Abstract
In the natural environment, plants are often bombarded by a combination of abiotic (such as drought, salt, heat or cold) and biotic (necrotrophic and biotrophic pathogens) stresses simultaneously. It is critical to understand how the various response pathways to these stresses interact with one another within the plants, and where the points of crosstalk occur which switch the responses from one pathway to another. Calcium sensors are often regarded as the first line of response to external stimuli to trigger downstream signaling. Abscisic acid (ABA) is a major phytohormone regulating stress responses, and it interacts with the jasmonic acid (JA) and salicylic acid (SA) signaling pathways to channel resources into mitigating the effects of abiotic stresses versus defending against pathogens. The signal transduction in these pathways are often carried out via GTP-binding proteins (G-proteins) which comprise of a large group of proteins that are varied in structures and functions. Deciphering the combined actions of these different signaling pathways in plants would greatly enhance the ability of breeders to develop food crops that can thrive in deteriorating environmental conditions under climate change, and that can maintain or even increase crop yield.
Keywords:
biotic stress; abiotic stress; ABA; JA; SA; calcium sensors; ethylene; G-proteins; crosstalk; plant hormones 1. Introduction
The individual effects of isolated abiotic and biotic stresses on plants and their physiological responses to each of them have been studied extensively under experimental conditions, along with the corresponding regulatory mechanisms at the genetic and molecular levels [1,2,3,4]. However, living in the natural environment, plants are constantly challenged with a combination of abiotic and biotic stresses [5,6]. The plant responses toward the combined impact of these stresses have been largely unknown. Upon the introduction of various external stimuli, different patterns of cytosolic calcium level fluctuations are induced within seconds [7]. Therefore, calcium (Ca2+) signaling has been recognized as the front line of signaling events. Intracellular Ca2+ level is closely influenced by abscisic acid (ABA) signaling, which is an important plant hormone with multiple roles in abiotic and biotic stress responses [8]. Besides ABA, GTP-binding proteins (G-proteins) are also important components for signal transduction [9]. In this review, we will discuss the crosstalks among the signaling pathways in response to abiotic and biotic stresses. The roles of stress-responsive signaling components, including Ca2+ sensors, ABA-mediated signaling events, and stress-responsive G-proteins, will be discussed to spotlight the convergent points between the regulatory pathways of abiotic stress and biotic stress responses.
2. Ca2+ Sensors
The fluctuations in cytosolic calcium level upon external stimuli are known as calcium waves, which are perceived by calcium sensors. Calcium sensors are proteins that bind to Ca2+ and change their conformations and functions upon binding. Therefore, calcium signaling is considered to be the translator of different stimuli, and the first step in initiating downstream responses. Several calcium sensors have been shown to play roles in both abiotic and biotic stresses. Therefore, calcium sensors have also been regarded as hubs in plant responses to concurrent abiotic and biotic stresses [10].
Calcium sensors are categorized into calmodulins (CaMs) or calmodulin-like proteins (CMLs), calcineurin B-like proteins (CBLs), calcium-dependent protein kinases (CPKs), and calcium/calmodulin-dependent protein kinases (CCaMKs) [11,12].
2.1. CaM/CML
The CaM protein consists of two pairs of elongation factor (EF)-hands, each made up of a helix-loop-helix motif forming a Ca2+-binding pocket. Within each pair, the two EF-hands are connected by an α-helix. Since each EF-hand pair forms a globular structure, the whole CaM protein consists of two globular domains. Upon the binding of Ca2+, the conformation of CaM changes to expose hydrophobic surfaces for binding to its targets [13]. CMLs are structurally similar to CaMs, in that they are also made up of EF-hands, and they have the ability to bind to Ca2+. However, CMLs have low sequence similarities with CaMs. The targets of CaMs/CMLs include promoters, enzymes such as kinases and phosphatases, transcription factors, and ion channels [13,14]. The sequence divergence between CaMs and CMLs reflects their divergent properties of Ca2+ binding and their specific targets. Several CaMs were found to be involved in abiotic stress, while some were found to be involved in biotic stress (Table 1).

Table 1.
Stress-responsive Ca2+ sensors in plants.
In tomato (Solanum lycopersicum), the expressions of SlCaM1, SlCaM2, SlCaM3, SlCaM4, SlCaM5, and SlCaM6 were reported to be induced by mechanical wounding and Botrytis cinerea infection [15]. Transgenic tomato overexpressing SlCaM2 had enhanced resistance to Botrytis cinerea infection, while transgenic tomato overexpressing the antisense SlCaM2 and thus having reduced expression of SlCaM2 was less resistant to the infection [15]. The expression levels of SlCaM1, SlCaM2, SlCaM3, SlCaM4, SlCaM5, and SlCaM6 could also be induced by salicylic acid (SA) and jasmonic acid (JA) [15]. In Arabidopsis thaliana, the mutation of AtCaM3 led to a reduced tolerance to heat stress [16]. In soybean, it was found that the expression levels of SCaM-4 and SCaM-5 in soybean cell suspensions were induced by Fuasrium solani infection [17]. The constitutive expression of SCaM-4 or SCaM-5 in tobacco led to enhanced resistance to infection by Phytophthora parasitica var. nicotianae [17].
For CMLs, in tomato, the expression of ShCML44 could be induced by cold, drought, mannitol, (osmotic stress), salt, ABA, methyl-jasmonate (MeJA), and ethephon (a plant growth regulator) [18]. In Arabidopsis thaliana, several CMLs were found to be involved in both abiotic and biotic stresses. The mutation of AtCML9 led to increased sensitivity to ABA, and enhanced tolerance to salt stress and water deficit [19]. On the other hand, the mutation of AtCML9 led to reduced resistance to Pseudomonas syringae pv. tomato (Pto) strain DC3000 infection [20]. The mutation of AtCML37 resulted in increased susceptibility to drought stress [21] and herbivory by down-regulating the JA pathway [22]. On the other hand, the mutation of AtCML42 led to enhanced resistance to herbivory but reduced tolerance of UV-B [23]. It was suggested that AtCML42 downregulates JA sensitivity [23]. Similarly, AtCML37 and AtCML42 were also shown to be regulators of the JA response pathway [21,22,23].
2.2. CBL
All CBL proteins have a conserved four-EF-hand calcium-binding domain, and they specifically interact with a family of protein kinases, namely, CBL-interacting protein kinases (CIPKs) [24]. In rapeseed (Brassica napus), the expression of BnCBL1 was induced by both salt and osmotic stress [25]. The overexpression of BnCBL1 in Arabidopsis conferred improved tolerance of low inorganic phosphate stress and salt stress [25]. In Arabidopsis thaliana, the overexpression of AtCBL5 resulted in improved tolerance of salt and drought stress [26]. The mutation of CBL10 in Arabidopsis thaliana led to a reduced tolerance of salt stress [27]. On the other hand, in tomato, the silencing of CBL10 led to improved resistance to Pto DC3000 infection [28].
2.3. CPK
The typical CPK protein consists of four functional domains including an N-terminal variable domain (N-VD), a protein kinase (PK) domain, an autoinhibitory junction (AJ), and a calcium-binding domain (CBD) made up of one to five EF-hands [29]. Upon binding to Ca2+, CPKs can then phosphorylate their specific targets [30]. The targets of CPKs include ion channels, ABA-responsive element binding factors (ABFs), and calcium ATPases (ACAs) [31]. In Arabidopsis thaliana, the mutation of CPK10 led to an enhanced tolerance of drought [32]. The expression of AtCPK3 in Arabidopsis thaliana protoplasts was induced by cold, salt, heat, hydrogen peroxide (H2O2), flagellin, and laminarin [33]. The mutation of AtCPK3 led to reduced tolerance of sodium chloride (NaCl) [33]. The overexpression of AtCPK6 led to improved tolerance of salt and drought stress [34]. It was also shown that AtCPK3 functioned together with AtCPK6 to regulate stomatal aperture when stimulated by ABA [35]. In maize, the overexpression of ZmCPK4 conferred tolerance to drought stress by increasing the sensitivity to ABA [36]. In rice, the expression of OsCPK9 was induced by ABA, NaCl, and polyethylene glycol (PEG) (osmotic stress). The overexpression of OsCPK9 conferred tolerance to drought stress [37]. OsCPK12 is involved in both abiotic and biotic stress. The overexpression of OsCPK12 promoted tolerance of salt stress, increased sensitivity to ABA and increased susceptibility to blast fungus (Magnaporthe grisea) by reducing the accumulation of reactive oxygen species (ROS) [38].
2.4. CCaMK
CCaMKs have a serine-threonine kinase domain, an autoinhibitory domain, a CaM-binding domain, and a neutral visinin-like Ca2+-binding domain consisting of three EF-hands. The binding of Ca2+ leads to the autophosphorylation of the protein and enhances its affinity to CaM, resulting in the activation of kinase activity [39]. Several transcription factors, such as CYCLOPS and ZmNAC84, were reported to be the targets of CCaMKs [40,41,42]. CYCLOPS, which is a transcriptional activator regulating symbiotic nodule development, was reported to be a phosphorylation target of CCaMK [42]. On the other hand, ZmCCaMK phosphorylates ZmNAC84 and mediates antioxidant defense [41]. In soybean, the expression of GsCBRLK was induced by cold, ABA, NaCl, and PEG [29]. The overexpression of GsCBRLK in Arabidopsis thaliana led to improved tolerance of NaCl stress and reduced sensitivity to ABA [29]. In tomato, the expression of SlCCaMK in the leaf was induced by Sclerotinia sclerotiorum infection but repressed by Pto DC3000/Xanthomonas oryzae pv. oryzae (Xoo) infection [29]. Knocking down SlCCaMK reduced resistance to S. scelrotiorum and Pto DC3000 in tomato [29]. In wheat, it was found that the expression of TaCCaMK was reduced by ABA, NaCl, and PEG [43]. The overexpression of TaCCaMK in Arabidopsis led to decreased sensitivity to ABA and improved tolerance of NaCl during seed germination, but increased sensitivity to NaCl, with heavier chlorosis in seedlings [43].
2.5. Calcium Sensors Involved in both Abiotic and Biotic Stresses
A lot of calcium sensors were found to be involved in stress responses in plants. Among the characterized calcium sensors, some of them were found to be involved in both abiotic and biotic stresses. Examples are AtCML37, AtCML42, AtCML9, and OsCPK12.
AtCML37 and AtCML42 are regulators of JA signaling. JA has been known for its dual role in both abiotic and biotic stress signaling, and the application of JA to plants protects them against both types of stresses [44]. JA has been known to play a protective role against cold and freezing, salinity, drought, and heat [45]. Upon infection by necrotrophic pathogens, attack by insects, and wounding events, JA activates plant defense responses by activating protective genes [46]. Therefore, calcium sensors that regulate the JA pathway are probable hubs of abiotic and biotic stress responses. In addition, among the stress-responsive calcium sensors, several are related to ABA sensitivity (Table 1). Like AtCML37 and AtCML42, AtCML9 is also involved in both abiotic and biotic stresses. It was reported that AtCML9 regulates ABA sensitivity [19]. Besides, OsCPK12 was also reported to play a dual role in abiotic and biotic stresses [38]. It is a positive regulator of salt stress by reducing ROS accumulation, but a negative regulator of defense responses against blast fungus as a result of its ROS-reducing activity which prevents the activation of defense-related genes. It was also proposed that the dual role of OsCPK12 was due to its effect on ABA signaling, which is a major event in both abiotic and biotic stresses. ABA and JA pathways are closely interconnected. The relation between ABA and JA signaling will be discussed in more detail in Section 3. The links among the calcium sensors, ABA and JA, strongly suggest the important roles played by calcium sensors in abiotic and biotic stress responses. Table 1 shows a list of stress-responsive calcium sensors found in plants.
3. ABA-Mediated Stress Responses
ABA is an important phytohormone that plays multiple roles in abiotic and biotic stresses. ABA closely interacts with several other stress-response hormones including JA, ethylene, and SA. In this section, the interactions between ABA and other stress hormones will be discussed.
3.1. The Interactions between ABA and JA Pathways in Response to Biotic and Abiotic Stresses
Upon contact with Pseudomonas syringae on the leaf, flg22, a 22-amino acid epitope on the flagellum of the bacterial cell and a typical pathogen-associated molecular pattern (PAMP), is recognized by the receptor, FLS2, on the leaf epidermal cells. The receptors then trigger SA- & ABA-mediated responses to close the stomata to prevent the pathogen from gaining entry into the plant [47]. A virulent strain of Pseudomonas syringae then injects the phytotoxin, coronatine (COR), which mimics the action of jasmonic acid–isoleucine (JA–ILeu), which is the active form of JA, into the host plant, in order to stimulate the re-opening of the stomata. COR, thus termed an effector, functions by stimulating the biosynthesis of JA via Jai1, which in turn induces JA–ILeu-inducible wound-responsive genes via the transcription factor, MYC2, while repressing PATHOGENESIS-RELATED (PR) genes [48,49,50]. Thus, the wounding response becomes activated at the expense of defense response.
In the absence of the virulence factor COR or actual wounding, induction of wounding-responsive genes is inhibited by the repression of MYC2 by jasmonate ZIM-domain (JAZ) proteins [51]. However, in the presence of COR or JA–ILeu, the F-Box protein, COI1, a component of the Skp1–Cul1–F-box protein (SCF) of the ubiquitin E3 ligase complex, recruits the JAZ protein to tag it for 26S proteasome-mediated degradation [52,53]. Thus the repression on JA-inducible wounding-responsive genes by JAZ protein is lifted by JA–ILeu or COR [54].
A small GTPase protein, NOG1-2, stimulates stomatal closure during abiotic stresses, and the nog1-2 mutant of Nicotiana benthamiana is more susceptible to the pathogen Pseudomonas syringae, due to its inability to close its stomata [55], proving its role in the re-closing of stomata opened by Pseudomonas syringae. NOG1-2-mediated stomatal closure is stimulated in the presence of COR, indicating that its function is dependent on JA–Ileu [48]. In Arabidopsis, JAZ9 is the protein that interacts with MYC2 and COI1 through its JA-associated (Jas) domain [56]. JAZ 9 also interacts with NOG1-2 through this same domain, and by binding to JAZ9, NOG1-2 interrupts the interaction between JAZ9 and COI1 and prevents JAZ9 from being targeted for degradation. Thus, JAZ9 can perform its role in inhibiting wounding response (i.e., stomatal re-opening) upon bacterial infection as part of the pathway in effector-triggered immunity (ETI) [48].
The commonly downregulated genes in nog1-2 and jaz mutants are often induced in response to ABA or drought, indicating the cooperative nature of these two proteins in inducing ABA-responsive genes [48]. Also, the nog1-2 mutant of Nicotiana benthamiana has reduced accumulation of ROS (necessary for ABA-mediated stomatal closure) in guard cells, even after external application of ABA, indicating the role of NOG1-2 in ABA-mediated stomatal closure [48]. Thus NOG1-2 acts as a point of crosstalk between the ABA and JA signaling pathways, since NOG1-2 is responsible for inducing stomatal closure as part of the ABA-dependent pathway during abiotic stress, while it causes the closing of stomata through the JA-Ileu-mediated pathway during pathogen infection [48]. Though the effect of both ABA and JA pathways is the same in terms of resulting in stomatal closure, the effects of these two hormones are opposite in the case of evoking wounding response.
The PDF1.2 defensin gene is induced upon fungal infection or MeJA challenge, but not by wounding or salicylic acid (SA) challenge, indicating that its induction is triggered only by pathogen attack but not by wounding [57]. PDF1.2 requires both JA and ethylene (ET) for its activation [57], or at least the overexpression of a downstream component of the ethylene signaling pathway, such as ERF-1 [58]. Constitutive expression of PDF1.2 occurs when ERF-1 is overexpressed both in the ethylene insensitive (ein2) and JA insensitive (coi1) mutants [58]. At the same time, its expression is upregulated by NOG1-2 in the presence of ABA [48]. This further shows the important position of NOG1-2 as a hub between the JA/ET and ABA signaling pathways. A summary of the interactions between the ABA and JA pathways is presented in Figure 1.

Figure 1.
The interactions between the abscisic acid (ABA) and jasmonic acid (JA) signaling pathways under biotic and abiotic stresses are mediated through NOG1-2.
3.2. The Interactions between ABA and Ethylene Pathways under Biotic and Abiotic Stresses
The plant ethylene receptor family consists of five members that function in conjunction with its negative regulator, CONSTITUTIVE TRIPLE RESPONSE1 (CTR1) [59,60]. CTR1 is a Raf-like protein that is associated with ethylene receptors in the endoplasmic membrane of Arabidopsis [61], and it is insensitive to ABA [60]. In the absence of ethylene, CTR1 phosphorylates and thus deactivates ETHYLENE-INSENSITIVE PROTEIN 2 (EIN2), and thus all the downstream signaling in the ethylene pathway [62]. The presence of ethylene results in the increase in the level of EIN2 by the deactivation of ETHYLENE INSENSITIVE3-BINDING F-BOX PROTEIN 1 and 2 (EBF1 and EBF2), which are responsible for the 26S proteosomal degradation of EIN2 [62,63]. Also, ethylene causes the deactivation of CTR1 [59], and therefore prevents the phosphorylation of EIN2. The C-terminal end of EIN2 is then cleaved and localized to the nucleus to activate the next component along the ethylene pathway, EIN3 [62]. The immediate target of EIN3 and the final component of the ethylene-initiated pathway in Arabidopsis is ETHYLENE RESPONSE FACTOR1 (ERF1), the constitutive expression of which elicits ethylene responses at both the transcriptional and the phenotypic levels [64]. ERF1 belongs to the Ethylene Response Factor (ERF) family of transcription factors, containing a single DNA-binding domain. Members of the ERF subfamily of the ERF family recognize the AGCCGCC motif (the GCC box) in the promoters of ethylene-inducible defense genes [65,66,67,68,69], while members of the DREB subfamily binds to the A/GCCGAC motif in the promoters of DEHYDRATION RESPONSIVE ELEMENT (DRE) genes in response to drought stress [65,70]. Among the members of the ERF subfamily in Arabidopsis, ERF1, 2 and 5 are activators, while ERF3 and 4 are repressors of gene transcription [68].
The biotic stress-inducible Arabidopsis ERF proteins are also found to be associated with various abiotic stresses such as salt, drought, cold, heat, and light [71,72,73]. AtERF6 induces many ROS-inducible genes, and thus ensures that the plant is protected against both biotic and abiotic stresses [74]. The ERF homologues in other plants are also found to be induced by biotic stresses or ethylene, and they can bind to the GCC box or DRE sequence in response to biotic and/or abiotic stresses.
Overexpression of ERF related genes such as, OsEREBP1 in rice and Tsi1 in tobacco, and ectopic expression of ERF genes such as JERF3 from tomato and GmERF3 from soybean in tobacco, induce the expression of PATHOGENESIS-RELATED (PR) genes and enhance the resistance to various biotic and abiotic stresses [75,76,77,78]. Ectopic expression of GmERF3 (from soybean) and JERF1 (from tomato) in tobacco, and overexpression of SlERF5 (in tomato), TaERF3 (in wheat), and OsEREBP1 (in rice), conferred resistance to drought, possibly by binding both the DRE promoter sequence along with the GCC box, which results in the upregulation of ABA biosynthesis genes and the accumulation of proline [71,75,78,79,80,81]. The rice homologue (OsWR1) of the Arabidopsis wax/cutin-synthesizing gene, WIN1/SHN1, is also an ERF gene. Overexpression of OsWR1 improves drought tolerance by inducing wax biosynthesis [82]. In Arabidopsis, ERF1-overexpression increases resistance to drought stress by upregulating the drought response-specific genes, GEA6 and LEA4-5, accumulating ABA and proline, and closing the stomata, whereby ERF1 binds only to the DRE promoter sequence, whereas ERF1 binds the GCC box exclusively during biotic stress responses [71]. AtERF4, which is known to be a transcriptional repressor in response to ethylene, causes the downregulation of ABA-responsive genes when constitutively expressed [83].
ERF1-overexpression also causes upregulation of the heat-inducible genes, HSP101, HSP70, and HSP23.6, and the transcription factor, AtHsfA3, responsible for inducing heat-responsive genes, possibly by binding to the DRE promoter sequence [71].
Though the overexpression of many ERF transcription factors, such as those found in soybean, tomato, wheat, and tobacco, has been shown to confer salt tolerance [76,77,78,79,80,81,84], most of the research is focused on the salt tolerance mechanism in Arabidopsis. In Arabidopsis, the ethylene signaling pathway component, EIN3, activates ETHYLENE AND SALT INDUCIBLE1 (ESE1), one of the salt- and ethylene-inducible genes of the ERF family in Arabidopsis. ESE1 confers salt tolerance by positively modulating the salt tolerance genes, COR15A and RD29A [85]. In a separate study, overexpression of AtERF1 caused the induction of salt-responsive genes, P5CS1, SRO5, GLP9, and ATOSM34, during salt stress by binding to the DRE sequence of the promoter [71].
ERFs from various species, including Arabidopsis, tomato, and wheat [81,86,87] are also found to confer cold tolerance, possibly by upregulating the COR gene in an ABA-independent pathway [88].
Many ERFs that confer abiotic stress tolerance are induced not only by ethylene, but also by the other phytohormones involved in biotic stress response pathways, such as SA and JA, and hence these could be the facilitators of crosstalks between biotic and abiotic stress response pathways [76,77,78,84,89]. Characterizations of these ERFs are summarized in Table 2, and the roles of ERFs in the response pathways to various biotic and abiotic stresses are charted in Figure 2.

Table 2.
Stress-responsive Ethylene Response Factors (ERFs).
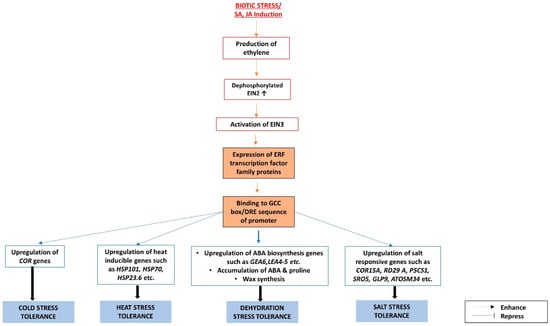
Figure 2.
The interactions between abscisic acid (ABA) and ethylene signaling pathways under biotic and abiotic stresses are mediated through Ethylene Response Factors (ERFs).
The Antagonistic Relationship between ABA and Ethylene Pathways
A unique ERF (ThERF1) from Tamarix hispida was found to negatively regulate drought tolerance by inhibiting superoxide dismutase (SOD) and peroxidase (POD) activities in transgenic Arabidopsis, which are both downstream components of the ABA-mediated stress response pathway [91]. It also negatively regulates salt tolerance by binding to the TTG motif of the promoter of salt-responsive genes with more affinity than to the GCC motif [94].
On the other hand, the ABA-INSENSITIVE4 (ABI4) protein binds to the promoters of the ethylene biosynthesis genes, ACS4 and ACS8, in Arabidopsis, and represses ethylene production in the presence of ABA [95]. One mutated allele of the ethylene signaling pathway component, ein2, caused higher ABA accumulation during germination, thus demonstrating the antagonistic relationship between ABA and ethylene [96].
3.3. The Interactions between ABA and SA under Biotic and Abiotic stresses
In response to drought stress, ABA promotes cuticular wax biosynthesis mediated by the transcription factor, MYB96, possibly by upregulating the enzyme, 3-ketoacyl-CoA synthase 2 (KCS2), involved in very-long-chain fatty acid biosynthesis in Arabidopsis [97]. MYB96 also increases drought tolerance by upregulating the RD22 gene, which decreases lateral root formation [98]. This phenomenon is accelerated in the super-active myb96-1d mutant, but it is reduced in the less-active myb96-1 mutant [98]. MYB96 also stimulates SA biosynthesis. SA-INDUCTION DEFICIENT 2 (SID2), the gene responsible for expressing isochorismate synthase, a key enzyme in the biosynthesis of SA and normally induced upon infection [99], is found to be upregulated in the super-active myb96-1d mutant. The myb96-1d mutant is more responsive to ABA-mediated signaling, and it also produces higher levels of SA and is more resistant to pathogens, compared to the wild type [100]. These mutant plants have impaired growth similar to those plants with higher levels of ABA. The dwarfism observed in these plants due to the increased ABA-mediated response can be negated by crossing with NahG plants [100], in which NahG encodes SA hydroxylase, which degrades SA [101]. This shows that the dwarfism resulting from response to ABA is actually mediated by SA. On the other hand, the stomatal closure in abiotic and biotic stresses is mediated by ABA and SA, respectively [102,103,104,105].
The ABA3 protein promotes ABA biosynthesis by activating aldehyde oxidase through sulfuration. The enzyme then catalyzes the final step of ABA biosynthesis by converting abscisic aldehyde to ABA [106]. ABA3 is also shown to induce SID2, as indicated by the absence of SID2 induction upon abiotic stress in the aba3 mutant [100].
SUMO (small ubiquitin-like modifier) E3 Ligase 1 (SIZ1) negatively regulates the ABA-responsive gene, ABI3, since the siz1 mutant accumulates a higher level of the ABI3 protein [107]. At the same time, it also negatively regulates SA levels, since the siz1 mutant has a higher accumulation of SA, higher expression levels of PR genes, and it exhibits greater resistance to bacterial infections and drought, and these effects can be reversed by introducing the NahG gene [108,109]. The interactions between the ABA and SA pathways are summarized in Figure 3.
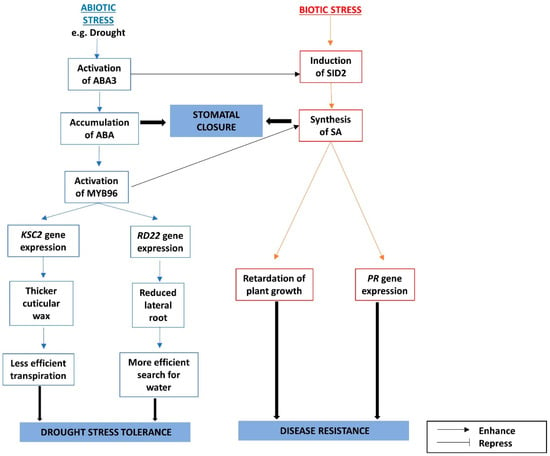
Figure 3.
The interactions between abscisic acid (ABA) and salicylic acid (SA) signaling pathways under biotic and abiotic stresses are mediated through MYB96.
The Crosstalks among ABA, SA, and Phospholipids under Biotic and Abiotic Stresses
Phospholipid pathways are divided into those that activate phospholipase C (PLC), and those that activate phospholipase D (PLD). Here we will focus on the PLD pathway, which is known to be involved in both abiotic and biotic stresses. There are different genes in the PLD gene family, such as PLDα, β, γ, δ, ε, and ζ, classified into subfamilies [110]. PLDs mediate phosphatidic acid (PA) production, which is a typical response to viral infection, e.g., in tobacco, likely via PLDα and β, since their presence is increased in the leaf upon infection [110,111,112]. PA facilitates viral replication, possibly by aiding in the formation of an active replicase complex [112]. PLDδ was found to provide a cell wall-based defense in Arabidopsis, together with PA, in the case of fungal [112,113] or bacterial [114] infection. PLDs are also reported to participate in abiotic stress responses. For example, PLD α1, α3, and δ cause stomatal closure by inducing the synthesis of PA and the activation of Mitogen-activated Protein Kinase 6 (MPK6), thus increasing tolerance to salt stress [115,116]. Salt increases the PA content in Arabidopsis, which was found to bind to MPK6 and increase its activity. The activation of MPK6 by PA is lost in the pldα1 mutant. MPK6 in turn activates a Na+/H+ antiporter via phosphorylation to confer salt tolerance [117]. PLDδ activity and the concentration of its catalytic product, phosphatidyl alcohols, increased upon dehydration stress; this was absent in Arabidopsis pldδ mutants [118]. PLDδ knockout mutants were found to be more sensitive to freezing stress, having less PA content, while overexpression increased the PA content, as well as conferring resistance [119].
Disassembly of microtubules is responsible for stomatal closure, which is inhibited in mutants that overexpress microtubules [120]. ABA, the phytohormone in the first line of response to drought and salt stress, activates PLD, which hydrolyzes membrane lipid to phosphatidyl choline (PC) [121], which is then hydrolyzed by PLDα1 to PA and choline [122]. PLDα1 and PA together depolymerize microtubules. This process is evidenced by the inability of ABA treatment to induce stomatal closure in pldα1 mutants, but co-treatment with ABA and microtubule-disassembling drugs (orzyline and propyzamide) results in stomatal closure in these mutants [123]. PLDα1-derived PA decreases the phosphatase activity of ABI1, one of the negative regulators of the ABA signaling pathway [124].
The intracellular increase in Ca2+ mediated by ABA [125] helps to activate PLDs by facilitating the binding of Ca2+ to the catalytic domain of PLDs [126]. The downstream production of ROS by activated PLDs further increases the intracellular Ca2+ concentration [121]. Ca2+ does not play any role downstream of PLDα1, as demonstrated by the fact that exogenous Ca2+ treatment could dis-assemble microtubules in guard cells, and this resulted in stomatal closure in wild-type Arabidopsis but not in the pldα1 mutant [123]. Treatment with exogenous ABA increases Ca2+ levels quickly in the wild type but slowly in the pldα1 mutant, demonstrating the role of PA produced by intact PLDα1. Treatment with PA increases Ca2+ concentrations more in the wild type than in the pldα1 mutant, showing the possibility that Ca2+ influx may occur both upstream and downstream of PLDα1 [123].. There may be a positive feedback loop, where Ca2+ produced by PA in turn induces PLDα1 to produce more PA. Exogenous treatment with Ca2+ increases the PLDα1 enzyme activity without increasing the protein production because the cellular concentration of PLDα1 remains the same [123].
The PLDα1-PA system stimulates NADPH oxidase activity, resulting in ROS production [127]. This is in agreement with the observation in Arabidopsis, where ROS production increased upon PA treatment [128].
During biotic stress, pathogen-associated molecular patterns (PAMPs) such as flg22 are recognized by the cell surface receptors of plants, initiating PAMP-triggered immunity (PTI). To circumvent this plant defense, bacteria and fungi produce effector proteins that disrupt PTI. In turn, plants produce intracellular resistance (R) proteins that can bind to pathogen-produced effector molecules and initiate effector-triggered immunity (ETI) [129]. Hence, a model is proposed, where upon pathogen infection, PAMPs activate PLD-mediated SA accumulation by the PA–ROS–SA pathway in two waves. The first peak of SA accumulation is rapid (0.5 to 2 h) during PTI and/or ETI. Accumulated SA activates PLD, which leads to a second wave of SA accumulation (after 2 to 10 h) by the PA–ROS–SA pathway, during ETI [130]. The crosstalks among SA, ABA, and phospholipid signaling are summarized in Figure 4.
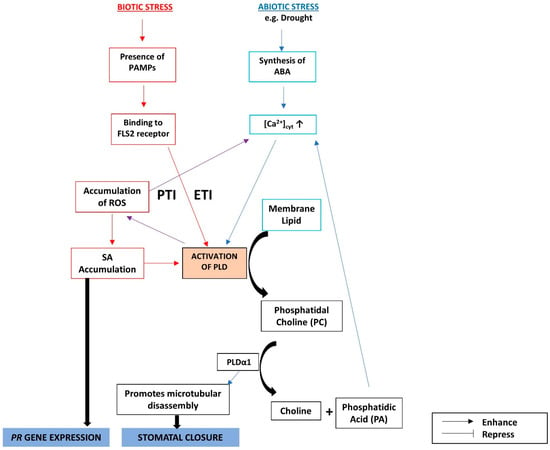
Figure 4.
The crosstalks among ABA, SA, and phospholipid signaling during biotic and abiotic stress responses act through PLDs. PTI: PAMP-triggered immunity; ETI: effector-triggered immunity.
4. Roles of G-proteins in Biotic and Abiotic Stress Responses
G-proteins are cellular molecules for relaying signals. The involvement of G-proteins in JA signaling has been discussed in Section 3 (Figure 1). Upon binding to GTP, G-proteins trigger the hydrolysis of GTP to GDP, and their conformations are changed to the active forms. The active G-proteins bind to downstream targets to modulate their functions. Many G-proteins are stress-responsive. Therefore, they have been regarded as important players in regulating signaling events upon stresses. G-proteins can be categorized into heterotrimeric G-proteins, unconventional G-proteins, and small G-proteins. Some G-proteins, such as the unconventional G-protein, YchF, have been reported to play dual roles in abiotic and biotic stresses.
4.1. Heterotrimeric G-proteins
A heterotrimeric G-protein typically comprises three subunits (α, β, and γ). In plants, a heterotrimeric G-protein consists of one α-subunit (AGA1), one β-subunit (AGB1), and three γ-subunits (AGG1, AGG2, and AGG3). It transduces signals from a G-protein-coupled receptor (GPCR) in response to environmental stimuli.
The Arabidopsis Gα subunit, namely GPA1, plays an important role in stomatal defense against bacterial pathogens. For example, growth of the coronatine-deficient P. syringae pv. tomato DC3118 is significantly increased in gpa1 mutant plants [131]. In addition, the rice d1 mutant, which is deficient in the Gα subunit, exhibits more severe disease symptoms when inoculated with Xanthomonas oryzae pv. oryzae, and it has significantly lowered production of the probenazole-inducible protein, PBZ1, which belongs to PR10 proteins and exhibits RNase activity, and thus induces cell death against necrotrophic pathogens, compared to the wild type [132].
On the other hand, using the bir1-1 mutant of Arabidopsis, the G-protein β-subunit (AGB1) was found to function downstream of the leucine-rich repeat receptor-like kinase (LRRR-RLK), SOBIR1, to initiate cell death [131]. In agb1 null-mutant plants, the induction of PAMP-triggered defense responses as mediated by three other RLKs: FLS2, EFR, and CERK1, are severely compromised, suggesting that AGB1 is a signaling component for plant immunity in common among pathways mediated by different RLKs [131].
Arabidopsis AGB1 also plays a role in the defense against the necrotrophic pathogens, Fusarium oxysporum f. sp. conglutinans and Alternaria brassicicola (isolate UQ4273). Using the agb1 mutant against these pathogens, AGB1-mediated signaling was found to suppress the induction of PR genes by SA, JA, ethylene, or ABA during the initial phase of the infection. However, with the use of double mutants, leaf necrosis resulting from the agb1-2 mutation was shown to be rescued by simultaneous coi1-21 and jin1-9 mutations. Thus, AGB1 works upstream of COI1 and ATMYC2 in JA signaling during the late phase of infection by necrotrophic pathogens [133].
For the Gγ subunits, AGG1 and AGG2 have redundant roles in Arabidopsis in response to elf18-induced oxidative bursts (a PTI response), although AGG1 seems to play the leading role. However, AGG3 is found to be involved in stomatal ion channel regulation and the development of reproductive organs, and it has no effect on the resistance against necrotrophic pathogens. Thus, AGG3 is probably not involved in PTI [131].
Heterotrimeric G-proteins also participate in abiotic stress responses. The Gα subunit from carrot seedlings were shown to exhibit lowered expression under heat stress and prolonged salt treatment [134,135] Meanwhile Gβ is found to be involved in salt stress tolerance only. Arabidopsis agb1-2 mutant exhibited lower cotyledon greening rates, fresh weight, root length, seedling germination rates, and survival rates [135].
4.2. Unconventional G-proteins
The unconventional G-proteins are unimolecular and are similar in size to the heterotrimeric Gα subunit. They are classified into extra-large G-protein, GPCR-type G-proteins (GTGs), and the Obg superfamily.
In the case of extra-large G-proteins, the Arabidopsis genome contains three genes encoding proteins with limited homology to Gα subunits, but more than twice the size of GPA1, and thus they were named extra-large G-proteins (XLGs) upon their discovery [136]. XLGs contain two distinct domains, an N-terminal cysteine-rich region and a C-terminal Gα-like domain [136]. The xlg mutants share some similar phenotypes with agb1 mutants, suggesting the existence of functional similarities between XLGs and Gβ. Similar to the agb1 mutants, xlg2 mutants displayed increased susceptibility to Pseudomonas syringae pv. tomato DC3000. XLG2 interacts directly with AGB1 and acts as a positive regulator in the salicylic acid pathway [137]
Another class of unconventional G-proteins, GPCR-type G-proteins (GTGs), has also been functionally characterized in Arabidopsis [138]. GTGs are multi-unit transmembrane G-proteins with a span topology reminiscent of G-protein-coupled receptors (GPCRs), but with nine predicted transmembrane spans instead of the seven that is characteristic of GPCRs. The Arabidopsis genome encodes two GTG proteins, GTG1 (At1g64990) and GTG2 (At4g27630), which have 90% sequence identity at the amino acid level. Both GTG1 and GTG2 interact with the Arabidopsis GPA1. The gtg1gtg2 double mutant is defective in a variety of classic ABA responses, including inhibition of seed germination, prevention of cotyledon greening, retardation of primary root elongation, induction of ABA-responsive genes such as RAB18, RD29B, DREB2A, DREB2B, and ERD10, and promotion of stomatal closure.
4.2.1. Obg Superfamily
The Obg superfamily is a TRAFAC class of P-loop GTPases that have a highly conserved amino acid sequence in various organisms from prokaryotes to eukaryotes, and are thus believed to be the descendants of ancient proteins. Some of them are found to play important roles in basic cellular processes, e.g., ribosome biogenesis, translation, and signal transduction. This superfamily includes the Obg, EngB, HflX, Era, TrmE, EngA, EngD/ YchF, Drg, and Nog1 G-protein families.
Obg/Era Family
Spo0B-associated GTP-binding (Obg) proteins was named as such because it was found to be downstream to the Spo0B gene in Bacillus subtilis. It is found to be essential for the viability of nearly all bacteria. However, its functions in plant are least to be studied. Rice OsTSV3 encodes a Spo0B-associated GTP-binding (Obg) protein was reported to exhibit functions related to cold stress responses. The cold-sensitive virescent tsv3 mutant of rice exhibits an albino phenotype at 20 °C, but develops normal green leaves at 32 °C before the three-leaf stage, and has a normal green phenotype at 20 °C after the 4-leaf stage. The expressions of genes associated with the biogenesis of the chloroplast ribosome 50S subunit in the tsv3 mutant were severely decreased in the three-leaf stage seedlings at 20 °C. This observation implied that the nuclear-encoded Obg G-protein member, TSV3, plays critical roles in chloroplast development at early developmental stages [139].
Drg Family
Developmentally regulated GTP-binding proteins (DRGs), being one of the family in the Obg superfamily, belongs to the Archaea-related type with Nog1. As suggested by the name, its functions are mainly related to development. However, current findings showed that it is also related to cold stress responses. Arabidopsis AtDRG1-3 is induced during seed drying and heat stress [140]. The expressed protein of AtDRG1-3 is localized not only in the cytoplasm, such as AtDRG1-1 and AtDRG1-2, but also in the nucleus. However, the function of AtDRG1-3, which is stress inducible, is not yet elucidated [140].
YchF Family
The YchFs are a group of ancient proteins, and their sequences are highly conserved among all kingdoms of life except Archaea [141]. The rice YchF1 protein (OsYchF1) was the first plant YchF protein, with its physiological functions characterized in relation to both biotic and abiotic stresses [141,142].
OsYchF1 is the interacting partner of a wounding-inducible protein, Oryza sativa GTPase-activating protein 1 (OsGAP1). OsGAP1 is a protein specific to higher plants, and it is induced during wounding in the bacterial blight-resistant rice line that expresses the R gene, Xa14, against bacterial blight. OsYchF1 is identified as its protein interacting partner through yeast two-hybrid experiments, in vitro pull-down and bimolecular fluorescence complementation (BiFC). OsGAP1 not only interacts with OsYchF1, but it also regulates OsYchF1 in several aspects. OsGAP1 alters the subcellular localization of OsYchF1, its RNA-binding affinity, and the substrate specificity of NTP hydrolysis [141].
More importantly, OsGAP1 is able to enhance plant resistance towards bacterial pathogens. OsGAP1-transgenic rice and Arabidopsis exhibit phenotypes with fewer disease lesions after inoculations with Xanthomonas oryzae pv. oryzae (Xoo) and Pseudomonas syringae pv. tomato DC3000 (Pto DC3000), respectively [143]. Besides having less severe disease symptoms, the OsGAP1-expressing transgenic plants also have lower pathogen titers from infected leaves, and higher induction of PR genes involved in the SA, JA, and ethylene pathways [143]. In addition, the ectopic expression of OsGAP1 in the Arabidopsis npr1-3 mutant failed to enhance resistance against Pto DC3000, and could not induce the above-mentioned PR genes, thus demonstrating that the function of OsGAP1 on biotic stress is upstream of NPR1, which is a well-known key regulator in biotic stress responses, and plays important roles as the convergence point of several signaling pathways, including the SA-dependent systemic acquired resistance, as well as the JA/ET-dependent induced systemic resistance [143].
However, the positive role of OsGAP1 in biotic stress responses function through its deactivation of OsYchF. Arabidopsis transgenic lines ectopically expressing OsYchF1 show increased susceptibility to Pto DC3000 [141]. Thus, it is speculated that OsGAP1 acts as a positive regulator in disease resistance pathways by stimulating the GTPase/ATPase activities of OsYchF1, and thus converting it into the GDP-/ADP-bound inactive form. Meanwhile, OsGAP1 also localizes OsYchF1 from the cytosol to the plasma membrane upon wounding. In addition, OsGAP1 interacts with OsYchF1 at its TGS domain, which is a ribonucleic acid-binding domain. OsYchF1 is able to bind the rice 26S rRNA, and OsGAP1 was found to compete with 26S rRNA for binding to OsYchF1 via in vitro pull-down experiments. Therefore, the function of OsYchF1 as a negative regulator for biotic stress is suppressed by OsGAP1 [141].
YchF1 and GAP1 proteins not only play roles in biotic stress, they are also involved in abiotic stress responses. The ectopic or over-expression of rice and Arabidopsis YchF1 proteins in transgenic Arabidopsis enhances the sensitivity of the plants towards salt stress. On the other hand, AtYchF1-knockdown mutant exhibits higher tolerance towards salt treatment when compared to the wild type. Meanwhile, transgenic Arabidopsis lines over-expressing AtGAP1 demonstrated higher salt tolerance when compared to the wild type while AtGAP1-knockdown lines showed lower salt tolerance [142]. Higher anti-oxidant activities were also found in the AtYchF1-knockdown mutant, AtGAP1-over-expressing lines, and OsGAP1-transgenic Arabidopsis lines. In addition, OsYchF1-transgenic tobacco BY-2 cells show higher levels of ROS generation and percentage of cell death under salt treatment [142].
Therefore, YchF1 acts as a negative regulator to suppress both biotic and abiotic stress responses under favorable growing conditions. When there are stresses present, GAP1 is induced, and the GAP1 protein would inactivate YchF1 and then trigger defense responses.
One of the special features of YchF1 is its lower nucleotide-binding specificity than typical GTPases; it can bind both GTP and ATP. By resolving the structure of OsYchF1 via crystallization, it is deduced that the G4 motif of OsYchF1 is responsible for the lowered specificity, and therefore its lower ability to use both ATP and GTP as a substrate. The canonical sequence of the G4 motif for GTP specificity is NKXD, while the sequence of the OsYchF1 G4 motif is NMSE. Mutation of the native noncanonical sequence of the G4 motif of OsYchF1 to the canonical sequence (i.e., mutating NMSE to NKSD) precludes the binding/hydrolysis of ATP by OsYchF1. Transgenic Arabidopsis over-expressing the mutated versions of OsYchF1-G4 and AtYchF1-G4 could only bind GTP but not ATP, and could no longer function as a negative regulator of plant defense responses. On the other hand, being only able to bind/hydrolyze GTP and not ATP still allows YchF1 to function as a negative regulator of abiotic stress responses. This study showcases the specific nature of the ATP-binding/hydrolyzing ability of YchF1 in determining its function in disease resistance [144].
4.3. Small G-proteins
In animals, there are two subfamilies within the Ras superfamily of small G-proteins: Ras and Rho. However, plants do not have Ras, but they contain Rho-like small G-proteins called RACs or ROPs.
The Arabidopsis genome encodes a family of 11 ROP small GTPases, while the rice genome comprises seven members and there are six ROPs in the barley genome [145,146].
The rice Rac protein (OsRAC1) is involved in defense against Magnaporthe grisea and elicitor (sphingolipid)-triggered responses. It positively regulates NADPH oxidase and cinnamoyl-CoA reductase, so as to increase ROS production and lignin biosynthesis, respectively [147].
Overexpression of a Ras-related small G-protein, RGPL, in Nicotiana tabacum results in higher production of SA and pathogenesis-related (PR) proteins, and increases resistance towards tobacco mosaic virus (TMV) [148].
In response to abiotic stress, Arabidopsis Rac1 (AtRac1) responds to ABA by disrupting the actin skeleton in guard cells, and this eventually leads to stomatal closure during drought conditions [149].
Transgenic rice and Arabidopsis lines overexpressing OsRAN2 exhibit increased sensitivity towards salt (NaCl) and osmotic stress (PEG6000), as well as exogenous ABA (10 μM). Meanwhile, transgenic rice lines with knocked-down OsRAN2 expressions showed the reverse phenotypes. OsRAN2 gene expression declines to around 20% to 40% of that in the wild type during the first 10 h of treatments, including salt stress, osmotic stress, and ABA treatment. Besides, both rice and Arabidopsis transgenic lines with higher or ectopic OsRAN2 expressions develop shorter and fewer roots, and smaller leaves under the aforementioned stress conditions. To conclude, the OsRAN2 protein is a negative regulator in abiotic stress responses [150].
On the other hand, OsRAN2 plays a positive role under cold stress. OsRAN2 was induced during cold stress, and transgenic lines over-expressing OsRAN2 showed higher survival rates under cold treatment, where over 50% of seedlings were able to grow and stay green after a two-week recovery period in normal greenhouse conditions when compared to the wild type, where only around 20% survived after the recovery period. OsRAN2 was found to maintain cell division via promoting the export of intra-nuclear tubulin at the end of mitosis, and it was thus able to maintain the normal nuclear envelope under cold stress [151].
A list of the different classes of G-proteins and their roles in abiotic and biotic stress responses is presented in Table 3.

Table 3.
G-proteins and their roles in biotic and abiotic stress responses.
5. Conclusions
Upon environmental challenges, a huge network of signaling events occur in the plant cell. Ca2+ sensors are responsible for decoding the natures of the stimuli, and the signals are then transduced through the appropriate stress hormone pathways to bring forth a series of physiological responses. Some major plant hormones, such as ABA and JA, are closely coordinated with Ca2+ signaling. Therefore, it has been speculated that ABA and JA are the convergent points between abiotic stresses and biotic stresses. This speculation is supported by studies showing the dual roles of Ca2+ sensors as both the initial stress signal detectors and also the regulators of ABA and JA signaling. At the same time, G-proteins are also known to play dual roles in abiotic stress and biotic stress responses, and it has been reported that G-proteins are also involved in the signaling pathways of plant hormones, such as JA. However, as G-proteins consist of so many different sub-groups (Table 3), their diverse behaviors could be an obstacle to fully understanding the roles of G-proteins in plant hormone signaling events. Nevertheless, the diverse natures of G-proteins also hint at the many possibilities of how G-proteins can regulate plant hormone signaling under biotic and abiotic stresses.
Author Contributions
Y.-S.K., M.S. and M.-Y.C. compiled the materials and wrote the first draft. H.-M.L. coordinated the writing effort and revised this manuscript. Y.-S.K. and H.-M.L. prepared the final version.
Funding
This work was supported by the Hong Kong Research Grants Council General Research Fund (14122715) and Area of Excellence Scheme (AoE/M-403/16).
Acknowledgments
Jee-Yan Chu copy-edited this manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ku, Y.; Yung, Y.; Li, M.; Wen, C.; Liu, X. Drought stress and tolerance in soybean. In A Comprehensive Survey of International Soybean Research—Genetics, Physiology, Agronomy and Nitrogen Relationships; Board, J.E., Ed.; IntechOpen: London, UK, 2013; p. 624. ISBN 978-953-51-0876-4. [Google Scholar]
- Phang, T.H.; Shao, G.; Lam, H.M. Salt tolerance in soybean. J. Integr. Plant Biol. 2008, 50, 1196–1212. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Vinocur, B.; Altman, A. Plant responses to drought, salinity and extreme temperatures: Towards genetic engineering for stress tolerance. Planta 2003, 218, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Mauch-Mani, B.; Mauch, F. The role of abscisic acid in plant-pathogen interactions. Curr. Opin. Plant Biol. 2005, 8, 409–414. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R. Abiotic stress, the field environment and stress combination. Trends Plant Sci. 2006, 11, 15–19. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, N.; Rivero, R.M.; Shulaev, V.; Blumwald, E.; Mittler, R. Abiotic and biotic stress combinations. New Phytol. 2014, 203, 32–43. [Google Scholar] [CrossRef] [PubMed]
- Trewavas, A. Le calcium, c’est la vie: Calcium makes waves. Plant Physiol. 1999, 120, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Edel, K.H.; Kudla, J. Integration of calcium and ABA signaling. Curr. Opin. Plant Biol. 2016, 33, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Nitta, Y.; Ding, P.; Zhang, Y. Heterotrimeric G proteins in plant defense against pathogens and ABA signaling. Environ. Exp. Bot. 2015, 114, 153–158. [Google Scholar] [CrossRef]
- Ranty, B.; Aldon, D.; Cotelle, V.; Galaud, J.-P.; Thuleau, P.; Mazars, C. Calcium sensors as key hubs in plant responses to biotic and abiotic stresses. Front. Plant Sci. 2016, 7, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Tuteja, N.; Mahajan, S. Calcium signaling network in plants: an overview. Plant Signal. Behav. 2007, 2, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Liese, A.; Romeis, T. Biochemical regulation of in vivo function of plant calcium-dependent protein kinases (CDPK). Biochim. Biophys. Acta—Mol. Cell Res. 2013, 1833, 1582–1589. [Google Scholar] [CrossRef] [PubMed]
- Perochon, A.; Aldon, D.; Galaud, J.P.; Ranty, B. Calmodulin and calmodulin-like proteins in plant calcium signaling. Biochimie 2011, 93, 2048–2053. [Google Scholar] [CrossRef] [PubMed]
- La Verde, V.; Dominici, P.; Astegno, A. Towards understanding plant calcium signaling through calmodulin-like proteins: A biochemical and structural perspective. Int. J. Mol. Sci. 2018, 19, 1331. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Yang, T.; Jurick II, W.M. Calmodulin gene expression in response to mechanical wounding and Botrytis cinerea infection in tomato fruit. Plants 2014, 3, 427–441. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Zhou, R.-G.; Gao, Y.-J.; Zheng, S.-Z.; Xu, P.; Zhang, S.-Q.; Sun, D.-Y. Molecular and genetic evidence for the key role of AtCaM3 in heat-shock signal transduction in Arabidopsis. Plant Physiol. 2009, 149, 1773–1784. [Google Scholar] [CrossRef] [PubMed]
- Heo, W.D.; Lee, S.H.; Kim, M.C.; Kim, J.C.; Chung, W.S.; Chun, H.J.; Lee, K.J.; Park, C.Y.; Park, H.C.; Choi, J.Y.; et al. Involvement of specific calmodulin isoforms in salicylic acid-independent activation of plant disease resistance responses. Proc. Natl. Acad. Sci. USA 1999, 96, 766–771. [Google Scholar] [CrossRef] [PubMed]
- Munir, S.; Liu, H.; Xing, Y.; Hussain, S.; Ouyang, B.; Zhang, Y.; Li, H.; Ye, Z. Overexpression of calmodulin-like (ShCML44) stress-responsive gene from Solanum habrochaites enhances tolerance to multiple abiotic stresses. Sci. Rep. 2016, 6, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Magnan, F.; Ranty, B.; Charpenteau, M.; Sotta, B.; Galaud, J.P.; Aldon, D. Mutations in AtCML9, a calmodulin-like protein from Arabidopsis thaliana, alter plant responses to abiotic stress and abscisic acid. Plant J. 2008, 56, 575–589. [Google Scholar] [CrossRef] [PubMed]
- Leba, L.-J.; Cheval, C.; Ortiz-Martín, I.; Ranty, B.; Beuzón, C.R.; Galaud, J.P.; Aldon, D. CML9, an Arabidopsis calmodulin-like protein, contributes to plant innate immunity through a flagellin-dependent signalling pathway. Plant J. 2012, 71, 976–989. [Google Scholar] [CrossRef] [PubMed]
- Scholz, S.S.; Reichelt, M.; Vadassery, J.; Mithöfer, A. Calmodulin-like protein CML37 is a positive regulator of ABA during drought stress in Arabidopsis. Plant Signal. Behav. 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- Scholz, S.S.; Vadassery, J.; Heyer, M.; Reichelt, M.; Bender, K.W.; Snedden, W.A.; Boland, W.; Mithöfer, A. Mutation of the Arabidopsis calmodulin-like protein CML37 deregulates the jasmonate pathway and enhances susceptibility to herbivory. Mol. Plant 2014, 7, 1712–1726. [Google Scholar] [CrossRef] [PubMed]
- Vadassery, J.; Reichelt, M.; Hause, B.; Gershenzon, J.; Boland, W.; Mithofer, A. CML42-mediated calcium signaling coordinates responses to Spodoptera herbivory and abiotic stresses in Arabidopsis. Plant Physiol. 2012, 159, 1159–1175. [Google Scholar] [CrossRef] [PubMed]
- Batistič, O.; Kudla, J. Plant calcineurin B-like proteins and their interacting protein kinases. Biochim. Biophys. Acta—Mol. Cell Res. 2009, 1793, 985–992. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Ren, F.; Zhou, L.; Wang, Q.Q.; Zhong, H.; Li, X.B. The Brassica napus Calcineurin B-Like 1/CBL-interacting protein kinase 6 (CBL1/CIPK6) component is involved in the plant response to abiotic stress and ABA signalling. J. Exp. Bot. 2012, 63, 6211–6222. [Google Scholar] [CrossRef] [PubMed]
- Cheong, Y.H.; Sung, S.J.; Kim, B.G.; Pandey, G.K.; Cho, J.S.; Kim, K.N.; Luan, S. Constitutive overexpression of the calcium sensor CBL5 confers osmotic or drought stress tolerance in Arabidopsis. Mol. Cells 2010, 29, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.G.; Waadt, R.; Cheong, Y.H.; Pandey, G.K.; Dominguez-Solis, J.R.; Schültke, S.; Lee, S.C.; Kudla, J.; Luan, S. The calcium sensor CBL10 mediates salt tolerance by regulating ion homeostasis in Arabidopsis. Plant J. 2007, 52, 473–484. [Google Scholar] [CrossRef] [PubMed]
- de la Torre, F.; Gutierrez-Beltran, E.; Pareja-Jaime, Y.; Chakravarthy, S.; Martin, G.B.; del Pozo, O. The tomato calcium sensor Cbl10 and its interacting protein kinase Cipk6 define a signaling pathway in plant immunity. Plant Cell 2013, 25, 2748–2764. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Ji, W.; Zhu, Y.; Gao, P.; Li, Y.; Cai, H.; Bai, X.; Guo, D. GsCBRLK, a calcium/calmodulin-binding receptor-like kinase, is a positive regulator of plant tolerance to salt and ABA stress. J. Exp. Bot. 2010, 61, 2519–2533. [Google Scholar] [CrossRef] [PubMed]
- Valmonte, G.R.; Arthur, K.; Higgins, C.M.; Macdiarmid, R.M. Calcium-dependent protein kinases in plants: Evolution, expression and function. Plant Cell Physiol. 2014, 55, 551–569. [Google Scholar] [CrossRef] [PubMed]
- Simeunovic, A.; Mair, A.; Wurzinger, B.; Teige, M. Know where your clients are: Subcellular localization and targets of calcium-dependent protein kinases. J. Exp. Bot. 2016, 67, 3855–3872. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.-J.; Wei, F.-J.; Wang, C.; Wu, J.-J.; Ratnasekera, D.; Liu, W.-X.; Wu, W.-H. Arabidopsis calcium-dependent protein kinase CPK10 functions in abscisic acid- and Ca2+-mediated stomatal regulation in response to drought stress. Plant Physiol. 2010, 154, 1232–1243. [Google Scholar] [CrossRef] [PubMed]
- Mehlmer, N.; Wurzinger, B.; Stael, S.; Hofmann-Rodrigues, D.; Csaszar, E.; Pfister, B.; Bayer, R.; Teige, M. The Ca2+-dependent protein kinase CPK3 is required for MAPK-independent salt-stress acclimation in Arabidopsis. Plant J. 2010, 63, 484–498. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Tian, Y.S.; Peng, R.H.; Xiong, A.S.; Zhu, B.; Jin, X.F.; Gao, F.; Fu, X.Y.; Hou, X.L.; Yao, Q.H. AtCPK6, a functionally redundant and positive regulator involved in salt/drought stress tolerance in Arabidopsis. Planta 2010, 231, 1251–1260. [Google Scholar] [CrossRef] [PubMed]
- Mori, I.C.; Murata, Y.; Yang, Y.; Munemasa, S.; Wang, Y.F.; Andreoli, S.; Tiriac, H.; Alonso, J.M.; Harper, J.F.; Ecker, J.R.; et al. CDPKs CPK6 and CPK3 function in ABA regulation of guard cell S-type anion- and Ca2+-permeable channels and stomatal closure. PLoS Biol. 2006, 4, 1749–1762. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Zhang, D.; Wang, L.; Pan, J.; Liu, Y.; Kong, X.; Zhou, Y.; Li, D. A maize calcium-dependent protein kinase gene, ZmCPK4, positively regulated abscisic acid signaling and enhanced drought stress tolerance in transgenic Arabidopsis. Plant Physiol. Biochem. 2013, 71, 112–120. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.; Hu, W.; Deng, X.; Zhang, Y.; Liu, X.; Zhao, X.; Luo, Q.; Jin, Z.; Li, Y.; Zhou, S.; et al. A rice calcium-dependent protein kinase OsCPK9 positively regulates drought stress tolerance and spikelet fertility. BMC Plant Biol. 2014, 14, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Asano, T.; Hayashi, N.; Kobayashi, M.; Aoki, N.; Miyao, A.; Mitsuhara, I.; Ichikawa, H.; Komatsu, S.; Hirochika, H.; Kikuchi, S.; et al. A rice calcium-dependent protein kinase OsCPK12 oppositely modulates salt-stress tolerance and blast disease resistance. Plant J. 2012, 69, 26–36. [Google Scholar] [CrossRef] [PubMed]
- Sathyanarayanan, P.V.; Cremo, C.R.; Poovaiah, B.W. Plant chimeric Ca2+/calmodulin-dependent protein kinase. Role of the neural visinin-like domain in regulating autophosphorylation and calmodulin affinity. J. Biol. Chem. 2000, 275, 30417–30422. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.-P.; Munyampundu, J.-P.; Xu, Y.-P.; Cai, X.-Z. Phylogeny of plant calcium and calmodulin-dependent protein kinases (CCaMKs) and functional analyses of tomato CCaMK in disease resistance. Front. Plant Sci. 2015, 6, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Yan, J.; Liu, W.; Liu, L.; Sheng, Y.; Sun, Y.; Li, Y.; Scheller, H.V.; Jiang, M.; Hou, X.; et al. Phosphorylation of a NAC transcription factor by a calcium/calmodulin-dependent protein kinase regulates abscisic acid-induced antioxidant defense in maize. Plant Physiol. 2016, 171, 1651–1664. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Katzer, K.; Lambert, J.; Cerri, M.; Parniske, M. CYCLOPS, a DNA-binding transcriptional activator, orchestrates symbiotic root nodule development. Cell Host Microbe 2014, 15, 139–152. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Li, A.; Zhao, Y.; Zhang, Z.; Zhu, Y.; Tan, X.; Geng, S.; Guo, H.; Zhang, X.; Kang, Z.; et al. Overexpression of a wheat CCaMK gene reduces ABA sensitivity of Arabidopsis thaliana during seed germination and seedling growth. Plant Mol. Biol. Report. 2011, 29, 681–692. [Google Scholar] [CrossRef]
- Ahmad, P.; Rasool, S.; Gul, A.; Sheikh, S.A.; Akram, N.A.; Ashraf, M.; Kazi, A.M.; Gucel, S. Jasmonates: Multifunctional roles in stress tolerance. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Kazan, K. Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci. 2015, 20, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Xie, D. Jasmonate in plant defence: Sentinel or double agent? Plant Biotechnol. J. 2015, 13, 1233–1240. [Google Scholar] [CrossRef] [PubMed]
- Schulze-Lefert, P.; Robatzek, S. Plant pathogens trick guard cells into opening the gates. Cell 2006, 126, 831–834. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Rojas, C.M.; Oh, S.; Kang, M.; Choudhury, S.R.; Lee, H.K.; Allen, R.D.; Pandey, S.; Mysore, K.S. Nucleolar GTP-binding protein 1-2 (NOG1-2) interacts with jasmonate-ZIMDomain protein 9 (JAZ9) to regulate stomatal aperture during plant immunity. Int. J. Mol. Sci. 2018, 19. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Thilmony, R.; Bender, C.L.; Schaller, A.; He, S.Y.; Howe, G.A. Virulence systems of Pseudomonas syringae pv. tomato promote bacterial speck disease in tomato by targeting the jasmonate signaling pathway. Plant J. 2003, 36, 485–499. [Google Scholar] [CrossRef] [PubMed]
- Boter, M.; Ruíz-Rivero, O.; Abdeen, A.; Prat, S. Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev. 2004, 18, 1577–1591. [Google Scholar] [CrossRef] [PubMed]
- Pauwels, L.; Barbero, G.F.; Geerinck, J.; Tilleman, S.; Grunewald, W.; Pérez, A.C.; Chico, J.M.; Bossche, R.V.; Sewell, J.; Gil, E.; et al. NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 2010, 464, 788–791. [Google Scholar] [CrossRef] [PubMed]
- Thines, B.; Katsir, L.; Melotto, M.; Niu, Y.; Mandaokar, A.; Liu, G.; Nomura, K.; He, S.Y.; Howe, G.A.; Browse, J. JAZ repressor proteins are targets of the SCFCOI1complex during jasmonate signalling. Nature 2007, 448, 661–665. [Google Scholar] [CrossRef] [PubMed]
- Chini, A.; Fonseca, S.; Fernández, G.; Adie, B.; Chico, J.M.; Lorenzo, O.; García-Casado, G.; López-Vidriero, I.; Lozano, F.M.; Ponce, M.R.; et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 2007, 448, 666–671. [Google Scholar] [CrossRef] [PubMed]
- Sheard, L.B.; Tan, X.; Mao, H.; Withers, J.; Ben-Nissan, G.; Hinds, T.R.; Kobayashi, Y.; Hsu, F.F.; Sharon, M.; Browse, J.; et al. Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 2010, 468, 400–407. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Senthil-Kumar, M.; Kang, M.; Rojas, C.M.; Tang, Y.; Oh, S.; Choudhury, S.R.; Lee, H.K.; Ishiga, Y.; Allen, R.D.; et al. The small GTPase, nucleolar GTP-binding protein 1 (NOG1), has a novel role in plant innate immunity. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.S.; Howe, G.A. A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM-Domain protein JAZ10 in Arabidopsis. Plant Cell 2009, 21, 131–145. [Google Scholar] [CrossRef] [PubMed]
- Manners, J.M.; Penninckx, I.A.M.A.; Vermaere, K.; Kazan, K.; Brown, R.L.; Morgan, A.; Maclean, D.J.; Curtis, M.D.; Cammue, B.P.A.; Broekaert, W.F. The promoter of the plant defensin gene PDF1.2 from Arabidopsis is systemically activated by fungal pathogens and responds to methyl jasmonate but not to salicylic acid. Plant Mol. Biol. 1998, 38, 1071–1080. [Google Scholar] [CrossRef] [PubMed]
- Lorenzo, O. ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 2003, 15, 165–178. [Google Scholar] [CrossRef] [PubMed]
- Shakeel, S.N.; Gao, Z.; Amir, M.; Chen, Y.F.; Rai, M.I.; Haq, N.U.; Schaller, G.E. Ethylene regulates levels of ethylene receptor/CTR1 signaling complexes in Arabidopsis thaliana. J. Biol. Chem. 2015, 290, 12415–12424. [Google Scholar] [CrossRef] [PubMed]
- Yasumura, Y.; Pierik, R.; Kelly, S.; Sakuta, M.; Voesenek, L.A.C.J.; Harberd, N.P. An ancestral role for CONSTITUTIVE TRIPLE RESPONSE1 proteins in both ethylene and abscisic acid signaling. Plant Physiol. 2015, 169, 283–298. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Chen, Y.F.; Randlett, M.D.; Zhao, X.C.; Findell, J.L.; Kieber, J.J.; Schaller, G.E. Localization of the Raf-like kinase CTR1 to the endoplasmic reticulum of Arabidopsis through participation in ethylene receptor signaling complexes. J. Biol. Chem. 2003, 278, 34725–34732. [Google Scholar] [CrossRef] [PubMed]
- Ju, C.; Yoon, G.M.; Shemansky, J.M.; Lin, D.Y.; Ying, Z.I.; Chang, J.; Garrett, W.M.; Kessenbrock, M.; Groth, G.; Tucker, M.L.; et al. CTR1 phosphorylates the central regulator EIN2 to control ethylene hormone signaling from the ER membrane to the nucleus in Arabidopsis. Proc. Natl. Acad. Sci. USA 2012, 109, 19486–19491. [Google Scholar] [CrossRef] [PubMed]
- An, F.; Zhao, Q.; Ji, Y.; Li, W.; Jiang, Z.; Yu, X.; Zhang, C.; Han, Y.; He, W.; Liu, Y.; et al. Ethylene-induced stabilization of ETHYLENE INSENSITIVE3 and EIN3-LIKE1 is mediated by proteasomal degradation of EIN3 binding F-box 1 and 2 that requires EIN2 in Arabidopsis. Plant Cell 2010, 22, 2384–2401. [Google Scholar] [CrossRef] [PubMed]
- Solano, R.; Stepanova, A.; Chao, Q.; Ecker, J.R. Nuclear events in ethylene signaling: A transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 1998, 12, 3703–3714. [Google Scholar] [CrossRef] [PubMed]
- Nakano, T.; Suzuki, K.; Fujimura, T.; Shinshi, H. Genome-wide analysis of the ERF gene family. Plant Physiol. 2006, 140, 411–432. [Google Scholar] [CrossRef] [PubMed]
- Ohmetakagi, M.; Shinshi, H. Ethylene-inducible DNA-bidning proteins that interact with an ethylene-responsive element. Plant Cell 1995, 7, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Sessa, G.; Meller, Y.; Fluhr, R. A GCC element and a G-box motif participate in ethylene-induced expression of the PRB-1b gene. Plant Mol. Biol. 1995, 28, 145–153. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, S.Y. Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 2000, 12, 393–404. [Google Scholar] [CrossRef] [PubMed]
- Shinshi, H.; Usami, S.; Ohme-Takagi, M. Identification of an ethylene-responsive region in the promoter of a tobacco class I chitinase gene. Plant Mol. Biol. 1995, 27, 923–932. [Google Scholar] [CrossRef] [PubMed]
- Sakuma, Y.; Liu, Q.; Dubouzet, J.G.; Abe, H.; Yamaguchi-Shinozaki, K.; Shinozaki, K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 2002, 290, 998–1009. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.-C.; Liao, P.-M.; Kuo, W.-W.; Lin, T.-P. The Arabidopsis ETHYLENE RESPONSE FACTOR1 regulates abiotic stress-responsive gene expression by binding to different cis-acting elements in response to different stress signals. Plant Physiol. 2013, 162, 1566–1582. [Google Scholar] [CrossRef] [PubMed]
- Dubois, M.; Skirycz, A.; Claeys, H.; Maleux, K.; Dhondt, S.; De Bodt, S.; Vanden Bossche, R.; De Milde, L.; Yoshizumi, T.; Matsui, M.; et al. ETHYLENE RESPONSE FACTOR6 acts as a central regulator of leaf growth under water-limiting conditions in Arabidopsis. Plant Physiol. 2013, 162, 319–332. [Google Scholar] [CrossRef] [PubMed]
- Seo, Y.J.; Park, J.B.; Cho, Y.J.; Jung, C.; Seo, H.S.; Park, S.K.; Nahm, B.H.; Song, J.T. Overexpression of the ethylene-responsive factor gene BrERF4 from Brassica rapa increases tolerance to salt and drought in Arabidopsis plants. Mol. Cells 2010, 30, 271–277. [Google Scholar] [CrossRef] [PubMed]
- Sewelam, N.; Kazan, K.; Thomas-Hall, S.R.; Kidd, B.N.; Manners, J.M.; Schenk, P.M. Ethylene response factor 6 is a regulator of reactive oxygen species signaling in Arabidopsis. PLoS ONE 2013, 8, e70289. [Google Scholar] [CrossRef] [PubMed]
- Jisha, V.; Dampanaboina, L.; Vadassery, J.; Mithöfer, A.; Kappara, S.; Ramanan, R. Overexpression of an AP2/ERF type transcription factor OsEREBP1 confers biotic and abiotic stress tolerance in rice. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- Park, J.M.; Park, C.J.; Lee, S.B.; Ham, B.K.; Shin, R.; Paek, K.H. Overexpression of the tobacco Tsi1 gene encoding an EREBP/AP2-type transcription factor enhances resistance against pathogen attack and osmotic stress in tobacco. Plant Cell 2001, 13, 1035–1046. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Huang, Z.; Chen, Q.; Zhang, Z.; Zhang, H.; Wu, Y.; Huang, D.; Huang, R. Ectopic overexpression of tomato JERF3 in tobacco activates downstream gene expression and enhances salt tolerance. Plant Mol. Biol. 2004, 55, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Chen, M.; Li, L.; Xu, Z.; Chen, X.; Guo, J.; Ma, Y. Overexpression of the soybean GmERF3 gene, an AP2/ERF type transcription factor for increased tolerances to salt, drought, and diseases in transgenic tobacco. J. Exp. Bot. 2009, 60, 3781–3796. [Google Scholar] [CrossRef] [PubMed]
- Rong, W.; Qi, L.; Wang, A.; Ye, X.; Du, L.; Liang, H.; Xin, Z.; Zhang, Z. The ERF transcription factor TaERF3 promotes tolerance to salt and drought stresses in wheat. Plant Biotechnol. J. 2014, 12, 468–479. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Seymour, G.B.; Lu, C.; Hu, Z.; Chen, X.; Chen, G. An ethylene response factor (ERF5) promoting adaptation to drought and salt tolerance in tomato. Plant Cell Rep. 2012, 31, 349–360. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Chen, X.; Ren, H.; Zhang, Z.; Zhang, H.; Wang, J.; Wang, X.C.; Huang, R. ERF protein JERF1 that transcriptionally modulates the expression of abscisic acid biosynthesis-related gene enhances the tolerance under salinity and cold in tobacco. Planta 2007, 226, 815–825. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wan, L.; Zhang, L.; Zhang, Z.; Zhang, H.; Quan, R.; Zhou, S.; Huang, R. An ethylene response factor OsWR1 responsive to drought stress transcriptionally activates wax synthesis related genes and increases wax production in rice. Plant Mol. Biol. 2012, 78, 275–288. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Tian, L.; Latoszek-Green, M.; Brown, D.; Wu, K. Arabidopsis ERF4 is a transcriptional repressor capable of modulating ethylene and abscisic acid responses. Plant Mol. Biol. 2005, 58, 585–596. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Zhang, Z.; Zhang, H.; Wang, X.-C.; Huang, R. Transcriptional modulation of ethylene response factor protein JERF3 in the oxidative stress response enhances tolerance of tobacco seedlings to salt, drought, and freezing. Plant Physiol. 2008, 148, 1953–1963. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Li, Z.; Quan, R.; Li, G.; Wang, R.; Huang, R. An AP2 domain-containing gene, ESE1, targeted by the ethylene signaling component EIN3 is important for the salt response in Arabidopsis. Plant Physiol. 2011, 157, 854–865. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Huang, R. Enhanced tolerance to freezing in tobacco and tomato overexpressing transcription factor TERF2/LeERF2 is modulated by ethylene biosynthesis. Plant Mol. Biol. 2010, 73, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Qi, L.; Liu, X.; Cai, S.; Xu, H.; Huang, R.; Li, J.; Wei, X.; Zhang, Z. The wheat ethylene response factor transcription factor PATHOGEN-INDUCED ERF1 mediates host responses to both the necrotrophic pathogen Rhizoctonia cerealis and freezing stresses. Plant Physiol. 2014, 164, 1499–1514. [Google Scholar] [CrossRef] [PubMed]
- Yi, S.Y.; Kim, J.-H.; Joung, Y.; Lee, S.; Kim, W.; Yu, S.H.; Choi, D. The pepper transcription factor CaPF1 confers pathogen and freezing tolerance in Arabidopsis. Plant Physiol. 2004, 136, 2862–2874. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Y.; Wang, Y.; Li, Y.; Lei, T.; Yan, F.; Su, L.; Li, X.; Zhao, Y.; Sun, X.; Li, J.; et al. Isolation and molecular characterization of GmERF7, a soybean ethylene-response factor that increases salt stress tolerance in tobacco. Gene 2013, 513, 174–183. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.-G.; Li, H.; Liu, J.-Y. Molecular characterization of three ethylene responsive element binding factor genes from cotton. J. Integr. Plant Biol. 2010. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Qin, L.; Liu, W.; Zhang, D.; Wang, Y. A novel ethylene-responsive factor from Tamarix hispida, ThERF1, is a GCC-box- and DRE-motif binding protein that negatively modulates abiotic stress tolerance in Arabidopsis. Physiol. Plant. 2014, 152, 84–97. [Google Scholar] [CrossRef] [PubMed]
- Serra, T.S.; Figueiredo, D.D.; Cordeiro, A.M.; Almeida, D.M.; Lourenço, T.; Abreu, I.A.; Sebastián, A.; Fernandes, L.; Contreras-Moreira, B.; Oliveira, M.M.; et al. OsRMC, a negative regulator of salt stress response in rice, is regulated by two AP2/ERF transcription factors. Plant Mol. Biol. 2013, 82, 439–455. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.J.; Chen, X.J.; Wu, X.L.; Ling, J.Q.; Xu, P. Overexpression of the AP2/EREBP transcription factor OPBP1 enhances disease resistance and salt tolerance in tobacco. Plant Mol. Biol. 2004, 55, 607–618. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, C.; Qin, L.; Liu, W.; Wang, Y. ThERF1 regulates its target genes via binding to a novel cis-acting element in response to salt stress. J. Integr. Plant Biol. 2015, 57, 838–847. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Yu, Y.; Li, S.; Wang, J.; Tang, S.; Huang, R. Abscisic acid antagonizes ethylene production through the ABI4-mediated transcriptional repression of ACS4 and ACS8 in Arabidopsis. Mol. Plant 2016, 9, 126–135. [Google Scholar] [CrossRef] [PubMed]
- Ghassemian, M.; Nambara, E.; Cutler, S.; Kawaide, H.; Kamiya, Y.; McCourt, P. Regulation of abscisic acid signaling by the ethylene response pathway in Arabidopsis. Plant Cell 2000, 12, 1117–1126. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, N.J.; Urwin, P.E. The interaction of plant biotic and abiotic stresses: From genes to the field. J. Exp. Bot. 2012, 63, 3523–3544. [Google Scholar] [CrossRef] [PubMed]
- Seo, P.J.; Xiang, F.; Qiao, M.; Park, J.-Y.; Lee, Y.N.; Kim, S.-G.; Lee, Y.-H.; Park, W.J.; Park, C.-M. The MYB96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis. Plant Physiol. 2009, 151, 275–289. [Google Scholar] [CrossRef] [PubMed]
- Wildermuth, M.C.; Dewdney, J.; Wu, G.; Ausubel, F.M. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 2001, 414, 562–565. [Google Scholar] [CrossRef] [PubMed]
- Seo, P.J.; Park, C.M. MYB96-mediated abscisic acid signals induce pathogen resistance response by promoting salicylic acid biosynthesis in Arabidopsis. New Phytol. 2010, 186, 471–483. [Google Scholar] [CrossRef] [PubMed]
- Friedrich, L.; Vernooij, B.; Gaffney, T.; Morse, A.; Ryals, J. Characterization of tobacco plants expressing a bacterial salicylate hydroxylase gene. Plant Mol. Biol. 1995, 29, 959–968. [Google Scholar] [CrossRef] [PubMed]
- Leckie, C.P.; McAinsh, M.R.; Allen, G.J.; Sanders, D.; Hetherington, A.M. Abscisic acid-induced stomatal closure mediated by cyclic ADP-ribose. Proc. Natl. Acad. Sci. USA 1998, 95, 15837–15842. [Google Scholar] [CrossRef] [PubMed]
- Munemasa, S.; Hauser, F.; Park, J.; Waadt, R.; Brandt, B.; Schroeder, J.I. Mechanisms of abscisic acid-mediated control of stomatal aperture. Curr. Opin. Plant Biol. 2015, 28, 154–162. [Google Scholar] [CrossRef] [PubMed]
- Hao, J.; Wang, X.; Dong, C.; Zhang, Z.; Shang, Q. Salicylic acid induces stomatal closure by modulating endogenous hormone levels in cucumber cotyledons. Russ. J. Plant Physiol. 2011, 58, 906–913. [Google Scholar] [CrossRef]
- Khokon, M.A.R.; Okuma, E.; Hossain, M.A.; Munemasa, S.; Uraji, M.; Nakamura, Y.; Mori, I.C.; Murata, Y. Involvement of extracellular oxidative burst in salicylic acid-induced stomatal closure in Arabidopsis. Plant Cell Environ. 2011, 34, 434–443. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Zeevaart, J.A.D. Overexpression of a 9-cis-epoxycarotenoid dioxygenase gene in Nicotiana plumbaginifolia increases abscisic acid and phaseic acid levels and enhances drought tolerance. Plant Physiol. 2002, 128, 544–551. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.I.; Kwak, J.S.; Song, J.T.; Seo, H.S. The E3 SUMO ligase AtSIZ1 functions in seed germination in Arabidopsis. Physiol. Plant. 2016, 158, 256–271. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Nam, J.; Park, H.C.; Na, G.; Miura, K.; Jin, J.B.; Yoo, C.Y.; Baek, D.; Kim, D.H.; Jeong, J.C.; et al. Salicylic acid-mediated innate immunity in Arabidopsis is regulated by SIZ1 SUMO E3 ligase. Plant J. 2007, 49, 79–90. [Google Scholar] [CrossRef] [PubMed]
- Miura, K.; Okamoto, H.; Okuma, E.; Shiba, H.; Kamada, H.; Hasegawa, P.M.; Murata, Y. SIZ1 deficiency causes reduced stomatal aperture and enhanced drought tolerance via controlling salicylic acid-induced accumulation of reactive oxygen species in Arabidopsis. Plant J. 2013, 73, 91–104. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Devaiah, S.P.; Zhang, W.; Welti, R. Signaling functions of phosphatidic acid. Prog. Lipid Res. 2006, 45, 250–278. [Google Scholar] [CrossRef] [PubMed]
- Kotel’nikova, I.M.; Nekrasov, E.V.; Krylov, A.V. Effect of tobacco mosaic virus on phospholipid content and phospholipase D activity in tobacco leaves. Russ. J. Plant Physiol. 2004, 51, 63–69. [Google Scholar] [CrossRef]
- Hyodo, K.; Taniguchi, T.; Manabe, Y.; Kaido, M.; Mise, K.; Sugawara, T.; Taniguchi, H.; Okuno, T. Phosphatidic acid produced by phospholipase D promotes RNA replication of a plant RNA virus. PLoS Pathog. 2015, 11. [Google Scholar] [CrossRef] [PubMed]
- Pinosa, F.; Buhot, N.; Kwaaitaal, M.; Fahlberg, P.; Thordal-Christensen, H.; Ellerstrom, M.; Andersson, M.X. Arabidopsis phospholipase D is involved in basal defense and nonhost resistance to powdery mildew fungi. Plant Physiol. 2013, 163, 896–906. [Google Scholar] [CrossRef] [PubMed]
- Johansson, O.N.; Fahlberg, P.; Karimi, E.; Nilsson, A.K.; Ellerström, M.; Andersson, M.X. Redundancy among phospholipase D isoforms in resistance triggered by recognition of the Pseudomonas syringae effector AvrRpm1 in Arabidopsis thaliana. Front. Plant Sci. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Bargmann, B.O.R.; Laxalt, A.M.; Riet, B.T.; van Schooten, B.; Merquiol, E.; Testerink, C.; Haring, M.A.; Bartels, D.; Munnik, T. Multiple PLDs required for high salinity and water deficit tolerance in plants. Plant Cell Physiol. 2009, 50, 78–89. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Lin, F.; Mao, T.; Nie, J.; Yan, M.; Yuan, M.; Zhang, W. Phosphatidic acid regulates microtubule organization by interacting with MAP65-1 in response to salt stress in Arabidopsis. Plant Cell 2012, 24, 4555–4576. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Nie, J.; Cao, C.; Jin, Y.; Yan, M.; Wang, F.; Liu, J.; Xiao, Y.; Liang, Y.; Zhang, W. Phosphatidic acid mediates salt stress response by regulation of MPK6 in Arabidopsis thaliana. New Phytol. 2010, 188, 762–773. [Google Scholar] [CrossRef] [PubMed]
- Katagiri, T.; Takahashi, S.; Shinozaki, K. Involvement of a novel Arabidopsis phospholipase D, AtPLD, in dehydration-inducible accumulation of phosphatidic acid in stress signalling. Plant J. 2001, 26, 595–605. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Li, M.; Zhang, W.; Welti, R.; Wang, X. The plasma membrane-bound phospholipase Ddelta enhances freezing tolerance in Arabidopsis thaliana. Nat. Biotechnol. 2004, 22, 427–433. [Google Scholar] [CrossRef] [PubMed]
- Marcus, A.I.; Moore, R.C.; Cyr, R.J. The role of microtubules in guard cell function. Plant Physiol. 2001, 125, 387–395. [Google Scholar] [CrossRef] [PubMed]
- Wang, X. Regulatory functions of phospholipase D and phosphatidic acid in plant growth, development, and stress responses. Plant Physiol. 2005, 139, 566–573. [Google Scholar] [CrossRef] [PubMed]
- Wang, X. Multiple forms of phospholipase D in plants: The gene family, catalytic and regulatory properties, and cellular functions. Prog. Lipid Res. 2000, 39, 109–149. [Google Scholar] [CrossRef]
- Jiang, Y.; Wu, K.; Lin, F.; Qu, Y.; Liu, X.; Zhang, Q. Phosphatidic acid integrates calcium signaling and microtubule dynamics into regulating ABA-induced stomatal closure in Arabidopsis. Planta 2014, 239, 565–575. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Qin, C.; Zhao, J.; Wang, X. Phospholipase D 1-derived phosphatidic acid interacts with ABI1 phosphatase 2C and regulates abscisic acid signaling. Proc. Natl. Acad. Sci. USA 2004, 101, 9508–9513. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.M.; Zhao, Z.; Assmann, S.M. Guard cells: A dynamic signaling model. Curr. Opin. Plant Biol. 2004, 7, 537–546. [Google Scholar] [CrossRef] [PubMed]
- Pappan, K.; Zheng, L.; Krishnamoorthi, R.; Wang, X. Evidence for and characterization of Ca2+ binding to the catalytic region of Arabidopsis thaliana phospholipase Dβ. J. Biol. Chem. 2004, 279, 47833–47839. [Google Scholar] [CrossRef] [PubMed]
- Sang, Y. Phospholipase D and phosphatidic acid-mediated generation of superoxide in Arabidopsis. Plant Physiol. 2001, 126, 1449–1458. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Gu, Y.; Lee, Y.Y.; Yang, Z.; Lee, Y.Y. Phosphatidic acid induces leaf cell death in Arabidopsis by activating the Rho-related small G protein GTPase-mediated pathway of reactive oxygen species generation. Plant Physiol. 2004, 134, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Xiao, S. Lipids in salicylic acid-mediated defense in plants: focusing on the roles of phosphatidic acid and phosphatidylinositol 4-phosphate. Front. Plant Sci. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Ding, P.; Sun, T.; Nitta, Y.; Dong, O.; Huang, X.; Yang, W.; Li, X.; Botella, J.R.; Zhang, Y. Heterotrimeric G proteins serve as a converging point in plant defense signaling activated by multiple receptor-like kinases. Plant Physiol. 2013, 161, 2146–2158. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, S.; Yang, G.; Hayashi, N.; Kaku, H.; Umemura, K.; Iwasaki, Y. Alterations by a defect in a rice G protein α subunit in probenazole and pathogen-induced responses. Plant Cell Environ. 2004, 27, 947–957. [Google Scholar] [CrossRef]
- Trusov, Y.; Sewelam, N.; Rookes, J.E.; Kunkel, M.; Nowak, E.; Schenk, P.M.; Botella, J.R. Heterotrimeric G proteins-mediated resistance to necrotrophic pathogens includes mechanisms independent of salicylic acid-, jasmonic acid/ethylene- and abscisic acid-mediated defense signaling. Plant J. 2009, 58, 69–81. [Google Scholar] [CrossRef] [PubMed]
- Asakura, Y.; Kurosaki, F. Cloning and expression of Dcga gene encoding alpha subunit of GTP-binding protein in carrot seedlings. Biol. Pharm. Bull. 2007, 30, 1800–1804. [Google Scholar] [CrossRef] [PubMed]
- Misra, S.; Wu, Y.; Venkataraman, G.; Sopory, S.K.; Tuteja, N. Heterotrimeric G-protein complex and G-protein-coupled receptor from a legume (Pisum sativum): Role in salinity and heat stress and cross-talk with phospholipase C. Plant J. 2007, 51, 656–669. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.R.J.; Assmann, S.M. Arabidopsis thaliana “extra-large GTP-binding protein” (AtXLG1): A new class of G-protein. Plant Mol. Biol. 1999, 40, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Li, G.J.; Ding, L.; Cui, X.; Berg, H.; Assmann, S.M.; Xia, Y. Arabidopsis extra large G-protein 2 (XLG2) interacts with the Gβ subunit of heterotrimeric G protein and functions in disease resistance. Mol. Plant 2009, 2, 513–525. [Google Scholar] [CrossRef] [PubMed]
- Pandey, A.; Sharma, M.; Pandey, G.K. Elucidation of Abiotic Stress Signaling in Plants; Springer: New York, NY, USA, 2015; ISBN 978-1-4939-2539-1. [Google Scholar]
- Lin, D.; Jiang, Q.; Ma, X.; Zheng, K.; Gong, X.; Teng, S.; Xu, J.; Dong, Y. Rice TSV3 encoding Obg-like GTPase protein is essential for chloroplast development during the early leaf stage under cold stress. G3 Genes Genomes Genet. 2017, 8, 253–263. [Google Scholar] [CrossRef]
- Suwastika, I.N.; Ohniwa, R.L.; Takeyasu, K.; Shiina, T. Plant Drg proteins are cytoplasmic small GTPase-Obg homologue. Procedia Environ. Sci. 2014, 20, 357–364. [Google Scholar] [CrossRef]
- Cheung, M.Y.; Xue, Y.; Zhou, L.; Li, M.W.; Sun, S.S.M.; Lam, H.M. An ancient P-loop GTPase in rice is regulated by a higher plant-specific regulatory protein. J. Biol. Chem. 2010, 285, 37359–37369. [Google Scholar] [CrossRef] [PubMed]
- Cheung, M.Y.; Li, M.W.; Yung, Y.L.; Wen, C.Q.; Lam, H.M. The unconventional P-loop NTPase OsYchF1 and its regulator OsGAP1 play opposite roles in salinity stress tolerance. Plant Cell Environ. 2013, 36, 2008–2020. [Google Scholar] [CrossRef] [PubMed]
- Cheung, M.-Y.; Zeng, N.-Y.; Tong, S.-W.; Li, W.-Y.F.; Xue, Y.; Zhao, K.-J.; Wang, C.; Zhang, Q.; Fu, Y.; Sun, Z.; et al. Constitutive expression of a rice GTPase-activating protein induces defense responses. New Phytol. 2008, 179, 530–545. [Google Scholar] [CrossRef] [PubMed]
- Cheung, M.-Y.; Li, X.; Miao, R.; Fong, Y.-H.; Li, K.-P.; Yung, Y.-L.; Yu, M.-H.; Wong, K.-B.; Chen, Z.; Lam, H.-M. ATP binding by the P-loop NTPase OsYchF1 (an unconventional G protein) contributes to biotic but not abiotic stress responses. Proc. Natl. Acad. Sci. USA 2016, 113, 2648–2653. [Google Scholar] [CrossRef] [PubMed]
- Nibau, C.; Wu, H.M.; Cheung, A.Y. RAC/ROP GTPases: “hubs” for signal integration and diversification in plants. Trends Plant Sci. 2006, 11, 309–315. [Google Scholar] [CrossRef] [PubMed]
- Schultheiss, H.; Preuss, J.; Pircher, T.; Eichmann, R.; Hückelhoven, R. Barley RIC171 interacts with RACB in planta and supports entry of the powdery mildew fungus. Cell. Microbiol. 2008, 10, 1815–1826. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, G.K.; Iwahashi, H.; Rakwal, R. Small GTPase “Rop”: Molecular switch for plant defense responses. FEBS Lett. 2003, 546, 173–180. [Google Scholar] [CrossRef]
- Sano, H.; Seo, S.; Orudgev, E.; Youssefian, S.; Ishizuka, K.; Ohashi, Y. Expression of the gene for a small GTP binding protein in transgenic tobacco elevates endogenous cytokinin levels, abnormally induces salicylic acid in response to wounding, and increases resistance to tobacco mosaic virus infection. Proc. Natl. Acad. Sci. USA 1994, 91, 10556–10560. [Google Scholar] [CrossRef] [PubMed]
- Lemichez, E.; Wu, Y.; Sanchez, J.P.; Mettouchi, A.; Mathur, J.; Chua, N.H. Inactivation of AtRac1 by abscisic acid is essential for stomatal closure. Genes Dev. 2001, 15, 1808–1816. [Google Scholar] [CrossRef] [PubMed]
- Zang, A.; Xu, X.; Neill, S.; Cai, W. Overexpression of OsRAN2 in rice and Arabidopsis renders transgenic plants hypersensitive to salinity and osmotic stress. J. Exp. Bot. 2010, 61, 777–789. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Xu, Y.; Wang, X.; Du, C.; Du, J.; Yuan, M.; Xu, Z.; Chong, K. OsRAN2, essential for mitosis, enhances cold tolerance in rice by promoting export of intranuclear tubulin and maintaining cell division under cold stress. Plant Cell Environ. 2011, 34, 52–64. [Google Scholar] [CrossRef] [PubMed]
- Zeng, W.; He, S.Y. A prominent role of the flagellin receptor FLAGELLIN-SENSING2 in mediating stomatal response to Pseudomonas syringae pv tomato DC3000 in Arabidopsis. Plant Physiol. 2010, 153, 1188–1198. [Google Scholar] [CrossRef] [PubMed]
- Ono, E.; Wong, H.L.; Kawasaki, T.; Hasegawa, M.; Kodama, O.; Shimamoto, K. Essential role of the small GTPase Rac in disease resistance of rice. Proc. Natl. Acad. Sci. USA 2001, 98, 759–764. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).