Abstract
The NAD+-dependent deacetylases Sirt1 and Sirt2 mediate cellular stress responses and are highly expressed in vascular endothelial cells. In contrast to the well-documented protective actions of Sirt1, the role of endothelial Sirt2 remains unknown. Using cDNA microarray and PCR validation, we examined global gene expression changes in response to Sirt2 knock down in primary human umbilical vein endothelial cells under oxidative stress. We found that Sirt2 knock down changed expression of 340 genes, which are mainly involved in cellular processes including actin binding, cellular amino acid metabolic process, transmembrane receptor protein serine/threonine kinase signaling, ferrous iron transport, protein transport and localization, cell morphogenesis, and functions associated with endosome membrane and the trans-Golgi network. These genes and associated functions were largely non-overlapping with those altered by Sirt1 knock down. Moreover, we showed that pharmacological inhibition of Sirt2 attenuated oxidant-induced cell toxicity in endothelial cells. These suggest that Sirt2 is functionally important in endothelial cells under oxidative stress, and may have a primarily distinct role as compared to Sirt1. Our results may provide a basis for future studies aiming to dissect the specific signaling pathway(s) that mediates specific Sirt2 functions in endothelial cells.
Abbreviations
| eNOS | nitric oxide synthase |
| NF-κB | nuclear factor-κB |
| HUVEC | human umbilical vein endothelial cell |
| siRNA | small interfering RNA |
| qPCR | quantitative polymerase chain reaction |
| GO | Gene Ontology |
| GPx | glutathione peroxidase |
| SOD | superoxide dismutase |
1. Introduction
Mammalian Sirt proteins (Sirt1 to Sirt7) are orthologues of the yeast SIR2 gene product, an NAD+-dependent class III histone deacetylase [1–4]. All Sirt proteins contain a conserved NAD+-dependent catalytic core domain of ~275 amino acids [3,4]. Among the seven Sirt proteins identified, Sirt1, 2, and 3 have the highest homology to yeast Sir2 and all exhibit specific protein deacetylase activity [5,6]. In addition to their important roles in aging and metabolic regulation, different studies have suggested that Sirt is also involved in modulating cardiovascular physiology and disease [1,7]. In the heart, for example, Sirt1 and Sirt3 have been implicated in promoting cardiomyocyte survival and preventing cardiac remodeling in response to different stress stimuli [8]. In blood vessels, activation of Sirt functions, especially those of Sirt1, is associated with multiple beneficial effects such as preventing vascular cell senescence, suppressing inflammation, decreasing cellular oxidative stress, and promoting vascular regeneration [1,7]. Moreover, Sirt may also exert cardiovascular protective actions by improving global glucose and lipid metabolism [8].
Endothelial cells have a pivotal role in maintaining the homeostasis of blood vessels. Endothelial dysfunction is recognized as a major cellular basis of the development of many cardiovascular diseases such as hypertension, atherosclerosis and heart failure [9]. Several lines of evidence have indicated that Sirt1 has an important role in modulating endothelial cell functions. In particular, Sirt1 physically interacts with and deacetylates endothelial nitric oxide synthase (eNOS), leading to enhanced eNOS activation [10]. Both in vitro and in vivo experiments revealed that activation of Sirt1 function led to increases in nitric oxide production and endothelium-dependent vasorelaxation, decreased inflammatory reactions in endothelial cells, and suppressed endothelial cell apoptosis and senescence [1,11–14].
Several Sirt members have pivotal roles in modulating cellular stress responses [2,6]. Under oxidative stress, Sirt1 exhibited broad cytoprotective effects in endothelial cells [7,15–19]. The molecular mechanisms by which Sirt1 produces these cytoprotective actions are not totally understood, while current evidence indicates that activation of the FoxO family members by Sirt1 is likely to be a major signaling route [2,6]. In contrast to Sirt1, the biological functions of Sirt2 in endothelial cells remain unknown [1]. Results from previous studies in non-endothelial cells indicate that the effects of Sirt2 on cell viability under stress are variable and appear to be cell type- and context-dependent [20–27]. Currently, research efforts have been made in the development of selective Sirt2 inhibitors, which may be used as novel chemotherapy agents [28]. Hence, it is important to determine whether and how Sirt2 is involved in modulating endothelial cell homeostasis under stress conditions. Like Sirt1, Sirt2 expression is also responsive to oxidative stress [24]. Moreover, Sirt2 and Sirt1 share a number of common substrates, including FoxO1, FoxO3, nuclear factor (NF)-κB, histone H3 and p300 [4,8,24,29–32]. These results prompted us to hypothesize that Sirt2 may also have critical functions in endothelial cells under oxidative stress. Therefore, in the present study we aim to examine the importance of endothelial Sirt2 on a systematic biology basis, by characterizing global gene expression changes after Sirt2 knock down in primary human umbilical vein endothelial cells (HUVECs) using mRNA microarray, an approach that has been used to delineate Sirt1 functions at the whole genome level [33,34].
2. Results and Discussion
To induce oxidative stress in cultured endothelial cells, we treated the cells with H2O2 at 300 μM for 6 h. Induction of a cellular stress response under this condition was demonstrated by the upregulation of detoxification enzymes glutathione peroxidase (GPx) and superoxide dismutases (SOD) as measured by qPCR (Figure 1A). We also confirmed that this treatment protocol did not induce obvious cytotoxic effects as assessed by a MTS cell viability assay (Figure 1B). A previous study demonstrated that Sirt2 expression was upregulated upon oxidant stimulation in adipocytes [24]. To clarify whether this is also the case in endothelial cells, we measured Sirt2 expression with qPCR in H2O2 challenged cells. We found that as compared to untreated cells, H2O2 increased Sirt2 expression by ~2 fold (data not shown). To demonstrate the intracellular localization of Sirt2 and in endothelial cells, we performed immunofluorescence staining. As shown in Figure 1C, the majority of Sirt2 showed a cytosolic localization, which was in contrast to Sirt1, which was mainly nuclear.
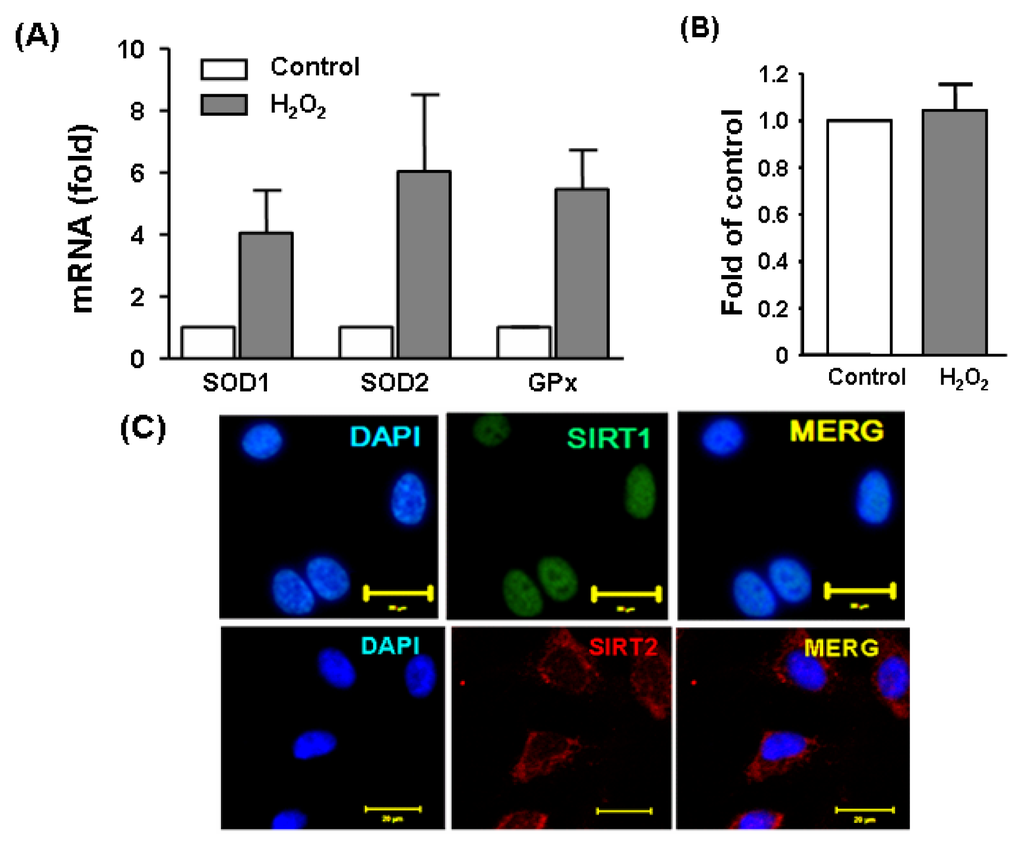
Figure 1.
(A) An oxidative stress response induced by H2O2 (300 μM for 6 h) in cultured human umbilical vein endothelial cells (HUVECs), as revealed by the upregulation of glutathione peroxidase (GPx) and superoxide dismutases (SOD) as measured by qPCR; (B) H2O2 treatment at 300 μM did not result in obvious cytotoxicity in the present experimental system. Cell viability was assessed with a MTS-based assay. Data are mean ± SEM, n = 3–4; (C) pseudo-colored immunofluorescence images showing the intracellular localization of Sirt1 and Sirt2 in untreated HUVEC. Nuclei were counter stained with DAPI (blue). Bar = 20 μm. MERG, merged image.
To clarify the functions of Sirt2 in endothelial cells at the genome level, we performed microarray experiments comparing the global gene expression profiles between control and Sirt2i cells. The efficiency of siRNA-mediated gene knock down was confirmed by qPCR and Western blot (Figure 2). We showed that under oxidative stress conditions, knock down of Sirt2 significantly changed the expression level of 340 genes, with 152 being upregulated and 188 downregulated (Figure 3A and Table 1). GO analysis of the Sirt2-sensitive genes showed that these genes were mainly involved in cellular processes related to actin binding, cellular amino acid metabolic process, transmembrane receptor protein serine/threonine kinase signaling pathways, ferrous iron transport, protein transport and localization, cell morphogenesis involved in differentiation, and functions associated with endosome membrane and the trans-Golgi network.
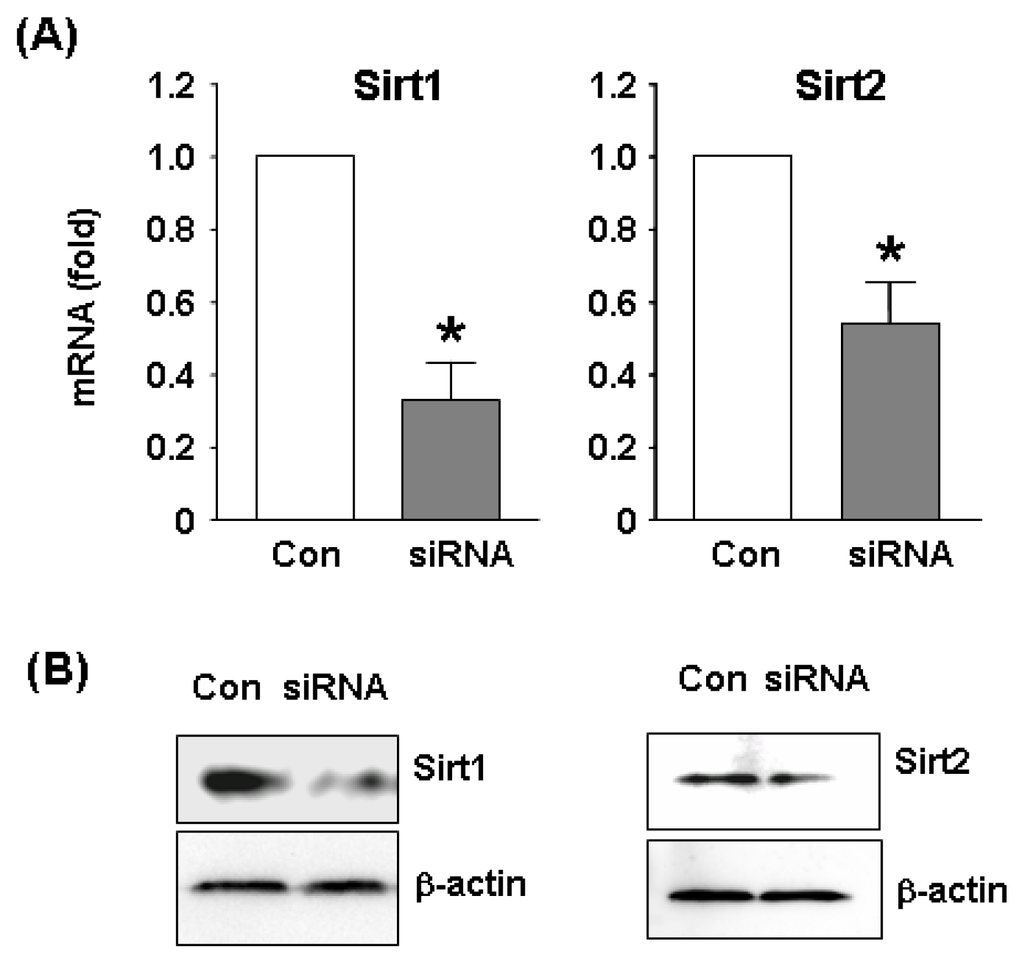
Figure 2.
(A) qPCR and (B) Western blot results showing gene silencing efficiency of siRNA sequences targeting Sirt1 or Sirt2 in H2O2-treated HUVECs. A non-specific siRNA was used as control. Data are mean ± SEM. * p < 0.05, student’s t-test, n = 4–5. Western blots were representative images from two independent experiments.
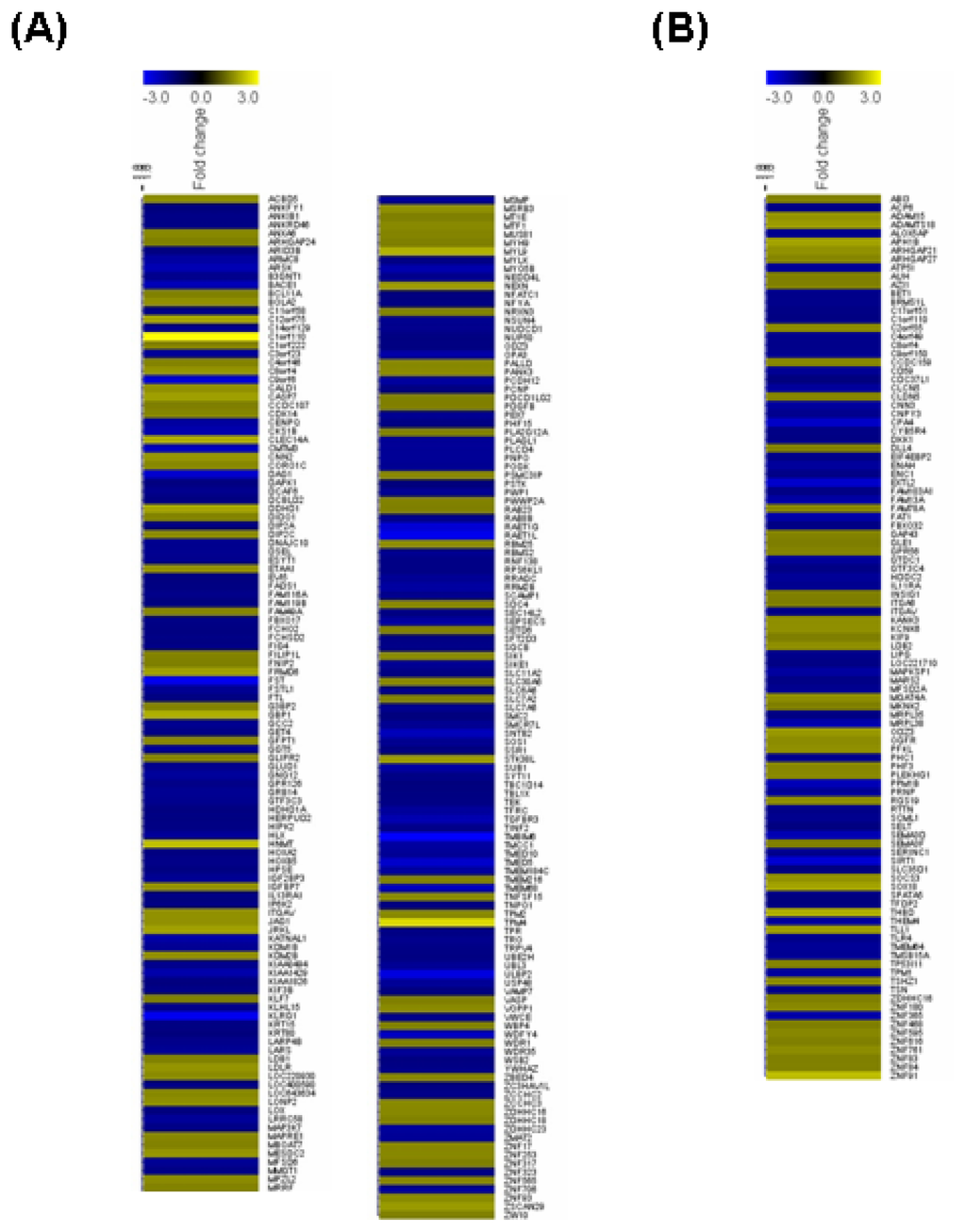
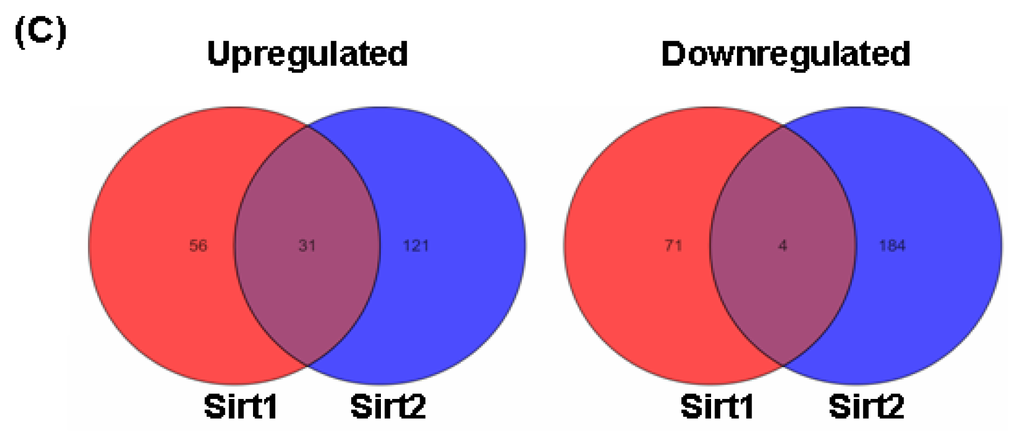
Figure 3.
Heat map diagrams illustrating the significantly changed (p < 0.05 with a fold change value >1.5 as compared to control cells) genes in HUVECs with gene silencing of (A) Sirt2 and (B) Sirt1, determined by Affymetrix Human Genome U219 Array (n = 3 biological replicates each); (C) Venn graphs showing the number of genes up- and downregulated by Sirt1 or Sirt2 gene silencing. A high-resolution version for Figure 3A,B is available online.

Table 1.
List of differentially expressed genes after Sirt1 or Sirt2 gene silencing (read the entire table column-wise).
To confirm that the altered gene expression after Sirt2 knock down was not caused by non-specific off target effects, we run a parallel experiment by knocking down Sirt1 using the same protocol. Sirt1 gene knock down induced significant alterations of expression of 162 genes (87 upregulated and 75 downregulated) (Figure 3B and Table 1). Among the upregulated genes with Sirt1i, only 31 (36%) overlapped with those changed in Sirt2i cells (20% of those changed in Sirt2i cells). Similarly, among the downregulated genes, only 4 (5%) overlapped with those changed in Sirt2i cells (2% of those changed in Sirt2i cells) (Figure 3C). GO analysis of the Sirt1-sensitive genes showed that these genes were mainly involved in cellular processes related to actin binding, ion binding, endoplasmic reticulum, cellular macromolecule biosynthetic process, cytoskeletal protein binding, and Golgi apparatus, of which the majority was distinct from those related to Sirt2. Further analysis of the differentially expressed genes in relation to disease processes with IPA software revealed that Sirt2-sensitive genes were enriched in categories including infectious disease, connective tissue disorders, developmental disorder, skeletal and muscular disorders, and cardiovascular disease. In comparison, Sirt1-sensitive genes were mainly enriched in categories including cardiovascular disease, inflammatory response, cancer, organismal injury and abnormalities, and connective tissue disorders. We also compared the two sets of cellular pathways significantly over-represented by Sirt1- or Sirt2-sensitive genes respectively, and found that the pathways affected by Sirt1 were primarily distinct from those affected by Sirt2 (Figure 4A). Moreover, IPA-Tox analysis revealed that Sirt1i and Sirt2i exhibited a discrete pattern of gene enrichment in categories of biological mechanisms that were related to toxicity responses (Figure 4B).
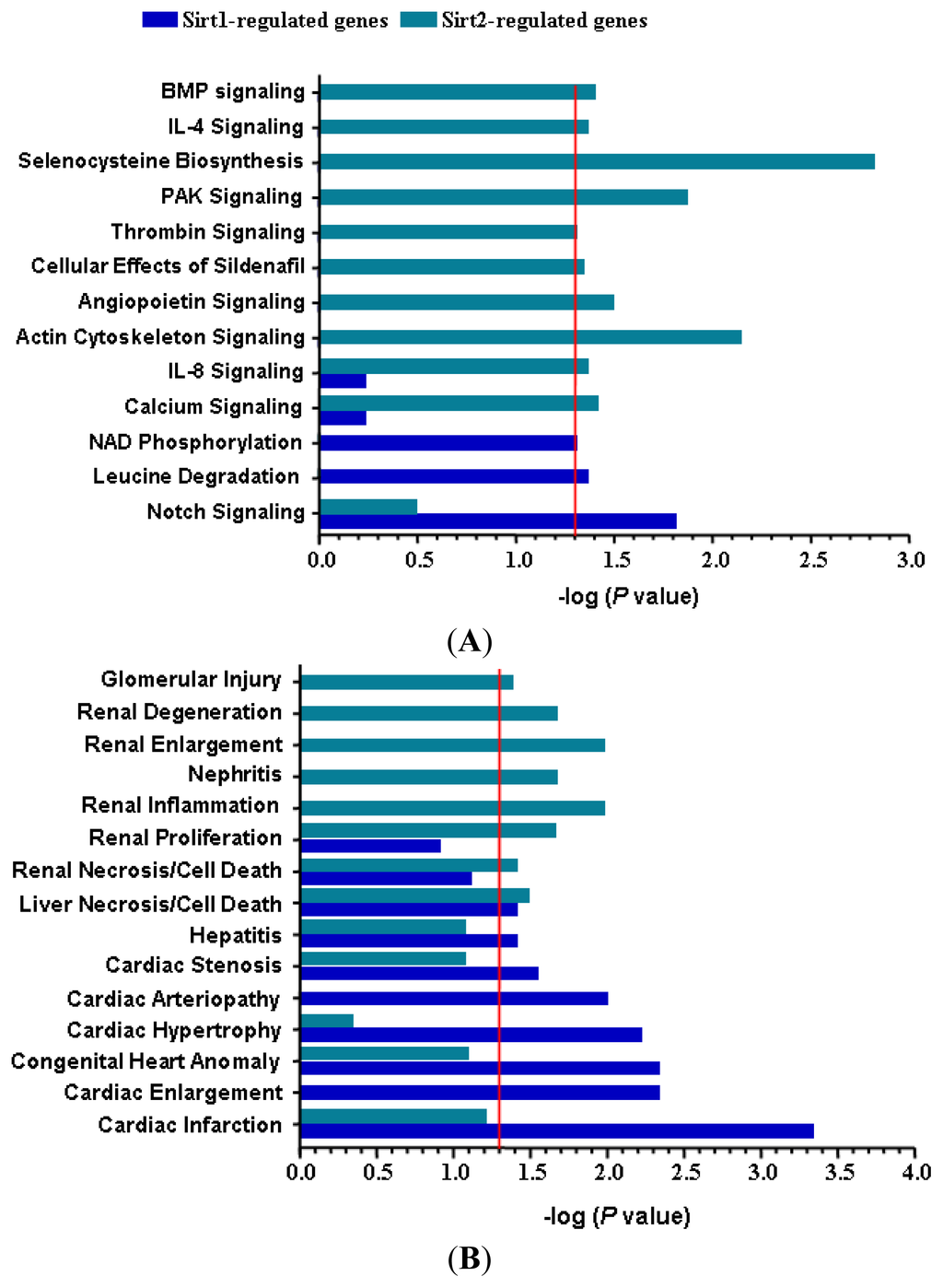
Figure 4.
Comparison of the potential categories of (A) intracellular pathways and (B) biological mechanisms related to toxicity responses that were significantly over-represented by Sirt1- or Sirt2-regulated genes respectively in stressed endothelial cells. The red line indicates the threshold of statistical significance. Functional gene enrichment analysis was performed with IPA software.
To validate our microarray data of Sirt2 effects on global gene expression, we carried out qPCR assays on selected genes including CALD1, CASP7, CNN2, RRAGC, ULBP2. We showed that Sirt2i induced upregulation CALD1, CASP7, CNN2 and downregulation of RRAGC, ULBP2 (Figure 5A). These changes were in accordance with the trend as detected by microarray (see Table 1). In contrast, expressions of these genes were not altered in Sirt1i cells (Figure 5B). To further clarify whether Sirt2 was functionally important in endothelial cells under stress, we treated HUVEC cells with a higher concentration (600 μM) of H2O2 for 2 h in the absence and presence of a selective Sirt2 inhibitor AGK2 (from Merck, Darmstadt, Germany) [35]. We found that pre-treatment with AGK2 (10 μM) attenuated H2O2-induced cell toxicity (Figure 6A), suggesting that under oxidative stress, activation of the Sirt2 pathway might have a detrimental effect on cell viability. In contrast, we showed that pre-treatment with the selective Sirt1 inhibitor EX-527 (10 μM) (from Merck) increased H2O2-induced cell toxicity (Figure 6B).
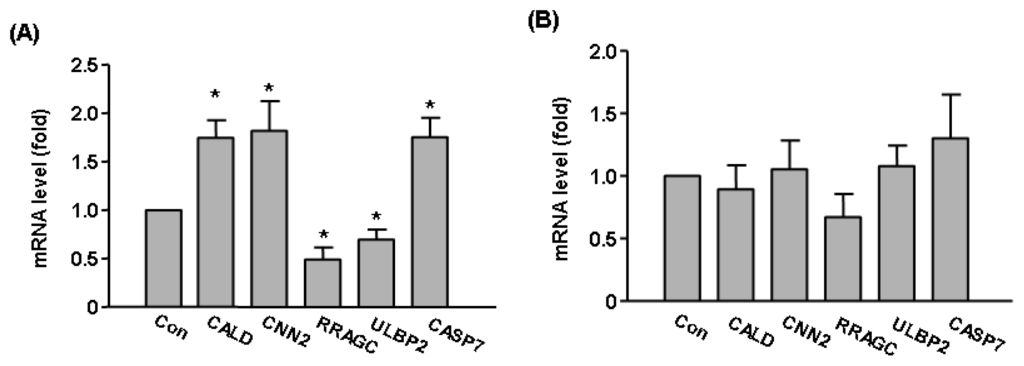
Figure 5.
Validation of microarray results with qPCR. The expression levels of CALD1, CASP7, CNN2, RRAGC, ULBP2 were measured in (A) Sirt2i cells and (B) Sirt1i cells in the presence of oxidant stress (H2O2 300 μM for 6 h). Gene expression levels were expressed as fold of control. Data are mean ± SEM. *p < 0.05 vs. Con, Student’s t-test, n = 3–4.
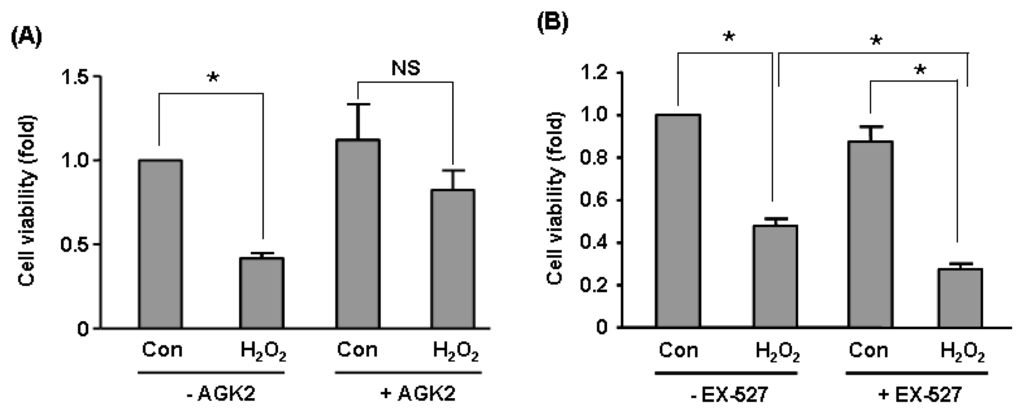
Figure 6.
Effects of (A) selective Sirt2 inhibitor AGK2 (10 μM) and (B) selective Sirt1 inhibitor EX-527 (10 μM) on H2O2-induced cell toxicity in HUVECs measured with MTS assay. Cells were treated with H2O2 (600 μM) for 2 h in the presence and absence of AGK2 or EX-527 pre-treatment. Data are mean ± SEM. *p < 0.05, one-way ANOVA, n = 4–6. NS: non-significant.
In the present study, we explored the functional importance of Sirt2 in endothelial cells under oxidative stress by measuring global gene expression changes in cells in which Sirt2 was knocked down. We found that Sirt2 gene knock down significantly altered the expression profile of 340 genes, which were involved in different cellular processes (see Table 1). We also confirmed the microarray data with qPCR for selected genes. Gene clustering analysis suggests that Sirt2-sensitive genes in endothelial cells may be involved in regulation of protein transport and localization, cellular amino acid metabolic process, and functions associated with endosome membrane and the trans-Golgi network. These functional annotations are in agreement with findings from cellular function studies showing that Sirt2 may have a pivotal role in modulating cell autophagy, an intracellular mechanism responsible for clearance of damaged proteins and organelles involving activation and mobilization of the endogenous membranous system [36]. Interestingly, a recent study demonstrated that overexpression of Sirt2 inhibited lysosome-mediated autophagic turnover and increased the sensitivity of cells to proteasomal stress-induced cytotoxicity [37]. Conversely, accumulation of ubiquitinated proteins and cytotoxicity in stressed cells were attenuated by Sirt2 knock down. These results indicate that a complex interaction between Sirt2 and autophagic process may be present. In line with these findings, we observed that pharmacological inhibition of autophagy in endothelial cells augmented H2O2-induced cell death [38]. Moreover, we found that inhibition of Sirt2 decreased H2O2-induced endothelial cytotoxicity (see below). Taken together, we propose that regulation of cellular autophagic processes might be a mechanistic link between Sirt2 and oxidative stress-induced injury in endothelial cells. In addition to the above-mentioned pathways, results from our gene function clustering analysis indicate that Sirt2-regularted genes may also be involved in actin binding, transmembrane receptor protein serine/threonine kinase signaling pathways, ferrous iron transport, and cell morphogenesis involved in differentiation.
Similar to Sirt1, Sirt2 has strong deacetylase activity, and may affect gene expression by modulating functions of multiple transcription factors and co-activators such as FoxO, NF-κB, p300, and histone [4–6,23,24,29–32]. However, the present gene profiling study showed that the potential intracellular pathways regulated by Sirt2 in stressed endothelial cells were primarily different from those regulated by Sirt1. Consistently, Sirt1- and Sirt2-sensitive genes were involved in distinct categories of diseases, for example inflammatory response, cancer, and organismal injury for Sirt2, and infectious disease, developmental disorder, skeletal and muscular disorders for Sirt1. As observed in neural cells [39], our data suggest that Sirt2 is likely to have a distinct functional role from Sirt1 in endothelial cells under stress conditions. Moreover, these data support that the observed gene expression changes in response to Sirt2 knock down are unlikely to be a result of non-specific off target effects of RNA interference.
The precise cellular functions of Sirt2 in endothelial cells remain largely unknown. A previous study has shown that Sirt2 may be implicated in mediating angiotensin II-induced endothelial cell migration via modulating α-tubulin acetylation and microtubule reorganization [40]. Consistent with this observation, we identified (and confirmed with qPCR) that Sirt2 knock down altered the expression of several genes involved in cytoskeletal organization, cell contraction and migration, such as CALD1 (caldesmon) and CNN2 (calponin) [41,42]. Moreover, we demonstrated that Sirt2 also affected expression of genes involved in modulating cell viability. This is exemplified by CASP7 (caspase 7), which is a master regulator of cell apoptosis, and RRAGC (Ras-related GTP binding C), which is involved in activation of the mTOR pathway [43].
To clarify the general role of Sirt2 in endothelial cells under oxidative stress, we challenged the cells with a high concentration of H2O2 and demonstrated that pharmacological inhibition of Sirt2 activity attenuated H2O2-induced cytotoxicity. This result is consistent with previous experiments in neural and cardiac cells showing that activation of Sirt2 promotes cell death, whereas knock down or inhibition of Sirt2 enhances cellular stress-tolerance [25,35]. Moreover, we confirmed that inhibitions of Sirt2 and Sirt1 had divergent effects on endothelial cell viability under H2O2-induced oxidative stress, an observation that was consistent with our microarray data revealing that there was only a small intersection between Sirt2- and Sirt1-sensitive genes in H2O2-challenged endothelial cells. Our experiments supported previous findings that Sirt1 exhibited profound cytoprotective effects in vascular endothelial cells in response to oxidative stress [7,15,16].
3. Experimental Section
3.1. Cell Culture
HUVECs were purchased from the American Type Culture Collection and maintained in ECM (from ScienCell Research Labortories, Carlsbad, CA, USA), supplemented with 10% fetal bovine serum, 1% ECG (endothelial cell growth supplement, ScienCell), and antibiotics (penicillin 100 U/mL, streptomycin 100 μg/mL). Cells were cultured at 37 °C with 5% CO2. Confluent cells were subcultured with 0.25% trypsin-EDTA, and cells of passage 3 to 5 were used for experimentation. Cell viability was assessed with the tetrazolium-based (MTS) assay using CellTiter 96 Aqueous kit (from Promega, Madison, WI, USA) according to the manufacturer’s direction.
3.2. RNA Interference
Small interfering RNA (siRNA) molecules targeting Sirt1 and Sirt2 were synthesized by GenePharma (Shanghai, China). For each target, 3 different siRNA sequences were tested with quantitative polymerase chain reaction (qPCR), and the one with highest efficacy was selected for following experiments. For siRNA transfection, cells were subcultured 24 h before treatment. Cells were incubated with siRNA (final concentration 30 nM) mixed with Lipofectamin RNAiMAX Reagent (Life Technologies, Carlsbad, CA, USA) for 6 h in antibiotic-free medium, and then changed to normal medium for additional 18 h.
3.3. Microarray Experiments and Data Processing
Cells were transfected with a control siRNA, Sirt2-specific siRNA (Sirt2i) or Sirt1-specific siRNA (Sirt1i). Three biological replicates were included for each group (hence a total of 9 arrays were analyzed). To induce oxidative stress, all transfected cells were treated with H2O2 at 300 μM for 6 h. Total RNA was isolated using TRIzol reagent (Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s protocol. RNA quality was tested with Bioanalyzer 2100 (Agilent, Santa Clara, CA, USA) and further purified with RNeasy Micro kit (Qiagen, Hilden, Germany). Microarray analysis was performed using Affymetrix Human Genome U219 Array, using standard labeling, hybridization and scanning protocols (ShanghaiBio Corporation, China). The raw data were processed and analyzed with GeneSpring GX software. Genes with a fold change of >1.5 and with a p value of <0.05 as compared to control were selected as differentially expressed genes. Gene Ontology (GO) functional annotation of the differentially regulated genes was carried out using DAVID Bioinformatics Resources 6.7 [44]. Further gene function clustering analysis was performed with IPA software (Ingenuity Systems, Redwood City, CA, USA).
3.4. Real-Time qPCR
Total RNA (500 ng) was reverse transcribed to cDNA using Prime Script RT reagent Kit (TaKaRa Biotechnology, Dalian, China). Real-time qPCR was performed with TaqMan gene expression assays primer-probe sets (Applied Biosystems, Carlsbad, CA, USA) or using a Sybr green-based master mix kit (SsoFast EvaGreen from Bio-Rad, Hercules, CA, USA) according to the manufacturer’s instructions. GAPDH or 18S was used as the housekeeping gene.
3.5. Fluorescent Immunocytochemistry
Cells grown on Lab-Tek II chamber slides (Nunc, Roskilde, Denmark) were fixed with cold methanol for 30 min, washed in PBS and blocked with 5% bovine serum albumin. Cells were incubated overnight with polyclonal anti-Sirt1 (1:200) (from Abcam, Cambridge, UK) or anti-Sirt2 (1:100) (from Millipore, Billerica, MA, USA). Immunofluorescent labeling was performed with DyLight594-conjugated donkey anti-rabbit IgG (1:400) (Jackson ImmunoResearch, West Grove, PA, USA). Cell nuclei were counter stained with DAPI. Images were captured using a Zeiss laser scanning confocal microscope (Zeiss LSM710, Oberkochen, Germany). Negative control experiments were performed using corresponding non-immune IgGs.
3.6. Western Blot
Total protein was resolved by 10% SDS-PAGE and transferred to nitrocellulose membranes. The membrane was blocked with 5% non-fat milk at room temperature for 1 h and then incubated with primary antibodies at 4 °C overnight. The blots were developed with ECL Prime reagents from GE Life Sciences (Piscataway, NJ, USA).
3.7. Data and Statistics
Microarray data were tested with Benjamini and Hochberg False Discovery Rate multiple testing correction. Other data were presented as mean ± SEM and tested with unpaired Student’s t-test or one-way ANOVA as appropriate, with a value of p < 0.05 being regarded as statistically significant. SPSS18.0 was used for statistical analysis.
4. Conclusions
In conclusion, to our knowledge this is the first genome-wide characterization of the gene expression profile in response to Sirt2 knockdown in endothelial cells. Sirt2-sensitive genes are involved in multiple cellular functions. Pharmacological inhibition of Sirt2 attenuated oxidant-induced endothelial cell death. These data suggest that Sirt2 is functionally important in endothelial cells under oxidative stress. Our results may provide a basis for future studies aiming to dissect the specific signaling pathway(s) that mediates specific Sirt2 functions in endothelial cells. Nevertheless, a limitation of the present study was that the microarray data did not provide direct evidence about the specific gene products that were involved in mediating the observed effects of Sirt2. Given the number of genes that are responsive to the changed Sirt2 level, it is likely that multiple mechanisms may be implicated in each specific biological function of Sirt2.
Acknowledgments
This research was partially supported by grants from the National 973 Basic Research Program of China (2010CB732605 for F.J.; 2012CB722406 for P.B.), National Natural Science Foundation of China (81070164 for F.J.; 81070076 for P.B.), and Shandong University graduate student independent innovation fund (21300070613085 for J.L.).
Conflict of Interest
The authors declare no conflict of interest.
References
- Haigis, M.C.; Sinclair, D.A. Mammalian sirtuins: Biological insights and disease relevance. Annu. Rev. Pathol 2010, 5, 253–295. [Google Scholar]
- Finkel, T.; Deng, C.X.; Mostoslavsky, R. Recent progress in the biology and physiology of sirtuins. Nature 2009, 460, 587–591. [Google Scholar]
- Saunders, L.R.; Verdin, E. Sirtuins: Critical regulators at the crossroads between cancer and aging. Oncogene 2007, 26, 5489–5504. [Google Scholar]
- Michan, S.; Sinclair, D. Sirtuins in mammals: Insights into their biological function. Biochem. J 2007, 404, 1–13. [Google Scholar]
- Bao, J.; Sack, M.N. Protein deacetylation by sirtuins: Delineating a post-translational regulatory program responsive to nutrient and redox stressors. Cell Mol. Life Sci 2010, 67, 3073–3087. [Google Scholar]
- Webster, B.R.; Lu, Z.; Sack, M.N.; Scott, I. The role of sirtuins in modulating redox stressors. Free Radic. Biol. Med 2012, 52, 281–290. [Google Scholar]
- Ota, H.; Eto, M.; Ogawa, S.; Iijima, K.; Akishita, M.; Ouchi, Y. SIRT1/eNOS axis as a potential target against vascular senescence, dysfunction and atherosclerosis. J. Atheroscler. Thromb 2010, 17, 431–435. [Google Scholar]
- Kelly, G.S. A review of the sirtuin system, its clinical implications, and the potential role of dietary activators like resveratrol: Part 2. Altern. Med. Rev 2010, 15, 313–328. [Google Scholar]
- Le Brocq, M.; Leslie, S.J.; Milliken, P.; Megson, I.L. Endothelial dysfunction: From molecular mechanisms to measurement, clinical implications, and therapeutic opportunities. Antioxid. Redox Signal 2008, 10, 1631–1674. [Google Scholar]
- Mattagajasingh, I.; Kim, C.S.; Naqvi, A.; Yamamori, T.; Hoffman, T.A.; Jung, S.B.; DeRicco, J.; Kasuno, K.; Irani, K. SIRT1 promotes endothelium-dependent vascular relaxation by activating endothelial nitric oxide synthase. Proc. Natl. Acad. Sci. USA 2007, 104, 14855–14860. [Google Scholar]
- Yang, L.; Zhang, J.; Yan, C.; Zhou, J.; Lin, R.; Lin, Q.; Wang, W.; Zhang, K.; Yang, G.; Bian, X.; et al. SIRT1 Regulates CD40 Expression Induced by TNF-alpha via NF-κB Pathway in Endothelial Cells. Cell Physiol. Biochem 2012, 30, 1287–1298. [Google Scholar]
- Xia, L.; Ding, F.; Zhu, J.H.; Fu, G.S. Resveratrol attenuates apoptosis of pulmonary microvascular endothelial cells induced by high shear stress and proinflammatory factors. Hum. Cell 2011, 24, 127–133. [Google Scholar]
- Ota, H.; Akishita, M.; Eto, M.; Iijima, K.; Kaneki, M.; Ouchi, Y. Sirt1 modulates premature senescence-like phenotype in human endothelial cells. J. Mol. Cell Cardiol 2007, 43, 571–579. [Google Scholar]
- Zu, Y.; Liu, L.; Lee, M.Y.; Xu, C.; Liang, Y.; Man, R.Y.; Vanhoutte, P.M.; Wang, Y. SIRT1 promotes proliferation and prevents senescence through targeting LKB1 in primary porcine aortic endothelial cells. Circ. Res 2010, 106, 1384–1393. [Google Scholar]
- Csiszar, A.; Labinskyy, N.; Podlutsky, A.; Kaminski, P.M.; Wolin, M.S.; Zhang, C.; Mukhopadhyay, P.; Pacher, P.; Hu, F.; de Cabo, R.; et al. Vasoprotective effects of resveratrol and SIRT1: Attenuation of cigarette smoke-induced oxidative stress and proinflammatory phenotypic alterations. Am. J. Physiol. Heart Circ. Physiol 2008, 294, H2721–H2735. [Google Scholar]
- Ota, H.; Eto, M.; Kano, M.R.; Ogawa, S.; Iijima, K.; Akishita, M.; Ouchi, Y. Cilostazol inhibits oxidative stress-induced premature senescence via upregulation of Sirt1 in human endothelial cells. Arterioscler. Thromb. Vasc. Biol 2008, 28, 1634–1639. [Google Scholar]
- Hou, J.; Wang, S.; Shang, Y.C.; Chong, Z.Z.; Maiese, K. Erythropoietin employs cell longevity pathways of SIRT1 to foster endothelial vascular integrity during oxidant stress. Curr. Neurovasc. Res 2011, 8, 220–235. [Google Scholar]
- Stein, S.; Schafer, N.; Breitenstein, A.; Besler, C.; Winnik, S.; Lohmann, C.; Heinrich, K.; Brokopp, C.E.; Handschin, C.; Landmesser, U.; et al. SIRT1 reduces endothelial activation without affecting vascular function in ApoE−/− mice. Aging (Albany N. Y.) 2010, 2, 353–360. [Google Scholar]
- Zhou, S.; Chen, H.Z.; Wan, Y.Z.; Zhang, Q.J.; Wei, Y.S.; Huang, S.; Liu, J.J.; Lu, Y.B.; Zhang, Z.Q.; Yang, R.F.; et al. Repression of P66Shc expression by SIRT1 contributes to the prevention of hyperglycemia-induced endothelial dysfunction. Circ. Res 2011, 109, 639–648. [Google Scholar]
- Nie, H.; Chen, H.; Han, J.; Hong, Y.; Ma, Y.; Xia, W.; Ying, W. Silencing of SIRT2 induces cell death and a decrease in the intracellular ATP level of PC12 cells. Int. J. Physiol. Pathophysiol. Pharmacol 2011, 3, 65–70. [Google Scholar]
- He, X.; Nie, H.; Hong, Y.; Sheng, C.; Xia, W.; Ying, W. SIRT2 activity is required for the survival of C6 glioma cells. Biochem. Biophys. Res. Commun 2011, 417, 468–472. [Google Scholar]
- Li, Y.; Matsumori, H.; Nakayama, Y.; Osaki, M.; Kojima, H.; Kurimasa, A.; Ito, H.; Mori, S.; Katoh, M.; Oshimura, M.; et al. SIRT2 down-regulation in HeLa can induce p53 accumulation via p38 MAPK activation-dependent p300 decrease, eventually leading to apoptosis. Genes Cells 2011, 16, 34–45. [Google Scholar]
- Liu, P.Y.; Xu, N.; Malyukova, A.; Scarlett, C.J.; Sun, Y.T.; Zhang, X.D.; Ling, D.; Su, S.P.; Nelson, C.; Chang, D.K.; et al. The histone deacetylase SIRT2 stabilizes Myc oncoproteins. Cell Death Differ 2013, 20, 503–514. [Google Scholar]
- Wang, F.; Nguyen, M.; Qin, F.X.; Tong, Q. SIRT2 deacetylates FOXO3a in response to oxidative stress and caloric restriction. Aging Cell 2007, 6, 505–514. [Google Scholar]
- Lynn, E.G.; McLeod, C.J.; Gordon, J.P.; Bao, J.; Sack, M.N. SIRT2 is a negative regulator of anoxia-reoxygenation tolerance via regulation of 14-3-3 zeta and BAD in H9c2 cells. FEBS Lett 2008, 582, 2857–2862. [Google Scholar]
- Liu, L.; Arun, A.; Ellis, L.; Peritore, C.; Donmez, G. Sirtuin 2 (SIRT2) enhances 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP)-induced nigrostriatal damage via deacetylating forkhead box O3a (Foxo3a) and activating Bim protein. J. Biol. Chem 2012, 287, 32307–32311. [Google Scholar]
- Luthi-Carter, R.; Taylor, D.M.; Pallos, J.; Lambert, E.; Amore, A.; Parker, A.; Moffitt, H.; Smith, D.L.; Runne, H.; Gokce, O.; et al. SIRT2 inhibition achieves neuroprotection by decreasing sterol biosynthesis. Proc. Natl. Acad. Sci. USA 2010, 107, 7927–7932. [Google Scholar]
- Zhang, Y.; Au, Q.; Zhang, M.; Barber, J.R.; Ng, S.C.; Zhang, B. Identification of a small molecule SIRT2 inhibitor with selective tumor cytotoxicity. Biochem. Biophys. Res. Commun 2009, 386, 729–733. [Google Scholar]
- Rothgiesser, K.M.; Erener, S.; Waibel, S.; Luscher, B.; Hottiger, M.O. SIRT2 regulates NF-kappaB dependent gene expression through deacetylation of p65 Lys310. J. Cell Sci 2010, 123, 4251–4258. [Google Scholar]
- Das, C.; Lucia, M.S.; Hansen, K.C.; Tyler, J.K. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature 2009, 459, 113–117. [Google Scholar]
- Black, J.C.; Mosley, A.; Kitada, T.; Washburn, M.; Carey, M. The SIRT2 deacetylase regulates autoacetylation of p300. Mol. Cell 2008, 32, 449–455. [Google Scholar]
- Bouras, T.; Fu, M.; Sauve, A.A.; Wang, F.; Quong, A.A.; Perkins, N.D.; Hay, R.T.; Gu, W.; Pestell, R.G. SIRT1 deacetylation and repression of p300 involves lysine residues 1020/1024 within the cell cycle regulatory domain 1. J. Biol. Chem 2005, 280, 10264–10276. [Google Scholar]
- Coussens, M.; Maresh, J.G.; Yanagimachi, R.; Maeda, G.; Allsopp, R. Sirt1 deficiency attenuates spermatogenesis and germ cell function. PLoS One 2008, 3, e1571. [Google Scholar]
- Potente, M.; Ghaeni, L.; Baldessari, D.; Mostoslavsky, R.; Rossig, L.; Dequiedt, F.; Haendeler, J.; Mione, M.; Dejana, E.; Alt, F.W.; et al. SIRT1 controls endothelial angiogenic functions during vascular growth. Genes Dev 2007, 21, 2644–2658. [Google Scholar]
- Outeiro, T.F.; Kontopoulos, E.; Altmann, S.M.; Kufareva, I.; Strathearn, K.E.; Amore, A.M.; Volk, C.B.; Maxwell, M.M.; Rochet, J.C.; McLean, P.J.; et al. Sirtuin 2 inhibitors rescue alpha-synuclein-mediated toxicity in models of Parkinson’s disease. Science 2007, 317, 516–519. [Google Scholar]
- De Oliveira, R.M.; Sarkander, J.; Kazantsev, A.G.; Outeiro, T.F. SIRT2 as a Therapeutic Target for Age-Related Disorders. Front Pharmacol 2012, 3, 82. [Google Scholar]
- Gal, J.; Bang, Y.; Choi, H.J. SIRT2 interferes with autophagy-mediated degradation of protein aggregates in neuronal cells under proteasome inhibition. Neurochem. Int 2012, 61, 992–1000. [Google Scholar]
- Liu, J.; Jiang, F. Shandong University, Jinan, Shandong Province, China. Personal communication, 2012. [Google Scholar]
- Pfister, J.A.; Ma, C.; Morrison, B.E.; D’Mello, S.R. Opposing effects of sirtuins on neuronal survival: SIRT1-mediated neuroprotection is independent of its deacetylase activity. PLoS One 2008, 3, e4090. [Google Scholar]
- Hashimoto-Komatsu, A.; Hirase, T.; Asaka, M.; Node, K. Angiotensin II induces microtubule reorganization mediated by a deacetylase SIRT2 in endothelial cells. Hypertens Res 2011, 34, 949–956. [Google Scholar]
- Numaguchi, Y.; Huang, S.; Polte, T.R.; Eichler, G.S.; Wang, N.; Ingber, D.E. Caldesmon-dependent switching between capillary endothelial cell growth and apoptosis through modulation of cell shape and contractility. Angiogenesis 2003, 6, 55–64. [Google Scholar]
- Tang, J.; Hu, G.; Hanai, J.; Yadlapalli, G.; Lin, Y.; Zhang, B.; Galloway, J.; Bahary, N.; Sinha, S.; Thisse, B.; et al. A critical role for calponin 2 in vascular development. J. Biol. Chem 2006, 281, 6664–6672. [Google Scholar]
- Dennis, M.D.; Baum, J.I.; Kimball, S.R.; Jefferson, L.S. Mechanisms involved in the coordinate regulation of mTORC1 by insulin and amino acids. J. Biol. Chem 2011, 286, 8287–8296. [Google Scholar]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc 2009, 4, 44–57. [Google Scholar]
Supplementary Files
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).