Y.; Ramsamooj, S.; Li, Q.; Johnson, J.L.; Yaron, T.M.; Sharra, K.; Cantley, L.C. Regulation of Folate and Methionine Metabolism by Multisite Phosphorylation of Human Methylenetetrahydrofolate Reductase. Sci. Rep. 2019, 9, 4190. [Google Scholar] [CrossRef] [PubMed]
- Pozo, N.; Zahonero, C.; Fernández, P.; Liñares, J.M.; Ayuso, A.; Hagiwara, M.; Pérez, A.; Ricoy, J.R.; Hernández-Laín, A.; Sepúlveda, J.M.; et al. Inhibition of DYRK1A Destabilizes EGFR and Reduces EGFR-Dependent Glioblastoma Growth. J. Clin. Investig. 2013, 123, 2475–2487. [Google Scholar] [CrossRef] [PubMed]
- Radhakrishnan, A.; Nanjappa, V.; Raja, R.; Sathe, G.; Puttamallesh, V.N.; Jain, A.P.; Pinto, S.M.; Balaji, S.A.; Chavan, S.; Sahasrabuddhe, N.A.; et al. A Dual Specificity Kinase, DYRK1A, as a Potential Therapeutic Target for Head and Neck Squamous Cell Carcinoma. Sci. Rep. 2016, 6, 36132. [Google Scholar] [CrossRef]
- Bai, Z.; Du, Y.; Cong, L.; Cheng, Y. The USP22 Promotes the Growth of Cancer Cells through the DYRK1A in Pancreatic Ductal Adenocarcinoma. Gene 2020, 758, 144960. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Wang, D.; Gao, Z.; Kan, H.; Qiu, F.; Chen, L.; Li, H. Licocoumarone Induces BxPC-3 Pancreatic Adenocarcinoma Cell Death by Inhibiting DYRK1A. Chem. Biol. Interact. 2020, 316, 108913. [Google Scholar] [CrossRef]
- Luna, J.; Boni, J.; Cuatrecasas, M.; Bofill-De Ros, X.; Núñez-Manchón, E.; Gironella, M.; Vaquero, E.C.; Arbones, M.L.; de la Luna, S.; Fillat, C. DYRK1A Modulates C-MET in Pancreatic Ductal Adenocarcinoma to Drive Tumour Growth. Gut 2019, 68, 1465–1476. [Google Scholar] [CrossRef]
- Li, L.; Wei, J.-R.; Song, Y.; Fang, S.; Du, Y.; Li, Z.; Zeng, T.-T.; Zhu, Y.-H.; Li, Y.; Guan, X.-Y. TROAP Switches DYRK1 Activity to Drive Hepatocellular Carcinoma Progression. Cell Death Dis. 2021, 12, 125. [Google Scholar] [CrossRef]
- Mauro, L.J.; Seibel, M.I.; Diep, C.H.; Spartz, A.; Perez Kerkvliet, C.; Singhal, H.; Swisher, E.M.; Schwartz, L.E.; Drapkin, R.; Saini, S.; et al. Progesterone Receptors Promote Quiescence & Ovarian Cancer Cell Phenotypes via DREAM in P53-Mutant Fallopian Tube Models. J. Clin. Endocrinol. Metab. 2021. [Google Scholar] [CrossRef]
- Iness, A.N.; Rubinsak, L.; Meas, S.J.; Chaoul, J.; Sayeed, S.; Pillappa, R.; Temkin, S.M.; Dozmorov, M.G.; Litovchick, L. Oncogenic B-Myb Is Associated with Deregulation of the DREAM-Mediated Cell Cycle Gene Expression Program in High Grade Serous Ovarian Carcinoma Clinical Tumor Samples. Front. Oncol. 2021, 11, 637193. [Google Scholar] [CrossRef]
- Jang, S.M.; Azebi, S.; Soubigou, G.; Muchardt, C. DYRK1A Phoshorylates Histone H3 to Differentially Regulate the Binding of HP1 Isoforms and Antagonize HP1-Mediated Transcriptional Repression. EMBO Rep. 2014, 15, 686–694. [Google Scholar] [CrossRef]
- Kim, J.-H.; Li, L.; Resar, L.M. Doubling up on Function: Dual-Specificity Tyrosine-Regulated Kinase 1A (DYRK1A) in B Cell Acute Lymphoblastic Leukemia. J. Clin. Investig. 2021, 131. [Google Scholar] [CrossRef]
- Lee, P.; Bhansali, R.; Izraeli, S.; Hijiya, N.; Crispino, J.D. The Biology, Pathogenesis and Clinical Aspects of Acute Lymphoblastic Leukemia in Children with Down Syndrome. Leukemia 2016, 30, 1816–1823. [Google Scholar] [CrossRef]
- Liu, A.; Zhang, B.; Zhao, W.; Tu, Y.; Wang, Q.; Li, J. MicroRNA-215-5p Inhibits the Proliferation of Keratinocytes and Alleviates Psoriasis-like Inflammation by Negatively Regulating DYRK1A and Its Downstream Signaling Pathways. Exp. Dermatol. 2020. [Google Scholar] [CrossRef]
- Deshmukh, V.; O’Green, A.L.; Bossard, C.; Seo, T.; Lamangan, L.; Ibanez, M.; Ghias, A.; Lai, C.; Do, L.; Cho, S.; et al. Modulation of the Wnt Pathway through Inhibition of CLK2 and DYRK1A by Lorecivivint as a Novel, Potentially Disease-Modifying Approach for Knee Osteoarthritis Treatment. Osteoarthr. Cartil. 2019, 27, 1347–1360. [Google Scholar] [CrossRef]
- Yazici, Y.; McAlindon, T.E.; Gibofsky, A.; Lane, N.E.; Lattermann, C.; Skrepnik, N.; Swearingen, C.J.; Simsek, I.; Ghandehari, H.; DiFrancesco, A.; et al. A Phase 2b Randomized Trial of Lorecivivint, a Novel Intra-Articular CLK2/DYRK1A Inhibitor and Wnt Pathway Modulator for Knee Osteoarthritis. Osteoarthr. Cartil. 2021. [Google Scholar] [CrossRef]
- Deshmukh, V.; Seo, T.; Lauren O’Green, A.; Ibanez, M.; Hofilena, B.; Sunil, K.; Stewart, J.; Dellamary, L.; Chiu, K.; Ghias, A.; et al. SM04755, a Small-Molecule Inhibitor of the Wnt Pathway, as a Potential Topical Treatment for Tendinopathy. J. Orthop. Res. 2020. [Google Scholar] [CrossRef]
- Kisaka, J.K.; Ratner, L.; Kyei, G.B. The Dual-Specificity Kinase DYRK1A Modulates the Levels of Cyclin L2 To Control HIV Replication in Macrophages. J. Virol. 2020, 94. [Google Scholar] [CrossRef] [PubMed]
- Booiman, T.; Loukachov, V.V.; van Dort, K.A.; van’t Wout, A.B.; Kootstra, N.A. DYRK1A Controls HIV-1 Replication at a Transcriptional Level in an NFAT Dependent Manner. PLoS ONE 2015, 10, e0144229. [Google Scholar] [CrossRef] [PubMed]
- Bol, S.M.; Moerland, P.D.; Limou, S.; van Remmerden, Y.; Coulonges, C.; van Manen, D.; Herbeck, J.T.; Fellay, J.; Sieberer, M.; Sietzema, J.G.; et al. Genome-Wide Association Study Identifies Single Nucleotide Polymorphism in DYRK1A Associated with Replication of HIV-1 in Monocyte-Derived Macrophages. PLoS ONE 2011, 6, e17190. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, S.T.; Hutterer, C.; Egilmezer, E.; Steingruber, M.; Milbradt, J.; Marschall, M.; Rawlinson, W.D. Human Cytomegalovirus Utilises Cellular Dual-Specificity Tyrosine Phosphorylation-Regulated Kinases during Placental Replication. Placenta 2018, 72, 10–19. [Google Scholar] [CrossRef]
- Dirmeier, S.; Dächert, C.; van Hemert, M.; Tas, A.; Ogando, N.S.; van Kuppeveld, F.; Bartenschlager, R.; Kaderali, L.; Binder, M.; Beerenwinkel, N. Host Factor Prioritization for Pan-Viral Genetic Perturbation Screens Using Random Intercept Models and Network Propagation. PLoS Comput. Biol. 2020, 16, e1007587. [Google Scholar] [CrossRef]
- Zhou, N.; Yuan, S.; Wang, R.; Zhang, W.; Chen, J.J. Role of Dual Specificity Tyrosine-Phosphorylation-Regulated Kinase 1B (Dyrk1B) in S-Phase Entry of HPV E7 Expressing Cells from Quiescence. Oncotarget 2015, 6, 30745–30761. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Saluja, T.S.; Kumar, V.; Agrawal, M.; Tripathi, A.; Meher, R.K.; Srivastava, K.; Gupta, A.; Singh, A.; Chaturvedi, A.; Singh, S.K. Mitochondrial Stress-Mediated Targeting of Quiescent Cancer Stem Cells in Oral Squamous Cell Carcinoma. Cancer Manag. Res. 2020, 12, 4519–4530. [Google Scholar] [CrossRef]
- Chen, H.; Shen, J.; Choy, E.; Hornicek, F.J.; Shan, A.; Duan, Z. Targeting DYRK1B Suppresses the Proliferation and Migration of Liposarcoma Cells. Oncotarget 2018, 9, 13154–13166. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, S.; He, Z.; Sun, F.; Huang, Y.; Ni, Q.; Wang, H.; Wang, Y.; Cheng, C. Dyrk1B Overexpression Is Associated with Breast Cancer Growth and a Poor Prognosis. Hum. Pathol. 2017, 66, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Gruber, W.; Hutzinger, M.; Elmer, D.P.; Parigger, T.; Sternberg, C.; Cegielkowski, L.; Zaja, M.; Leban, J.; Michel, S.; Hamm, S.; et al. DYRK1B as Therapeutic Target in Hedgehog/GLI-Dependent Cancer Cells with Smoothened Inhibitor Resistance. Oncotarget 2016, 7, 7134–7148. [Google Scholar] [CrossRef]
- Tandon, V.; de la Vega, L.; Banerjee, S. Emerging Roles of DYRK2 in Cancer. J. Biol. Chem. 2020. [Google Scholar] [CrossRef]
- Mehnert, M.; Ciuffa, R.; Frommelt, F.; Uliana, F.; van Drogen, A.; Ruminski, K.; Gstaiger, M.; Aebersold, R. Multi-Layered Proteomic Analyses Decode Compositional and Functional Effects of Cancer Mutations on Kinase Complexes. Nat. Commun. 2020, 11, 3563. [Google Scholar] [CrossRef]
- Moreno, R.; Banerjee, S.; Jackson, A.W.; Quinn, J.; Baillie, G.; Dixon, J.E.; Dinkova-Kostova, A.T.; Edwards, J.; de la Vega, L. The Stress-Responsive Kinase DYRK2 Activates Heat Shock Factor 1 Promoting Resistance to Proteotoxic Stress. Cell Death Differ. 2020. [Google Scholar] [CrossRef]
- Banerjee, S.; Wei, T.; Wang, J.; Lee, J.J.; Gutierrez, H.L.; Chapman, O.; Wiley, S.E.; Mayfield, J.E.; Tandon, V.; Juarez, E.F.; et al. Inhibition of Dual-Specificity Tyrosine Phosphorylation-Regulated Kinase 2 Perturbs 26S Proteasome-Addicted Neoplastic Progression. Proc. Natl. Acad. Sci. USA 2019, 116, 24881–24891. [Google Scholar] [CrossRef] [PubMed]
- Koike, C.; Okudela, K.; Matsumura, M.; Mitsui, H.; Suzuki, T.; Arai, H.; Kataoka, T.; Ishikawa, Y.; Umeda, S.; Tateishi, Y.; et al. Frequent DYRK2 Gene Amplification in Micropapillary Element of Lung Adenocarcinoma—An Implication in Progression in EGFR-Mutated Lung Adenocarcinoma. Histol. Histopathol. 2020, 18294. [Google Scholar] [CrossRef]
- Park, C.S.; Lacorazza, H.D. DYRK2 Controls a Key Regulatory Network in Chronic Myeloid Leukemia Stem Cells. Exp. Mol. Med. 2020, 52, 1663–1672. [Google Scholar] [CrossRef]
- Park, C.S.; Lewis, A.H.; Chen, T.J.; Bridges, C.S.; Shen, Y.; Suppipat, K.; Puppi, M.; Tomolonis, J.A.; Pang, P.D.; Mistretta, T.-A.; et al. A KLF4-DYRK2-Mediated Pathway Regulating Self-Renewal in CML Stem Cells. Blood 2019, 134, 1960–1972. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Zhang, L.; Wang, D.; Bao, Y.; Liu, C.; Xu, Z.; Huang, W.; Cheng, C. Regulation of Glioma Cells Migration by DYRK2. Neurochem. Res. 2017, 42, 3093–3102. [Google Scholar] [CrossRef] [PubMed]
- Kumamoto, T.; Yamada, K.; Yoshida, S.; Aoki, K.; Hirooka, S.; Eto, K.; Yanaga, K.; Yoshida, K. Impairment of DYRK2 by DNMT1-mediated Transcription Augments Carcinogenesis in Human Colorectal Cancer. Int. J. Oncol. 2020, 56, 1529–1539. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama-Mashima, S.; Yogosawa, S.; Kanegae, Y.; Hirooka, S.; Yoshida, S.; Horiuchi, T.; Ohashi, T.; Yanaga, K.; Saruta, M.; Oikawa, T.; et al. Forced Expression of DYRK2 Exerts Anti-Tumor Effects via Apoptotic Induction in Liver Cancer. Cancer Lett. 2019, 451, 100–109. [Google Scholar] [CrossRef]
- Wozniak, J.M.; Silva, T.A.; Thomas, D.; Siqueira-Neto, J.L.; McKerrow, J.H.; Gonzalez, D.J.; Calvet, C.M. Molecular Dissection of Chagas Induced Cardiomyopathy Reveals Central Disease Associated and Druggable Signaling Pathways. PLoS Negl. Trop. Dis. 2020, 14, e0007980. [Google Scholar] [CrossRef]
- Bogacheva, O.; Bogachev, O.; Menon, M.; Dev, A.; Houde, E.; Valoret, E.I.; Prosser, H.M.; Creasy, C.L.; Pickering, S.J.; Grau, E.; et al. DYRK3 Dual-Specificity Kinase Attenuates Erythropoiesis during Anemia. J. Biol. Chem. 2008, 283, 36665–36675. [Google Scholar] [CrossRef]
- Reed, K.S.M.; Ulici, V.; Kim, C.; Chubinskaya, S.; Loeser, R.F.; Phanstiel, D.H. Transcriptional Response of Human Articular Chondrocytes Treated with Fibronectin Fragments: An in Vitro Model of the Osteoarthritis Phenotype. Osteoarth. Cartil. 2021, 29, 235–247. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.-C.; Veeraraghavan, J.; Tan, Y.; Kim, J.-A.; Wang, X.; Loo, S.K.; Lee, S.; Hu, Y.; Wang, X.-S. A Novel Neoplastic Fusion Transcript, RAD51AP1-DYRK4, Confers Sensitivity to the MEK Inhibitor Trametinib in Aggressive Breast Cancers. Clin. Cancer Res. 2021, 27, 785–798. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Phoa, A.F.; Abbassi, R.H.; Hoque, M.; Reekie, T.A.; Font, J.S.; Ryan, R.M.; Stringer, B.W.; Day, B.W.; Johns, T.G.; et al. Structural Optimization and Pharmacological Evaluation of Inhibitors Targeting Dual-Specificity Tyrosine Phosphorylation-Regulated Kinases (DYRK) and CDC-like Kinases (CLK) in Glioblastoma. J. Med. Chem. 2017, 60, 2052–2070. [Google Scholar] [CrossRef]
- Hutterer, C.; Milbradt, J.; Hamilton, S.; Zaja, M.; Leban, J.; Henry, C.; Vitt, D.; Steingruber, M.; Sonntag, E.; Zeitträger, I.; et al. Inhibitors of Dual-Specificity Tyrosine Phosphorylation-Regulated Kinases (DYRK) Exert a Strong Anti-Herpesviral Activity. Antiviral Res. 2017, 143, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Dominguez, D.; Tsai, Y.-H.; Weatheritt, R.; Wang, Y.; Blencowe, B.J.; Wang, Z. An Extensive Program of Periodic Alternative Splicing Linked to Cell Cycle Progression. eLife 2016, 5. [Google Scholar] [CrossRef]
- Uzor, S.; Zorzou, P.; Bowler, E.; Porazinski, S.; Wilson, I.; Ladomery, M. Autoregulation of the Human Splice Factor Kinase CLK1 through Exon Skipping and Intron Retention. Gene 2018, 670, 46–54. [Google Scholar] [CrossRef]
- Hartmann, A.M.; Rujescu, D.; Giannakouros, T.; Nikolakaki, E.; Goedert, M.; Mandelkow, E.-M.; Gao, Q.S.; Andreadis, A.; Stamm, S. Regulation of Alternative Splicing of Human Tau Exon 10 by Phosphorylation of Splicing Factors. Mol. Cell. Neurosci. 2001, 18, 80–90. [Google Scholar] [CrossRef]
- Glatz, D.C.; Rujescu, D.; Tang, Y.; Berendt, F.J.; Hartmann, A.M.; Faltraco, F.; Rosenberg, C.; Hulette, C.; Jellinger, K.; Hampel, H.; et al. The Alternative Splicing of Tau Exon 10 and Its Regulatory Proteins CLK2 and TRA2-BETA1 Changes in Sporadic Alzheimer’s Disease. J. Neurochem. 2006, 96, 635–644. [Google Scholar] [CrossRef]
- Li, H.; Cui, X.; Hu, Q.; Chen, X.; Zhou, P. CLK3 Is A Direct Target of MiR-144 And Contributes To Aggressive Progression In Hepatocellular Carcinoma. Onco Targets Ther. 2019, 12, 9201–9213. [Google Scholar] [CrossRef]
- Bowler, E.; Porazinski, S.; Uzor, S.; Thibault, P.; Durand, M.; Lapointe, E.; Rouschop, K.M.A.; Hancock, J.; Wilson, I.; Ladomery, M. Hypoxia Leads to Significant Changes in Alternative Splicing and Elevated Expression of CLK Splice Factor Kinases in PC3 Prostate Cancer Cells. BMC Cancer 2018, 18, 355. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Lin, M.; Feng, X.; Ma, F.; Zhu, Y.; Liu, X.; Qu, C.; Sui, H.; Sun, B.; Zhu, A.; et al. Targeting CLK3 Inhibits the Progression of Cholangiocarcinoma by Reprogramming Nucleotide Metabolism. J. Exp. Med. 2020, 217. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Yang, H.; Zhang, W.; Liang, Z.; Huang, Q.; Xu, G.; Zhen, X.; Zheng, L.T. Clk1-Regulated Aerobic Glycolysis Is Involved in Glioma Chemoresistance. J. Neurochem. 2017, 142, 574–588. [Google Scholar] [CrossRef]
- Liu, B.; Kong, X.; Wang, R.; Xin, C. CLK2 Promotes Occurrence and Development of Non-Small Cell Lung Cancer. J. BUON 2021, 26, 58–64. [Google Scholar]
- Sako, Y.; Ninomiya, K.; Okuno, Y.; Toyomoto, M.; Nishida, A.; Koike, Y.; Ohe, K.; Kii, I.; Yoshida, S.; Hashimoto, N.; et al. Development of an Orally Available Inhibitor of CLK1 for Skipping a Mutated Dystrophin Exon in Duchenne Muscular Dystrophy. Sci. Rep. 2017, 7, 46126. [Google Scholar] [CrossRef]
- Artarini, A.; Meyer, M.; Shin, Y.J.; Huber, K.; Hilz, N.; Bracher, F.; Eros, D.; Orfi, L.; Keri, G.; Goedert, S.; et al. Regulation of Influenza A Virus MRNA Splicing by CLK1. Antiviral Res. 2019, 168, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Xu, L.-J.; Lian, W.-W.; Pang, X.-C.; Jia, H.; Liu, A.-L.; Du, G.-H. Anti-Influenza Effect and Action Mechanisms of the Chemical Constituent Gallocatechin-7-Gallate from Pithecellobium Clypearia Benth. Acta Pharmacol. Sin. 2018, 39, 1913–1922. [Google Scholar] [CrossRef]
- Zu, M.; Li, C.; Fang, J.-S.; Lian, W.-W.; Liu, A.-L.; Zheng, L.-S.; Du, G.-H. Drug Discovery of Host CLK1 Inhibitors for Influenza Treatment. Molecules 2015, 20, 19735–19747. [Google Scholar] [CrossRef] [PubMed]
- An, J.; Nakajima, T.; Shibata, H.; Arimura, T.; Yasunami, M.; Kimura, A. A Novel Link of HLA Locus to the Regulation of Immunity and Infection: NFKBIL1 Regulates Alternative Splicing of Human Immune-Related Genes and Influenza Virus M Gene. J. Autoimmun. 2013, 47, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Karlas, A.; Machuy, N.; Shin, Y.; Pleissner, K.-P.; Artarini, A.; Heuer, D.; Becker, D.; Khalil, H.; Ogilvie, L.A.; Hess, S.; et al. Genome-Wide RNAi Screen Identifies Human Host Factors Crucial for Influenza Virus Replication. Nature 2010, 463, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.; Xu, S.; Deyanat-Yazdi, G.; Peng, S.X.; Barnes, L.A.; Narla, R.K.; Tran, T.; Mikolon, D.; Ning, Y.; Shi, T.; et al. Synthetic Lethal Strategy Identifies a Potent and Selective TTK and CLK1/2 Inhibitor for Treatment of Triple-Negative Breast Cancer with a Compromised G1-S Checkpoint. Mol. Cancer Ther. 2018, 17, 1727–1738. [Google Scholar] [CrossRef] [PubMed]
- Wong, R.; Balachandran, A.; Mao, A.Y.; Dobson, W.; Gray-Owen, S.; Cochrane, A. Differential Effect of CLK SR Kinases on HIV-1 Gene Expression: Potential Novel Targets for Therapy. Retrovirology 2011, 8, 47. [Google Scholar] [CrossRef] [PubMed]
- Bidinosti, M.; Botta, P.; Krüttner, S.; Proenca, C.C.; Stoehr, N.; Bernhard, M.; Fruh, I.; Mueller, M.; Bonenfant, D.; Voshol, H.; et al. CLK2 Inhibition Ameliorates Autistic Features Associated with SHANK3 Deficiency. Science 2016, 351, 1199–1203. [Google Scholar] [CrossRef]
- Iwai, K.; Yaguchi, M.; Nishimura, K.; Yamamoto, Y.; Tamura, T.; Nakata, D.; Dairiki, R.; Kawakita, Y.; Mizojiri, R.; Ito, Y.; et al. Anti-Tumor Efficacy of a Novel CLK Inhibitor via Targeting RNA Splicing and MYC-Dependent Vulnerability. EMBO Mol. Med. 2018, 10. [Google Scholar] [CrossRef]
- Salvador, F.; Gomis, R.R. CLK2 Blockade Modulates Alternative Splicing Compromising MYC-Driven Breast Tumors. EMBO Mol. Med. 2018, 10. [Google Scholar] [CrossRef]
- Riggs, J.R.; Nagy, M.; Elsner, J.; Erdman, P.; Cashion, D.; Robinson, D.; Harris, R.; Huang, D.; Tehrani, L.; Deyanat-Yazdi, G.; et al. The Discovery of a Dual TTK Protein Kinase/CDC2-Like Kinase (CLK2) Inhibitor for the Treatment of Triple Negative Breast Cancer Initiated from a Phenotypic Screen. J. Med. Chem. 2017, 60, 8989–9002. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Kim, J.H.; Carver, K.; Su, Y.; Weremowicz, S.; Mulvey, L.; Yamamoto, S.; Brennan, C.; Mei, S.; Long, H.; et al. CLK2 Is an Oncogenic Kinase and Splicing Regulator in Breast Cancer. Cancer Res. 2015, 75, 1516–1526. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Piao, Y.; Thomas, C.; Fuller, G.N.; de Groot, J.F. Cdc2-like Kinase 2 Is a Key Regulator of the Cell Cycle via FOXO3a/P27 in Glioblastoma. Oncotarget 2016, 7, 26793–26805. [Google Scholar] [CrossRef]
- Park, S.Y.; Mittal, S.; Dong, J.; Jeong, K.; Martinez-Ledesma, E.; Piao, Y.; Khan, S.; Henry, V.; Verhaak, R.G.; Majd, N.; et al. Depletion of CLK2 Sensitizes Glioma Stem-like Cells to PI3K/MTOR and FGFR Inhibitors. Am. J. Cancer Res. 2020, 10, 3765–3783. [Google Scholar]
- Tam, B.Y.; Chiu, K.; Chung, H.; Bossard, C.; Nguyen, J.D.; Creger, E.; Eastman, B.W.; Mak, C.C.; Ibanez, M.; Ghias, A.; et al. The CLK Inhibitor SM08502 Induces Anti-Tumor Activity and Reduces Wnt Pathway Gene Expression in Gastrointestinal Cancer Models. Cancer Lett. 2020, 473, 186–197. [Google Scholar] [CrossRef] [PubMed]
- Murai, A.; Ebara, S.; Sasaki, S.; Ohashi, T.; Miyazaki, T.; Nomura, T.; Araki, S. Synergistic Apoptotic Effects in Cancer Cells by the Combination of CLK and Bcl-2 Family Inhibitors. PLoS ONE 2020, 15, e0240718. [Google Scholar] [CrossRef]
- Solyakov, L.; Halbert, J.; Alam, M.M.; Semblat, J.-P.; Dorin-Semblat, D.; Reininger, L.; Bottrill, A.R.; Mistry, S.; Abdi, A.; Fennell, C.; et al. Global Kinomic and Phospho-Proteomic Analyses of the Human Malaria Parasite Plasmodium Falciparum. Nat. Commun. 2011, 2, 565. [Google Scholar] [CrossRef]
- Agarwal, S.; Kern, S.; Halbert, J.; Przyborski, J.M.; Baumeister, S.; Dandekar, T.; Doerig, C.; Pradel, G. Two Nucleus-Localized CDK-like Kinases with Crucial Roles for Malaria Parasite Erythrocytic Replication Are Involved in Phosphorylation of Splicing Factor. J. Cell Biochem. 2011, 112, 1295–1310. [Google Scholar] [CrossRef]
- Kern, S.; Agarwal, S.; Huber, K.; Gehring, A.P.; Strödke, B.; Wirth, C.C.; Brügl, T.; Abodo, L.O.; Dandekar, T.; Doerig, C.; et al. Inhibition of the SR Protein-Phosphorylating CLK Kinases of Plasmodium Falciparum Impairs Blood Stage Replication and Malaria Transmission. PLoS ONE 2014, 9, e105732. [Google Scholar] [CrossRef]
- Alam, M.M.; Sanchez-Azqueta, A.; Janha, O.; Flannery, E.L.; Mahindra, A.; Mapesa, K.; Char, A.B.; Sriranganadane, D.; Brancucci, N.M.B.; Antonova-Koch, Y.; et al. Validation of the Protein Kinase PfCLK3 as a Multistage Cross-Species Malarial Drug Target. Science 2019, 365. [Google Scholar] [CrossRef] [PubMed]
- Mahindra, A.; Janha, O.; Mapesa, K.; Sanchez-Azqueta, A.; Alam, M.M.; Amambua-Ngwa, A.; Nwakanma, D.C.; Tobin, A.B.; Jamieson, A.G. Development of Potent PfCLK3 Inhibitors Based on TCMDC-135051 as a New Class of Antimalarials. J. Med. Chem. 2020, 63, 9300–9315. [Google Scholar] [CrossRef] [PubMed]
- Mahmud, F.; Lee, P.C.; Abdul Wahab, H.; Mustaffa, K.M.F.; Leow, C.H.; Azhar, R.; Lai, N.S. Plasmodium Falciparum Protein Kinase as a Potential Therapeutic Target for Antimalarial Drugs Development. Trop. Biomed. 2020, 37, 822–841. [Google Scholar] [CrossRef] [PubMed]
- Kii, I.; Sumida, Y.; Goto, T.; Sonamoto, R.; Okuno, Y.; Yoshida, S.; Kato-Sumida, T.; Koike, Y.; Abe, M.; Nonaka, Y.; et al. Selective Inhibition of the Kinase DYRK1A by Targeting Its Folding Process. Nat. Commun. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Bain, J.; McLauchlan, H.; Elliott, M.; Cohen, P. The Specificities of Protein Kinase Inhibitors: An Update. Biochem. J. 2003, 371, 199–204. [Google Scholar] [CrossRef]
- Grabher, P.; Durieu, E.; Kouloura, E.; Halabalaki, M.; Skaltsounis, L.A.; Meijer, L.; Hamburger, M.; Potterat, O. Library-Based Discovery of DYRK1A/CLK1 Inhibitors from Natural Product Extracts. Planta Med. 2012, 78, 951–956. [Google Scholar] [CrossRef]
- Debdab, M.; Renault, S.; Lozach, O.; Meijer, L.; Paquin, L.; Carreaux, F.; Bazureau, J.-P. Synthesis and Preliminary Biological Evaluation of New Derivatives of the Marine Alkaloid Leucettamine B as Kinase Inhibitors. Eur. J. Med. Chem. 2010, 45, 805–810. [Google Scholar] [CrossRef]
- Deshmukh, V.; Hu, H.; Barroga, C.; Bossard, C.; Kc, S.; Dellamary, L.; Stewart, J.; Chiu, K.; Ibanez, M.; Pedraza, M.; et al. A Small-Molecule Inhibitor of the Wnt Pathway (SM04690) as a Potential Disease Modifying Agent for the Treatment of Osteoarthritis of the Knee. Osteoarth. Cartil. 2018, 26, 18–27. [Google Scholar] [CrossRef]
- Yazici, Y.; McAlindon, T.E.; Gibofsky, A.; Lane, N.E.; Clauw, D.; Jones, M.; Bergfeld, J.; Swearingen, C.J.; DiFrancesco, A.; Simsek, I.; et al. Lorecivivint, a Novel Intraarticular CDC-like Kinase 2 and Dual-Specificity Tyrosine Phosphorylation-Regulated Kinase 1A Inhibitor and Wnt Pathway Modulator for the Treatment of Knee Osteoarthritis: A Phase II Randomized Trial. Arthritis Rheumatol. 2020, 72, 1694–1706. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).