Abstract
The function of a wheat starch regulator 1 (TaRSR1) in regulating the synthesis of grain storage starch was determined using the barley stripe mosaic virus—virus induced gene-silencing (BSMV-VIGS) method in field experiments. Chlorotic stripes appeared on the wheat spikes infected with barley stripe mosaic virus-virus induced gene-silencing- wheat starch regulator 1 (BSMV-VIGS-TaRSR1) at 15 days after anthesis, at which time the transcription levels of the TaRSR1 gene significantly decreased. Quantitative real-time PCR was also used to measure the transcription levels of 26 starch synthesis-related enzyme genes in the grains of BSMV-VIGS-TaRSR1-silenced wheat plants at 20, 27, and 31 days after anthesis. The results showed that the transcription levels of some starch synthesis-related enzyme genes were markedly induced at different sampling time points: TaSSI, TaSSIV, TaBEIII, TaISA1, TaISA3, TaPHOL, and TaDPE1 genes were induced at each of the three sampling time points and TaAGPS1-b, TaAGPL1, TaAGPL2, TaSSIIb, TaSSIIc, TaSSIIIb, TaBEI, TaBEIIa, TaBEIIb, TaISA2, TaPHOH, and TaDPE2 genes were induced at one sampling time point. Moreover, both the grain starch contents, one thousand kernel weights, grain length and width of BSMV-VIGS-TaRSR1-infected wheat plants significantly increased. These results suggest that TaRSR1 acts as a negative regulator and plays an important role in starch synthesis in wheat grains by temporally regulating the expression of specific starch synthesis-related enzyme genes.
1. Introduction
Starch is the major storage carbohydrate reserve in the endosperm of cereals, including rice, wheat, and maize, and provides about 80% of the calories consumed by humans [1]. Plant starch is composed of two different glucose polymers: amylose and amylopectin. The former is a linear polymer composed of α-1,4-glucosidic chains, whereas the latter consists of a highly branched glucan with α-1,6 glucosidic bonds that connect linear chains [2]. Amylose is synthesized by adenosine diphosphate glucose pyrophosphorylase (AGPase, EC 2.7.7.27) and granule-bound starch synthase (GBSS, EC 2.4.1.21), whereas amylopectin is catalyzed by the coordinated actions of AGPase, soluble starch synthase (SS, EC 2.4.1.21), starch branching enzyme (BE, EC 2.4.1.18), starch debranching enzyme (DBE) (isoamylase, ISA, EC 3.2.1.68; pullulanase, PUL, EC 3.2.1.41), disproportionating enzyme (DPE, EC 2.4.1.25), and phosphorylase (PHO, EC 2.4.1.1) [2]. AGPase catalyzes adenosine diphosphate glucose (ADP-Glc) to glucose-1-phosphate (G-1-P) and provides substrates for amylose and amylopectin synthesis. GBSS is a key enzyme involved in amylose synthesis, whereas SS, BE, and DBE function together with distinct roles to catalyze amylopectin synthesis [2,3]. In starch biosynthesis, PHO and DPE are believed to be involved in the initiation steps, elongating α-1,4-linked glucan polymers using G-1-P as substrate, although their precise mechanisms remain unclear [2,4].
Most starch synthesis-related enzymes have multiple subunits or isoforms in higher plants, and the number of subunits or isoforms for each enzyme is plant species-specific [5]. In the rice genome, 11 and 3 isozymes of SS and BE, respectively, have been identified [5]; in the maize genome, 9 and 4 isozymes of SS and BE, respectively, have been identified [6]; and in wheat, rice, and maize, 26, 29, and 32 starch synthesis-related enzyme genes, respectively, have been identified [5,6,7]. Temporal and spatial coordination of these starch synthesis-related enzymes may be important for converting photosynthetic products to starch and grain storage organs. To some extent, these genes are differentially expressed among plant species because there are some differences in the functional properties of starch [8]. Comprehensive transcription profiles of starch synthesis-related enzyme genes in rice, barley, maize and wheat have been determined, and of them, key genes that play essential roles in endosperm starch synthesis have been identified [5,6,7,9].
Previous studies have focused on identifying the expression profiles and effects of single starch synthesis-related enzyme genes in starch biosynthesis [2]; however, the molecular machinery regulating the expression of these genes remains unclear despite the fact that this is a finely regulated process. Transcription factors play important roles in plant growth, development, and abiotic and biotic stress responses because they specifically bind to cis-acting elements in the promoter region of target genes [10]. Most of the transcription factors isolated from higher plants are involved in abiotic and biotic stress responses, whereas few have been associated with starch synthesis [11,12,13].
Bread wheat (Triticum aestivum L.), one of the major staple crops for the human diet, is an essential component of the global food security [14]. It has a large, complex, and allohexaploid genome consisting of A, B, and D subgenomes (2n = 6x = 42, AABBDD; 2n is the number of chromosomes in each somatic cell and 6× is the basic chromosome); therefore, each wheat gene potentially exists as a trio of A, B, and D homoeoloci [15]. However, there are complicated regulatory mechanisms within the cells of an allopolyploid genome, and through genetic or epigenetic modifications, these mechanisms can orchestrate the complex intergenomic gene expression [15,16]. The genetic or epigenetic modifications have brought genomic asymmetry toward a diploid-like mode of expression, because either of mutation, elimination or repression of most genes that, in most cases, confine the activity of sets of genes to only one genome [17]. With the recent release and annotation of the bread wheat genome, it is now possible to expand better characterize its growth, development, and responses to biotic and abiotic stresses [18,19].
The euAPETALA2 (euAP2) group of the APETALA2/ethylene-responsive element binding protein (AP2/EREBP) family is characterized by the APETALA2 (AP2) domain, and some members of this group function in the boundaries of floral organ identity, floral meristem identity, or the control of floral organ number [20,21,22,23]. In our previous study, transcription levels of Rice Starch Regulator 1 (RSR1), an euAP2 group transcription factor, was significantly and negatively associated with the transcription levels of 11 starch synthesis-related enzyme genes during the wheat grain-filling period, suggesting that TaRSR1 may negatively regulate the expression of some starch synthesis-related enzyme genes in wheat grains [7]. In bread wheat, particle bombardment and co-cultivation with Agrobacterium tumefaciens have been used to explore the function of candidate genes. However, the disadvantages of these two methods include multiple copy insertions, cultivar-specificity, time consumption, and low transformation efficiency; this species may be the last cereal to be genetically transformed [23,24]. The barley stripe mosaic virus–virus induced gene silencing (BSMV-VIGS) was recently developed as an attractive tool for the rapid generation of gene knockdown phenotypes in plants. The technique mainly exploits the BSMV-mediated antiviral defense mechanism of plants to study the function of endogenous genes based on homology-dependent gene silencing. It has played an important role in the functional identification of genes in bread wheat because it is not required for plant transformation and can potentially accelerate the characterization of target genes [25,26]. In this study, BSMV-VIGS method was used to further verify the above-mentioned hypothesis. Our results indicate that TaRSR1 may negatively and temporally regulate the expression of some starch synthesis-related enzyme genes, thereby supporting its regulatory role in wheat starch synthesis.
2. Results
2.1. Phenotypes of Barley Stripe Mosaic Virus-Wheat Starch Regulator 1 (BSMV-TaRSR1) Infected Wheat Plants
Our previous study indicated that, in bread wheat, starch synthesis-related enzymes are encoded by 26 genes: AGPase (TaAGPS1-a, TaAGPS1-b, TaAGPS2, TaAGPL1, and TaAGPL2), GBSS (TaGBSSI and TaGBSSII), SS (TaSSI, TaSSIIa, TaSSIIb, TaSSIIc, TaSSIIIa, TaSSIIIb and TaSSIV), BE (TaBEI, TaBEIIa, TaBEIIb and TaBEIII), DBE (TaISA1, TaISA2, TaISA3 and TaPUL), PHO (TaPHOL and TaPHOH), and DPE (TaDPE1 and TaDPE2). Fu and Xue [22] reported that the transcription levels of OsRSR1 were almost opposite those of 15 rice starch synthesis-related enzyme genes (OsAGPS1, OsAGPS2, OsAGPL1, OsAGPL2, OsGBSSI, OsSSI, OsSSIIa, OsSSIIIa, OsBEI, OsBEIIb, OsISA1, OsISA2, OsISA3, OsPHOL, and OsPUL), and in an rsr1 mutant, these genes were distinctly upregulated during the early stage (6 days) of the grain-filling period when grain starch contents significantly increased. Our previous data indicated that the transcription levels of TaRSR1 were significantly negatively correlated with those of 11 starch synthesis-related enzyme genes (TaAGPS1-a, TaAGPL1, TaGBSSI, TaSSI, TaSSIIa, TaSSIV, TaBEI, TaBEIIb, TaPUL, TaPHOL, and TaDPE1) during the entire grain-filling period (35 days) of bread wheat, suggesting that (similar to OsRSR1) TaRSR1 negatively regulates the expression of these genes and affects grain starch synthesis. However, there may be some differences (e.g., target genes, regulatory stages) in the regulatory mechanisms of RSR1 involved in starch synthesis between bread wheat and rice [7]. In this study, the function of TaRSR1 in the starch synthesis of wheat grains was further explored.
Higher plants including important crops are grown under field conditions with natural variable factors (e.g., temperature, light illumination and humidity). Thus, field experiments with variable growth factors may more accurately measure the function of candidate genes than controlled environments with designed growth factors [27]. The BSMV-VIGS method was previously used to explore the function of candidate genes of cereal crops under controlled environments [13,28,29]. Here, this method was used in field experiments to explore the function of TaRSR1. Humidity is a major growth factor that affects the efficiency of the BSMV-VIGS method [30]. To increase humidity, wheat plants grown under field conditions were covered with an arch plastic shed at the heading stage, and wheat plants in the shed were sprayed with distilled water at 1 d before inoculation. Thus, relative humidity in the arch plastic shed was close to 85.0%, which was suitable for BSMV-VIGS inoculation [30].
When a fragment of a plant gene is inserted into virus induced gene-silencing (VIGS) vector, a recombinant virus is formed, and after a plant host is infected, this recombinant virus can induce post-transcriptional gene silencing targeting both the virus RNA and homologous endogenous plant RNA sequences for degradation. Only plant barley stripe mosaic virus (BSMV), a tri-partite RNA genome comprising RNAα, RNAβ and RNAγ, has so far been developed into the VIGS vector, and there appears chlorosis induced by BSMV virus on the inoculated tissues [31,32]. In VIGS experimental design, at least one negative control VIGS construct containing GFP or β-glucuronidase gene (GUS) should be included, and appearance of chlorosis on the inoculated tissues suggests that BSMV virus is inoculated and the target genes are silenced by VIGS in plant host [33]. In this study, a fragment (284 bp) of the TaRSR1 gene (GenBank accession no. JX473823) was isolated (Figure S1), the BSMV-derived TaRSR1 silencing vector was constructed, which was used to inoculate the spikes of 45 plants grown in the arch plastic shed during the heading stage under field conditions. BSMV-derived green fluorescent protein (GFP)-silenced wheat plants were used as the control. Our data indicated that BSMV-VIGS-induced chlorosis appeared in the middle of the wheat spikes at 15 days after anthesis, after which it extended throughout the entire spikes and persisted until the mature grain stage (Figure 1A). This suggested that TaRSR1 was successfully silenced in these wheat spikes. Of the 45 BSMV-TaRSR1-infected or BSMV-GFP-infected wheat spikes, 25 and 22 showed chlorosis with silencing efficiencies of 55.6% and 48.9% (average of 52.5%), respectively (Figure 1B).
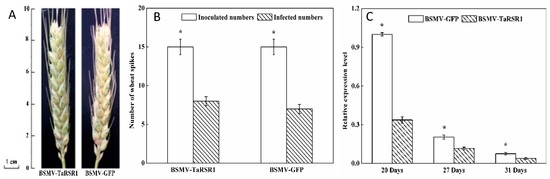
Figure 1.
Phenotypes (A); number (B); and TaRSR1 gene transcription levels (C) of the BSMV-TaRSR1-infected and BSMV-GFP-infected wheat spikes. Appearance of chlorosis on the inoculated wheat spikes at 22 days after anthesis suggests that BSMV virus has been infected and the target genes are successfully silenced by VIGS in this tissue. Efficiency of the BSMV-VIGS method was based on chlorosis phenotype, and the average values of the number of VIGS infection from 15 plants were considered as one replicate and three independent biological replicates were performed. Transcription levels at 20, 27 and 31 days after anthesis were measured by qPCR using Actin gene as internal control. Each value is the mean ± standard deviation of three independent biological replicates. Asterisks indicate significant differences (p < 0.05).
2.2. Transcription Levels of TaRSR1 in BSMV-TaRSR1 Infected Wheat Plants
These qualitative phenotypic results were further confirmed by quantitative analysis of the expression levels of the TaRSR1 gene using quantitative real-time PCR (qPCR) at 20, 27, and 31 days after anthesis. Wheat β-actin gene was used as an internal control. Similar data were obtained using the glyceraldehyde 3-phosphate dehydrogenase (GAPDH) gene as another internal control, which is shown in Figure S2. Our results showed that transcription levels of the TaRSR1 gene in the grains of BSMV-TaRSR1 infected spikes were markedly decreased by 43.5%–66.3% at the three sampling time points (Figure 1C). The degree of suppression was in accordance with previous data in wheat [34], thereby supporting the fact that the TaRSR1 gene is silenced in the grains of wheat plants grown under field conditions. In this study, the efficiency (52.5%) of the BSMV-VIGS method used in wheat plants under field conditions was far lower than that (90%) in experiments performed by Ma et al. [28] under greenhouse conditions. This lower efficiency may be due to the variable growth parameters under field conditions including humidity, light illumination, and temperature during the reproductive period of BSMV virus inoculation [30,35].
2.3. Starch Contents and One Thousand Kernel (1000-Kernel) Weights in BSMV-TaRSR1 Infected Wheat Plants
The starch contents, 1000-kernel weights, grain length and width in grains of BSMV-TaRSR1-infected wheat spikes were 30.0%, 19.5%, 14.8% and 8.2% higher, respectively, than those in BSMV-GFP-infected wheat spikes (Figure 2). These results suggest that TaRSR1 silencing resulted in enhanced starch synthesis in wheat grains; thus TaRSR1 may act as a negative regulator of starch synthesis, thereby playing a crucial role in the regulation of starch metabolism in bread wheat. The transcription levels of 26 starch synthesis-related enzyme genes in grains of BSMV-TaRSR1 and BSMV-GFP infected wheat spikes at the three sampling time points were also measured by qPCR, with the Actin gene used as an internal control (Figure 3, Figure 4 and Figure 5). Similar results were obtained using the GAPDH gene as another internal control, as shown in Figure S3.
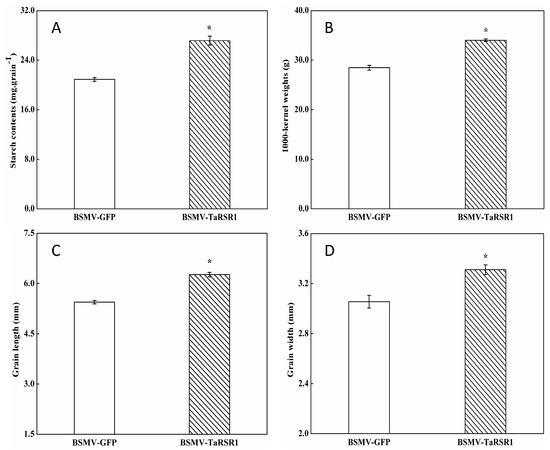
Figure 2.
Matured grain starch contents (A); 1000-kernel weights (B); length (C); and width (D) of BSMV-TaRSR1-infected and BSMV-GFP-infected wheat spikes. Each value is the mean ± standard deviation of three independent biological replicates. Asterisks indicate significant differences (p < 0.05).
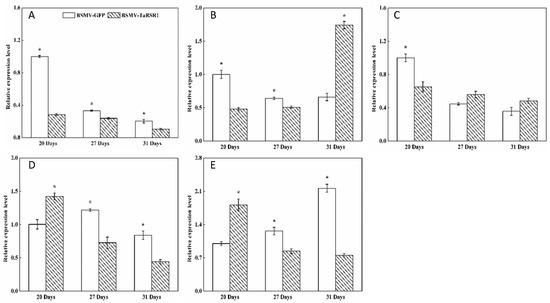
Figure 3.
Transcription levels of the AGPase starch synthesis-related enzyme genes in wheat grains of BSMV-TaRSR1-infected and BSMV-GFP-infected spikes at 20, 27 and 31 days after anthesis. Transcription levels were measured by qPCR using Actin gene as internal control. (A–E), transcription levels of TaAGPS1-a, TaAGPS1-b, TaAGPS2, TaAGPL1 and TaAGPL2 genes, respectively. Each value is the mean ± standard deviation of three independent biological replicates. Asterisks indicate significant differences (p < 0.05).
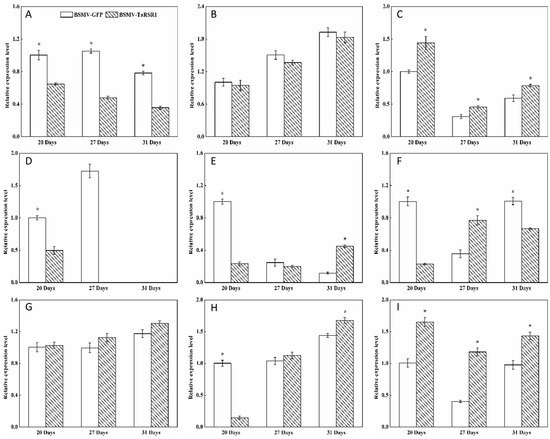
Figure 4.
Transcription levels of the GBSS and SS starch synthesis-related enzyme genes in wheat grains of BSMV-TaRSR1-infected and BSMV-GFP-infected spikes at 20, 27 and 31 days after anthesis. Transcription levels were measured by qPCR using Actin gene as internal control. (A–I), transcription levels of TaGBSSI, TaGBSSII, TaSSI, TaSSIIa, TaSSIIb, TaSSIIc, TaSSIIIa, TaSSIIIb and TaSSIV genes, respectively. Each value is the mean ± standard deviation of three independent biological replicates. Asterisks indicate significant differences (p < 0.05).
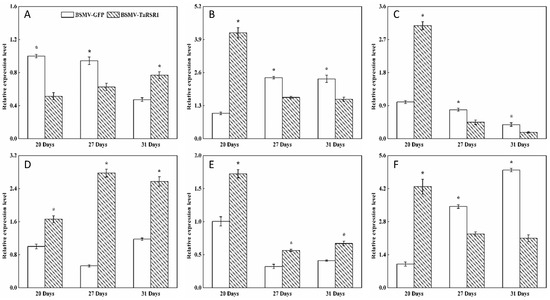
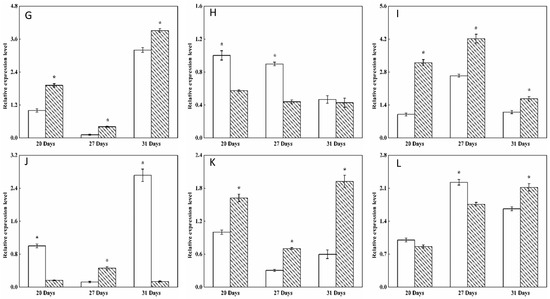
Figure 5.
Transcription levels of the branching enzyme (BE), debranching enzyme (DBE), phosphorylase (PHO) and disproportionating enzyme (DPE) starch synthesis-related enzyme genes in wheat grains of BSMV-TaRSR1-infected and BSMV-GFP-infected spikes at 20, 27 and 31 days after anthesis. Transcription levels were measured by qPCR using Actin gene as internal control. (A–L), transcription levels of TaBEI, TaBEIIa, TaBEIIb, TaBEIII, TaISA1, TaISA2, TaISA3, TaPUL, TaPHOL, TaPHOH, TaDPE1 and TaDPE2 genes, respectively. Each value is the mean ± standard deviation of three independent biological replicates. Asterisks indicate significant differences (p < 0.05).
2.4. Transcription Levels of 26 Starch Synthesis-Related Enzyme Genes in BSMV-TaRSR1 Infected Wheat Plants
Some transcription factors can temporally regulate the expression of target genes [36,37]. Our results showed that, in grains of BSMV-TaRSR1-infected wheat spikes, transcription levels of TaAGPS1-a (Figure 3A), TaAGPS2 (Figure 3C), TaGBSSI (Figure 4A), TaSSIIa (Figure 4D), and TaPUL (Figure 5H) genes were strongly downregulated or unchanged at one or more sampling time points. The transcription levels of TaGBSSII (Figure 4B) and TaSSIIIa (Figure 4G) did not show significant changes; however, TaSSI (Figure 4C), TaSSIV (Figure 4I), TaBEIII (Figure 5D), TaISA1 (Figure 5E), TaISA3 (Figure 5G), TaPHOL (Figure 5I), and TaDPE1 (Figure 5K) genes were markedly upregulated at each of the three sampling time points. TaAGPL1 (Figure 3D), TaAGPL2 (Figure 3E), TaBEIIa (Figure 5B), TaBEIIb (Figure 5C), and TaISA2 (Figure 5F) genes at 20 days; TaSSIIc (Figure 4F) and TaPHOH (Figure 5J) at 27 days; and TaAGPS1-b (Figure 3B), TaSSIIb (Figure 4E), TaSSIIIb (Figure 4H), TaBEI (Figure 5A), and TaDPE2 (Figure 5L) genes at 31 days were markedly induced, but their transcription levels were repressed or unchanged at the other two sampling time points. These results suggest that TaRSR1 negatively and temporally regulates the expression of several starch synthesis-related enzyme genes (TaAGPS1-b, TaAGPL1, TaAGPL2, TaSSI, TaSSIIb, TaSSIIc, TaSSIIIb, TaSSIV, TaBEI, TaBEIIa, TaBEIIb, TaBEIII, TaISA1, TaISA2, TaISA3, TaPHOL, TaPHOH, TaDPE1, and TaDPE2) and plays a role in wheat starch synthesis. The decreased transcription levels of these starch synthesis-related enzyme genes at one or more sampling time points may be due to the increased transcription of their isoform genes, or these genes may be positively regulated by TaRSR1; however, further studies are required. Similar expression patterns of starch synthesis-related enzyme genes have been reported in rice overexpressing the OsbZIP58 transcription factor [38].
The number (19) of regulated starch synthesis-related enzyme genes in the grains of BSMV-TaRSR1-infected wheat spikes was determined (Figure 3, Figure 4 and Figure 5), and found to be greater than that (11) of starch synthesis-related enzyme genes whose transcription levels showed a negative relationship with TaRSR1 [7]. In addition, only seven starch synthesis-related enzyme genes (TaAGPL1, TaSSI, TaSSIV, TaBEI, TaBEIIb, TaPHOL, and TaDPE1) had similar expression profiles in both experiments. The differences between these two studies may mainly be because the BSMV-VIGS method used in this study more accurately determined the function of target genes than the correlation analysis used in our previous study [7]. In addition, differences may be partially due to the different sampling time points between this study (21, 27, and 31 days after anthesis) and previous studies (5, 10, 15, 20, 25, 30, and 35 days after anthesis).
2.5. Putative Mechanism of TaRSR1 Regulation in Higher Plants
In higher plants, APETALA2/ethylene-responsive element binding protein (AP2/EREBP) is one of the largest families of transcription factors [39]. The AP2/EREBP superfamily is divided into AP2, ethylene-responsive transcription factor, and related ABI3/VP1 families [40,41]. The RSR1 transcription factor belongs to the AP2 group of the AP2/EREBP family, which is characterized by an AP2 domain [20]. AP2/EREBP can recognize the GCAC(A/G)N(A/T)TCCC(A/G)ANG(C/T), DRE, GCC, JERE, CAACA, ACGT, and ACTCAT boxes in the promoters of target genes [38,42,43,44,45,46]. These boxes have been identified in the promoters of some upregulated starch synthesis-related enzyme genes in grains of BSMV-TaRSR1-infected wheat spikes. For example, ACGT and ACTCAT boxes were found in the promoters of GBSSI and BEI genes of rice [38,45], and AGPS1, SSI, SSIIIa, and ISA1 genes of maize, respectively [46]. Therefore, TaRSR1 may directly and negatively regulate the expression of some starch synthesis-related enzyme genes.
3. Materials and Methods
3.1. Plant Materials
Bread wheat cultivar (Bainong 207, a high-yield wheat cultivar developed by Henan University of Science and Technology, Xinxiang, China) was grown in the Agricultural and Experimental Farm of Henan Agricultural University (Zhengzhou, China, N34°9′, E113°6′) during the wheat growing season (October 2014 to May 2015). Type of soil, organic matter, nitrogen, phosphorus, and potassium, fertilizer application, and planting management were reported in our previous study [8]. Seeds were sown on 16 October 2014, and the plant density was adjusted to 150 plants/m2 at the three-leaf stage. The plot dimension was 4 m × 8 m, and plots were separated by a ridge (20 cm in width). Wheat plants with a 15 m2 area (length 5.0 m × width 3.0 m) were covered with an arch plastic shed (3.0 m width × 1.5 m height × 0.5 m arch height) during the heading stage, and light transmittance of the polyvinylchlorid (PVC) film (Auspicious dragon import and export trade company, Zhejiang, China) was 75%. Ninety wheat spikes with the same anthesis date (18 April 2015) in the arch plastic shed, which was built with concrete pillars and bamboo poles, were marked. At 1 day before inoculation, the surrounding PVC plastic film was covered with soil to prevent water loss and increase humidity in the shed. Developing grains in the middle of BSMV-infected spikes were sampled at 20, 27, and 31 days after anthesis at 11:00 in the morning. The sampled grains were rapidly frozen in liquid nitrogen for 2 min and then stored at −80 °C for transcription analysis of starch synthesis-related enzyme genes. During the mature stage, grains of BSMV-infected wheat plants were harvested and dried to a constant weight at 70 °C in the oven for starch determination.
3.2. Construction of BSMV-Derived Vectors
Because of asymmetric expression patterns of two or more homoeoalleles in allopolyploid plants [17], only one copy (Genome location: 1DS) of TaRSR1 could be expressed in grains of bread wheat in our previous study [7]. In this study, the full-length cDNA sequence of this copy, its conserved cDNA fragment (284 bp) used to construct the BSMV-TaRSR1 vector, and the other two copies (1AS and 1BS) in bread wheat A and B subgenomes are indicated in Figure S1. Because there are high similarities in cDNA or protein sequences among three or more copies of a gene in allohexaploid bread wheat, VIGS method usually results in the simultaneous silencing of three copies for the targeted gene in bread wheat [47]. In this study, similarly, the chosen cDNA fragment of TaRSR1 (1DS) for VIGS silencing also had high similarities (99.1%) to those of the other two copies (Figure S1), and thus, all three copies of TaRSR1 gene could be simultaneously silenced in this study. The forward and reverse primers were 5′-CTAGCTAGCGCCTGCAACTCCACCATG-3′ and 5′-CTAGCTAGCCGGACGATGACGACGAGA-3′, respectively. The amplified cDNA sequence of the TaRSR1 gene fragment was sequenced by Applied Biosystems and the correct TaRSR1 sequence was cloned into the BSMV plasmid using an Nhe I site. The schematic construction of recombinant BSMV-TaRSR1 vector is described in Figure S4. Similar to a study by Pacak et al. [48], GFP was used as a control in this study.
3.3. In Vitro Transcription of Viral RNAs and Plant Inoculation
The in vitro transcription of viral RNAs was performed as described by Ma et al. [49]. The RNA-α, RNA-β, and RNA-γ-derivative clones were linearized with Mlu I and Spe I, respectively. RNA synthesis was performed using the RiboMAX™ Large Scale RNA Production System-T7 kit (Promega, Fitchburg, WI, USA) according to the manufacturer’s instructions. The RNA-α, RNA-β, and RNA-γ (or its derivative) transcripts were mixed in a 1:1:1 ratio, and subsequently diluted with nine volumes of DEPC water. In addition, 12 volumes of 2× GKP buffer were added to the diluted transcript mixture for subsequent inoculation. To increase humidity, wheat plants in the shed were sprayed with distilled water until it ran off at 1 day before inoculation. Virus inoculation was performed on the spike of wheat plants during the heading stage and was accomplished by gently rubbing the spike surface five times from the central section of wheat spikes to the tip and then from base to tip. Wheat spikes were inoculated with BSMV-TaRSR1 or BSMV-GFP transcript mixtures with a gloved finger (Figure S5). A total of 45 plants were inoculated with BSMV-TaRSR1 or BSMV-GFP, respectively, and 20 µL BSMV-TaRSR1 or BSMV-GFP transcript mixtures were used for each spike. After inoculation, BSMV-inoculated spikes were fog-sprayed with nuclease-free water and covered with plastic film to maintain high relative humidity (85.0%) for 24 h, and wheat plants remained covered with the arch plastic shed for 7 days and two sides were opened during the daytime due to high temperatures and closed at night to maintain high humidity.
The manufacturer’s instructions were followed and treated (Takara Biotechnology (Dalian) Co., Ltd., Dalian, China) to remove contaminating. First-strand cDNAs were synthesized from 2 µg of total RNA using Super-Script II reverse transcriptase (Invitrogen, Carlsbad, CA, USA).
3.4. Determination of Transcription Levels of TaRSR1 and 26 Starch Synthesis-Related Enzyme Genes
Total RNA was extracted from the grains of BSMV-TaRSR1-infected or BSMV-GFP-infected wheat spikes with TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and genomic DNA was removed with RNase-free DNase I. The first strand cDNA was synthesized from 2 µg of total RNA using the Revert Aid™ First Strand cDNA Synthesis Kit (Fermentas, Burlington, ON, Canada). Transcription levels of TaRSR1 and 26 starch synthesis-related enzyme genes were measured using the qPCR method, which was performed on a Light Cycler 480 Real-Time PCR System (Roche Diagnostics Ltd., West Sussex, UK) using an SYBR Premix Ex Taq (Perfect Real Time) Kit (Takara Biotechnology Co., Ltd., Dalian, China). Each reaction consisted of 10 µL SYBR Green Supermix (2×), 1 µL diluted cDNA, 0.5 µL forward primer, and 0.5 µL reserve primer, in a total volume of 20 µL. The relative transcription levels of each starch synthesis-related enzyme gene were calculated using the 2−ΔΔCt method, with wheat glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (GenBank accession no. EF592180) and β-actin (GenBank accession no. AB181991) genes as the internal controls. All of primers used for qPCR are indicated in Table S1.
3.5. Assays on Wheat Grains Parameters
Starch contents were measured according to the methods of Zhao et al. [50]. Grain length and width were determined by calculating ten grains from each plant. The 1000-kernel weight was determined by counting four replicates of 100-grain samples independently on an electronic balance. Fifty seeds were analyzed for seed size, and data are presented as means ± SD.
3.6. Statistical Analysis
Data were statistically analyzed using Student’s t-test with SPSS Statistics 17.0 (SPSS Inc., Chicago, IL, USA) and significant differences among group means was tested by using Duncan’s multiple range test at p < 0.05 level. The averaged values of three spikes were considered one replicate and three independent biological replicates were performed. All of the recorded values represent the means of the results from three independent biological replicates.
4. Conclusions
In this study, the TaRSR1 transcription factor was silenced in grains of wheat plants using the BSMV-VIGS method under field conditions. In BSMV-TaRSR1-infected wheat plants, starch contents, 1000-kernel weights, grain length and width markedly increased in mature grains. In addition, the transcription levels of 19 starch synthesis-related enzyme genes were significantly upregulated at one or more sampling time points in the grains of the infected wheat plants. These results indicate that TaRSR1 acts as a negative regulator and temporally regulates the expression of some starch synthesis-related enzyme genes in bread wheat.
Supplementary Materials
Supplementary materials can be found at www.mdpi.com/1422-0067/17/9/1557/s1.
Acknowledgments
The BSMV vectors (BSMV-α, BSMV-β and γ-GFP) were kindly provided by Daowen Wang (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing, China). This study was financially supported by the National Natural Science Foundation of China (31571575), the National Transgenic Major Project (2016ZX08002-003-04), the Program for Science & Technology Innovation Talents in Universities of Henan Province (15HASIT029), the Independent Item of the National Key Laboratory of Wheat and Maize Crop Science (SKL2014ZH-03), and the Open Item of the State Key Laboratory of Crop Stress Biology for Arid Areas (CSBAA2015010).
Author Contributions
Guoyu Liu and Yufang Wu performed the main experimental work, Mengjun Xu, Tian Gao and Lina Wang participated in data analysis, Pengfei Wang and Tiancai Guo designed the experiments. Guozhang Kang and Guoyu Liu wrote this manuscript, and Tiancai Guo and Guozhang Kang provided the financial support. All authors have read and approved the final manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Stamova, B.S.; Laudencia-Chingcuanco, D.; Beckles, D.M. Transcriptomic analysis of starch biosynthesis in the developing grain of hexaploid wheat. Int. J. Plant Genom. 2009, 2009, 407426. [Google Scholar] [CrossRef] [PubMed]
- Jeon, J.S.; Ryoo, N.; Hahn, T.R.; Walia, H.; Nakamura, Y. Starch biosynthesis in cereal endosperm. Plant Physiol. Biochem. 2010, 48, 383–392. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Chen, Y.; Tong, C.; Huang, Y.; Xu, F.; Li, K.; Corke, H.; Sun, M.; Bao, J. Association mapping of starch physicochemical properties with starch synthesis-related gene markers in nonwaxy rice (Oryza Sativa L.). Mol. Breed. 2014, 34, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Satoh, H.; Shibahara, K.; Tokunaga, T.; Nishi, A.; Tasaki, M.; Hwang, S.K.; Okita, T.W.; Kaneko, N.; Fujita, N.; Yoshida, M.; et al. Mutation of the plastidial a-glucan phosphorylase gene in rice affects the synthesis and structure of starch in the endosperm. Plant Cell 2008, 20, 1833–1849. [Google Scholar] [CrossRef] [PubMed]
- Ohdan, T.; Francisco, P.B.J.; Sawada, T.; Hirose, T.; Terao, T.; Satoh, H.; Nakamura, Y. Expression profiling of genes involved in starch synthesis in sink and source organs of rice. J. Exp. Bot. 2005, 56, 3229–3244. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.B.; Pan, X.X.; Jiang, H.W.; Wu, G.J. Comparison of the starch synthesis genes between maize and rice: Copies, chromosome location and expression divergence. Theor. Appl. Genet. 2009, 119, 815–825. [Google Scholar] [CrossRef] [PubMed]
- Kang, G.; Xu, W.; Liu, G.; Peng, X.; Guo, T. Comprehensive analysis on the transcript levels of starch synthesis genes and RSR1 transcript factor in endosperm of wheat. Genome 2013, 56, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Kang, G.; Peng, X.; Wang, L.; Yang, Y.; Wang, C.; Zhu, Y.; Guo, T. Ultrastructural observation of mesophyll cells and temporal expression profiles of the genes involved in transitory starch metabolism in flag leaves of wheat after anthesis. Physiol. Plant 2015, 153, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Radchuk, V.V.; Borisjuk, L.; Sreenivasulu, N.; Merx, K.; Mock, H.P.; Rolletschek, H.; Wobus, U.; Weschke, W. Spatiotemporal profiling of starch biosynthesis and degradation in the developing barley grain. Plant Physiol. 2009, 150, 190–204. [Google Scholar] [CrossRef] [PubMed]
- Tombuloglu, H.; Kekec, G.; Sakcali, M.S.; Unver, T. Transcriptome-wide identification of R2R3-MYB transcription factors in barley with their boron responsive expression analysis. Mol. Genet. Genom. 2013, 288, 141–155. [Google Scholar] [CrossRef] [PubMed]
- Quan, R.D.; Hu, S.J.; Zhang, Z.L.; Zhang, H.W.; Zhang, Z.J.; Huang, R.F. Overexpression of an ERF transcription factor TSRF1 improves rice drought tolerance. Plant Biotechnol. J. 2010, 8, 476–488. [Google Scholar] [CrossRef] [PubMed]
- Morran, S.; Eini, O.; Pyvovarenko, T.; Parent, B.; Singh, R.; Ismagul, A.; Eliby, S.; Shirley, N.; Langridge, P.; Lopato, S. Improvement of stress tolerance of wheat and barley by modulation of expression of DREB/CBF factor. Plant Biotechnol. J. 2011, 9, 230–249. [Google Scholar] [CrossRef] [PubMed]
- Rong, W.; Qi, L.; Wang, A.; Ye, X.; Du, L.; Liang, H.; Xin, Z.; Zhang, Z. The ERF transcription factor TaERF3 promotes tolerance to salt and drought stresses in wheat. Plant Biotechnol. J. 2014, 12, 468–479. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, M.; Bonnett, D.; Chapman, S.C.; Furbank, R.T.; Manes, Y.; Mather, D.E.; Parry, M.A.J. Raising yield potential of wheat. I. Overview of a consortium approach and breeding strategies. J. Exp. Bot. 2011, 62, 439–453. [Google Scholar] [CrossRef] [PubMed]
- Feldman, M.; Levy, A.A.; Fahima, T.; Korol, A. Genomic asymmetry in allopolyploid plants: Wheat as a model. J. Exp. Bot. 2012, 63, 5045–5059. [Google Scholar] [CrossRef] [PubMed]
- Pfeifer, M.; Kugler, K.G.; Sandve, S.R.; Zhan, B.; Rudi, H.; Hvidsten, T.R.; Mayer, K.F.; Olsen, O.A.; International Wheat Genome Sequencing Consortium. Genome interplay in the grain transcriptome of hexaploid bread wheat. Science 2014, 345, 1250091. [Google Scholar] [CrossRef] [PubMed]
- Flagel, L.E.; Chen, L.; Chaudhary, B.; Wendel, J.F. Coordinated and fine-scale control of homoeologous gene expression in allotetraploid cotton. J. Hered. 2009, 100, 487–490. [Google Scholar] [CrossRef] [PubMed]
- International Wheat Genome Sequencing Consortium. A chromosome-based draft sequence of the hexaploid bread wheat genome. Science 2014, 345, 1251788. [Google Scholar]
- Pingault, L.; Choulet, F.; Alberti, A.; Glover, N.; Wincker, P.; Feuillet, C.; Paux, E. Deep transcriptome sequencing provides new insights into the structural and functional organization of the wheat genome. Genome Biol. 2015, 16, 29. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Soltis, P.S.; Wall, K.; Soltis, D.E. Phylogeny and domain Evolution in the APETALA2-likegenefamily. Mol. Biol. Evol. 2006, 23, 107–120. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.Y.; Lee, J.W.; Moon, S.; Park, S.Y.; An, G. The rice heterochronic gene SUPERNUMERARY BRACT regulates the transition from spikelet meristem to floral meristem. Plant J. 2006, 49, 64–78. [Google Scholar] [CrossRef] [PubMed]
- Fu, F.F.; Xue, H.W. Coexpression analysis identifies rice starch regulator1, a rice AP2/EREBP family transcription factor, as a novel rice starch biosynthesis regulator. Plant Physiol. 2010, 154, 927–938. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Sparks, C.; Amoah, B.; Jones, H.D. Factors influencing successful Agrobacterium-mediated genetic transformation of wheat. Plant Cell Rep. 2003, 21, 659–668. [Google Scholar] [PubMed]
- Zhao, T.; Zhao, S.; Chen, H.; Zhao, Q.; Hu, Z.; Hou, B.; Xia, G. Transgenic wheat frogeny resistant to powdery mildew generated by Agrobacterium inoculumto the basal portion of wheat seedling. Plant Cell Rep. 2006, 25, 1199–1204. [Google Scholar] [CrossRef] [PubMed]
- Bhullar, N.K.; Keller, B. Virus induced gene silencing in wheat: A review. In Advances in Biotechnology; Bentham Science Publishers Ltd.: Hilversum, The Netherlands, 2010; pp. 185–203. [Google Scholar]
- Tufan, H.A.; Stefanato, F.L.; McGrann, G.R.D.; MacCormack, R.; Boyd, L.A. The barley stripe mosaic virus system used for virus-induced gene silencing in cereals differentially affects susceptibility to fungal pathogens in wheat. J. Plant Physiol. 2011, 168, 990–994. [Google Scholar] [CrossRef] [PubMed]
- Pfleeger, T.; Olszyk, D.; Lee, E.H.; Plocher, M. Comparing effects of low levels of herbicides on greenhouse and field-grown potatoes (Solanum tuberosum L.), soybeans (Glycine max L.), and peas (Pisum sativum L.). Environ. Toxicol. Chem. 2011, 30, 455–468. [Google Scholar] [CrossRef] [PubMed]
- Ma, M.; Yan, Y.; Huang, L.; Chen, M.; Zhao, H. Virus-induced gene-silencing in wheat spikes and grains and its application in functional analysis of HMW-GS-encoding genes. BMC Plant Biol. 2012, 12, 141. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Li, H.; Chen, Y.; Zheng, Q.; Li, B.; Li, Z. TaSCL14, a novel wheat (Triticum aestivum L.) GRAS gene, regulates plant growth, photosynthesis, tolerance to photooxidative stress, and senescence. J. Genet. Genom. 2015, 42, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Renner, T.; Bragg, J.; Driscoll, H.E.; Jackson, A.O.; Specht, C.D. Virus-induced gene silencing in the Culinary Ginger (Zingiber officinale): An effective mechanism for down-regulating gene expression in tropical monocots. Mol. Plant 2009, 2, 1084–1094. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, P.M.; Wang, M.B.; Lough, T. Gene silencing as an adaptive defence against viruses. Nature 2001, 411, 834–842. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.S.; Rudd, J.J.; Kanyuka, K. Virus induced gene silencing (VIGS) for functional analysis of wheat genes involved in Zymoseptoria tritici susceptibility and resistance. Fungal Genet. Biol. 2015, 79, 84–88. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.S.; Hammond-Kosack, K.E.; Kanyuka, K. Barley stripe mosaic virus-mediated tools for investigating gene function in cereal plants and their pathogens: Virus-induced gene silencing, host-mediated gene silencing, and virus-mediated overexpression of heterologous proteins. Plant Physiol. 2012, 160, 582–590. [Google Scholar] [CrossRef] [PubMed]
- Scofield, S.R.; Huang, L.; Brandt, A.S.; Gill, B.S. Development of a virus-induced gene-silencing system for hexaploid wheat and its use in functional analysis of the lr21-mediated leaf rust resistance pathway. Plant Physiol. 2005, 138, 2165–2173. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Chen, X.; Zhao, H.; Yu, S.; Long, H.; Deng, G.; Pan, Z.; Yu, M. The impacts of BSMV on vegetative growth and water status in hulless barley (Hordeum vulgarevar nudum) in VIGS study. Acta Soc. Bot. Pol. 2015, 84, 43–51. [Google Scholar] [CrossRef]
- Pearce, S.; Tabbita, F.; Cantu, D.; Buffalo, V.; Avni, R.; Vazquez-Gross, H.; Zhao, R.; Conley, C.J.; Distelfeld, A.; Dubcovksy, J. Regulation of Zn and Fe transporters by the GPC1 gene during early wheat monocarpic senescence. BMC Plant Biol. 2014, 14, 368. [Google Scholar] [CrossRef] [PubMed]
- Sato, H.; Todaka, D.; Kudo, M.; Mizoi, J.; Kidokoro, S.; Zhao, Yu.; Shinozaki, K.; Yamaguchi-Shinozaki, K. The Arabidopsis transcriptional regulator DPB3-1 enhances heat stress tolerance without growth retardation in rice. Plant Biotechnol. J. 2016, 1–12. [Google Scholar]
- Wang, J.C.; Xu, H.; Zhu, Y.; Liu, Q.Q.; Cai, X.L. OsbZIP58, a basic leucine zipper transcription factor, regulates starch biosynthesis in rice endosperm. J. Exp. Bot. 2013, 64, 3453–3466. [Google Scholar] [CrossRef] [PubMed]
- Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 2012, 1819, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Nakano, T.; Suzuki, K.; Fujimura, T.; Shinshi, H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 2006, 140, 411–432. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.L.; Lv, H.; Li, L.; Liu, J.; Mu, S.H.; Li, X.P.; Gao, J. Genome-wide analysis of the AP2/ERF transcription factors family and the expression patterns of DREB genes in Moso Bamboo (Phyllostachys edulis). PLoS ONE 2015, 10, e0126657. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.L. AP2/EREBP: A special transcription factor family in plant. J. Qinghai Norm. Univ. 2006, 3, 80–83, In Chinese with English Abstract. [Google Scholar]
- Gong, W.; He, K.; Covington, M.; Dinesh-Kumar, S.P.; Snyder, M.; Harme, S.L.; Zhu, Y.X.; Deng, X.W. The development of protein microarrays and their applications in DNA-protein and protein-protein interaction analyses of Arabidopsis transcription factors. Mol. Plant 2008, 1, 27–41. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Je, J.Y.; Song, C.; Hwang, J.E.; Lim, C.O. A proximal promoter region of Arabidopsis DREB2C confers tissue-specific expression under heat stress. J. Integr. Plant Biol. 2012, 54, 640–651. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Xie, D.L.; Wang, Z.Y.; Hong, M.M. Interaction of rice bZIP protein REB with the 5′-upstream region of both rice sbe1 gene and waxy gene. Chin. Sci. Bull. 2002, 47, 310–314. [Google Scholar] [CrossRef]
- Chen, J.; Yi, Q.; Cao, Y.; Wei, B.; Zheng, L.J.; Xiao, Q.L.; Xie, Y.; Gu, Y.; Li, Y.P.; Huang, H.H.; et al. ZmbZIP91 regulates expression of starch synthesis-related genes by binding to ACTCAT elements in their promoters. J. Exp. Bot. 2016, 67, 1327–1338. [Google Scholar] [CrossRef] [PubMed]
- Ma, M.; Wang, Q.; Li, Z.; Cheng, H.; Li, Z.; Liu, X.; Song, W.; Appels, R.; Zhao, H. Expression of TaCYP78A3, a gene encoding cytochrome P450CYP78A3 protein in wheat (Triticum aestivum L.), affects seed size. Plant J. 2015, 83, 312–325. [Google Scholar] [CrossRef] [PubMed]
- Pacak, A.; Geisler, K.; Jørgensen, B.; Barciszewska-Pacak, M.; Nilsson, L.; Nielsen, T.H.; Johansen, E.; Grønlund, M.; Jakobsen, I.; Albrechtsen, M. Investigations of barley stripe mosaic virus as a gene silencing vector in barley roots and in Brachypodium distachyon and oat. Plant Methods 2010, 6, 26. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.Z.; Liu, G.Q.; Li, C.W.; Kang, G.Z.; Guo, T.C. Identification of the TaBTF3 gene in wheat (Triticum aestivum L.) and the effect of its silencing on wheat chloroplast, mitochondria and mesophyll cell development. Biochem. Biophys. Res. Commun. 2012, 426, 608–614. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Dai, T.; Jiang, D.; Cao, W. Effects of high temperature on key enzymes involved in starch and protein formation in grains of two wheat cultivars. J. Agron. Crop. Sci. 2008, 194, 47–54. [Google Scholar] [CrossRef]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).