Correlates with Vaccine Protective Capacity and COVID-19 Disease Symptoms Identified by Serum Proteomics in Vaccinated Individuals
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design with Serum Samples from Different Cohorts
2.2. Serum Proteomics
2.3. Quality Control of Proteomics Data
2.4. Data Analysis
2.5. Antibody Neutralization Test
2.6. Antibody Levels against HBV and Zika Virus
2.7. Human Autoantibody General Survey Microarray
3. Results and Discussion
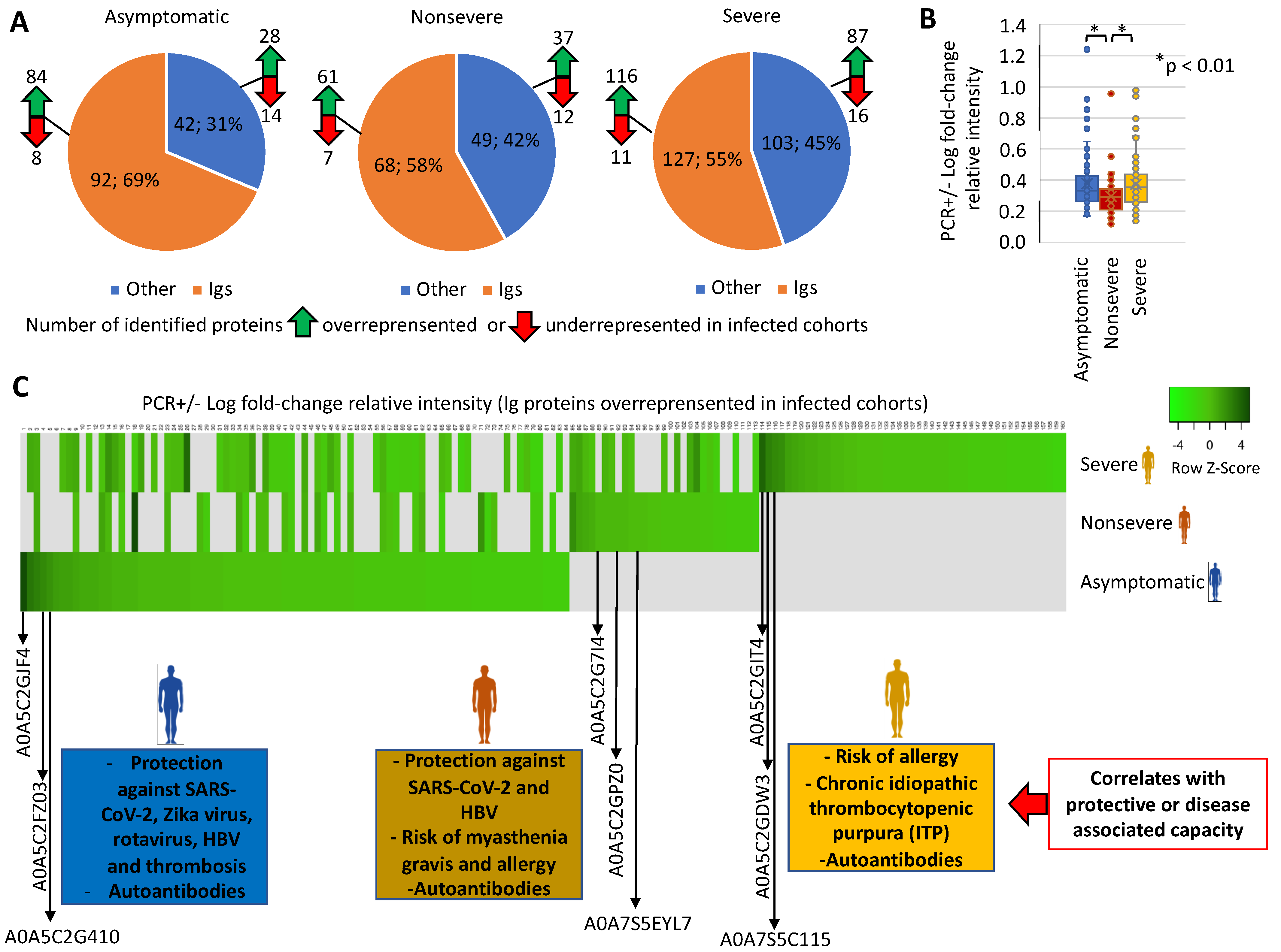
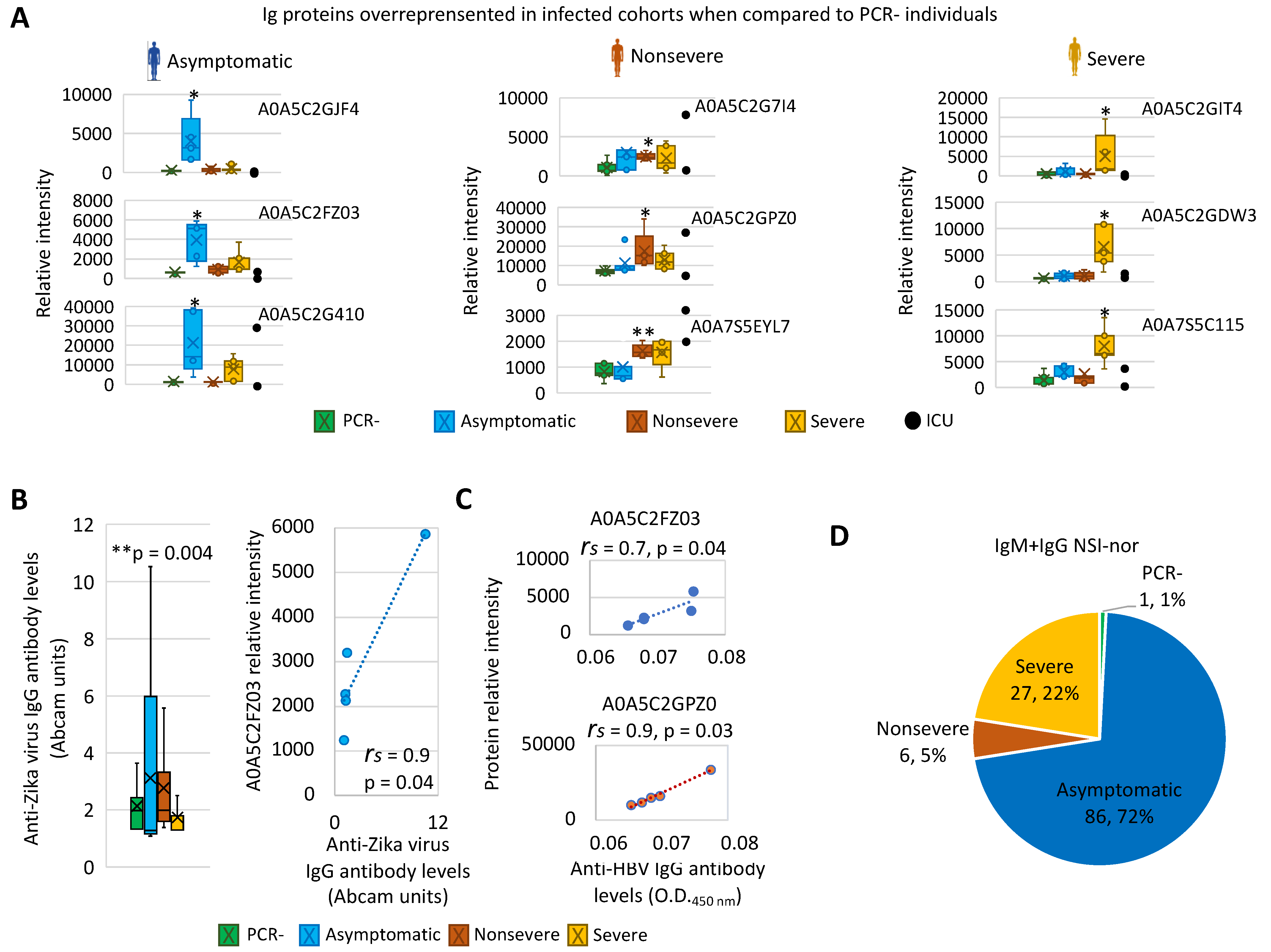
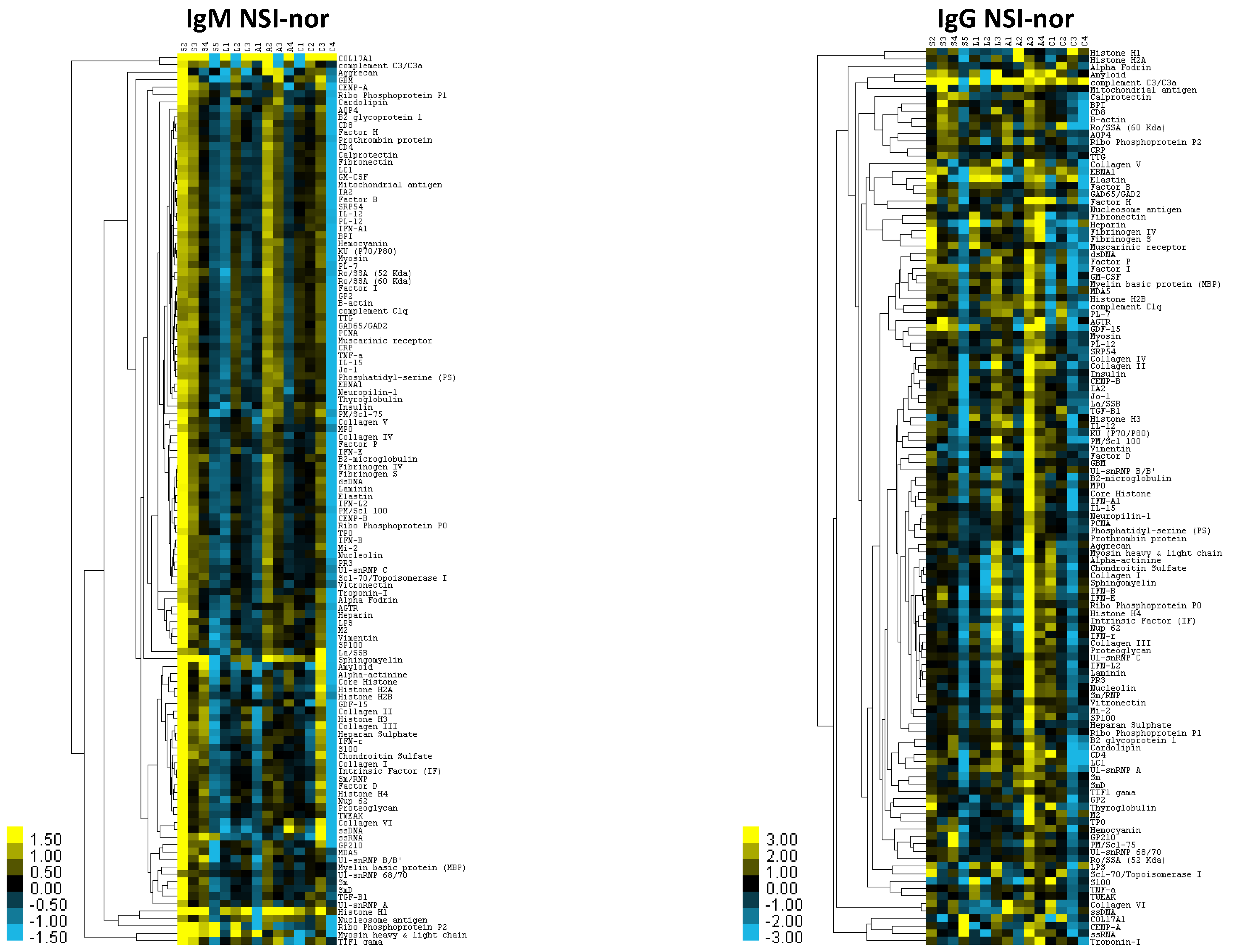
3.1. Characterization of Immunoglobulin Protein Profiles and Correlation with Protective- or Disease-Associated Capacity
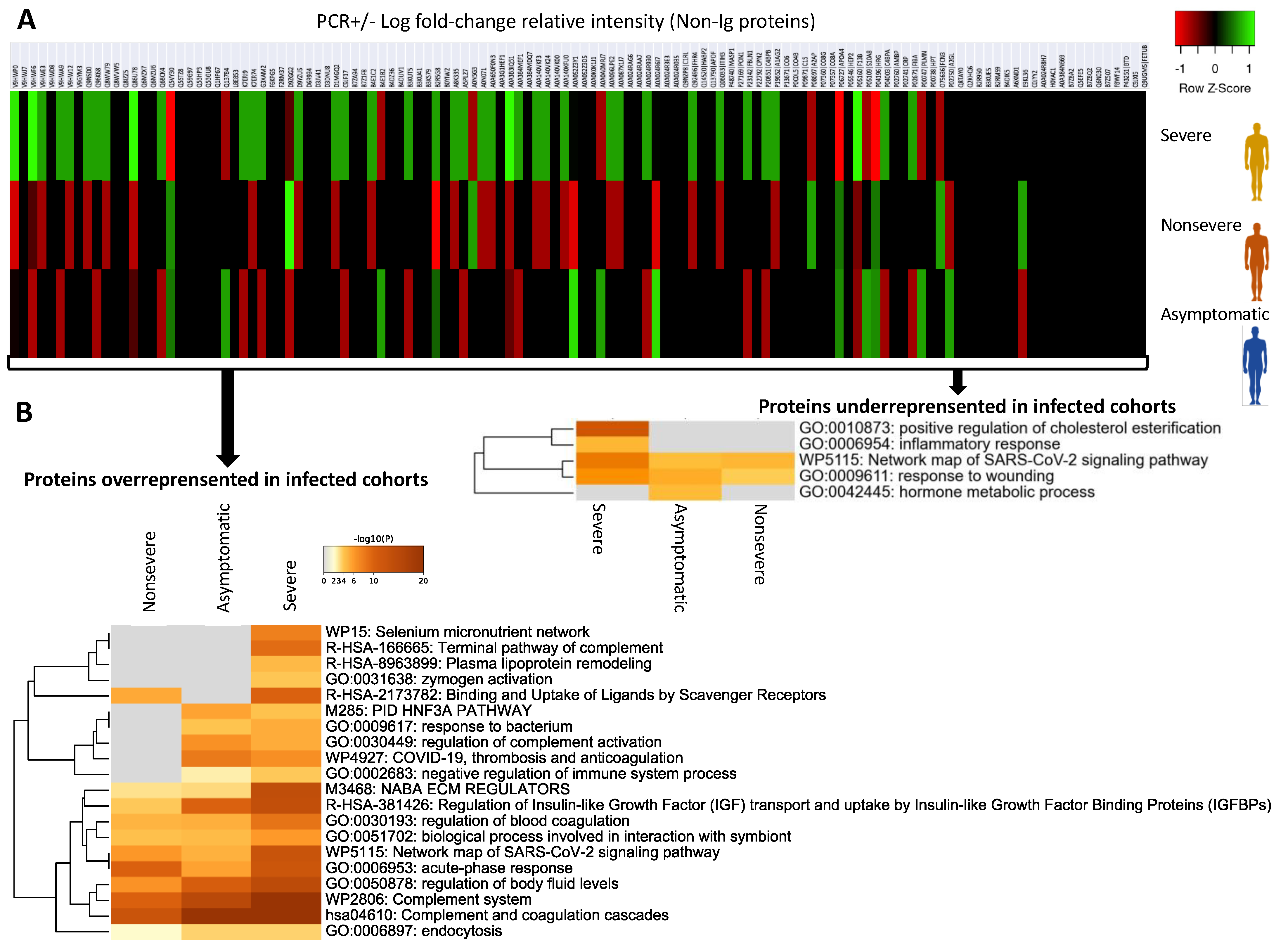
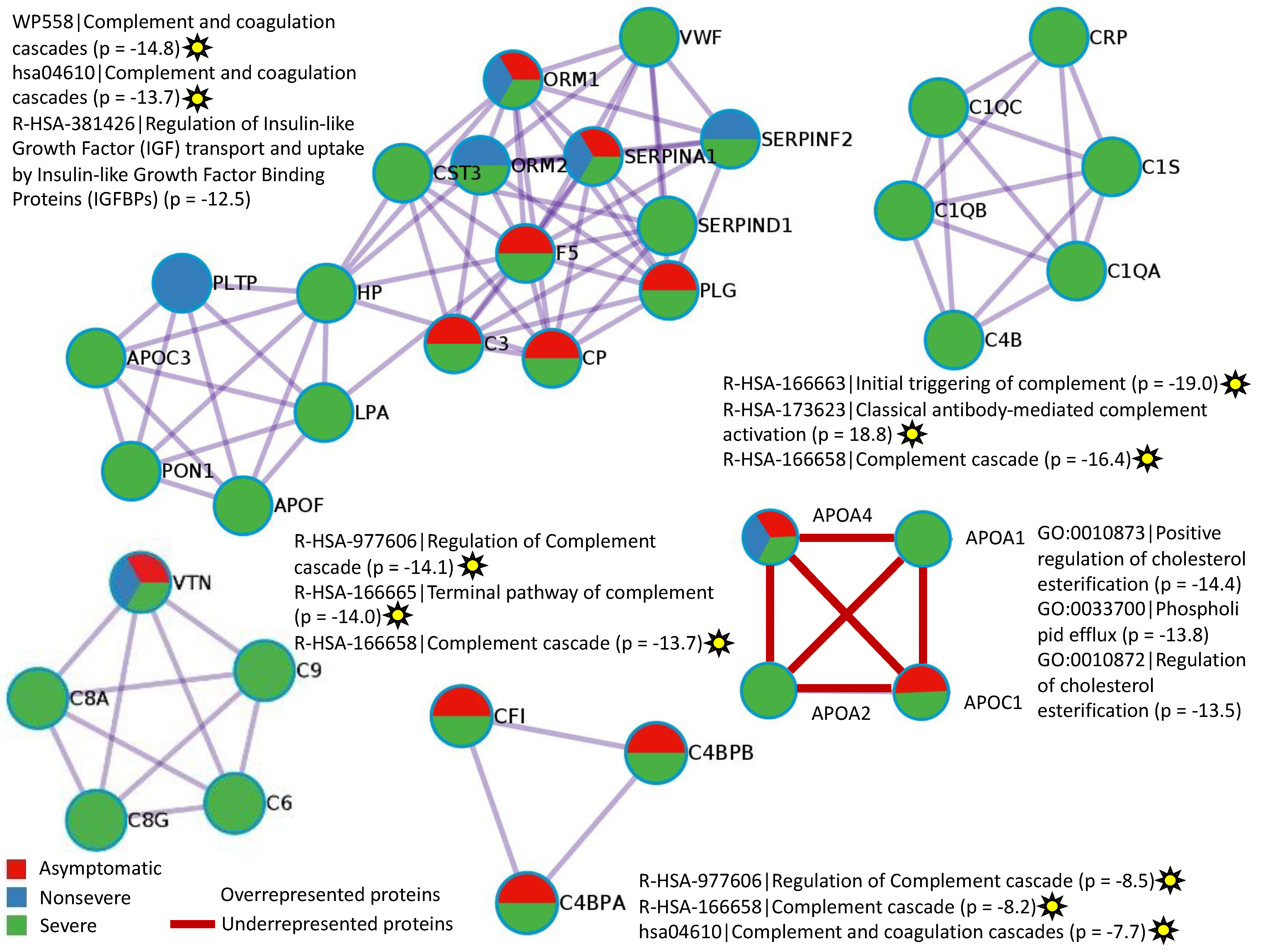
3.2. Characterization of Non-Ig Protein Profiles and Correlation with COVID-19
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Adam, D. The pandemic’s true death toll: Millions more than official counts. Nature 2022, 601, 312–315. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, J.K.; Yadav, R.; Chaudhary, P.K.; Maurya, A.; Kant, N.; Rugaie, O.A.; Haokip, H.R.; Yadav, D.; Roshan, R.; Prasad, R.; et al. Insights into COVID-19 Vaccine development based on immunogenic structural proteins of SARS-CoV-2, host immune responses, and herd immunity. Cells 2021, 10, 2949. [Google Scholar] [CrossRef] [PubMed]
- Ceacareanu, A.C.; Wintrob, Z.A.P. Summary of COVID-19 vaccine-related reports in the vaccine adverse event reporting system. J. Res. Pharm. Pract. 2021, 10, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Adesokan, A.; Obeid, M.A.; Lawal, A.F. SARS-CoV-2: Vaccinology and emerging therapeutics; challenges and future developments. Ther. Deliv. 2022, 13, 117–183. [Google Scholar] [CrossRef]
- Mohapatra, R.K.; Kuppili, S.; Kumar Suvvari, T.; Kandi, V.; Behera, A.; Verma, S.; Kudrat-E-Zahan Biswal, S.K.; Al-Noor, T.H.; El-Ajaily, M.M.; Sarangi, A.K.; et al. SARS-CoV-2 and its variants of concern including Omicron: Looks like a never ending pandemic. Chem. Biol. Drug Des. 2022, 99, 769–788. [Google Scholar] [CrossRef]
- Li, J.; Wang, Y.; Liu, Y.; Zhang, Z.; Zhai, Y.; Dai, Y.; Wu, Z.; Nie, X.; Du, L. Polymorphisms and mutations of ACE2 and TMPRSS2 genes are associated with COVID-19: A systematic review. Eur. J. Med. Res. 2022, 27, 26. [Google Scholar] [CrossRef]
- Amiri-Dashatan, N.; Koushki, M.; Rezaei-Tavirani, M. Mass spectrometry-based proteomics research to fight COVID-19: An expert review on hopes and challenges. OMICS 2022, 26, 19–34. [Google Scholar] [CrossRef]
- Villar, M.; Urra, J.M.; Rodríguez-Del-Río, F.J.; Artigas-Jerónimo, S.; Jiménez-Collados, N.; Ferreras-Colino, E.; Contreras, M.; Fernández de Mera, I.G.; Estrada-Peña, A.; Gortázar, C.; et al. Characterization by quantitative serum proteomics of immune-related prognostic biomarkers for COVID-19 symptomatology. Front. Immunol. 2021, 12, 730710. [Google Scholar] [CrossRef]
- Kunik, V.; Ashkenazi, S.; Ofran, Y. Paratome: An online tool for systematic identification of antigen-binding regions in antibodies based on sequence or structure. Nucleic Acids Res. 2012, 40, W521–W524. [Google Scholar] [CrossRef]
- D’Angelo, S.; Ferrara, F.; Naranjo, L.; Erasmus, M.F.; Hraber, P.; Bradbury, A.R.M. Many routes to an Antibody Heavy-Chain CDR3: Necessary, yet insufficient, for specific binding. Front. Immunol. 2018, 9, 395. [Google Scholar] [CrossRef] [Green Version]
- Taeschler, P.; Cervia, C.; Zurbuchen, Y.; Hasler, S.; Pou, C.; Tan, Z.; Adamo, S.; Raeber, M.E.; Bächli, E.; Rudiger, A.; et al. Autoantibodies in COVID-19 correlate with antiviral humoral responses and distinct immune signatures. Allergy 2022. [Google Scholar] [CrossRef] [PubMed]
- Uchida, M.; Tashiro-Itoh, T.; Matsuda, Y.; Ishihara, K.; Asakura, H. Autoantibodies against a 210 kDa glycoprotein of the nuclear pore complex as a prognostic marker in patients with primary biliary cirrhosis. J. Gastroenterol. Hepatol. 1998, 13, 257–265. [Google Scholar]
- Yeung, M.L.; Teng, J.L.L.; Jia, L.; Zhang, C.; Huang, C.; Cai, J.P.; Zhou, R.; Chan, K.H.; Zhao, H.; Zhu, L.; et al. Soluble ACE2-mediated cell entry of SARS-CoV-2 via interaction with proteins related to the renin-angiotensin system. Cell 2021, 184, 2212–2228.e12. [Google Scholar] [CrossRef]
- Miedema, J.; Schreurs, M.; van der Sar-van der Brugge, S.; Paats, M.; Baart, S.; Bakker, M.; Hoek, R.; Dik, W.A.; Endeman, H.; Van Der Velden, V.; et al. Antibodies against Angiotensin II Receptor Type 1 and Endothelin A Receptor are associated with an unfavorable COVID19 disease course. Front. Immunol. 2021, 12, 684142. [Google Scholar] [CrossRef]
- Niu, L.; Wittrock, K.N.; Clabaugh, G.C.; Srivastava, V.; Cho, M.W. A structural landscape of neutralizing antibodies against SARS-CoV-2 Receptor Binding Domain. Front. Immunol. 2021, 12, 647934. [Google Scholar] [CrossRef] [PubMed]
- Bastard, P. Why do people die from COVID-19? Science 2022, 375, 829–830. [Google Scholar] [CrossRef]
- Schiaffini, R.; Campana, A.; Deodati, A.; Peschiaroli, E.; Lanzillotta, M.F.; Fierabracci, A. SARS-CoV-2 infection as possible downstream disease precipitator in autoantibody-positive insulin-dependent diabetes mellitus: A case report. Ital. J. Pediatr. 2022, 48, 33. [Google Scholar] [CrossRef] [PubMed]
- Dotan, A.; David, P.; Arnheim, D.; Shoenfeld, Y. The autonomic aspects of the post-COVID19 syndrome. Autoimmun. Rev. 2022, 21, 103071. [Google Scholar] [CrossRef] [PubMed]
- Juanes-Velasco, P.; Landeira-Viñuela, A.; García-Vaquero, M.L.; Lecrevisse, Q.; Herrero, R.; Ferruelo, A.; Góngora, R.; Corrales, F.; Rivas, J.L.; Lorente, J.A.; et al. SARS-CoV-2 Infection Triggers Auto-Immune Response in ARDS. Front. Immunol. 2022, 13, 732197. [Google Scholar] [CrossRef]
- Afzali, B.; Noris, M.; Lambrecht, B.N.; Kemper, C. The state of complement in COVID-19. Nat. Rev. Immunol. 2022, 22, 77–84. [Google Scholar] [CrossRef]
- Ilias, I.; Diamantopoulos, A.; Botoula, E.; Athanasiou, N.; Zacharis, A.; Tsipilis, S.; Jahaj, E.; Vassiliou, A.G.; Vassiliadi, D.A.; Kotanidou, A.; et al. COVID-19 and Growth Hormone/Insulin-Like Growth Factor 1: Study in critically and non-critically ill patients. Front. Endocrinol. 2021, 12, 644055. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Yin, C.; Wang, J.; Yang, M.; Ma, H.; Jin, G.; Song, M.; Hu, Z.; Shen, H.; Hang, D. Pre-diagnostic circulating concentrations of insulin-like growth factor-1 and risk of COVID-19 mortality: Results from UK Biobank. Eur. J. Epidemiol. 2021, 36, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Yoon, I.S.; Park, H.; Kwak, H.W.; Woo Jung, Y.; Nam, J.H. Macrophage-derived insulin-like growth factor-1 affects influenza vaccine efficacy through the regulation of immune cell homeostasis. Vaccine 2017, 35, 4687–4694. [Google Scholar] [CrossRef]
- Weller, R.; Hueging, K.; Brown, R.J.P.; Todt, D.; Joecks, S.; Vondran, F.W.R.; Pietschmann, T. Hepatitis C virus strain-dependent usage of Apolipoprotein E modulates assembly efficiency and specific infectivity of secreted virions. J. Virol. 2017, 91, e00422-17. [Google Scholar] [CrossRef] [PubMed]
- Hubacek, J.A. Effects of selected inherited factors on susceptibility to SARS-CoV-2 infection and COVID-19 progression. Physiol. Res. 2021, 70, S125–S134. [Google Scholar] [CrossRef]
- Nuñez, E.; Orera, I.; Carmona-Rodríguez, L.; Paño, J.R.; Vázquez, J.; Corrales, F.J. Mapping the Serum Proteome of COVID-19 Patients; Guidance for Severity Assessment. Biomedicines 2022, 10, 1690. [Google Scholar] [CrossRef]
- Ulloque-Badaracco, J.R.; Hernandez-Bustamante, E.A.; Herrera-Añazco, P.; Benites-Zapata, V.A. Prognostic value of apolipoproteins in COVID-19 patients: A systematic review and meta-analysis. Travel Med. Infect. Dis. 2021, 44, 102200. [Google Scholar] [CrossRef]
- Cervin, M.; Anderson, R. Modulation of coronavirus-mediated cell fusion by homeostatic control of cholesterol and fatty acid metabolism. J. Med. Virol. 1991, 35, 142–149. [Google Scholar] [CrossRef]
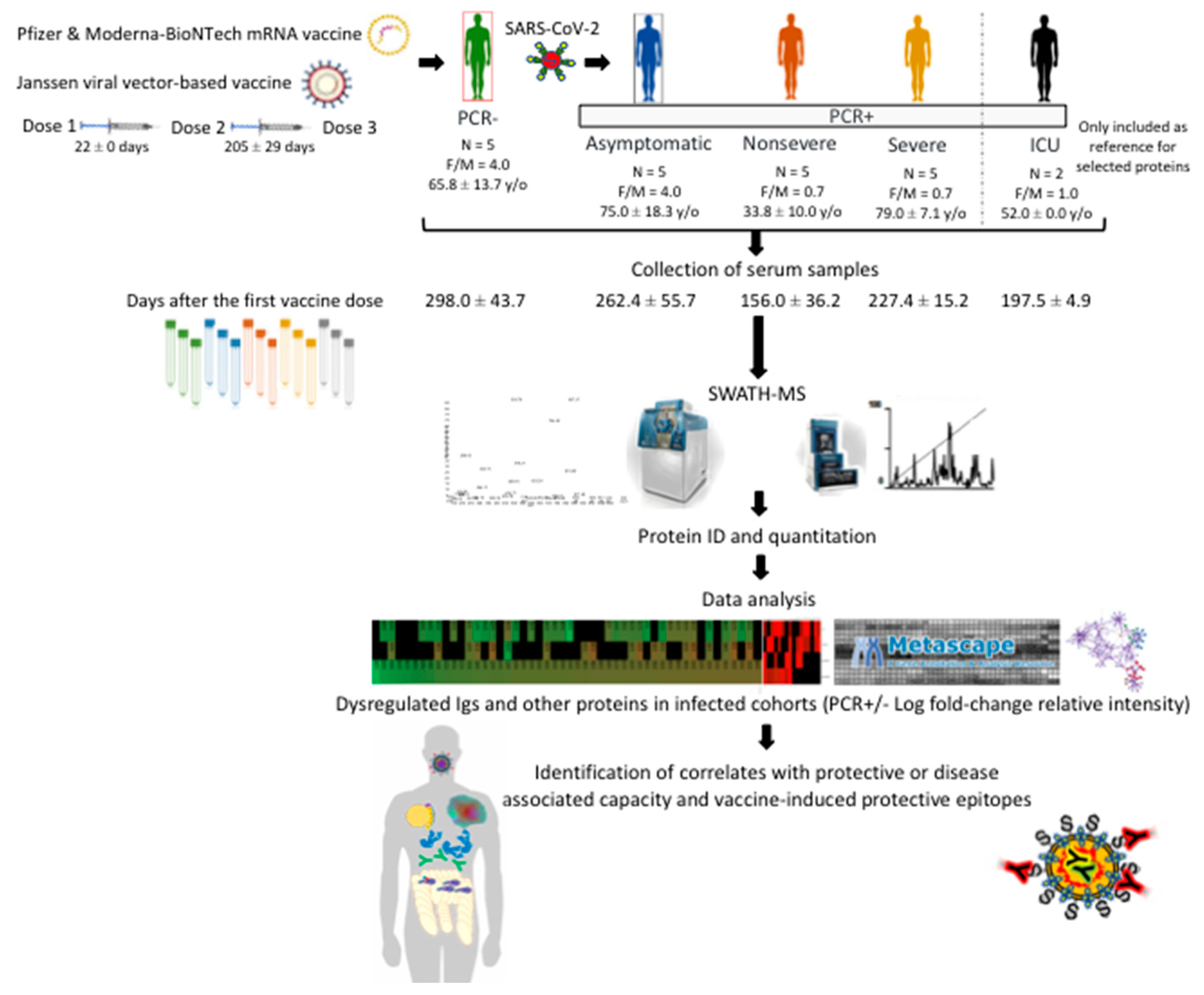

| Lab, Proteome ID | Age (y/o) | Sex | Sample Date (*) | Vaccine Doses | SARS-CoV-2-Neutralizing Antibodies | Dates of Vaccination |
|---|---|---|---|---|---|---|
| Cohort PCR– | ||||||
| 16107663, C1 | 79 | F | 15.12.2021 (247 days) | Pfizer Pfizer Moderna | 95.2% | 13.04.2021 11.05.2021 11.11.2021 |
| 16107723, C2 | 54 | M | 15.12.2021 (323 days) | Pfizer Pfizer | 32.0% | 27.01.2021 22.02.2021 |
| 39385248, C3 | 60 | F | 15.12.2021 (323 days) | Pfizer Pfizer Moderna | 95.3% | 27.01.2021 22.02.2021 30.11.2021 |
| 39385665, C4 | 82 | F | 15.12.2021 (255 days) | Pfizer Pfizer Pfizer | 92.4% | 05.04.2021 26.04.2021 08.11.2021 |
| 39386122, C5 | 54 | F | 16.12.2021 (342 days) | Pfizer Pfizer | 2.3% (negative) | 09.01.2021 30.01.2021 |
| Cohort PCR+ Asymptomatic | ||||||
| 16107241, A1 | 84 | F | 09.12.2021 (255 days) | Pfizer Pfizer | 39.4% | 30.03.2021 20.04.2021 |
| 1433003, A2 | 88 | F | 12.12.2021 (272 days) | Pfizer Pfizer Pfizer | 95.6% | 16.03.2021 06.04.2021 29.10.2021 |
| 161082999, A3 | 89 | F | 23.12.2021 (351 days) | Pfizer Pfizer Pfizer | 51.4% | 07.01.2021 28.01.2021 04.10.2021 |
| 1437141, A4 | 68 | M | 26.12.2021 (229 days) | Pfizer Pfizer Pfizer | 94.1% | 12.05.2021 04.06.2021 24.11.2021 |
| 88403647, A5 | 46 | F | 30.12.2021 (205 days) | Pfizer Pfizer | 70.2% | 09.06.2021 30.06.2021 |
| Cohort PCR+ Non-severe Hospital Discharge. Symptoms: fever, cough | ||||||
| 1429191, L1 | 41 | F | 28.11.2021 (170 days) | Pfizer | 95.6% | 12.06.2021 |
| 1433753, L2 | 19 | M | 14.12.2021 (140 days) | Pfizer Pfizer | 75.6% | 28.07.2021 18.08.2021 |
| 1433789, L3 | 25 | F | 14.12.2021 (103 days) | Pfizer | 45.0% | 03.09.2021 |
| 1435300, L4 | 26 | M | 20.12.2021 (168 days) | Pfizer Pfizer | 62.8% | 06.07.2021 27.07.2021 |
| 1435504, L5 | 47 | M | 20.12.2021 (199 days) | Pfizer Pfizer | 96.1% | 05.06.2021 26.06.2021 |
| Cohort PCR+ Severe Hospitalized. Symptoms: pneumonia, diarrhea, body weakness | ||||||
| 16105221, S1 | 79 | M | 10.11.2021 (209 days) | Pfizer Pfizer | 7.3% (negative) | 16.04.2021 07.05.2021 |
| 16106123, S2 | 77 | F | 22.11.2021 (223 days) | Pfizer Pfizer | 94.6% | 14.04.2021 05.05.2021 |
| 1431680, S3 | 91 | M | 08.12.2021 (251 days) | Pfizer Pfizer Pfizer | 61.7% | 03.03.2021 24.03.2021 03.11.2021 |
| 1432590, S4 | 73 | M | 10.12.2021 (229 days) | Pfizer Pfizer | 95.5% | 26.04.2021 17.05.2021 |
| 1434692, S5 | 75 | F | 17.12.2021 (225 days) | Pfizer Pfizer Moderna | 92.9% | 07.05.2021 28.05.2021 09.12.2021 |
| Cohort PCR+ ICU. Symptoms: severe bilateral pneumonia with acute respiratory distress | ||||||
| 1432410, U1 | 52 | F | 10.12.2021 (194 days) | Janssen Moderna | 95.0% | 31.05.2021 23.11.2021 |
| 1434573, U2 | 52 | M | 17.12.2021 (201 days) | Janssen Moderna | 94.7% | 31.05.2021 23.11.2021 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Villar, M.; Urra, J.M.; Artigas-Jerónimo, S.; Mazuecos, L.; Contreras, M.; Vaz-Rodrigues, R.; Rodríguez-del-Río, F.J.; Gortázar, C.; de la Fuente, J. Correlates with Vaccine Protective Capacity and COVID-19 Disease Symptoms Identified by Serum Proteomics in Vaccinated Individuals. Molecules 2022, 27, 5933. https://doi.org/10.3390/molecules27185933
Villar M, Urra JM, Artigas-Jerónimo S, Mazuecos L, Contreras M, Vaz-Rodrigues R, Rodríguez-del-Río FJ, Gortázar C, de la Fuente J. Correlates with Vaccine Protective Capacity and COVID-19 Disease Symptoms Identified by Serum Proteomics in Vaccinated Individuals. Molecules. 2022; 27(18):5933. https://doi.org/10.3390/molecules27185933
Chicago/Turabian StyleVillar, Margarita, José Miguel Urra, Sara Artigas-Jerónimo, Lorena Mazuecos, Marinela Contreras, Rita Vaz-Rodrigues, Francisco J. Rodríguez-del-Río, Christian Gortázar, and José de la Fuente. 2022. "Correlates with Vaccine Protective Capacity and COVID-19 Disease Symptoms Identified by Serum Proteomics in Vaccinated Individuals" Molecules 27, no. 18: 5933. https://doi.org/10.3390/molecules27185933
APA StyleVillar, M., Urra, J. M., Artigas-Jerónimo, S., Mazuecos, L., Contreras, M., Vaz-Rodrigues, R., Rodríguez-del-Río, F. J., Gortázar, C., & de la Fuente, J. (2022). Correlates with Vaccine Protective Capacity and COVID-19 Disease Symptoms Identified by Serum Proteomics in Vaccinated Individuals. Molecules, 27(18), 5933. https://doi.org/10.3390/molecules27185933