Abstract
An amphipathic α-helical peptide, Hp1404, was isolated from the venomous gland of the scorpion Heterometrus petersii. Hp1404 exhibits antimicrobial activity against methicillin-resistant Staphylococcus aureus but is cytotoxic. In this study, we designed antimicrobial peptides by substituting amino acids at the 14 C-terminal residues of Hp1404 to reduce toxicity and improve antibacterial activity. The analog peptides, which had an amphipathic α-helical structure, were active against gram-positive and gram-negative bacteria, particularly multidrug-resistant Acinetobacter baumannii, and showed lower cytotoxicity than Hp1404. N-phenyl-1-naphthylamine uptake and DisC3-5 assays demonstrated that the peptides kill bacteria by effectively permeating the outer and cytoplasmic membranes. Additionally, the analog peptides inhibited biofilm formation largely than Hp1404 at low concentrations. These results suggest that the analog peptides of Hp1404 can be used as therapeutic agents against A. baumannii infection.
1. Introduction
Acinetobacter baumannii is a rod-shaped gram-negative bacterium and among the major causes of opportunistic nosocomial infections, known as ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, A. baumannii, Pseudomonas aeruginosa and Enterobacter species). ESKAPE pathogens are highly antibiotic-resistant and difficult to treat, accounting for the majority of nosocomial and community-acquired infections and show high mortality rates [1,2]. The World Health Organization recently cited antibacterial resistance as one of the three most important problems facing human health and reported that the number of multidrug-resistant (MDR) A. baumannii infections has increased rapidly, posing a public health threat worldwide [3,4,5,6]. To treat A. baumannii infection, antibiotics such as β-lactams or carbapenems with a broad spectrum are used as first-line options, but strains that have recently gained carbapenem resistance have emerged. Polymyxin or tigecycline is used to treat carbapenem-resistant A. baumannii [7], but most strains are multidrug resistant, and thus treatment options are very limited [8]. Multidrug-resistant A. baumannii causes opportunistic infections, including septicemia, pneumonia, endocarditis, skin and wound infections and urinary tract infections following hospitalization of patients with severe illness [9,10]. This increased bacterial resistance to antibiotics is attributed to A. baumannii’s ability to form biofilms.
Biofilms are communities of microorganisms attached to a solid surface and produce a matrix known as extracellular polymeric substances (EPS) [11]. EPS are composed mostly of polysaccharides, glycoproteins, glycolipids and extracellular DNA [12,13,14,15]. The matrix acts to protect bacteria, enabling them to survive in difficult-to-grow environments. Therefore, enhanced resistance or resistance to antibiotics and other antimicrobial agents compared to plankton bacteria is a typical feature of biofilms. Most bacterial infections are thought to be related to bacterial biofilms, and many cases of device-related infections have been reported. Device-related infections typically occur in medical devices such as catheters, contact lenses, mechanical heart valves and artificial joints [16]. Therefore, the development of new antimicrobial drugs against these multidrug-resistant pathogens is needed; antimicrobial peptides (AMPs) are considered an alternative to antibiotics.
Organisms such as humans, animals and plants are exposed to various microbial at-tacks; they protect themselves from these threats by producing antimicrobial peptides (AMPs) via the innate immune system [17,18]. AMPs, which have cationic and amphipathic properties, consist of fewer than 50 amino acids and show potent activity against bacteria, fungi, yeast and viruses [19,20,21]. AMPs also exert anticancer activities and regulate inflammatory responses and wound healing [22,23,24]. Killing of bacteria leads to membrane destruction by causing the formation of pores or micelles and inhibition of protein synthesis or DNA replication by binding to RNA and DNA [25]. Despite the outstanding properties of AMPs, they exhibit disadvantages such as hemolytic activity against red blood cells and sensitivity to salt. Since antimicrobial peptides have the disadvantage of being sensitive to a salt environment, it is important to develop peptides that maintain their antibacterial activity in the presence of salts [26]. AMPs that can overcome these shortcomings would be promising candidates for treating bacterial infectious diseases [27].
In a previous study, the new amphipathic peptide Hp1404 was identified from the venomous gland cDNA library of the scorpion Heterometrus petersii [28]. Hp1404 consists of 14 amino acids, has a net charge of + 1 and exhibits an α-helical structure in a bacterial membrane-mimicking environment. Hp1404 has been reported to have potential antibac-terial activity, including against only gram-positive bacteria, such as Staphylococcus aureus. However, Hp1404 has a relatively cytotoxicity in mammalian cells and a high level of hemolysis of red blood cells.
Hence, the purpose of this study has been to design a peptide less toxic than its parent Hp1404 peptide and to investigate the antibacterial activity of the newly designed antimicrobial peptide on multidrug-resistant strains. We designed analog peptides by replacing the position 14 amino acid with different amino acids. Antimicrobial activity against gram-positive, gram-negative and multidrug-resistant bacteria of the designed analog peptides was tested, and their mechanism of action was examined. Furthermore, we investigated the activity of peptides against A. baumannii strains and their ability to inhibit and eliminate bacterial biofilms. Our findings suggest that the newly designed peptides have the potential to be promising candidates as antimicrobial agents against A. baumannii strains.
2. Results
2.1. Peptide Design and Characterization
The analog peptides were designed based on the peptide Hp1404 by substituting the F residue with six different amino acids (A, K, V, L, I and W). Wheel diagram analysis showed that hydrophobic and hydrophilic amino acids formed an amphipathic structure (Figure 1). The peptide sequence, retention time, molecular weight, hydrophobicity and net charge of Hp1404 and its analog peptides are listed in Table 1. The hydrophobicity values of Hp1404, Hp1404-A, Hp1404-K, Hp1404-V, Hp1404-L, Hp1404-I and Hp1404-W were 0.686, 0.580, 0.487, 0.645, 0.679, 0.686 and 0.719, respectively. The parent peptide and five analog peptides showed a net charge of +1, whereas Hp1404-K had a net charge of +2.
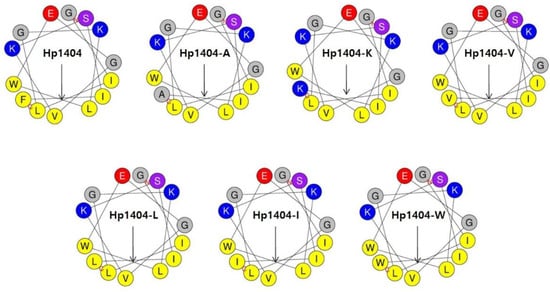
Figure 1.
Wheel diagram of Hp1404 and its analog peptides. Wheel diagram was obtained from the HeliQuest site. Negatively charged residue is shown in red, positively charged residues are shown in blue, hydrophobic residues are shown in yellow and certain polar residues are shown in purple. Arrows indicate helical hydrophobic moments.

Table 1.
Sequence and physicochemical properties of Hp1404 and its analog peptides.
2.2. Secondary Structure of the Peptides
Circular dichroism (CD) spectroscopy was performed to determine the secondary structure of the peptides in various environments. This analysis was performed in 10 mM SP buffer (aqueous environment), bacterial membrane-mimicking solvents such as 30 mM sodium dodecyl sulfate (SDS; negatively charged environment) and 50% 2, 2, 2-trifluoroethanol (TFE; hydrophobic environment). As shown in Figure 2, in the aqueous environment, all peptides displayed a random coil conformation, as indicated by the minimum peak at 198 nm. In the presence of 30 mM SDS and 50% TFE, the parent peptide and all analog peptides had an α-helical conformation, as demonstrated by minimum peaks at 208 and 222 nm. These results showed that the analog peptides affected the bacterial membrane by forming an α-helix structure.
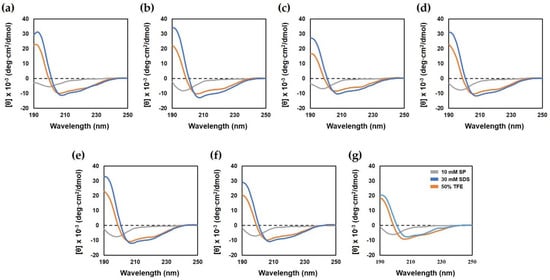
Figure 2.
Circular dichroism spectra of Hp1404 and its analog peptides. The secondary structure of peptides was measured in 10 mM SP buffer, 50% TFE and 30 mM SDS by CD spectrometry. (a) Hp1404, (b) Hp1404-A, (c) Hp1404-K, (d) Hp1404-V, (e) Hp1404-L (f) Hp1404-I and (g) Hp1404-W peptide concentration was fixed at 40 μM.
2.3. Antimicrobial Activity
The antimicrobial activity of Hp1404 and its analog peptides against gram-positive and gram-negative bacteria was determined using the two-fold broth microdilution method. The results are summarized in Table 2. The parent peptide, Hp1404, displayed minimum inhibitory concentration at 3.13 ranging from 12.5 μM against gram-positive and gram-negative bacteria. All four analog peptides (except Hp1404-A and Hp1404-K) showed broad-spectrum antimicrobial activities, whereas Hp1404-A and Hp1404-K exhibited weak effects towards only a few of these tested strains. Hp1404-L and Hp1404-I exhibited a minimum inhibitory concentration (MIC) of 3.13 μM against A. baumannii.

Table 2.
Antimicrobial activity of peptides against gram-positive and gram-negative bacteria.
In addition, these analog peptides were effective against multidrug-resistant A. baumannii strains, with Hp1404-L and Hp1404-W showing the same activity as the parent peptide (Table 3). Thus, the analog peptides have excellent antibacterial activity against A. baumannii.

Table 3.
Antimicrobial activity of peptides and antibiotics against A. baumannii strains isolated from patients.
2.4. Cytotoxicity Assay
We evaluated the hemolytic activity of the peptides against mouse erythrocytes. All peptides with amino acid substitutions showed lower hemolysis than Hp1404 (Figure 3a). At the highest tested concentration of 50 μM, Hp1404, Hp1404-L and Hp1404-W showed hemolytic activities of 49.2%, 45.8% and 15.9%, respectively. However, Hp1404-A, Hp1404-K and Hp1404-V showed no hemolytic activity. The parent peptide Hp1404 showed hemolytic activity of 41.7%, similar to the rate at 50 μM, whereas Hp1404-L, Hp1404-I and Hp1404-W induced 21.3%, 0% and 1.7% hemolysis at 25 μM. Melittin, used as a positive control, showed 42% hemolytic activity at concentrations up to 1.56 μM, indicating that it is toxic.
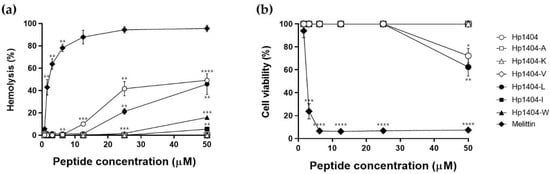
Figure 3.
Hemolytic activity and cytotoxicity of peptides. (a) Hemolytic activity of peptides against 8% mouse red blood cells. (b) Cytotoxicity of peptides against HaCaT cells determined by MTT assay. Data are presented as the mean ± SEM of three independent experiments. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001 versus the control group.
To confirm the cytotoxicity of Hp1404 and its analog peptides toward human keratinocyte HaCaT cells, treatment with various peptide concentrations after 24 h was measured by MTT assay. Similar to the hemolytic activity, cell viability was lowest after treatment with Hp1404 and Hp1404-L with values of 72.3% and 62.4%, respectively, against HaCaT cells at 50 μM. Cell viability was 100% after treatment with Hp1404-A, Hp1404-V, Hp1404-I and Hp1404-W (Figure 3b). Compared to melittin, analog peptides showed significantly lower toxicity. Cell viability was high after treatment with all analog peptides, indicating that they were not cytotoxic.
2.5. Calcein Leakage Assay
As shown in Figure 4a,b, to evaluate the toxicity of the peptides via their effects on the cell membranes, large unilamellar vesicles (LUVs) containing calcein were used in a leakage assay. Upon treatment with various concentrations of peptides at LUVs, calcein was released in a concentration-dependent manner. For Hp1404-A and Hp1404-K, phosphatidylcholine: cholesterol (PC:CH) liposomes (mimicking human erythrocyte membrane) and PC:CH: sphingomyelin (SM) liposomes (mimicking mammalian cell membrane) did not release calcein, indicating no toxicity. Following treatment with melittin, which was used as a positive control, regardless of the concentration, the fluorescence intensity of calcein was released up to 1500 arbitrary units (a.u.). Hp1404-V, Hp1404-I, Hp1404-L and Hp1404-W caused similar calcein leakage as the parent peptide Hp1404 and was less toxic than melittin. Furthermore, calcein leakage was assessed using a fluorescence-labeled organism bioimaging instrument. As shown in Figure 4c, the analog peptides (Hp1404-V, Hp1404-I, Hp1404-L and Hp1404-W) caused lower leakage activity in liposomes compared to the positive control melittin and parent peptide Hp1404, and no fluorescence was observed for Hp1404-A and Hp1404-K. Based on these results, the analog peptides are not highly toxic towards mammalian cell membranes.
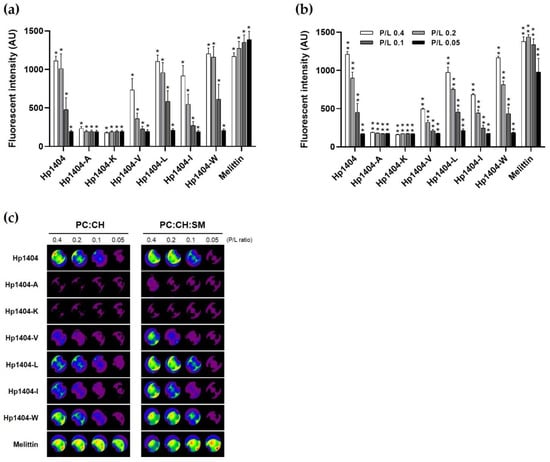
Figure 4.
Calcein leakage assay. (a) PC:CH (1:1, w/w) and (b) PC:CH:SM (1:1:1, w/w) LUV emission intensity after treatment with the peptides. (c) Visualization of calcein release from calcein-loaded LUVs using a fluorescence-labeled organism bioimaging instrument. Data are presented as the mean ± SEM of three independent experiments using one-way ANOVA. * p < 0.05, ** p < 0.01.
2.6. Salt Stability
To evaluate the effect of a salt-containing environment on the antibacterial peptide, peptides were assessed by MIC assay in the presence of 150 mM NaCl. As shown in Figure 5, the MIC values of Hp1404, Hp1404-L and Hp1404-W for A. baumannii KCTC 2508 increased from 3.13 to 6.25 μM and Hp1404-I and Hp1404-V increased from 3.13 to 12.5 μM and 6.25 to 25 μM, respectively, reducing the antibacterial activity. In all resistant strains, Hp1404 and analog peptides has antibacterial activity at 4-fold the MIC value.
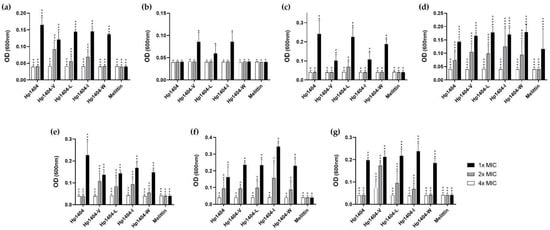
Figure 5.
Antibacterial activity of peptides in salt environments. Salt stability of peptides were measured in the presence of 150 mM NaCl against A. baumannii. (a) A. baumannii KCTC 2508, (b) A. baumannii 409081, (c) A. baumannii 719705, (d) A. baumannii 907233, (e) A. baumannii #2, (f) A. baumannii #3 and (g) A. baumannii #4. Data are presented as the mean ± SEM of three independent experiments using one-way ANOVA. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001.
2.7. Time-Killing Assay
To measure bacterial mortality over time caused by the peptides, we performed time-kill analysis. The peptides, except for Hp1404-I and Hp1404-V at 1x MIC, quickly killed the bacteria within 60 min. However, at 4× MIC, analog peptides killed bacteria within 10 min, similar to the parent peptide Hp1404. Based on these results, both Hp1404 and its analog peptides killed bacteria in a short time (Figure 6).
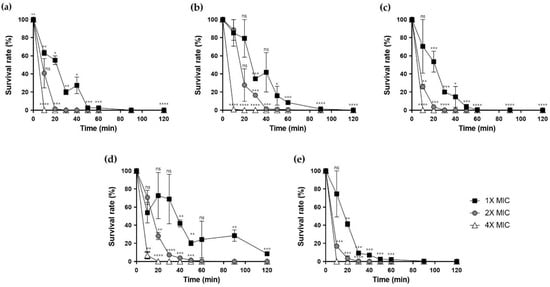
Figure 6.
Time-killing assay. Peptides were prepared at concentrations of 1×, 2× and 4× MIC and treated against A. baumannii KCTC 2508. (a) Hp1404, (b) Hp1404-V, (c) Hp1404-L, (d) Hp1404-I and (e) Hp1404-W. Data are presented as the mean ± SEM of three independent experiments. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001 versus the control group. ns = Not statistically significant.
2.8. Biofilm Inhibition
We quantified biofilm formation in a crystal violet staining assay. The absorbance values of the standard strain and six clinically isolated strains were measured, and the highest formation rate of the four strains was used to confirm biofilm inhibition ability. As shown in Figure 7a, Hp1404 and its 4 analog peptides (Hp1404-V, Hp1404-L, Hp1404-I and Hp1404-W) inhibited A. baumannii KCTC 2508 biofilm formation at 6.25, 12.5, 6.25, 6.25 and 6.25 μM, respectively. The antibiotics meropenem and polymyxin B also inhibited biofilm formation at 6.25 μM. For A. baumannii #3 and 409081, Hp1404 inhibited biofilm formation at 6.25 μM and its 4 analog peptides inhibited this formation at 12.5 μM. Meropenem did not affect biofilm inhibition at less than 25 μM, and polymyxin B showed inhibition at 6.25 μM (Figure 7b,d). In A. baumannii #4, Hp1404, Hp1404-V, Hp1404-L, Hp1404-I and Hp1404-W inhibited biofilm formation at 6.25, 12.5, 6.25, 12.5 and 12.5 μM, respectively. Additionally, Hp1404, Hp1404-V, Hp1404-L, Hp1404-I, and Hp1404-W showed biofilm inhibition effects at 6.25, 12.5, 6.25, 12.5 and 12.5 μM, respectively, in A. baumannii 719705. Similarly, for A. baumannii #3 and 719705, meropenem exhibited no biofilm inhibition activity and polymyxin B showed activity at 6.25 μM (Figure 7c,e).
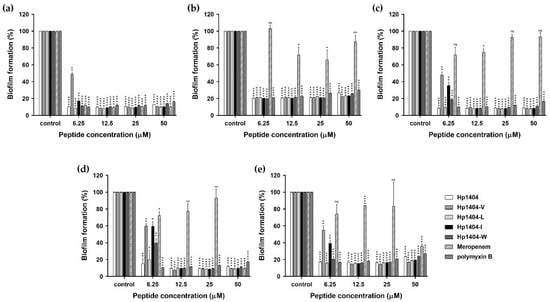
Figure 7.
Anti-biofilm activity of peptides and antibiotics. The biofilm mass was measured in a crystal violet assay at wavelength 595 nm. (a) A. baumannii KCTC 2508, (b) A. baumannii 409081, (c) A. baumannii 719705 and (d) A. baumannii #3, and (e) A. baumannii #4. Data are presented as the mean ± SEM of three independent experiments. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001 versus the control group. ns = Not statistically significant.
2.9. Biofilm Reduction and Visualization
We performed a crystal violet staining assay to evaluate the biofilm reduction ability of the peptides and antibiotics (Figure 8a–c). In A. baumannii KCTC 2508, Hp1404, Hp1404-V, Hp1404-L, Hp1404-I and Hp1404-W showed reduced biofilm activity at 50 μM, with reductions of 74.06%, 79.50%, 84.72%, 83.82% and 79.90%, respectively. Hp1404 and its analog peptides (Hp1404-V, Hp1404-L, Hp1404-I and Hp1404-W) reduced biofilm formation by up to 80% in the baumannii #3 at 100, 50, 50, 50 and 50 μM, respectively. For A. baumannii 409081, Hp1404, Hp1404-V, Hp1404-L, Hp1404-I and Hp1404-W peptides of 50 μM reduced biofilm formation by 34.52%, 53.23%, 64.76%, 72.68% and 65.08%, respectively. All analog peptides removed the biofilm at a lower concentration compared to Hp1404. Polymyxin B showed lower MIC values than the peptides and strong biofilm inhibition effects, but the ability to remove formed biofilm was low.
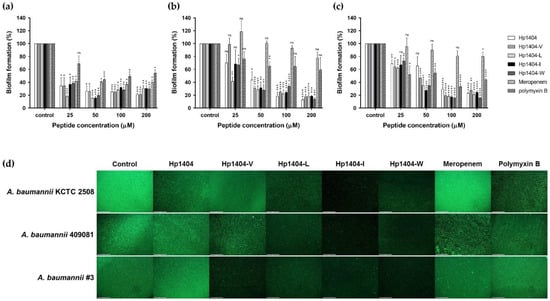
Figure 8.
Biofilm reduction ability of peptides and antibiotics. The biofilm reduction rate was determined by crystal violet staining. (a) A. baumannii KCTC 2508, (b) A. baumannii #3 and (c) A. baumannii 409081. (d) Representative fluorescence images of A. baumannii strains stained with SYTO 9 (green) and evaluated using an EVOS™ FL Auto 2 Imaging System. Scale bar = 200 μm. Data are presented as the mean ± SEM of three independent experiments. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001 versus the control group. ns = Not statistically significant.
The biofilm reduction ability was confirmed by visualizing A. baumannii biofilms using an EVOS™ FL Auto 2 imaging system (Figure 8d). The biofilm was treated with 50 μM Hp1404, its analog peptides and meropenem or polymyxin B. Treatment with Hp1404-I and Hp1404-W, decreased SYTO9 fluorescence by more than Hp1404 in all strains. In contrast, meropenem or polymyxin B did not decrease biofilm formation of A. baumannii, including the strains isolated from patients.
2.10. EPS Determination
EPS are a complex mixture of carbohydrates, DNA and proteins in biofilms. We measured the total carbohydrate content in EPS using the phenol-sulfuric acid method. After the formation of biofilm, treatment with 50 μM peptides or antibiotics resulted in reduced EPS production (Figure 9). For the standard strain A. baumannii KCTC 2508, the control showed 0.4 μg/mL EPS production. After treatment with the peptides, EPS production decreased from 0.1 to 0.25 μg/mL. Antibiotics reduced this value from 0.12 to 0.15 μg/mL, which is less than that after peptide treatment. Antibiotic-resistant strains also reduced EPS production. Thus, the peptides and antibiotics caused similar reductions in EPS production.
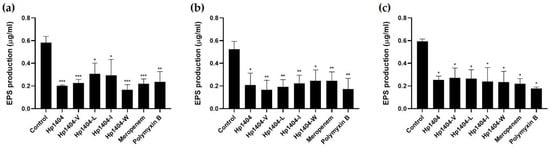
Figure 9.
Effect of Hp1404 and its analog peptides on biofilm reduction and EPS production in A. baumannii strains. (a) A. baumannii KCTC 2508, (b) A. baumannii #3 and (c) A. baumannii 409081. Data are presented as the mean ± SEM of three independent experiments. * p < 0.05, ** p < 0.01, *** p < 0.001 versus the control group.
2.11. NPN Uptake
The increase in fluorescence due to N-phenyl-1-naphthylamine (NPN) in the outer membrane was measured to determine the outer membrane permeability of the peptides. NPN fluorescence increases when the bacterial outer membrane is disrupted, and hydrophobic regions are exposed by the action of the peptide. As shown in Figure 10, Hp1404 and its analog peptides induced NPN fluorescence in A. baumannii cells in time -and concentration-dependent manners. The fluorescence intensity of 4× MIC Hp1404 and its analog peptides (Hp1404-V, Hp1404-L, Hp1404-I and Hp1404-W) for 30 min reached approximately 600, 650, 540, 530 and 490 arbitrary units (a.u.), respectively.
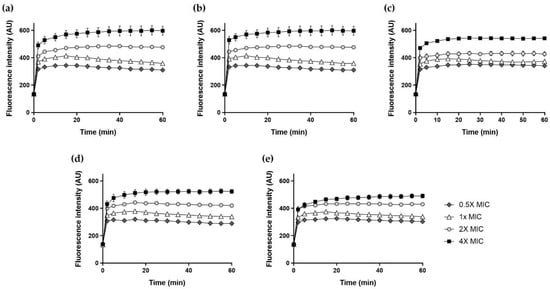
Figure 10.
Outer membrane permeabilization of A. baumannii KCTC 2508 determined in NPN uptake assay. The increase of fluorescence intensity measured for 30 min at excitation wavelength of 350 nm and emission wavelength of 420 nm. (a) Hp1404, (b) Hp1404-V, (c) Hp1404-L (d) Hp1404-I and (e) Hp1404-W. Data are presented as the mean ± SEM of three independent experiments.
2.12. DisC3-5
We assessed whether Hp1404 and its analog peptides induced membrane depolarization using DisC3-5, which accumulates in intact cytoplasmic membrane cells and fluoresces when the membrane is disrupted by the peptides. All peptides produced time- and dose-dependent effects (Figure 11). Hp1404 and four analog peptides (Hp1404-V, Hp1404-L, Hp1404-I and Hp1404-W) at 4× MIC increased the fluorescence intensity to almost 210, 200, 200, 220 and 260 arbitrary units (a.u.), respectively. Analog peptides induced cytoplasmic membrane depolarization comparable to that of Hp1404. DisC3-5 fluorescence increased immediately to the highest level after addition of Hp1404. Hp1404 and its analog peptides caused rapid membrane depolarization, and Hp1404-W showed higher fluorescence over time.
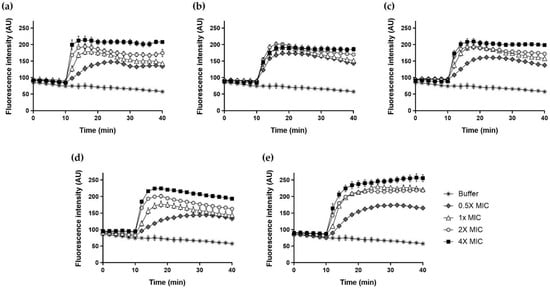
Figure 11.
Cytoplasmic membrane depolarization of A. baumannii determined using membrane potential sensitive dye DisC3-5. The release of DisC3-5 was measured at an excitation wavelength of 622 nm and emission wavelength of 670 nm. (a) Hp1404, (b) Hp1404-V, (c) Hp1404-L (d) Hp1404-I and (e) Hp1404-W. Data are presented as the mean ± SEM of three independent experiments.
3. Discussion
The venom of various animal species is the source of biologically active molecules with attractive candidates for the development of novel therapeutics [29]. Antimicrobial peptides (AMPs) are bioactive molecules and notable sources of known antibacterial activity found in scorpions, ants, bees, snakes, wasps and spiders. AMPs isolated from scorpions have been reported to effectively kill multidrug-resistant pathogens and possess anticancer activity but are slightly toxic [30]. Modifying AMPs could improve the antibacterial effect while reducing cytotoxicity, thereby overcoming the barriers preventing their development as therapeutic agents. Thus, new peptide analogs of Heterometrus petersii scorpion AMPs have been designed and synthesized, and their antimicrobial spectrum and hemolytic/cytotoxic activity have been determined.
In the current study, we predicted that by replacing various amino acids in Hp1404, the peptides would show increased hydrophobicity and reduced toxicity. Six analog peptides of Hp1404 were designed by replacing phenylalanine (Phe, F) with an electrically charged amino acid, lysine (Lys, K) [31] and hydrophobic amino acids, alanine (Ala, A) [32], valine (Val, V) [33], leucine (Leu, L) [34], isoleucine (Ile, I) [35] and tryptophan (Trp, W) [36]. The net charge was increased by substituting the charged amino acid lysine, and hydrophobicity was similar to or increased by substitution with hydrophobic amino acids.
We examined the antimicrobial activity of the parent peptide Hp1404 and its six analog peptides against gram-positive and gram-negative bacteria. Hp1404 and four analog peptides (Hp1404-W, Hp1404-L, Hp1404-I and Hp1404-W) displayed antimicrobial activity, with excellent activity against A. baumannii, including clinical strains resistant to antibiotics. Particularly, the peptides substituted with Lue and Trp maintained antibacterial activity. Hp1404 and melittin were toxic towards mammalian cells, but the toxicity of the analog peptides decreased and cell viability increased by 80% even at a concentration 4-fold the active concentration, and the analogs exhibited stable antibacterial activity in a high-salt environment. Hp1404-L maintained antibacterial activity but showed toxicity similar to that of the parent Hp1404 peptide, while Hp1404-W showed no toxicity. For use as therapeutic agents, peptides need to have low toxicity and the ability to maintain the activity in a salt environment [37]. Cations such as Na+ interfere with the electrostatic interaction between the peptide and bacterial membrane, thereby reducing antimicrobial activity. Therefore, we determined whether each peptide maintains antibacterial activity in the presence of Na+; Hp1404-L and Hp1404-W showed excellent activity along with the parent peptide Hp1404. Thus, the analog peptides show potential for treating A. baumannii infections and overcome the limitations of Hp1404.
AMPs display random coil structures in their native forms; in bacterial membrane-mimicking environments, they display α-helix, β-sheet or loop structures which act on the bacterial membrane [38]. It is known that AMPs such as melittin [39], magainin2 [40], cecropins [41] and LL37 [42] adopt α-helix structures. Hp1404 has been reported to form amphipathic α-helix when adsorbed to the bilayer lipid membrane of the bacterial membrane [28]. The structures of the analog peptides were confirmed under different conditions by CD spectroscopy. As a result, the parent peptide and its analog peptides formed a random coil in aqueous solution, and an α-helical structure was adopted in membrane-mimicking environments. The high amphiphilic and strong helix properties of the peptides are associated with their antimicrobial activity in high-salt environments (Figure 5). The helix structure and positive charges are strongly bound to negatively charged phospholipid bacterial membranes and efficiently exert their effects on them. We investigated the mechanism of action of outer membrane permeability and cytoplasmic membrane depolarization using NPN and DisC3-5. Peptides showed an increased fluorescence intensity of NPN and DiSC3-5 at each active concentration. This is consistent with the findings of studies on the mechanisms of action of AMP such as LL37 and melittin that act by destroying bacterial membranes. These results suggest that the peptides interact with the bacterial membrane to exert antibacterial effects by causing outer and inner membrane permeability in A. baumannii. Therefore, we observed that owing to this mechanism of action, analog peptides killed the bacteria in short time and thus showed potential as therapeutic agents.
Biofilm formation increases antibiotic resistance compared to strains in the planktonic state, and there are concerns that the effect of the peptide may be reduced by the production of polysaccharides. Microorganisms attach to surfaces such as medical devices and produce a polymer matrix containing extracellular DNA, polysaccharides and glycoproteins that cause infection [43,44]. The tolerance of these biofilms is increased by endogenous oxidative stress, upregulation of the effluent pump and common tolerance mechanisms such as beta-lactamase [45]. To assess biofilm inhibition activity, we used Hp1404, four analog peptides and two conventional antibiotics. The parent and its analog peptides showed similar levels of inhibition of biofilm formation. Meropenem inhibited biofilm formation of the standard strain A. baumannii KCTC 2508 but did not inhibit that of drug-resistant A. baumannii strains. Polymyxin B inhibited biofilm formation of A. baumannii, including that of resistant strains. For biofilm eradication, analog peptides were more effective than Hp1404, with Hp1404-I showing the strongest effects. Polymyxin B was superior for inhibiting biofilm formation but exhibited a significantly lower eradication ability than the peptides. Furthermore, the effect of the peptides on EPS secreted by the biofilm was confirmed. Analog peptides reduced EPS production to a level similar as that of Hp1404. Thus, analog peptides of Hp1404 may be useful alternatives to antibiotics.
In summary, we designed Hp1404 analogs with reduced toxicity by substituting the amino acid in position 14. The analog peptides showed activity against gram-positive and gram-negative bacteria. We determined the activity of these peptides against multidrug-resistant A. baumannii and their anti-biofilm effects. The toxicity of the analog peptides was lower, and the anti-biofilm effect was better than that of Hp1404. Thus, these newly designed peptides are promising candidates as antimicrobial agents against A. baumannii strains.
4. Materials and Methods
4.1. Materials
SDS, TFE, thiazolyl blue tetrazolium bromide (MTT), NPN, 3,3′-dipropylthiadicarbocyanine iodide (DisC3-5), SYTOX green, propidium iodide (PI), dimethyl sulfoxide, meropenem and polymyxin B were purchased from Sigma-Aldrich (St. Louis, MO, USA). Vancomycin was purchased from LPS solution (Daejeon, South Korea).
4.2. Microorganisms
Staphylococcus aureus ATCC 25923, S. aureus ATCC 29213, P. aeruginosa ATCC 27853, Escherichia coli ATCC 25922 and Salmonella typhimurium ATCC 14028 strains were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). Listeria monocytogenes KCTC 3710, Bacillus cereus KCTC 1012, K. pneumoniae KCTC 2208 and A. baumannii KCTC 2508 were obtained from the Korean Collection for Type Cultures (KRIBB, Daejeon, South Korea). Clinical isolates of A. baumannii strains were collected from Eulji University Hospital (Seoul, South Korea).
4.3. Peptide Design and Characterization
The analog peptides were designed by substituting amino acids in Hp1404. The three-dimensional structures of Hp1404 and its analog peptides were constructed using Mobyle@RPBS (http://mobyle.rpbs.univ-paris-diderot.fr/cgi-bin/portal.py#welcome accessed on 14 August 2020). Helical wheel diagrams and the hydrophobicity of the peptides were obtained using the HeliQuest site (http://heliquest.ipmc.cnrs.fr, accessed on 14 August 2020).
4.4. Peptide Synthesis
The peptides were synthesized using the solid-phase-9-fluorenylmethoxycarbonyl (Fmoc) method, as reported previously [46], on a Rink amide 4-methylbenzhydrylamine resin using a Liberty microwave peptide synthesizer (CEM, Matthews, NY, USA). The following chemicals were used as linkage reagents: 0.45 M 2-(1H-benzotriazole-1-yil)-1,1,3,3-tetramethyluronium hexafluorophosphate (HBTU) diluted in dimethylformamide (DMF), 0.1 M N-hydroxybenzotriazole (HOBt) diluted in piperidine/DMF and 2 M N, N-diisopropylethylamine (DIEA) diluted in N-methylpyrrolidone (NMP). After washing with dichloromethane (DCM), cleavage was performed by incubating for 2 h at 25 °C in a trifluoroacetic acid (TFA) solution containing water, phenol and triisopropylsilane. The crude peptide was precipitated by dilution with ice-cold diethyl ether, and then spread on the tube wall and dried. After resuspension in water at 25 °C, the peptide was purified by reversed-phase high-performance liquid chromatography (RP-HPLC) on a Jupiter C18 column (4.6 × 250 mm, 300 Å, 5 µm; Phenomenex, Torrance, CA, USA). The molecular weights of the peptides were confirmed using matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry (Kratos Analytical Inc., Chestnut Ridge, NY, USA). The peptides were dissolved in deionized water (DI H2O) and stored at −20 °C. In this study, all the peptides used were >95% pure.
4.5. CD Spectroscopy
CD spectra were measured to determine the secondary structure of the peptide, which was changed by the bacterial membrane-mimicking environment. The peptides were used at a concentration of 40 μM. The secondary structure of the peptides was investigated in 10 mM SP buffer (pH 7.2), 30 mM SDS and 50% TFE. Spectra were investigated at wavelengths ranging from 190 to 250 nm using a quartz cell with a 1.0-mm path length on a JASCO 810 spectropolarimeter (Jasco, Tokyo, Japan) [47].
4.6. Antibacterial Activity Test
Hp1404 and its analog peptides activity against both gram-positive and gram-negative bacterial strains were tested using the broth microdilution method [48]. Briefly, S. aureus (ATCC 25923, ATCC 29213) and B. cereus KCTC 1012 were cultured overnight at 37 °C in tryptic soy broth (TSB). P. aeruginosa ATCC 27853, K. pneumoniae KCTC 2208, A. baumannii KCTC 2508 and S. typhimurium ATCC 14028 were cultured overnight at 37 °C in nutrient broth (NB). E. coli ATCC 25922 was cultured overnight at 37 °C in luria bertani (LB). L. monocytogenes KCTC 3710 was cultured overnight at 37 °C in brain heart infusion (BHI). The cultures were diluted with fresh medium at a density of 2 × 105 CFU/mL. Peptides were serially diluted in sodium phosphate buffer (pH 7.2) to a final concentration of 25 μM. In a 96-well plate, 50-μL bacterial aliquots were mixed with 50 μL peptide solution. After incubation at 37 °C for 18 h, the MIC was determined using a plate reader at 600 nm. The experiment was performed in triplicate.
4.7. Salt Stability Assay
To determine whether antibacterial activity was affected by the presence of salts, experiments were conducted as previously reported [49]. We determined the MICs of NB supplemented with 150 mM NaCl. MIC assays were performed as described above. The assay was performed in triplicate.
4.8. Hemolytic Assay
The hemolytic activity of the peptides against mouse erythrocytes was determined using red blood cells [50]. Fresh mouse red blood cells were washed three times and resuspended in PBS to attain an 8% dilution. Peptides were serially diluted to a final concentration of 50 μM. Next, 100 µL peptide solution was added to 100 μL of mouse red blood cells, wrapped with aluminum foil and incubated for 1 h at 37 °C. The mixture was then centrifuged at 1500× g for 10 min. The supernatant was transferred into a new 96-well plate, and the absorbance of the supernatant was measured at 414 nm. As positive and negative controls, PBS and 0.1% Triton X-100 in mouse red blood cells were used. Hemolysis was evaluated according to the following equation:
Hemolysis (%) = [(Abs414 nm in the peptide solution − Abs414 nm in PBS)/(Abs414 nm in 0.1% Triton X-100 − Abs414 nm in PBS)] × 100.
4.9. Cytotoxicity Assay
The cytotoxicity of the peptides against mammalian cells was calculated using the MTT assay. Human keratinocyte HaCaT cells were incubated in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum and 1% penicillin at 37 °C with 5% CO2. HaCaT cells were added at 2 × 105 cells/mL to DMEM supplemented with 10% fetal bovine serum and placed in 96-well plates. The plates were incubated overnight at 37 °C in a CO2 incubator. Peptides were serially diluted two-fold in DMEM, added to the cell cultures and incubated for 24 h at 37 °C. MTT solution (5 mg/mL) was added to each well and incubated for 1 h at 37 °C. The media was discarded, and the formazan crystals were dissolved in 100 μL dimethyl sulfoxide. The absorbance was measured using a Versa-Max microplate ELISA reader (Molecular Devices, Sunnyvale, CA, USA) at 570 and 650 nm. As a negative control, 0.1% Triton X-100 was used.
4.10. Time-Killing Assay
A time-killing assay of the peptides against A. baumannii KCTC 2508 was performed. The bacteria were grown to mid-log phase in NB medium, with cell density of A. baumannii KCTC 2508 of 2 × 105 CFU/mL. The peptides were used to treat the bacteria at concentrations of 1×, 2× and 4× MIC and incubated at 37 °C with shaking for 0–180 min. The reactive aliquots were plated on NB agar and incubated at 37 °C for 24 h, and surviving bacterial cells were counted. The assay was conducted in triplicate.
4.11. Calcein Leakage Assay
Peptide-induced membrane permeability was monitored be measuring calcein dye emission in calcein-loaded LUVs [49]. We prepared the lipid mixture, and PC:CH (2:1, w/w) and PC:CH:SM liposomes (1:1:1, w/w) were prepared with 70 mM calcein buffer. A Sephadex G-50 column was used to separate calcein-loaded liposomes from free calcein dye. The LUVs (10 μM) were mixed with various concentrations of the peptide on a 96-well black plate. The fluorescence intensity was measured at excitation and emission wavelengths of 480 and 520 nm, respectively.
4.12. Biofilm Inhibition Assay
Acinetobacter baumannii KCTC 2508, #3, #4, 409081 and 719705 were incubated overnight at 37°C to mid-log phase in NB medium. Bacterial cells (5 × 105 CFU/mL) were suspended in NB containing 0.2% glucose. A final concentration of 25 μM of peptides and antibiotics (meropenem and polymyxin B) was prepared with the bacterial suspensions in 96-well tissue culture plates and incubated at 37 °C for 24 h. The culture media was carefully removed, and biofilms were fixed with 100% methanol for 15 min and then stained with 0.1% crystal violet for 1 h and rinsed three times with distilled water. After air-drying, the biofilm mass was dissolved in 100 μL of 95% ethanol and measured at 595 nm using a microplate reader [51].
4.13. Biofilm Reduction Assay
Acinetobacter baumannii KCTC 2508, #3 and 409081 cells were cultured in NB at 37 °C. Bacterial cells were diluted to 5 × 105 CFU/mL in fresh NB with 0.2% glucose in a 96-well plate and incubated for 24 h. After incubation, the wells were washed with PBS to remove non-adherent cells, and serially diluted peptides and antibiotics (final concentration 200 μM) were treated for 24 h. A crystal violet assay was performed, and biofilm formation was measured as described above.
4.14. Live Biofilm Viability Assay
An A. baumannii biofilm viability assay was performed by staining with Syto9 dye [52]. Acinetobacter baumannii strains were diluted to approximately 5 × 105 CFU/mL and incubated for 24 h. Biofilms of A. baumannii strains were washed with PBS and treated with 25 and 50 μM peptides and antibiotics for 24 h. Biofilms were stained with SYTO 9 diluted in PBS and the biofilm was visualized using an EVOS™ FL Auto 2 Imaging System (Thermo Fisher Scientific, Waltham, MA, USA).
4.15. EPS Determination
The total carbohydrate content in EPS was measured using the phenol-sulfuric acid method with glucose as a standard [53]. Briefly, biofilms of A. baumannii strains were incubated for 24 h, washed with PBS and treated with 50 μM peptides or antibiotics. After 24 h, 100 μL EPS solution was mixed with 100 μL 5% phenol. Next, 500 μL sulfuric acid was added, and the mixture was incubated at room temperature for 1 h in the dark. The mixture was cooled on ice for 15 min and centrifuged 10,000× g for 10 min. The supernatant was transferred to a 96-well plate. Absorbance was read at 490 nm with a microplate reader.
4.16. NPN Uptake Assay
The hydrophobic fluorescent dye NPN was used to investigate whether the peptides induced outer membrane permeability [54,55]. Briefly, A. baumannii KCTC 2508 was grown in NB at 37 °C, washed three times and diluted to an OD600 of 0.2, in 5 mM HEPES buffer (pH 7.3). Diluted bacterial cells (100 μL) were added to 96-well black microplates and mixed with 50 μL of 10 μM NPN (dissolved in 95% ethanol), and background fluorescence was measured. Next, 0.5×, 1×, 2× and 4× MIC of peptides were added, and the increase in fluorescence caused by the peptides was recorded for 30 min. HEPES buffer (5 mM) was used as a negative control. The excitation and emission wavelengths were 350 and 420 nm, respectively.
4.17. Inner Membrane Depolarization Using DisC3-5
The inner membrane depolarization in the presence of peptides was determined using a membrane potential-dependent probe, DisC3-5. Briefly, mid-log phase A. baumannii KCTC 2508 cells were washed three times with 5 mM HEPES buffer (pH 7.3) containing 20 mM glucose. Next, 100 µL bacterial suspension (OD600 = 0.05) and 50 μL of a final concentration 1 μM DisC3-5 (in 5 mM HEPES with 20 mM glucose and 0.1 M KCl) mixtures were added to 96-well black microplates and incubated for 60 min until dye uptake reached a maximum. The peptides (0.5×, 1×, 2× and 4× MIC of peptides) were treated at 50 μL, and the change in fluorescence intensity was monitored for 30 min at excitation and emission wavelengths of 622 and 670 nm, respectively [56].
4.18. Statistical Analysis
All experiments were performed in triplicates in three independent experiments. Data are presented as the mean ± SEM. Statistical analysis of the results was performed with two-tailed Student’s t-test and one-way analysis of variance using GraphPad Prism version 8. 0 (La Jolla, CA, USA). The results were considered significant at * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001.
5. Conclusions
We designed analog peptides (Hp1404-V, Hp1404-L, Hp1404-I and Hp1404-W) that are less toxic than the parent peptide Hp1404 but have the same antibacterial activity against A. baumannii strains. Furthermore, the peptides maintained antibacterial activity under high-salt conditions and effectively inhibited biofilm formation and elimination. The analog peptides showed an α-helical structure in a bacterial membrane-mimicking environment and the same mechanism of action as the parent peptide. Overall, Hp1404-V, Hp1404-L, Hp1404-I and Hp1404-W are potential therapeutic agents against A. baumannii strains.
Author Contributions
Y.P. planned the research project. M.J.H. and M.K.K. performed the experiments, analyses, prepared the figures and wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by a National Research Foundation of Korea (NRF) grant funded by the Korean Government (No. 2019R1A2B5B03070330, NRF-2017M3A9E4077206) and Institute for Information and Communications Technology Promotion (IITP) grant funded by the Korean government (MSIT) (No. 2017-0-01714, Development of Antimicrobial Peptide using Deep Learning).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable for studies that do not involve humans.
Data Availability Statement
Data is contained within the article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Pfalzgraff, A.; Brandenburg, K.; Weindl, G. Antimicrobial Peptides and Their Therapeutic Potential for Bacterial Skin Infections and Wounds. Front. Pharmacol. 2018, 9, 281. [Google Scholar] [CrossRef] [PubMed]
- Pletzer, D.; Mansour, S.C.; Hancock, R.E.W. Synergy between conventional antibiotics and anti-biofilm peptides in a murine, sub-cutaneous abscess model caused by recalcitrant ESKAPE pathogens. PLoS Pathog. 2018, 14, e1007084. [Google Scholar] [CrossRef] [PubMed]
- Kengkla, K.; Kongpakwattana, K.; Saokaew, S.; Apisarnthanarak, A.; Chaiyakunapruk, N. Comparative efficacy and safety of treatment options for MDR and XDR Acinetobacter baumannii infections: A systematic review and network meta-analysis. J. Antimicrob. Chemother. 2018, 73, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Mendelson, M.; Matsoso, M.P. The World Health Organization global action plan for antimicrobial resistance. SAMJ S. Afr. Med. J. 2015, 105, 325. [Google Scholar] [CrossRef]
- World Health Organization. The Evolving Threat of Antimicrobial Resistance: Options for Action: Executive Summary. 2012. Available online: https://apps.who.int/iris/handle/10665/75389 (accessed on 17 September 2012).
- World Health Organization. Worldwide Country Situation Analysis: Response to Antimicrobial Resistance; WHO/HSE/PED/AIP/2015.1; WHO: Geneva, Switzerland, 29 April 2015; ISBN 978-92-4-156494-6. [Google Scholar]
- Kanafani, Z.; Kanj, S. Acinetobacter Infection: Treatment and Prevention. Available online: https://www.uptodate.com/contents/acinetobacterinfection-treatment-and-prevention (accessed on 16 June 2015).
- Deveci, Ö.; Dal, T.T.R.; Bozkurt, F.; Tekin, A.; Dayan, S. Carbapenem resistance in Acinetobacter baumannii: Where is it heading. Infez. Med. 2013, 21, 211–215. [Google Scholar]
- Dijkshoorn, L.; Nemec, A.; Seifert, H. An increasing threat in hospitals: Multidrug-resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 2007, 5, 939–951. [Google Scholar] [CrossRef]
- Fournier, P.E.; Richet, H. The epidemiology and control of Acinetobacter baumannii in health care facilities. Clin. Infect. Dis. 2006, 42, 692–699. [Google Scholar] [CrossRef]
- Flemming, H.C.; Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, S. Biofilms: An emergent form of bacterial life. Nat. Rev. Microbiol. 2016, 14, 563–575. [Google Scholar] [CrossRef]
- Donlan, R.M. Biofilms: Microbial life on surfaces. Emerg. Infect. Dis. 2002, 8, 881–890. [Google Scholar] [CrossRef]
- Flemming, H.C.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef]
- Jiao, Y.; Cody, G.D.; Harding, A.K.; Wilmes, P.; Schrenk, M.; Wheeler, K.E.; Banfield, J.F.; Thelen, M.P. Characterization of extracellular polymeric substances from acidophilic microbial biofilms. Appl. Environ. Microbiol. 2010, 76, 2916–2922. [Google Scholar] [CrossRef] [PubMed]
- Flemming, H.C.; Neu, T.R.; Wozniak, D.J. The EPS matrix: The “house of biofilm cells”. J. Bacteriol. 2007, 189, 7945–7947. [Google Scholar] [CrossRef]
- Jamal, M.; Ahmad, W.; Andleeb, S.; Jalil, F.; Imran, M.; Nawaz, M.A.; Hussain, T.; Ali, M.; Rafiq, M.; Kamil, M.A. Bacterial biofilm and associated infections. J. Chin. Med Assoc. 2018, 81, 7–11. [Google Scholar] [CrossRef] [PubMed]
- Mirski, T.; Niemcewicz, M.; Bartoszcze, M.; Gryko, R.; Michalski, A. Utilisation of peptides against microbial infections—A review. Ann. Agric. Environ. Med. 2017, 25, 205–210. [Google Scholar] [CrossRef]
- Pasupuleti, M.; Schmidtchen, A.; Malmsten, M. Antimicrobial peptides: Key components of the innate immune system. Crit. Rev. Biotechnol. 2012, 32, 143–171. [Google Scholar] [CrossRef] [PubMed]
- Carnicelli, V.; Lizzi, A.; Ponzi, A.; Amicosante, G.; Bozzi, A.; Di Giulio, A. Interaction between antimicrobial peptides (AMPs) and their primary target, the biomembranes. In Microbial Pathogens and Strategies for Combating Them: Science, Technology and Education; Méndez-Vilas, A., Ed.; Formatex Research Center: Badajoz, Spain, 2013; pp. 1123–1134. [Google Scholar]
- Bergaoui, I.; Zairi, A.; Tangy, F.; Aouni, M.; Selmi, B.; Hani, K. In vitro antiviral activity of dermaseptin S4 and derivatives from amphibian skin against herpes simplex virus type 2. J. Med Virol. 2013, 85, 272–281. [Google Scholar] [CrossRef] [PubMed]
- De Cesare, G.B.; Cristy, S.A.; Garsin, D.A.; Lorenz, M.C. Antimicrobial Peptides: A New Frontier in Antifungal Therapy. Mbio 2020, 11. [Google Scholar] [CrossRef]
- Cho, E.; Lee, J.K.; Park, E.; Seo, C.H.; Luchian, T.; Park, Y. Antitumor activity of HPA3P through RIPK3-dependent regulated necrotic cell death in colon cancer. Oncotarget 2018, 9, 7902–7917. [Google Scholar] [CrossRef]
- Brandl, K.; Plitas, G.; Mihu, C.N.; Ubeda, C.; Jia, T.; Fleisher, M.; Schnabl, B.; DeMatteo, R.P.; Pamer, E.G. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature 2008, 455, 804–807. [Google Scholar] [CrossRef]
- Han, H.M.; Ko, S.; Cheong, M.J.; Bang, J.K.; Seo, C.H.; Luchian, T.; Park, Y. Myxinidin2 and myxinidin3 suppress inflammatory responses through STAT3 and MAPKs to promote wound healing. Oncotarget 2017, 8, 87582–87597. [Google Scholar] [CrossRef]
- Le, C.-F.; Fang, C.-M.; Sekaran, S.D. Intracellular targeting mechanisms by antimicrobial peptides. Antimicrob. Agents Chemother. 2017, 61. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Zhou, L.; Lakshminarayanan, R.; Beuerman, R. Multivalent antimicrobial peptides as therapeutics: Design principles and structural diversities. Int. J. Pept. Res. Ther. 2010, 16, 199–213. [Google Scholar] [CrossRef] [PubMed]
- Rios, A.C.; Moutinho, C.G.; Pinto, F.C.; Del Fiol, F.S.; Jozala, A.; Chaud, M.V.; Vila, M.M.; Teixeira, J.A.; Balcão, V.M. Alternatives to overcoming bacterial resistances: State-of-the-art. Microbiol. Res. 2016, 191, 51–80. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Xu, X.; Meng, L.; Zhang, Q.; Cao, L.; Li, W.; Wu, Y.; Cao, Z. Hp1404, a new antimicrobial peptide from the scorpion Heterometrus petersii. PLoS ONE 2014, 9, e97539. [Google Scholar] [CrossRef]
- Primon-Barros, M.; José Macedo, A. Animal Venom Peptides: Potential for New Antimicrobial Agents. Curr. Top. Med. Chem. 2017, 17, 1119–1156. [Google Scholar] [CrossRef]
- Harrison, P.L.; Abdel-Rahman, M.A.; Miller, K.; Strong, P.N. Antimicrobial peptides from scorpion venoms. Toxicon 2014, 88, 115–137. [Google Scholar] [CrossRef]
- Jiang, Z.; Vasil, A.I.; Hale, J.D.; Hancock, R.E.; Vasil, M.L.; Hodges, R.S. Effects of net charge and the number of positively charged residues on the biological activity of amphipathic α-helical cationic antimicrobial peptides. Pept. Sci. 2008, 90, 369–383. [Google Scholar] [CrossRef]
- Taniguchi, M.; Takahashi, N.; Takayanagi, T.; Ikeda, A.; Ishiyama, Y.; Saitoh, E.; Kato, T.; Ochiai, A.; Tanaka, T. Effect of substituting arginine and lysine with alanine on antimicrobial activity and the mechanism of action of a cationic dodecapeptide (CL (14-25)), a partial sequence of cyanate lyase from rice. Pept. Sci. 2014, 102, 58–68. [Google Scholar] [CrossRef]
- Perumal, P.; Pandey, V.P. Antimicrobial peptides: The role of hydrophobicity in the alpha helical structure. J. Pharm. Pharmacogn. Res. 2013, 1, 39–53. [Google Scholar]
- Chen, Y.; Guarnieri, M.T.; Vasil, A.I.; Vasil, M.L.; Mant, C.T.; Hodges, R.S. Role of peptide hydrophobicity in the mechanism of action of α-helical antimicrobial peptides. Antimicrob. Agents Chemother. 2007, 51, 1398–1406. [Google Scholar] [CrossRef]
- Sun, J.; Xia, Y.; Li, D.; Du, Q.; Liang, D. Relationship between peptide structure and antimicrobial activity as studied by de novo designed peptides. Biochim. Biophysica Acta (BBA) Biomembr. 2014, 1838, 2985–2993. [Google Scholar] [CrossRef] [PubMed]
- Bi, X.; Wang, C.; Ma, L.; Sun, Y.; Shang, D. Investigation of the role of tryptophan residues in cationic antimicrobial peptides to determine the mechanism of antimicrobial action. J. Appl. Microbiol. 2013, 115, 663–672. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Dong, N.; Wang, Z.; Ma, Z.; Zhang, L.; Ma, Q.; Shan, A. Design of imperfectly amphipathic α-helical antimicrobial peptides with enhanced cell selectivity. Acta Biomater. 2014, 10, 244–257. [Google Scholar] [CrossRef]
- Liu, Y.; Xia, X.; Xu, L.; Wang, Y. Design of hybrid β-hairpin peptides with enhanced cell specificity and potent anti-inflammatory activity. Biomaterials 2013, 34, 237–250. [Google Scholar] [CrossRef]
- Dempsey, C.E. The actions of melittin on membranes. Biochim. Biophys. Acta (BBA) Rev. Biomembr. 1990, 1031, 143–161. [Google Scholar] [CrossRef]
- Kim, M.K.; Kang, N.H.; Ko, S.J.; Park, J.; Park, E.; Shin, D.W.; Kim, S.H.; Lee, S.A.; Lee, J.I.; Lee, S.H. Antibacterial and antibiofilm activity and mode of action of magainin 2 against drug-resistant Acinetobacter baumannii. Int. J. Mol. Sci. 2018, 19, 3041. [Google Scholar] [CrossRef]
- Steiner, H. Secondary structure of the cecropins: Antibacterial peptides from the moth Hyalophora cecropia. FEBS Lett. 1982, 137, 283–287. [Google Scholar] [CrossRef]
- Henzler Wildman, K.A.; Lee, D.-K.; Ramamoorthy, A. Mechanism of lipid bilayer disruption by the human antimicrobial peptide, LL-37. Biochemistry 2003, 42, 6545–6558. [Google Scholar] [CrossRef]
- Veerachamy, S.; Yarlagadda, T.; Manivasagam, G.; Yarlagadda, P.K. Bacterial adherence and biofilm formation on medical implants: A review. Proc. Inst. Mech. Eng. Part H J. Eng. Med. 2014, 228, 1083–1099. [Google Scholar] [CrossRef] [PubMed]
- Watnick, P.; Kolter, R. Biofilm, city of microbes. J. Bacteriol. 2000, 182, 2675–2679. [Google Scholar] [CrossRef] [PubMed]
- Høiby, N.; Bjarnsholt, T.; Givskov, M.; Molin, S.; Ciofu, O. Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents 2010, 35, 322–332. [Google Scholar] [CrossRef] [PubMed]
- Fields, G.B.; Noble, R.L. Solid phase peptide synthesis utilizing 9-fluorenylmethoxycarbonyl amino acids. Int. J. Pept. Protein Res. 1990, 35, 161–214. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.K.; Park, S.C.; Hahm, K.S.; Park, Y. A helix-PXXP-helix peptide with antibacterial activity without cytotoxicity against MDRPA-infected mice. Biomaterials 2014, 35, 1025–1039. [Google Scholar] [CrossRef] [PubMed]
- Wiegand, I.; Hilpert, K.; Hancock, R.E. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008, 3, 163. [Google Scholar] [CrossRef]
- Kim, M.K.; Kang, H.K.; Ko, S.J.; Hong, M.J.; Bang, J.K.; Seo, C.H.; Park, Y. Mechanisms driving the antibacterial and antibiofilm properties of Hp1404 and its analogue peptides against multidrug-resistant Pseudomonas aeruginosa. Sci. Rep. 2018, 8, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.G.; Kim, H.N.; Park, Y.; Kim, H.K.; Choi, B.H.; Choi, C.H.; Hahm, K.S. Design of novel analogue peptides with potent antibiotic activity based on the antimicrobial peptide, HP (2-20), derived from N-terminus of Helicobacter pylori ribosomal protein L1. Biochim. Biophys. Acta 2002, 1598, 185–194. [Google Scholar] [CrossRef]
- Mohamed, M.F.; Hamed, M.I.; Panitch, A.; Seleem, M.N. Targeting methicillin-resistant Staphylococcus aureus with short salt-resistant synthetic peptides. Antimicrob. Agents Chemother. 2014, 58, 4113–4122. [Google Scholar] [CrossRef]
- Rasamiravaka, T.; Vandeputte, O.M.; Pottier, L.; Huet, J.; Rabemanantsoa, C.; Kiendrebeogo, M.; Andriantsimahavandy, A.; Rasamindrakotroka, A.; Stévigny, C.; Duez, P.; et al. Pseudomonas aeruginosa Biofilm Formation and Persistence, along with the Production of Quorum Sensing-Dependent Virulence Factors, Are Disrupted by a Triterpenoid Coumarate Ester Isolated from Dalbergia trichocarpa, a Tropical Legume. PLoS ONE 2015, 10, e0132791. [Google Scholar] [CrossRef]
- Gowrishankar, S.; Sivaranjani, M.; Kamaladevi, A.; Ravi, A.V.; Balamurugan, K.; Karutha Pandian, S. Cyclic dipeptide cyclo(l-leucyl-l-prolyl) from marine Bacillus amyloliquefaciens mitigates biofilm formation and virulence in Listeria monocytogenes. Pathog. Dis. 2016, 74, ftw017. [Google Scholar] [CrossRef]
- Helander, I.M.; Mattila-Sandholm, T. Fluorometric assessment of gram-negative bacterial permeabilization. J. Appl. Microbiol. 2000, 88, 213–219. [Google Scholar] [CrossRef]
- Loh, B.; Grant, C.; Hancock, R.E. Use of the fluorescent probe 1-N-phenylnaphthylamine to study the interactions of aminoglycoside antibiotics with the outer membrane of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 1984, 26, 546–551. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Park, S.C.; Yoon, M.Y.; Hahm, K.S.; Park, Y. C-terminal amidation of PMAP-23: Translocation to the inner membrane of Gram-negative bacteria. Amino Acids 2011, 40, 183–195. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).