Abstract
Gastric cancer (GC) is a common malignant neoplasm worldwide and one of the main cause of cancer-related deaths. Despite some advances in therapies, long-term survival of patients with advanced disease remains poor. Different types of classification have been used to stratify patients with GC for shaping prognosis and treatment planning. Based on new knowledge of molecular pathways associated with different aspect of GC, new pathogenetic classifications for GC have been and continue to be proposed. These novel classifications create a new paradigm in the definition of cancer biology and allow the identification of relevant GC genomic subsets by using different techniques such as genomic screenings, functional studies and molecular or epigenetic characterization. An improved prognostic classification for GC is essential for the development of a proper therapy for a proper patient population. The aim of this review is to discuss the state-of-the-art on combining histological and molecular classifications of GC to give an overview of the emerging therapeutic possibilities connected to the latest discoveries regarding GC.
1. Introduction
Gastric cancer (GC) is the fifth malignant neoplasm worldwide and the third cause of cancer-related deaths [1]. Despite some advances in therapies for GC, long-term survival of patients with advanced disease is poor. GC is a multifactorial disease in which both genetic and environmental factors are involved. Historically, different types of classification have been used to shape prognosis and plan treatment [2,3,4,5,6]. Proposed in 1965, the Laurén system was widely used in GC classification for half a century, which was very useful in evaluating the natural history of GC carcinogenesis. Based on pathological morphology, the Laurén system divides GC into intestinal (G-INT), diffuse (G-DIF) and mixed GC (G-Mix). An improved prognostic classification for GC is essential for the development of a proper therapy for patients. Therefore, based on new knowledge of molecular pathways, new pathogenetic classifications for GC have been proposed. The aim of this review is to update molecular classifications of GC to give an overview of the emerging therapeutic possibilities
2. Histological and Molecular Classifications of GC
Based on the gene expression profile for GC cell lines and patients’ tissue, Tan et al. [7] classified GC into two intrinsic genomic subtypes that overlapped with the histological Lauren’s classification. The G-INT subtype and the G-DIF are related to intestinal and diffuse histology, respectively. The two intrinsic subtypes have distinct patterns of gene expression.
In the G-INT subtype, genes associated with the carbohydrate and protein metabolism (FUT2) and cell adhesion (LGALS4, CDH17) are upregulated. The FUT2 gene codes for the galactoside 2-alpha-l-fucosyltransferase 2 enzyme affecting the Lewis blood group involved in Helicobacter pylori (H. pylori) infection; the LGALS4 gene codes the galectin 4 implicated in the modulation of the interaction between cell-cell and cell-matrix and the peptide transporter cadherin-17 coded by the CDH17 gene.
Instead, in the G-DIF subtype, genes related to cell proliferation (AURKB) and fatty acid metabolism (ELOVL5) are upregulated. The AURKB gene codes for the Aurora B kinase that functions in the attachment of the mitotic spindle to the centromere, and the ELOVL5 gene encodes the elongation of the very long chain fatty acids protein. The prognosis of G-DIF tumour type is poor, and the response to chemotherapy is reduced compared to those of the G-INT type. In vitro, G-INT cell lines are more sensitive to 5-FU and oxaliplatin than G-DIF lines, which result in being more sensitive to cisplatin [7,8]. There were many more other molecular studies based on the Laurén classification [9,10,11,12].
A molecular classification for GC, independent of the histological Laurent classification, was made in 2013 by Singapore Researchers. They categorized GC into three main types: [13] a proliferative profile associated with a high genomic instability and TP53 gene mutation, a metabolic profile associated with a higher anaerobic glycolysis and resulting in tumour cells more sensitive to 5-FU therapy and a mesenchymal stem cell profile with a high capacity for self-renewal, immunomodulation and tissue regeneration showing a sensitivity to PIK3CA-mTOR pathway inhibitors.
Soon after, The Cancer Genome Atlas (TCGA) research group categorized GC into four main groups by introducing new technologies of large-scale genome sequencing analyses [14]: Epstein-Barr virus (EBV)-positive cancers (9% of all GC) characterized by DNA hypermethylation, a high frequency of PIK3CA mutations and PDL1/PDL2 overexpression, microsatellite instable (MSI, 22%) tumours, showing a very high number of mutations and DNA methylation sites and chromosome instable tumours (CIN, 50%) mainly coding for alteration in tyrosine kinase receptors and genome stable tumours (GS, 20%).
In 2015, by using similar multi-platform molecular approaches, the Asian Cancer Research Group (ACRG) developed a novel molecular classification for GC based on a pre-defined set of genetic pathways relevant to the biology of GC, including epithelial-mesenchymal transition (EMT), microsatellite instability, cytokine signaling and P53 activity [15]. The ACRG classification included four subtypes [16]: an MSI subtype (22.7%), a mesenchymal group microsatellite stable (MSS)/EMT (15.3%) based on the evidence of epithelial-to-mesenchymal transition, a microsatellite stable TP53-positive subtype MSS/TP53+ (26.3%) and a microsatellite stable TP53-negative subtype MSS/TP53− (35.7%), according to the presence/absence of P53 mutations. By using this approach, the MSI subtype had the best prognosis, while the MSS/EMT subtype had the worst one. The former occurred predominantly at an early stage in the distal part of the stomach and showed mainly an intestinal histology (according to Lauren’s classification); the latter occurred at an advanced stage, at a younger age and with a diffuse histology (>80%) including a large set of signet ring cell carcinomas seeding in the peritonea with malignant ascites (64.1% vs. 15–24% in the other subtypes) and showed loss of CDH1 expression. Given the earlier stage of diagnosis, MSI and MSS/TP53− patients also had the best overall survival and when recurrence occurs, this was generally limited to liver metastasis (about 20%). EBV infection was more frequent in the MSS/TP53 active group.
In ACRG, the correlation between molecular classification and prognosis was validated using the TCGA [14] and the Gastric Cancer Project ′08 Singapore datasets [16]. As shown in Table 1, the ACRG subtypes show a significant overlap with the TCGA subtypes, and this confirms the association between better survival and the MSI subtype [17]. However, the overlap is only partial and probably due to the differences in the patient population (Korea in ACRG and USA and Western Europe in TCGA), tumour sampling and technical platforms used. Nonetheless, these novel classifications created a new paradigm in the definition of GC, although some limitations persist:

Table 1.
Key characteristics of The Cancer Genome Atlas (TCGA) and the Asian Cancer Research Group (ACRG) molecular classifications of gastric cancer (GC). MSI, microsatellite instable; CIN, chromosome instable; GS, genome stable; EGJ, esophagogastric junction; MSS, microsatellite stable.
- these classifications are based on a highly complex methodology, which is not always available in every laboratory;
- they lack a prospective validation on a large scale;
- they have striking differences in epidemiology, underlying molecular mechanisms and prognosis;
- their prognostic power is decreased by limited follow-up of patients;
- none of them takes into account the active, non-malignant stromal cells
3. Integrated Molecular Signatures to Discriminate Intestinal and Diffuse Histological GC Subtypes
Previous findings indicated that diffuse and intestinal GC might be two distinct diseases with different molecular bases, aetiologies, epidemiologies and, thus, response to therapies. A recent study based on a population of 300 GC identified 40 genes specifically expressed in diffuse or intestinal GC [12] and three genes associated with the patients’ prognosis, namely EFEMP1 and FRZB in G-DIF and KRT23 in G-INT. The products of the former are an extracellular matrix glycoprotein and a secreted protein regulating bone development and influencing the Wnt/beta-catenin pathway. The latter encodes for a member of the keratin family, which regulates epithelial cell structures.
In the last year, a nine-gene signature, including two negative impact factors (NR1I2 and LGALSL) and seven positive ones (C1ORF198, CST2, LAMP5, FOXS1, CES1P1, MMP7 and COL8A1), was proposed to predict the outcome of GC, and the model was able to predict patients’ outcome in terms of survival and recurrence, clustering GC cases into low-risk and high-risk groups [18].
Although molecular characterizations have identified the gene signature for prognosis in GC, today, signatures are still inadequate for accurate patient therapy. Identifying new tumour markers or constructing gene models is still the focus of many research works and studies.
4. TCGA Classification of GC and Related Signaling Pathways
4.1. EBV-Related GC
EBV-positive GC is one of the four subtypes of GC, as defined by TCGA, found in 9% of GC and characterised by high EBV burden [13]. EBV-positive tumours were more frequent in men (81% of the cases) and mainly occurred in the upper part of the stomach. In addition, EBV-positive GC was more prevalent in younger patients compared to older subjects (Figure 1). The histology of EBV-related GC is moderately- to poorly-differentiated adenocarcinoma, often accompanied by dense lymphocytic infiltration [19,20,21,22]. In this subtype were identified pathways related to the elevated expression of programmed death ligands 1 and 2 (PD-L1 and PD-L2), phosphatidylinositol-4,5-bisphosphate 3-kinase, the catalytic subunit α (PIK3CA) mutation and Janus kinase 2 (JAK2) amplification.
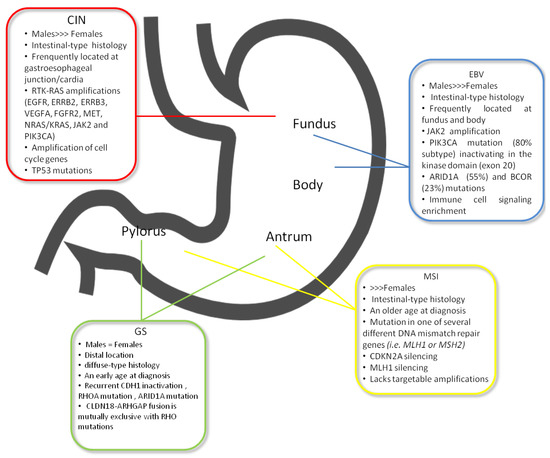
Figure 1.
The most relevant clinic-pathological and molecular features of TCGA subtypes.
PD-L1 helps neoplastic cells to escape from antitumoral immune response, by binding to PD-1, which is expressed on cytotoxic T-cells [23,24,25]. In the literature PD-L1, expressed on cancer cells or tumour infiltrating immune cells, has emerged as a prognostic factor in GC, but its specific role in EBV-related GC has not yet been described [26,27,28,29,30]. In a recent study [31] focusing on EBV-related GC, the expression of PD-1/PDL-1 on immune and neoplastic cells, respectively, was directly related to diffuse histology (according to Lauren’s classification) and depth of tumour invasion. Therefore, targeted immune therapy against the PD-L1/PD-1 axis could be effective in this subtype. Pembrolizumab, a highly specific monoclonal antibody targeting the PD-1 receptor, showed an overall response rate of 22% in a cohort of patients previously treated with chemotherapy [32]. Subsequently, PD-L1 expression in at least 1% of neoplastic cells from paraffin-embedded tissue was significantly related to response to this drug [33]. Another anti-immune strategy, already employed in melanoma, targeting both the PD-1/PD-L1 and the CTLA/B7 axis, is under evaluation in several clinical trials [34].
The PI3K family of intracellular kinases is involved in cell survival, proliferation, differentiation and migration [35]. In GC, the PI3K/AKT/mTOR pathway is frequently activated and associated with nodal metastasis: in 35–80% of GC cases, PI3KCA is overexpressed [27,28,29], and in 40–82% of GC cases, phosphorylation of AKT is described [36,37,38,39,40]. The EBV and MSI molecular subtypes of GC show alterations in PIK3CA, in 80% and 42% of cases, respectively [14]. However, molecular mechanisms responsible for sensitivity to PI3K inhibitors are not clearly defined, and the potential use of this drug category in advanced GC is still in the preclinical stage. [41]. In GC, the PIK3CA mutation could be predictive of response to everolimus and AKT inhibitors [42,43]. It is hypothesised that AKT affects the BCL2 protein and the NF-κB pathway. PI3K may also induce upregulation of the chemo-resistance proteins, MDR1/Pgp, BCL2 and XIAP, and downregulation of the expression of BAX and caspase 3. In vitro, in tumour tissues of GC patients, AKT activation and PTEN loss were associated with increased resistance to multiple chemotherapeutic agents (5-FU, doxorubicin, mitomycin C and cisplatin) [44]. Similarly, in GC cell lines, a combination of PI3K and AKT inhibitors with chemotherapy agents has successfully attenuated chemotherapeutic resistance [45,46].
The JAK/STAT signaling pathway has been identified in several types of tumours, including GC, and especially in the EBV-subtype [47,48]. The phosphorylation and subsequent activation of JAK2 lead to STAT activation by phosphorylation and activation of downstream gene expression involved in cell proliferation and apoptosis arrest [49]. Therefore, JAK2 inhibitors may also represent a potential therapeutic treatment for solid tumours, such as GC, despite them being primarily studied in inflammatory and myeloproliferative disorders [50]. Ruxolitinib, a JAK1 and JAK2 inhibitor, in combination with capecitabine has demonstrated preliminary efficacy in pancreatic cancer and, in combination with regorafenib, and is currently under evaluation in colorectal cancer (ClinicalTrials.gov identifier: NCI02119676) [51]. However, there are no trials ongoing in GC.
4.2. GC with MSI
Microsatellite instability (MSI) is the hallmark of the MSI subtype according to TGCA classification. MSI represents 15–30% of all GCs, is more frequently associated with intestinal histology and usually arises in the mucosa of the antrum, mainly in females at an older age [14,52,53] (Figure 1). MSI is a change that occurs in the DNA of certain cells (such as tumour cells) in which the number of repeats of microsatellites (short, repeated sequences of DNA) is different than the number of repeats in the DNA when it was inherited. The cause of MSI may be a defect in the ability to repair mistakes made when DNA is copied in the cell, determined by mutations in one of several different DNA mismatch repair genes (i.e., MLH1 or MSH2) [54]. The principal mechanism causing MMR deficiency in this GC subtype relies on different MMR genes probably involved in MSI-high (MSI-H) sporadic GC without MLH1 hypermethylation [55,56]. Zhu et al. in a meta-analysis showed a significant reduction of mortality in patients with MSI-H compared with MSI-L (low) or microsatellite stable (MSS) cases [57]. In the MRC MAGIC trial, the relationship between MMRd, MSI and survival in patients with resectable GC randomised to surgery alone or perioperative chemotherapy has been examined. MSI status and MLH1 deficiency had a positive prognostic role in patients treated with surgery alone, while a negative prognostic effect was established in patients treated with chemotherapy [55]. In contrast to MSI in colorectal cancer, in MSI GC, alterations in PIK3CA, ERBB3, ERB22 and EGFR genes, along with major histocompatibility complex I are known [14,53], whereas BRAF V600E mutations have never been found [14]. In MSI-positive colorectal cancer, pembrolizumab has shown objective response and progression-free survival rates of 40% and 78%, respectively [58]. Both MSI and EBV subtypes have been associated with a more favourable prognosis and are, therefore, detected in lower percentage in the metastatic setting, with subsequent difficult case finding in clinical trial design [59,60].
4.3. GC with CIN
The largest group, CIN subtype, accounts for approximately 50% of GCs, and its most frequent location is in the esophagogastric junction (EGJ)/cardia, as established by the TCGA study [14] (Figure 1). CIN GC with an intestinal type histology is associated with copy number gains of chromosomes 8q, 17q and 20q, whereas gains at 12q and 13q are more related to diffuse histology [61,62]. The effect of these alterations is the loss or gain of function of oncogenes and tumour suppressor genes [63]. In the CIN subtype, some specific mutations are frequently found, i.e., in the TP53 gene and receptor tyrosine kinases (RTKs), as well as amplifications of cell cycle genes (cyclin E1, cyclin D1 and cyclin-dependent kinase 6) and of the gene that encodes the ligand vascular endothelial growth factor A (VEGFA) [14,64]. Furthermore, HER2, BRAF, epidermal growth factor (EGFR), MET, FGFR2 and RAS mutations have been discovered in the CIN subtype [14,65] (Figure 2).

Figure 2.
The most relevant targetable pathways in GC.
The most frequent genetic alteration of this subtype, along with their respective targeted drugs, is detailed in Table 2.

Table 2.
Gene alteration and their respective targeted drugs.
4.4. Genomic Stable (GS) GC
The GS subgroup included all tumours that did not fulfil appropriate criteria for inclusion in one of the other groups [14]. Patients included in this subgroup represent nearly 20% of all GC, usually show diffuse histology, have a diagnosis at an earlier age (median 59 years), distal localization and occurring equally in males and females (Figure 1). Several subtype-specific molecular changes have been described for GS tumours. The principal somatic genomic alterations observed in GS gastric tumours involve CDH1, ARID1A and RHOA and are described in Table 2. Moreover, an additional translocation (between CLDN18 and ARHGAP26) involved in cell motility was later identified [14].
4.5. Patient-Derived Preclinical Models of GC
The lack of effective preclinical models of human tumours, reflecting the complexity and heterogeneity of cancer, has consistently limited the development of targeted drugs. In vitro and in vivo models are available: cancer cell lines; cell line xenograft mouse models (PDX), created transplanting human neoplastic fresh tissue into immunodeficient mice and organoids, which are three-dimensionally cultured tissues, mimicking human tissues [110,111,112,113,114,115,116,117,118,119,120,121,122]. Their advantages and disadvantages are summarised in Table 3.

Table 3.
Patient-derived preclinical models of GC: advantages and disadvantages.
Stem cell-derived gastric organoids have proven to be effective models of gastric cancer pathogenesis: H. pylori-activated c-Met by its virulence factor cytotoxin-associated gene A and induced a two-fold increase in epithelial cell proliferation [123]. Furthermore, epithelial dysplasia was found in gastric organoids, and adenocarcinoma quickly developed in mice having mutations in KRAS or P53 [124]. Murine epithelial-mesenchymal organoids were used also to successfully replicate hereditary GC, with short hairpin RNA knockdown of TGFBR2 [125]. The fundamental role of RHOA function in mediating anoikis in diffuse-type GC was demonstrated also in mouse organoids [126].
4.6. Role of microRNAs in Signaling Pathways of GC
MicroRNAs (miRNAs) are short, approximately 22 nucleotides in length that play key roles in the regulation of gene expression [127]. Accumulating evidence indicates that miRNAs play an important role in regulating cancer-related genes. They contribute to GC as oncogenes or tumour suppressors by inhibiting either directly or indirectly the expression of target genes, some of which are involved in signaling pathways [128]. Phosphatase and tensin homologue (PTEN) functions as a tumour suppressor by counteracting PI3K signaling [129]. miRNA-221/222 has been found to be a modulator of PTEN: by antisense or overexpression strategies, it directly affects PTEN expression [130]. PTEN is also a target gene of miRNA-21 that increases the proliferation and invasion of GC cells. A similar effect is displayed by miRNA-214 [131].
miRNA-375 is one of the most downregulated miRNAs in GC, by directly targeting PDK1, a kinase that phosphorylates Akt. Ectopic expression of miRNA-375 reduces cell viability by inducing the caspase-dependent apoptotic pathway [132]. Instead, miRNA-143 regulates the function of GC cells in the PI3K/Akt pathway because its gene target is Akt itself [133]. Down-expression of miR-181c stimulates KRAS expression and may have an important role in GC [134]. It was found that miRNA-29s could influence the Ras/Raf/MEK/ERK pathway, which acts on cell cycle progression by induction of cell cycle regulatory proteins such as CDKs and cyclins. miRNA-29c inhibits protein expression/phosphorylation of Cdc42 [135,136]. Feng et al. demonstrated that CDK6 is regulated by miRNA-107 [137]. Its expression is significantly decreased in GC, and its re-expression significantly decreases proliferation. In GC, miRNA-206 modulates downstream target cyclin D2, involved in proliferation [138]. miRNA-106b and miRNA-93 could be upregulated in GC and be downstream targets of the oncogenic transcription factor E2F1, which make the tumour-suppressive function of transforming growth factor-β less effective [139]. E2F1 is a gene target of miRNA-331-3p and miRNA-106a, modulating the G1/s transition [140,141]. miRNA-331-3p is a tumour suppressor, whereas miRNA-106a promotes tumour growth. A group of researchers demonstrated that the p21 family of CDK inhibitors was suppressed by miRNA-106b-93-25 and miRNA-222-221 clusters. In particular, miRNA-25 targets p57 through the 3′-untranslated region; miRNA-106b and miRNA-93 control p21, whereas p27 and p57 are downregulated by miRNA-222 and miRNA-221 [142]. miRNA-148a has as direct target, p27, so by suppressing p27 expression, it may promote gastric cell proliferation [143]. miRNA-196a, when highly expressed, is associated with clinic-pathological parameters, such as tumour size, poor pT stage, pN stage and patients’ overall survival times. In vitro and in vivo, a downregulation of miRNA-196a suppresses gastric cancer proliferation by targeting p27kip [144]. Previous research has shown that miRNA-375, by targeting the JAK2 oncogene, may act as a tumour suppressor and regulate GC cell proliferation [139]. Moreover, miRNA-135 by targeting JAK2 may repress p-STAT3 activation, reduce cyclin D1 Bcl-xL expression and inhibit cell proliferation [47].
4.7. Clinical Implications of Tissue miRNAs in GC
Tissue-based GC-related miRNA biomarkers are listed in Table 4, focusing particularly on their application as diagnostic and prognostic indicators [145,146,147,148,149,150,151,152,153,154,155,156,157,158,159]. Dysregulated expression of miRNA can play an oncogenic or tumour-suppressor role. In fact, they can regulate different signal pathways, targeting genes involved in cell migration, angiogenesis and cell proliferation. Table 5 summarises specific miRNA targeting pathways described above [160,161,162,163,164,165,166,167].

Table 4.
Diagnostic and prognostic role of tissue-based GC-related miRNAs.

Table 5.
Expression and deregulation of miRNAs in gastric cancer.
5. Conclusions
The recent molecular research on GC has generated large amounts of data that are currently not integrated into clinical practice.
However, they may be of help in the design of future clinical trials aiming to personalise treatment in several ways: (i) by identifying the driving pathways of tumour growth; (ii) by discovering potential drugs targeting such pathways; (iii) by finding predictable mechanisms of resistance and strategies to overcome them.
It must be emphasised that each targetable molecular alteration/pathway is not specific to a distinct subtype of GC; therefore, molecular subgroups alone are not sufficient to assign a patient to a clinical trial. On the contrary, molecular characterization of patients is useful to select a small population to be screened for protocol-eligible molecular aberrations. The implementation of GC research and the molecular classification of patients in clinical trials may be important to select the most appropriate therapies in GC. The hope is that combining histological and molecular classification will be supportive of GC therapeutics and prognosis, but also in the near future for new non-invasive diagnostic approaches such as to identify specific GC biomarker subtypes from circulating nucleic acid or tumour cells.
Conflicts of Interest
The author declares no conflicts of interest.
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2016. CA Cancer J. Clin. 2016, 66, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Borrmann, R. Geschwulste des margens. In Handbuch spez pathol anat und histo; Henke, F., Lubarsch, O., Eds.; Springer: Berlin, Germany, 1926; pp. 864–871. [Google Scholar]
- Siewert, J.R.; Stein, H.J. Classification of adenocarcinoma of the oesophagogastric junction. Br. J. Surg. 1998, 85, 1457–1459. [Google Scholar] [CrossRef] [PubMed]
- Lauren, P. The two histological main types of gastric carcinoma: Diffuse and so called intestinal-type carcinoma: An attempt at a histo-clinical classification. Acta Pathol. Microbiol. Scand. 1965, 64, 31–49. [Google Scholar] [CrossRef] [PubMed]
- Lauwers, G.Y.; Carneiro, F.; Graham, D.Y. Gastric carcinoma. In WHO Classification of Tumours of the Digestive System, 4th ed.; Bosman, F.T., Carneiro, F., Hruban, R.H., Theise, N.D., Eds.; World Health Organization: Lyon, France, 2010; Volume 3, pp. 48–58, ISBN-13 9789283224327. [Google Scholar]
- Kajitani, T. The general rules for the gastric cancer study in surgery and pathology I: Clinical classification. Jpn. J. Surg. 1981, 1, 127–139. [Google Scholar]
- Tan, I.B.; Ivanova, T.; Lim, K.H.; Ong, C.W.; Deng, N.; Lee, J.; Tan, S.H.; Wu, J.; Lee, M.H.; Ooi, C.H.; et al. Intrinsic subtypes of gastric cancer, based on gene expression pattern, predict survival and respond differently to chemotherapy. Gastroenterology 2011, 141, 476–485. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.Y.; Cheong, J.H. Beyond precision surgery: Molecularly motivated precision care for gastric cancer. Eur. J. Surg. Oncol. 2017, 43, 856–864. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Cho, Y.S.; Lee, G.K.; Lee, S.; Kim, K.W.; Jho, S.; Kim, H.M.; Hong, S.H.; Hwang, J.H.; Kim, S.Y.; et al. Genomic profile analysis of diffuse-type gastric cancers. Genome Biol. 2014, 15, R55. [Google Scholar] [CrossRef] [PubMed]
- Tanabe, S.; Aoyagi, K.; Yokozaki, H.; Sasaki, H. Gene expression signatures for identifying diffuse-type gastric cancer associated with epithelial-mesenchymal transition. Int. J. Oncol. 2014, 44, 1955–1970. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.; Bang, S.; Lee, S.; Kim, S.; Jung, Y.; Lee, C.; Choi, K.; Lee, S.G.; Lee, K.; Lee, Y.; et al. Expression Profiling and Subtype-Specific Expression of stomach cancer. Cancer Res. 2003, 63, 8248–8255. [Google Scholar] [PubMed]
- Min, L.; Zhao, Y.; Zhu, S.; Qiu, X.; Cheng, R.; Xing, J.; Shao, L.; Guo, S.; Zhang, S. Integrated Analysis Identifies Molecular Signatures and Specific Prognostic Factors for Different Gastric Cancer Subtypes. Transl. Oncol. 2017, 10, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Lei, Z.; Tan, I.B.; Das, K.; Deng, N.; Zouridis, H.; Pattison, S.; Chua, C.; Feng, Z.; Guan, Y.K.; Ooi, C.H.; et al. Identification of molecular subtypes of gastric cancer with different responses to PI3-kinase inhibitors and 5-fluorouracil. Gastroenterology 2013, 145, 554–565. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014, 513, 202–209. [Google Scholar] [CrossRef]
- Benita, Y.; Cao, Z.; Giallourakis, C.; Li, C.; Gardet, A.; Xavier, R.J. Gene enrichment profiles reveal T-cell development, differentiation, and lineage-specific transcription factors including ZBTB25 as a novel NF-AT repressor. Blood 2010, 115, 5376–5384. [Google Scholar] [CrossRef] [PubMed]
- Ooi, C.H.; Ivanova, T.; Wu, J.; Lee, M.; Tan, I.B.; Tao, J.; Ward, L.; Koo, J.H.; Gopalakrishnan, V.; Zhu, Y.; et al. Oncogenic pathway combinations predict clinical prognosis in gastric cancer. PLoS Genet. 2009, 5, e1000676. [Google Scholar] [CrossRef] [PubMed]
- Cristescu, R.; Lee, J.; Nebozhyn, M.; Kim, K.M.; Ting, J.C.; Wong, S.S.; Liu, J.; Yue, Y.G.; Wang, J.; Yu, K.; et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat. Med. 2015, 21, 449–456. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Chen, G.; Wang, Q.; Lu, W.; Xu, M. Identification and validation of a prognostic 9-genes expression signature for gastric cancer. Oncotarget 2017, 10, 73826–73836. [Google Scholar] [CrossRef] [PubMed]
- Shinozaki-Ushiku, A.; Kunita, A.; Fukayama, M. Update on Epstein–Barr virus and gastric cancer [review]. Int. J. Oncol. 2015, 46, 1421–1434. [Google Scholar] [CrossRef] [PubMed]
- Abe, H.; Kaneda, A.; Fukayama, M. Epstein–Barr virus associated gastric carcinoma: Use of host cell machineries and somatic gene mutations. Pathobiology 2015, 82, 212–223. [Google Scholar] [CrossRef] [PubMed]
- Fukayama, M.; Hino, R.; Uozaki, H. Epstein–Barr virus and gastric carcinoma: Virus–host interactions leading to carcinoma. Cancer Sci. 2008, 99, 1726–1733. [Google Scholar] [CrossRef] [PubMed]
- Song, H.J.; Srivastava, A.; Lee, J.; Kim, Y.S.; Kim, K.M.; Ki Kang, W.; Kim, M.; Kim, S.; Park, C.K.; Kim, S. Host inflammatory response predicts survival of patients with Epstein–Barr virus-associated gastric carcinoma. Gastroenterology 2010, 139, 84–92.e2. [Google Scholar] [CrossRef] [PubMed]
- Iwai, Y.; Ishida, M.; Tanaka, Y.; Okazaki, T.; Honjo, T.; Minato, N. Involvement of PD- L1 on tumor cells in the escape from host immune system and tumor immunotherapy by PD-L1 blockade. Proc. Natl. Acad. Sci. USA 2002, 99, 12293–12297. [Google Scholar] [CrossRef] [PubMed]
- Blank, C.; Gajewski, T.F.; Mackensen, A. Interaction of PD-L1 on tumor cells with PD-1 on tumor-specific T cells as a mechanism of immune evasion:implications for tumor immunotherapy. Cancer Immunol. Immunother. 2005, 54, 307–314. [Google Scholar] [CrossRef] [PubMed]
- Francisco, L.M.; Sage, P.T.; Sharpe, A.H. The PD-1 pathway in tolerance and autoimmunity. Immunol. Rev. 2010, 236, 219–242. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Qiu, M.; Jin, Y.; Li, B.; Wang, X.; Yan, S.; Xu, R.; Yang, D. Programmed cell death ligand 1 (PD-L1) expression on gastric cancer and its relationship with clinicopathologic factors. Int. J. Clin. Exp. Pathol. 2015, 8, 11084–11091. [Google Scholar] [PubMed]
- Qing, Y.; Li, Q.; Ren, T.; Xia, W.; Peng, Y.; Liu, G.L.; Luo, X.Y.; Dai, X.Y.; Zhou, S.F.; Wang, D. Upregulation of PD-L1 and APE1 is associated with tumorigenesis and poor prognosis of gastric cancer. Drug Des. Devel. Ther. 2015, 9, 901–909. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.W.; Nam, K.H.; Ahn, S.H.; Park, D.J.; Kim, H.H.; Kim, S.H.; Chang, H.; Lee, J.O.; Kim, Y.J.; Lee, H.S.; et al. Prognostic implications of immunosuppressive protein expression in tumors as well as immune cell infiltration within the tumor microenvironment in gastric cancer. Gastric Cancer 2016, 19, 42–52. [Google Scholar] [CrossRef] [PubMed]
- Thompson, E.D.; Zahurak, M.; Murphy, A.; Cornish, T.; Cuka, N.; Abdelfatah, E.; Yang, S.; Duncan, M.; Ahuja, N.; Taube, J.M.; et al. Patterns of PD-L1 expression and CD8 T cell infiltration in gastric adenocarcinomas and associated immune stroma. Gut 2017, 66, 794–801. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.X.; Wang, X.S.; Wang, Y.F.; Hu, X.C.; Yan, J.Q.; Zhang, Y.L.; Wang, W.; Yang, R.J.; Feng, Y.Y.; Gao, S.G.; et al. Prognostic significance of PD-L1 expression in patients with gastric cancer in East Asia: A meta-analysis. OncolTargets Ther. 2016, 9, 2649–2654. [Google Scholar] [CrossRef]
- Abe, H.; Kunita, A.; Yamashita, H.; Seto, Y.; Fukayama, M. Overexpression and gene amplification of PD-L1 in cancer cells and PD-L1+ immune cells in Epstein-Barr virus-associated gastric cancer: The prognostic implications. Mod. Pathol. 2017, 30, 427–439. [Google Scholar] [CrossRef]
- Shankaran, V.; Muro, K.; Bang, Y.; Geva, R.; Catenacci, D.; Gupta, S.; Eder, J.P.; Berger, R.; Loboda, A.; Albright, A.; et al. Correlation of gene expression signatures and clinical outcomes in patients with advanced gastric cancer treated with pembrolizumab (MK-3475). J. Clin. Oncol. 2015, 33, 3026. [Google Scholar] [CrossRef]
- Bang, Y.; Im, S.; Lee, K.; Cho, J.; Song, E.; Lee, K.; Kim, Y.H.; Park, J.O.; Chun, H.G.; Zang, D.Y.; et al. Randomized, double-blind phase II trial with prospective classification by ATM protein level to evaluate the efficacy and tolerability of olaparib plus paclitaxel in patients with recurrent or metastatic gastric cancer. J. Clin. Oncol. 2015, 33, 3858–3865. [Google Scholar] [CrossRef] [PubMed]
- Fontana, E.; Smyth, E.C. Novel targets in the treatment of advanced gastric cancer: A perspective review. Ther. Adv. Med. Oncol. 2016, 8, 113–125. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.F.; Zhou, Z.X.; Chen, J.H.; Yi, G.; Chen, H.G.; Ba, M.C.; Lin, S.Q.; Qi, Y.C. Up-regulation of PIK3CA promotes metastasis in gastric carcinoma. World J. Gastroenterol. 2010, 16, 4986–4991. [Google Scholar] [CrossRef] [PubMed]
- Tapia, O.; Riquelme, I.; Leal, P.; Sandoval, A.; Aedo, S.; Weber, H.; Letelier, P.; Bellolio, E.; Villaseca, M.; Garcia, P.; et al. The PI3K/AKT/mTOR pathway is activated in gastric cancer with potential prognostic and predictive significance. Virchows Arch. 2014, 465, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Ye, B.; Jiang, L.; Xu, H.; Zhou, D.; Li, Z. Expression of PI3K/AKT pathway in gastric cancer and its blockade suppresses tumor growth and metastasis. Int. J. Immunopathol. Pharmacol. 2012, 25, 627–636. [Google Scholar] [CrossRef] [PubMed]
- Cinti, C.; Vindigni, C.; Zamparelli, A.; Sala, D.; Epistolato, M.; Marrelli, D.; Cevenini, G.; Tosi, P. Activated Akt as an indicator of prognosis in gastric cancer. Virchows Arch. 2008, 453, 449–455. [Google Scholar] [CrossRef] [PubMed]
- Sangawa, A.; Shintani, M.; Yamao, N.; Kamoshida, S. Phosphorylation status of Akt and caspase-9 in gastric and colorectal carcinomas. Int. J. Clin. Exp. Pathol. 2014, 7, 3312–3317. [Google Scholar] [PubMed]
- Welker, M.E.; Kulik, G. Recent syntheses of PI3K/Akt/mTOR signaling pathway inhibitors. Bioorg. Med. Chem. 2013, 2, 4063–4091. [Google Scholar] [CrossRef] [PubMed]
- Janku, F.; Tsimberidou, A.M.; Garrido-Laguna, I.; Wang, X.; Luthra, R.; Hong, D.S.; Naing, A.; Falchook, G.S.; Moroney, J.W. PIK3CA mutations in patients with advanced cancers treated with PI3K/AKT/mTOR axis inhibitors. Mol. Cancer. Ther. 2011, 10, 558–565. [Google Scholar] [CrossRef] [PubMed]
- Davies, B.R.; Greenwood, H.; Dudley, P.; Crafter, C.; Yu, D.H.; Zhang, J.; Li, J.; Gao, B.; Ji, Q.; Maynard, J.; et al. Preclinical pharmacology of AZD5363, an inhibitor of AKT: Pharmacodynamics, antitumor activity, and correlation of monotherapy activity with genetic background. Mol. Cancer Ther. 2012, 11, 873–887. [Google Scholar] [CrossRef] [PubMed]
- Li, V.; Wong, C.; Chan, T.; Chan, A.S.; Zhao, W.; Chu, K.M.; So, S.; Chen, X.; Yuen, S.T.; Leung, S.Y. Mutations of PIK3CA in gastric adenocarcinoma. BMC Cancer 2005, 5, 29. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.G.; Ai, Y.W.; Yu, L.L.; Zhou, X.D.; Liu, J.; Li, J.H.; Xu, X.M.; Liu, S.; Chen, J.; Liu, F.; et al. Phosphoinositide 3-kinase/Akt pathway plays an important role in chemoresistance of gastric cancer cells against etoposide and doxorubicin induced cell death. Int. J. Cancer 2008, 122, 433–443. [Google Scholar] [CrossRef] [PubMed]
- Oki, E.; Kakeji, Y.; Tokunaga, E.; Nishida, K.; Koga, T.; Egashira, A.; Morita, M.; Maehara, Y. Impact of PTEN/AKT/ PI3K signal pathway on the chemotherapy for gastric cancer. J. Clin. Oncol. 2006, 24, 4034. [Google Scholar] [CrossRef]
- Im, S.; Lee, K.; Nam, E. Potential prognostic significance of p185HER2 overexpression with loss of PTEN expression in gastric carcinomas. Tumori 2005, 91, 513–521. [Google Scholar] [PubMed]
- Wu, H.; Huang, M.; Cao, P.; Wang, T.; Shu, Y.; Liu, P. MiR-135a targets JAK2 and inhibits gastric cancer cell proliferation. Cancer Biol. Ther. 2012, 13, 281–288. [Google Scholar] [CrossRef] [PubMed]
- Brooks, A.J.; Dai, W.; O’Mara, M.L.; Abankwa, D.; Chhabra, Y.; Pelekanos, R.A.; Gardon, O.; Tunny, K.A.; Blucher, K.M.; Morton, C.J.; et al. Mechanism of activation of protein kinase JAK2 by the growth hormone receptor. Science 2014, 344, 1249783. [Google Scholar] [CrossRef] [PubMed]
- Levine, R.L.; Wadleigh, M.; Cools, J.; Ebert, B.L.; Wernig, G.; Huntly, B.J.; Boggon, T.J.; Wlodarska, I.; Clark, J.J.; Moore, S.; et al. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell 2005, 7, 387–397. [Google Scholar] [CrossRef] [PubMed]
- Buchert, M.; Burns, C.; Ernst, M. Targeting JAK kinase in solid tumors: Emerging opportunities and challenges. Oncogene 2016, 35, 939–951. [Google Scholar] [CrossRef] [PubMed]
- Hurwitz, H.; Uppal, N.; Wagner, S.; Bendell, J.; Beck, J.; Wade, S.; Nemunaitis, J.J.; Stella, P.J.; Pipas, J.M.; Wainberg, Z.A.; et al. A randomized doubleblind phase 2 study of ruxolitinib (RUX) or placebo (PBO) with capecitabine (CAPE) as second-line therapy in patients (pts) with metastatic pancreatic cancer (mPC). J. Clin. Oncol. 2015, 32, 4000. [Google Scholar] [CrossRef]
- Pedrazzani, C.; Corso, G.; Velho, S.; Leite, M.; Pascale, V.; Bettarini, F.; Marrelli, D.; Seruca, R.; Roviello, F. Evidence of tumor micro satellite instability in gastric cancer with familial aggregation. Fam. Cancer 2009, 8, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Velho, S.; Fernandes, M.S.; Leite, M.; Figueiredo, C.; Seruca, R. Causes and consequences of microsatellite instability in gastric carcinogenesis. World J. Gastroenterol. 2014, 20, 16433–16442. [Google Scholar] [CrossRef] [PubMed]
- Chung, D.C.; Rustgi, A.K. DNA mismatch repair and cancer. Gastroenterology 1995, 109, 1685–1699. [Google Scholar] [CrossRef]
- Smyth, E.C.; Wotherspoon, A.; Peckitt, C.; Nankivell, M.G.; Eltahir, Z.; Wilson, S.H.; de Castro, D.G.; Okines, A.F.C.; Langley, R.E.; Cunningham, D. Correlation between mismatch repair deficiency (MMRd), microsatellite instability (MSI) and survival in MAGIC. J. Clin. Oncol. 2016, 15, 4064. [Google Scholar] [CrossRef]
- Pinto, M.; Wu, Y.; Mensink, R.G.; Cirnes, L.; Seruca, R.; Hofstra, R.M. Somatic mutations in mismatch repair genes in sporadic gastric carcinomas are not a cause but a consequence of the mutator phenotype. Cancer Genet. Cytogenet. 2008, 180, 110–114. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Li, Z.; Wang, Y.; Zhang, C.; Liu, Y.; Qu, X. Microsatellite instability and survival in gastric cancer: A systematic review and meta-analysis. Mol. Clin. Oncol. 2015, 3, 699–705. [Google Scholar] [CrossRef] [PubMed]
- Le, D.; Uram, J.; Wang, H.; Bartlett, B.; Kemberling, H.; Eyring, A.; Skora, A.D.; Luber, B.S.; Azad, N.S.; Laheru, D.; et al. PD-1 blockade in tumors with mismatch-repair deficiency. N. Engl. J. Med. 2015, 372, 2509–2520. [Google Scholar] [CrossRef] [PubMed]
- Camargo, M.C.; Kim, W.H.; Chiaravalli, A.M.; Kim, K.M.; Corvalan, A.H.; Matsuo, K.; Yu, J.; Sung, J.J.; Herrera-Goepfert, R.; Meneses-Gonzalez, F.; et al. Improved survival of gastric cancer with tumour Epstein-Barr virus positivity: An international pooled analysis. Gut 2014, 63, 236–243. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.Y.; Bae, J.M.; An, J.Y.; Kwon, I.G.; Cho, I.; Shin, H.B.; Eiji, T.; Aburahmah, M.; Kim, H.I.; Cheong, J.H.; et al. Is microsatellite instability a prognostic marker in gastric cancer? A systematic review with meta-analysis. J. Surg. Oncol. 2014, 110, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Giam, M.; Rancati, G. Aneuploidy and chromosomal instability in cancer: A jackpot to chaos. Cell Div. 2015, 10, 3. [Google Scholar] [CrossRef] [PubMed]
- Chia, N.Y.; Tan, P. Molecular classification of gastric cancer. Ann. Oncol. 2016, 27, 763–769. [Google Scholar] [CrossRef] [PubMed]
- Aprile, G.; Giampieri, R.; Bonotto, M.; Bittoni, A.; Ongaro, E.; Cardellino, G.G.; Graziano, F.; Giuliani, F.; Fasola, G.; Cascinu, S.; et al. The challenge of targeted therapies for gastric cancer patients: The beginning of a long journey. Expert Opin. Investig. Drugs 2014, 23, 925–942. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Xu, X.Y.; Zhou, P.H. Emerging molecular classifications and therapeutic implications for gastric cancer. Chin. J. Cancer 2016, 35, 49. [Google Scholar] [CrossRef] [PubMed]
- Tan, P.; Yeoh, K.G. Genetics and Molecular Pathogenesis of Gastric Adenocarcinoma. Gastroenterology 2015, 149, 1153–1162.e3. [Google Scholar] [CrossRef] [PubMed]
- Gravalos, C.; Jimeno, A. HER2 in gastric cancer: A new prognostic factor and a novel therapeutic target. Ann. Oncol. 2008, 19, 1523–1529. [Google Scholar] [CrossRef] [PubMed]
- Bang, Y.J.; Van Cutsem, E.; Feyereislova, A.; Chung, H.C.; Shen, L.; Sawaki, A.; Lordick, F.; Ohtsu, A.; Omuro, Y.; Satoh, T.; et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): A phase 3, open-label, randomised controlled trial. Lancet 2010, 376, 687–697. [Google Scholar] [CrossRef]
- Hecht, J.R.; Bang, Y.J.; Qin, S.K.; Chung, H.C.; Xu, J.M.; Park, J.O.; Jeziorski, K.; Shparyk, Y.; Hoff, P.M.; Sombrero, A.; et al. Lapatinib in Combination with Capecitabine Plus Oxaliplatin in Human Epidermal Growth Factor Receptor 2-Positive Advanced or Metastatic Gastric, Esophageal, or Gastroesophageal Adenocarcinoma: TRIO-013/LOGiC—A Randomized Phase III Trial. J. Clin. Oncol. 2016, 34, 443–451. [Google Scholar] [CrossRef] [PubMed]
- Baselga, J.; Cortés, J.; Kim, S.B.; Im, S.A.; Hegg, R.; Im, Y.H.; Roman, L.; Pdrini, J.L.; Pienkowki, T.; Kontt, A.; et al. Pertuzumab plus trastuzumab plus docetaxel for metastatic breast cancer. N. Engl. J. Med. 2012, 366, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Hecht, J.R.; Bang, Y.J.; Qin, S.K.; Chung, H.C.; Xu, J.M.; Park, J.O.; Jeziorski, K.; Shparyk, Y.; Hoff, P.M.; Sombrero, A.; et al. Lapatinib in combination with capecitabine plus oxaliplatin (CapeOx) in HER2 positive advanced or metastatic gastric (A/MGC), esophageal (EAC), or astroesophageal (GEJ) adenocarcinoma: The logic trial. J. Clin. Oncol. 2013, 31, LBA4001. [Google Scholar] [CrossRef]
- Satoh, T.; Xu, R.H.; Chung, H.C.; Sun, G.P.; Doi, T.; Xu, J.M.; Tsuji, A.; Omuro, Y.; Li, J.; Wang, J.W.; et al. Lapatinib plus paclitaxel versus paclitaxel alone in the second-line treatment of HER2-amplified advanced gastric cancer in Asian populations: TyTAN-a randomized, phase III study. J. Clin. Oncol. 2014, 32, 2039–2049. [Google Scholar] [CrossRef] [PubMed]
- Deva, S.; Baird, R.; Cresti, N.; Garcia-Corbacho, J.; Hogarth, L.; Frenkel, E.; Kawaguchi, K.; Arimura, A.; Donaldson, K.; Posner, J.; et al. Phase I expansion of S-222611, a reversible inhibitor of EGFR and HER2, in advanced solid tumors, including patients with brain metastases. J. Clin. Oncol. 2015, 33, 2511. [Google Scholar] [CrossRef]
- Lee, J.Y.; Hong, M.; Kim, S.T.; Park, S.H.; Kang, W.K.; Kim, K.M.; Lee, J. The impact of concomitant genomic alterations on treatment outcome for trastuzumab therapy in HER2-positive gastric cancer. Sci. Rep. 2015, 5, 9289. [Google Scholar] [CrossRef] [PubMed]
- Zuo, Q.; Liu, J.; Zhang, J.; Wu, M.; Guo, L.; Liao, W. Development of trastuzumab-resistant human gastric carcinoma cell lines and mechanisms of drug resistance. Sci. Rep. 2015, 5, 11634. [Google Scholar] [CrossRef] [PubMed]
- Piro, G.; Carbone, C.; Cataldo, I.; Di Nicolantonio, F.; Giacopuzzi, S.; Aprile, G.; Simionato, F.; Boschi, F.; Zanotto, M.; Mina, M.M.; et al. An FGFR3 Autocrine Loop Sustains Acquired Resistance to Trastuzumab in Gastric Cancer Patients. Clin. Cancer Res. 2016, 22, 6164–6175. [Google Scholar] [CrossRef] [PubMed]
- Arienti, C.; Zanoni, M.; Pignatta, S.; Del Rio, A.; Carloni, S.; Tebaldi, M.; Tedaldi, G.; Tesei, A. Preclinical evidence of multiple mechanisms underlying trastuzumab resistance in gastric cancer. Oncotarget 2016, 7, 18424–18439. [Google Scholar] [CrossRef] [PubMed]
- White, C.D.; Brown, M.D.; Sacks, D.B. IQGAPs in cancer: A family of scaffold proteins underlying tumorigenesis. FEBS Lett. 2009, 583, 1817–1824. [Google Scholar] [CrossRef] [PubMed]
- Walch, A.; Seidl, S.; Hermannstädter, C.; Rauser, S.; Deplazes, J.; Langer, R.; von Weyhern, C.H.; Sarbia, M.; Busch, R.; Feith, M.; et al. Combined analysis of Rac1, IQGAP1, Tiam1 and E-cadherin expression in gastric cancer. Mod. Pathol. 2008, 21, 544–552. [Google Scholar] [CrossRef] [PubMed]
- Khoury, H.; Naujokas, M.A.; Zuo, D.; Sangwan, V.; Frigault, M.M.; Petkiewicz, S.; Dankort, D.L.; Muller, W.J.; Park, M. HGF converts ErbB2/Neu epithelial morphogenesis to cell invasion. Mol. Biol. Cell 2005, 16, 550–561. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.T.; Kim, H.; Liska, D.; Gao, S.; Christensen, J.G.; Weiser, M.R. MET activation mediates resistance to lapatinib inhibition of HER2- amplified gastric cancer cells. Mol. Cancer Ther. 2012, 11, 660–669. [Google Scholar] [CrossRef] [PubMed]
- De Silva, N.; Schulz, L.; Paterson, A.; Qain, W.; Secrier, M.; Godfrey, E.; Cheow, H.; O’Donovan, M.; Lao-Sirieix, P.; Jobanputra, M.; et al. Molecular effects of Lapatinib in the treatment of HER2 overexpressing oesophago-gastric adenocarcinoma. Br. J. Cancer 2015, 113, 1305–1312. [Google Scholar] [CrossRef] [PubMed]
- Lordick, F.; Kang, Y.; Chung, H.; Salman, P.; Oh, S.; Bodoky, G.; Kurteva, G.; Volovat, C.; Moiseyenko, V.M.; Gorbunova, V.; et al. Capecitabine and cisplatin with or without cetuximab for patients with previously untreated advanced gastric cancer (EXPAND): A randomised, open-label phase 3 Trial. Lancet Oncol. 2013, 14, 490–499. [Google Scholar] [CrossRef]
- Waddell, T.; Chau, I.; Cunningham, D.; Gonzalez, D.; Okines, A.F.; Okines, C.; Wotherspoon, A.; Saffert, C.; Middleton, G.; Wadsley, J.; et al. Epirubicin, oxaliplatin, and capecitabine with or without panitumumab for patients with previously untreated advanced oesophagogastric cancer (REAL3): A randomised, open-label phase 3 trial. Lancet Oncol. 2013, 14, 481–489. [Google Scholar] [CrossRef]
- Dragovich, T.; Mccoy, S.; Fenoglio-Preiser, C.; Wang, J.; Benedetti, J.; Baker, A.F.; Hackett, C.B.; Urba, S.G.; Zaner, K.S.; Blanke, C.D.; et al. Phase II trial of erlotinib in gastroesophageal junction and gastric adenocarcinomas: SWOG 0127. J. Clin. Oncol. 2006, 24, 4922–4927. [Google Scholar] [CrossRef] [PubMed]
- Dutton, S.J.; Ferry, D.R.; Blazeby, J.M.; Abbas, H.; Dahle-Smith, A.; Mansoor, W.; Thompson, J.; Harrison, M.; Chatteriee, A.; Falk, S.; et al. Gefitinib for oesophageal cancer progressing after chemotherapy (COG): A phase 3, multicentre, double-blind, placebocontrolled randomised trial. Lancet Oncol. 2014, 15, 894–904. [Google Scholar] [CrossRef]
- Ha, S.Y.; Lee, J.; Kang, S.Y.; Do, I.G.; Ahn, S.; Park, J.O; Kang, W.K.; Choi, M.G.; Sohn, T.S.; Bae, J.M.; et al. MET overexpression assessed by new interpretation method predicts gene amplification and poor survival in advanced gastric carcinomas. Mod. Pathol. 2013, 26, 1632–1641. [Google Scholar] [CrossRef] [PubMed]
- Scagliotti, G.V.; Novello, S.; Von Pawel, J. The emerging role of MET/HGF inhibitors in oncology. Cancer Treat. Rev. 2013, 39, 793–801. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, D.; Tebbutt, N.; Davidenko, I.; Murad, A.; Al-Batran, S.; Ilson, D.; Tjulandin, S.; Gotovkin, E.; Karaszewska, B.; Bondarenko, I.; et al. Phase III, randomized, double-blind, multicenter, placebo (P)-controlled trial of rilotumumab (R) plus epirubicin, cisplatin and capecitabine (ECX) as first-line therapy in patients (pts) with advanced MET-positive (pos) gastric or gastroesophageal junction (G/GEJ) cancer: RILOMET-1 study. J. Clin. Oncol. 2015, 33, 4000. [Google Scholar] [CrossRef]
- Shah, M.; Bang, Y.; Lordick, F.; Tabernero, J.; Chen, M.; Hack, S.; Phan, S.; Shames, D.S.; Cunningham, D. Metgastric: A phase III study of onartuzumab plus mFOLFOX6 in patients with metastatic HER2-negative (HER2-) and METpositive (MET+) adenocarcinoma of the stomach or gastroesophageal junction (GEC). J. Clin. Oncol. 2015, 33, 4012. [Google Scholar] [CrossRef]
- Iveson, T.; Donehower, R.; Davidenko, I.; Tjulandin, S.; Deptala, A.; Harrison, M.; Nirni, S.; Lakshmaiah, K.; Thomas, A.; Jiang, Y.; et al. Rilotumumab in combination with epirubicin, cisplatin, and capecitabine as first-line treatment for gastric or oesophagogastric junction adenocarcinoma: An open-label, dose de-escalation phase 1b study and a double-blind, randomised phase 2 study. Lancet Oncol. 2014, 15, 1007–1018. [Google Scholar] [CrossRef]
- Chen, J.; Zhou, S.J.; Zhang, Y.; Zhang, G.Q.; Zha, T.Z.; Feng, Y.Z.; Zhang, K. Clinicopathological and prognostic significance of galectin-1 and vascular endothelial growth factor expression in gastric cancer. World J. Gastroenterol. 2013, 19, 2073–2079. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.J.; Kim, J.G.; Sohn, S.K.; Chae, Y.S.; Moon, J.H.; Kim, S.N.; Bae, H.I.; Chung, H.Y.; Yu, W. No association of vascular endothelial growth factor-A (VEGF-A) and VEGF-C expression with survival in patients with gastric cancer. Cancer Res. Treat. 2009, 41, 218–223. [Google Scholar] [CrossRef] [PubMed]
- Deguchi, K.; Ichikawa, D.; Soga, K.; Watanabe, K.; Kosuga, T.; Takeshita, H.; Konishi, H.; Morimura, R.; Tsujiura, M.; Komatsu, S.; et al. Clinical significance of vascular endothelial growth factors C and D and chemokine receptor CCR7 in gastric cancer. Anticancer Res. 2010, 30, 2361–2366. [Google Scholar] [PubMed]
- Gou, H.F.; Chen, X.C.; Zhu, J.; Jiang, M.; Yang, Y.; Cao, D.; Hou, M. Expressions of COX-2 and VEGF-C in gastric cancer: Correlations with lymphangiogenesis and prognostic implications. J. Exp. Clin. Canc. Res. 2011, 30, 14. [Google Scholar] [CrossRef] [PubMed]
- Ohtsu, A.; Shah, M.A.; Van Cutsem, E.; Rha, S.Y.; Sawaki, A.; Park, S.R.; Lim, H.Y.; Yamada, Y.; Wu, J.; Langer, B.; et al. Bevacizumab in combination with chemotherapy as first-line therapy in advanced gastric cancer: A randomized, double-blind, placebo-controlled phase III study. J. Clin. Oncol. 2011, 29, 3968–3976. [Google Scholar] [CrossRef] [PubMed]
- Van Cutsem, E.; de Haas, S.; Kang, Y.K.; Ohtsu, A.; Tebbutt, N.C.; Ming, X.J.; Peng Yong, W.; Langer, B.; Delmar, P.; Scherer, S.J.; et al. Bevacizumab in combination with chemotherapy as first-line therapy in advanced gastric cancer: A biomarker evaluation from the AVAGAST randomized phase III trial. J. Clin. Oncol. 2012, 30, 2119–2127. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, C.S.; Tomasek, J.; Yong, C.J.; Dumitru, F.; Passalacqua, R.; Goswami, C.; Safran, H.; dos Santos, L.V.; Aprile, G.; Ferry, D.R.; et al. Ramucirumab monotherapy for previously treated advanced gastric or gastro-oesophageal junction adenocarcinoma (REGARD): An international, randomised, multicentre, placebocontrolled, phase 3 trial. Lancet 2014, 383, 31–39. [Google Scholar] [CrossRef]
- Li, J.; Qin, S.; Xu, J.; Guo, W.; Xiong, J.; Bai, Y.; Sun, G.; Yang, Y.; Wang, L.; Xu, N.; et al. Apatinib for chemotherapy-refractory advanced metastatic gastric cancer: Results from a randomized, placebocontrolled, parallel-arm, phase II trial. J. Clin. Oncol. 2013, 31, 3219–3225. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.Y.; Noh, S.H.; Cheong, J.H. Molecular Dimensions of Gastric Cancer: Translational and Clinical Perspectives. J. Pathol. Transl. Med. 2016, 50, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Corso, G.; Marrelli, D.; Pascale, V.; Vindigni, C.; Roviello, F. Frequency of CDH1 germline mutations in gastric carcinoma coming from high- and low-risk areas: Metanalysis and systematic review of the literature. BMC Cancer 2012, 12, 8. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.C.; Shen, C.Y.; Wu, H.S.; Hsieh, T.Y.; Chan, D.C.; Chen, C.J.; Yu, J.C.; Yu, C.P.; Harn, H.J.; Chen, P.J.; et al. Mechanisms inactivating the gene for E-cadherin in sporadic gastric carcinomas. World J. Gastroenterol. 2006, 12, 2168–2173. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wu, W.K.; Xing, R.; Wong, S.H.; Liu, Y.; Fang, X.; Zhang, Y.; Wang, M.; Wang, J.; Li, L.; et al. Distinct Subtypes of Gastric Cancer Defined by Molecular Characterization Include Novel Mutational Signatures with Prognostic Capability. Cancer Res. 2016, 76, 1724–1732. [Google Scholar] [CrossRef] [PubMed]
- Weissman, B.; Knudsen, K.E. Hijacking the chromatin remodeling machinery: Impact of SWI/SNF perturbations in cancer. Cancer Res. 2009, 69, 8223–8230. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.D.; Chen, Y.B.; Pan, K.; Wang, W.; Chen, S.P.; Chen, J.G.; Zhao, J.J.; Lv, L.; Pan, Q.Z.; Li, Y.Q.; et al. Decreased expression of the ARID1A gene is associated with poor prognosis in primary gastric cancer. PLoS ONE 2012, 7, e40364. [Google Scholar] [CrossRef] [PubMed]
- Shang, X.; Marchioni, F.; Evelyn, C.R.; Sipes, N.; Zhou, X.; Seibel, W.; Wortman, M.; Zheng, Y. Small-molecule inhibitors targeting G-protein-coupled Rho guanine nucleotide exchange factors. Proc. Natl. Acad. Sci. USA 2013, 110, 3155–3160. [Google Scholar] [CrossRef] [PubMed]
- Shang, X.; Marchioni, F.; Sipes, N.; Evelyn, C.R.; Jerabek-Willemsen, M.; Duhr, S.; Seibel, W.; Wortman, M.; Zheng, Y. Rational design of small molecule inhibitors targeting RhoA subfamily Rho GTPases. Chem. Biol. 2012, 19, 699–710. [Google Scholar] [CrossRef] [PubMed]
- Türeci, O.; Koslowski, M.; Helftenbein, G.; Castle, J.; Rohde, C.; Dhaene, K.; Seitz, G.; Sahin, U. Claudin-18 gene structure, regulation, and expression is evolutionary conserved in mammals. Gene 2011, 481, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Yao, F.; Kausalya, J.P.; Sia, Y.Y.; Teo, A.S.; Lee, W.H.; Ong, A.G.; Zhang, Z.; Tan, J.H.; Li, G.; Bertrand, D.; et al. Recurrent Fusion Genes in Gastric Cancer: CLDN18-ARHGAP26 Induces Loss of Epithelial Integrity. Cell Rep. 2015, 12, 272–285. [Google Scholar] [CrossRef] [PubMed]
- Al Batran, S.E.; Schuler, M.H.; Zvirbule, Z.; Manikhas, G.; Lordick, F.; Rusyn, A.; Vynnyk, Y.; Vynnychenko, I.; Fadeeva, N.; Nechaeva, M.; et al. FAST: An international, multicenter, randomized, phase II trial of epirubicin, oxaliplatin, and capecitabine (EOX) with or without IMAB362, a first-in-class anti-CLDN18.2 antibody, as firstline therapy in patients with advanced CLDN18.2 gastric and gastroesophageal junction (GEJ) adenocarcinoma. J. Clin. Oncol. 2016, 34, LBA4001. [Google Scholar] [CrossRef]
- Hidalgo, M.; Amant, F.; Biankin, AV.; Budinskà, E.; Byrne, A.T.; Caldas, C.; Clarke, R.B.; de Jong, S.; Jonkers, J.; Mælandsmo, G.M.; et al. Patient-derived xenograft models: An emerging platform for translational cancer research. Cancer Discov. 2014, 4, 998–1013. [Google Scholar] [CrossRef] [PubMed]
- Hausser, H.J.; Brenner, R.E. Phenotypic instability of Saos-2 cells in long-term culture. Biochem. Biophys. Res. Commun. 2005, 333, 216–222. [Google Scholar] [CrossRef] [PubMed]
- Gillet, J.P.; Calcagno, A.M.; Varma, S.; Marino, M.; Green, L.J.; Vora, M.I.; Patel, C.; Orina, J.N.; Eliseeva, T.A.; Singal, V.; et al. Redefining the relevance of established cancer cell lines to the study of mechanisms of clinical anti-cancer drug resistance. Proc. Natl. Acad. Sci. USA 2011, 108, 18708–18713. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, T.; Kubota, T.; Watanabe, M.; Kitaijma, M.; Hoffman, R.M. Orthotopic transplantation of histologically intact clinical specimens of stomach cancer to nude mice: Correlation of metastatic sites in mouse and individual patient donors. Int. J. Cancer 1993, 53, 608–612. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, T.; Fu, X.; Kubota, T.; Watanabe, M.; Kitajima, M.; Hoffman, R.M. Nude mouse metastatic models of human stomach cancer constructed using orthotopic implantation of histologically intact tissue. Cancer Res. 1993, 53, 1204–1208. [Google Scholar] [PubMed]
- Zhang, L.; Yang, J.; Cai, J.; Song, X.; Deng, J.; Huang, H.; Chen, D.; Yang, M.; Wery, J.P.; Li, S.; et al. A subset of gastric cancers with EGFR amplification and overexpression respond to cetuximab therapy. Sci. Rep. 2013, 3, 2992. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Tian, T.; Li, Z.; Tang, Z.; Wang, L.; Wu, J.; Li, Y.; Dong, B.; Li, Y.; Dong, B.; et al. Establishment and characterization of patient-derived tumor xenograft using gastroscopic biopsies in gastric cancer. Sci. Rep. 2015, 5, 8542. [Google Scholar] [CrossRef] [PubMed]
- Lau, W.M.; Teng, E.; Chong, H.S.; Lopez, K.A.; Tay, A.Y.; Salto-Tellez, M.; Shabbir, A.; So, J.B.; Shan, S.L. CD44v8-10 is a cancer-specific marker for gastric cancer stem cells. Cancer Res. 2014, 74, 2630–2641. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Korn, J.M.; Ferretti, S.; Monahan, J.E.; Wang, Y.; Singh, M.; Zhang, C.; Schnell, C.; Yang, G.; Zhang, Y.; et al. High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat. Med. 2015, 21, 1318–1325. [Google Scholar] [CrossRef] [PubMed]
- Park, H.; Cho, S.Y.; Kim, H.; Na, D.; Han, J.Y.; Chae, J.; Park, C.; Park, O.K.; Min, S.; Kang, J.; et al. Genomic alterations in BCL2L1 and DLC1 contribute to drug sensitivity in gastric cancer. Proc. Natl. Acad. Sci. USA 2015, 112, 12492–12497. [Google Scholar] [CrossRef] [PubMed]
- Dedhia, P.H.; Bertaux-Skeirik, N.; Zavros, Y.; Spence, J.R. Organoid models of human gastrointestinal development and disease. Gastroenterology 2016, 150, 1098–1112. [Google Scholar] [CrossRef] [PubMed]
- Hill, D.R.; Spence, J.R. Gastrointestinal organoids: Understanding the molecular basis of the host-microbe interface. Cell Mol. Gastroenterol Hepatol. 2017, 3, 138–149. [Google Scholar] [CrossRef] [PubMed]
- van de Wetering, M.; Francies, H.E.; Francis, J.M.; Bounova, G.; Iorio, F.; Pronk, A.; van Houdt, W.; van Gorp, J.; Taylor-Weiner, A.; Kester, L.; et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 2015, 161, 933–945. [Google Scholar] [CrossRef] [PubMed]
- McCracken, K.W.; Catá, E.M.; Crawford, C.M.; Sinagoga, K.L.; Schumacher, M.; Rocjich, B.E.; Tsai, Y.H.; Mayhew, C.N.; Spence, J.R.; Zavros, Y.; et al. Modelling human development and disease in pluripotent stemcell-derived gastric organoids. Nature 2014, 516, 400–404. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Nadauld, L.; Ootani, A.; Corney, D.C.; Pai, R.K.; Gevaert, O.; Cantrell, M.A.; Rack, P.G.; Neal, J.T.; Chan, C.W.; et al. Oncogenic transformation of diverse gastrointestinal tissues in primary organoid culture. Nat. Med. 2014, 20, 769–777. [Google Scholar] [CrossRef] [PubMed]
- Nadauld, L.D.; Garcia, S.; Natsoulis, G.; Bell, J.M.; Miotke, L.; Hopmans, E.S.; Xu, H.; Pai, R.K.; Palm, C.; Regan, J.F.; et al. Metastatic tumor evolution and organoid modeling implicate TGFBR2 as a cancer driver in diffuse gastric cancer. Genome Biol. 2014, 15, 428. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Yuen, S.T.; Xu, J.; Lee, S.P.; Yan, H.H.; Shi, S.T.; Siu, H.C.; Deng, S.; Chu, K.M.; Law, S.; et al. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat. Genet. 2014, 46, 573–582. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Radmark, O.; Kim, S. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef] [PubMed]
- Ruan, K.; Fang, X.; Ouyang, G. MicroRNAs: Novel regulators in the hallmarks of human cancer. Cancer Lett. 2009, 285, 116–126. [Google Scholar] [CrossRef] [PubMed]
- Kim, R.H.; Mak, T.W. Tumours and tremors: How PTEN regulation underlies both. Br. J. Cancer 2006, 94, 620–624. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.-Z.; Han, L.; Zhang, A.-L.; Fu, Y.-C.; Yue, X.; Wang, G.-X.; Jia, Z.-F.; Pu, P.-Y.; Zhang, Q.-Y.; Kang, C.-S. MicroRNA221 and microRNA-222 regulate gastric carcinoma cell proliferation and radioresistance by targeting PTEN. BMC Cancer 2010, 10, 367. [Google Scholar] [CrossRef]
- Zhang, B.G.; Li, J.F.; Yu, B.Q.; Zhu, Z.G.; Liu, B.Y.; Yan, M. microRNA-21 promotes tumor proliferation and invasion in gastric cancer by targeting PTEN. Oncol. Rep. 2012, 27, 1019–1026. [Google Scholar] [CrossRef] [PubMed]
- Tsukamoto, Y.; Nakada, C.; Noguchi, T.; Tanigawa, M.; Nguyen, L.T.; Uchida, T.; Hijina, T.; Matsuura, K.; Fujioka, T.; Seto, M.; et al. MicroRNA-375 is downregulated in gastric carcinomas and regulates cell survival by targeting PDK1 and 14-3-3zeta. Cancer Res. 2010, 70, 2339–2349. [Google Scholar] [CrossRef] [PubMed]
- Takagi, T.; Iio, A.; Nakagawa, Y.; Naoe, T.; Tanigawa, N.; Akao, Y. Decreased expression of microRNA-143 and -145 in human gastric cancers. Oncology 2009, 77, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, Y.; Akiyama, Y.; Otsubo, T.; Shimada, S.; Yuasa, Y. Involvement of epigenetically silenced microRNA-181c in gastric carcinogenesis. Carcinogenesis 2010, 31, 777–784. [Google Scholar] [CrossRef] [PubMed]
- Lang, N.; Liu, M.; Tang, Q.L.; Chen, X.; Liu, Z.; Bi, F. Effects of microRNA-29 family members on proliferation and invasion of gastric cancer cell lines. Chin. J. Cancer 2010, 29, 603–610. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.T., Jr.; McCubrey, J.A. The Raf/MEK/ERK signal transduction cascade as a target for chemotherapeutic intervention in leukemia. Leukemia 2002, 16, 486–507. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Xie, Y.; Zhang, H.; Wu, Y. miR-107 targets cyclin-dependent kinase 6 expression, induces cell cycle G1 arrest and inhibits invasion in gastric cancer cells. Med. Oncol. 2012, 29, 856–863. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liu, X.; Jin, H.; Guo, X.; Xia, L.; Chen, Z.; Bai, M.; Liu, J.; Shang, X.; Wu, K.; et al. MiR-206 inhibits gastric cancer proliferation in part by repressing CyclinD2. Cancer Lett. 2013, 332, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Xu, Y.; Zhang, W.; Deng, Y.; Si, M.; Du, Y.; Yao, H.; Liu, X.; Ke, Y.; Si, J.; et al. MiR-375 frequently downregulated in gastric cancer inhibits cell proliferation by targeting JAK2. Cell Res. 2010, 20, 784–793. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Guo, L.; Ji, J.; Zhang, J.; Chen, X.; Cai, Q.; Li, J.; Gu, Q.; Liu, B.; Zhu, Z.; et al. miRNA-331-3p directly targets E2F1 and induces growth arrest in human gastric cancer. Biochem. Biophys. Res. Commun. 2010, 398, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Xiao, B.; Guo, J.; Miao, Y.; Jiang, Z.; Huan, R.; Zhang, Y.; Li, D.; Zhong, J. Detection of miR-106a in gastric carcinoma and its clinical significance. Clin. Chim. Acta 2009, 400, 97–102. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.K.; Yu, J.; Han, T.S.; Park, S.Y.; Namkoong, B.; Kim, D.H.; Hur, K.; Yoo, M.W.; Lee, H.J.; Yang, H.K.; et al. Functional links between clustered microRNAs: Suppression of cell-cycle inhibitors by microRNA clusters in gastric cancer. Nucleic Acids Res. 2009, 37, 1672–1681. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.L.; Peng, Z.; Yang, X.; Fan, K.J.; Ye, H.; Li, Z.H.; Wang, Y.; Xu, X.L.; Li, J.; Wang, Y.L.; et al. miR-148a promoted cell proliferation by targeting p27 in gastric cancer cells. Int. J. Biol. Sci. 2011, 7, 567–574. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Liu, X.H.; Li, J.H.; Yang, J.S.; Zhang, E.B.; Yin, D.D.; Liu, Z.L.; Zhou, J.; Ding, Y.; Li, S.Q.; et al. MiR-196a is upregulated in gastric cancer and promotes cell proliferation by downregulating p27(kip1). Mol. Cancer Ther. 2012, 11, 842–852. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.H.; Wu, C.W.; Li, A.F.; Chi, C.W.; Lin, W.C. miR-21 microRNA expression in human gastric carcinomas and its clinical association. Anticancer Res. 2008, 28, 907–911. [Google Scholar] [PubMed]
- Motoyama, K.; Inoue, H.; Mimori, K.; Tanaka, F.; Kojima, K.; Uetake, H.; Sugihara, K.; Mori, M. Clinicopathological and prognostic significance of PDCD4 and microRNA-21 in human gastric cancer. Int. J. Oncol. 2010, 36, 1089–1095. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Zhao, Y.; Guo, G.; Li, W.; Zhu, E.D.; Luo, X.; Mao, X.H.; Zou, Q.M.; Yu, P.W.; Zuo, Q.F.; et al. Plasma microRNAs, miR-223, miR-21 and miR-218, as novel potential biomarkers for gastric cancer detection. PLoS ONE 2012, 7, e41629. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Guo, J.; Li, D.; Xiao, B.; Miao, Y.; Jiang, Z.; Zhuo, H. Down-regulation of miR-31 expression in gastric cancer tissues and its clinical significance. Med. Oncol. 2010, 27, 685–689. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Luo, F.; Li, Q.; Xu, M.; Feng, D.; Zhang, G.; Wu, W. Identification of new aberrantly expressed miRNAs in intestinal-type gastric cancer and its clinical significance. Oncol. Rep. 2011, 26, 1431–1439. [Google Scholar] [CrossRef] [PubMed]
- Katada, T.; Ishiguro, H.; Kuwabara, Y.; Kimura, M.; Mitui, A.; Mori, Y.; Ogawa, R.; Harata, K.; Fujii, Y. microRNA expression profile in undifferentiated gastric cancer. Int. J. Oncol. 2009, 34, 537–542. [Google Scholar] [CrossRef] [PubMed]
- Bandres, E.; Bitarte, N.; Arias, F.; Agorreta, J.; Fortes, P.; Agirre, X.; Zarate, R.; Diaz-Gonzalez, J.A.; Ramirez, N.; Sola, J.J.; et al. microRNA-451 regulates macrophage migration inhibitory factor production and proliferation of gastrointestinal cancer cells. Clin. Cancer Res. 2009, 15, 2281–2290. [Google Scholar] [CrossRef] [PubMed]
- Gong, J.; Li, J.; Wang, Y.; Liu, C.; Jia, H.; Jiang, C.; Wang, Y.; Luo, M.; Zhao, H.; Dong, L.; et al. Characterization of microRNA-29 family expression and investigation of their mechanistic roles in gastric cancer. Carcinogenesis 2014, 35, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.S.; Yang, X.H.; Chen, X.; Wang, X.D.; Hua, J.; Zhou, D.L.; Zhou, B.; Song, Z.S. MicroRNA-106b in cancer-associated fibroblasts from gastric cancer promotes cell migration and invasion by targeting PTEN. FEBS Lett. 2014, 588, 2162–2169. [Google Scholar] [CrossRef] [PubMed]
- Nishida, N.; Mimori, K.; Fabbri, M.; Yokobori, T.; Sudo, T.; Tanaka, F.; Shibata, K.; Ishii, H.; Doki, Y.; Mori, M. MicroRNA-125a-5p is an independent prognostic factor in gastric cancer and inhibits the proliferation of human gastric cancer cells in combination with trastuzumab. Clin. Cancer Res. 2011, 17, 2725–2733. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Zhang, C.; Huang, B.; Li, H.; Zhang, R.; Huang, Y.; Wang, J. Downregulation of microRNA-206 is a potent prognostic marker for patients with gastric cancer. Eur. J. Gastroenterol Hepatol. 2013, 25, 953–957. [Google Scholar] [CrossRef] [PubMed]
- Volinia, S.; Calin, G.A.; Liu, C.G.; Ambs, S.; Cimmino, A.; Petrocca, F.; Visone, R.; Iorio, M.; Roldo, C.; Ferracin, M.; et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA 2006, 103, 2257–2261. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, Y.; Zhang, Y.; Ding, J.; Wu, K.; Fan, D. Survival prediction of gastric cancer by a seven-microRNA signature. Gut 2010, 59, 579–585. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, Y.; Zhang, H.; Liu, X.; Gong, T.; Li, M.; Sun, L.; Ji, G.; Shi, Y.; Han, Z.; et al. miRNA-223 promotes gastric cancer invasion and metastasis by targeting tumor suppressor EPB41L3. Mol. Cancer Res. 2011, 9, 824–833. [Google Scholar] [CrossRef] [PubMed]
- Ueda, T.; Volinia, S.; Okumura, H.; Shimizu, M.; Taccioli, C.; Rossi, S.; Alder, H.; Liu, C.G.; Oue, N.; Yasui, W.; et al. Relation between microRNA expression and progression and prognosis of gastric cancer: A microRNA expression analysis. Lancet Oncol. 2010, 11, 136–146. [Google Scholar] [CrossRef]
- Kogo, R.; Mimori, K.; Tanaka, F.; Komune, S.; Mori, M. Clinical significance of miR-146a in gastric cancer cases. Clin. Cancer Res. 2011, 17, 4277–4284. [Google Scholar] [CrossRef] [PubMed]
- Bou Kheir, T.; Futoma-Kazmierczak, E.; Jacobsen, A.; Krogh, A.; Bardram, L.; Hother, C.; Gronbaek, K.; Federspiel, B.; Lund, A.H.; Friis-Hansen, L. miR-449 inhibits cell proliferation and is down-regulated in gastric cancer. Mol. Cancer 2011, 10, 29. [Google Scholar] [CrossRef] [PubMed]
- Tsai, M.M.; Wang, C.S.; Tsai, C.Y.; Huang, H.W.; Chi, H.C.; Lin, Y.H.; Lu, P.H. Potential diagnostic, prognostic and therapeutic targets of microRNAs in human gastric cancer. Int. J. Mol. Sci. 2016, 17. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Guo, F.; Wang, Y.; Lv, Y.; Huo, B.; Wang, L.; Liu, W. MicroRna-200c regulates the sensitivity of chemotherapy of gastric cancer SGC7901/DDP cells by directly targeting RhoE. Pathol. Oncol. Res. 2014, 20, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.H.; Swiderski, P.M.; Kaplan, B.E.; Takao, M.; Yasui, A.; Shen, B.; Pfeifer, G.P. Processing of UV damage in vistro by FEN-1 proteins aas part of an alternative DNA excision repair pathway. Biochemistry 1999, 38, 4809–4817. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.B.; Li, Q.L.; Hu, J.F.; Zhang, Q.; Xie, J.P.; Deng, L. miR-124 inhibits growth and invasion of gastric cancer by targeting ROCK1. Asian Pac. J. Cncer Prev. 2014, 15, 6543–6546. [Google Scholar] [CrossRef]
- Xu, Y.; Huang, Z.; Liu, Y. Reduced miR-125a-5p expression is associated with gastric carcinogenesis through the targeting of E2F3. Mol. Med. Rep. 2014, 10, 2601–2608. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Pu, J.; Qi, T.; Qi, M.; Li, D.; Xiang, X.; Huang, K.; Tong, Q. miRNA-145 targets v-ets erythroblastosis virus E26 oncogene homolog 1 to suppress the invasion, metastasis, and angiogenesi of gastric cancer cell. Mol. Cancer Res. 2013, 11, 182–193. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).