Abstract
Malus hupehensis belongs to the Malus genus (Rosaceae) and is an indigenous wild crabapple of China. This species has received more and more attention, due to its important medicinal, and excellent ornamental and economical, values. In this study, the whole chloroplast (cp) genome of Malus hupehensis, using a Hiseq X Ten sequencing platform, is reported. The M. hupehensis cp genome is 160,065 bp in size, containing a large single copy region (LSC) of 88,166 bp and a small single copy region (SSC) of 19,193 bp, separated by a pair of inverted repeats (IRs) of 26,353 bp. It contains 112 genes, including 78 protein-coding genes (PCGs), 30 transfer RNA genes (tRNAs), and four ribosomal RNA genes (rRNAs). The overall nucleotide composition is 36.6% CG. A total of 96 simple sequence repeats (SSRs) were identified, most of them were found to be mononucleotide repeats composed of A/T. In addition, a total of 49 long repeats were identified, including 24 forward repeats, 21 palindromic repeats, and four reverse repeats. Comparisons of the IR boundaries of nine Malus complete chloroplast genomes presented slight variations at IR/SC boundaries regions. A phylogenetic analysis, based on 26 chloroplast genomes using the maximum likelihood (ML) method, indicates that M. hupehensis clustered closer ties with M. baccata, M. micromalus, and M. prunifolia than with M. tschonoskii. The availability of the complete chloroplast genome using genomics methods is reported here and provides reliable genetic information for future exploration on the taxonomy and phylogenetic evolution of the Malus and related species.
1. Introduction
Chloroplasts are important organelles involved in photosynthesis, supplying the indispensable energy for plant growth and development. The chloroplast genome typically has a quadripartite organization, with a LSC region, a SSC region and two identical copies of IR regions [1]. In angiosperms, the most complete cp genome sizes range from 120 to 160 kb [2]. Apart from its quadripartite structure, about 100–130 genes were included in chloroplast genome, and therefore the performance in their composition and arrangement are very conservative [3]. The chloroplast DNA shows maternal inheritance in most plant species, less recombination and has a slow rate of evolution, which is substantially different from the nuclear genome [4] that has been widely applied in evolutionary relationships at the taxonomic level in plants. The cp DNA genome Sequencing can support knowledge for researching the molecular evolutionary, RNA Editing, population genetics, and transplastomic studies [5,6,7,8,9]. With the development of next-generation sequencing technologies, provides a cost-effective means and efficiently get complete chloroplast genome information, which can contribute to the resolution of species relationships. Moreover, the comparative analysis of chloroplast genomes can contribute to a theoretical basis for a phylogenetic status study [10,11].
Malus Miller is an economically important genus of about 62 species (http://www.theplantlist.org/1.1/browse/A/Rosaceae/Malus/). The genus Malus Miller (Rosaceae) are widely found in the Northern Hemisphere temperate zone [12]. About 30 to 35 species of the Malus genus are widely distributed in China [13]. Species of the Malus genus are well known for their leaves, flower and fruit, which have great value in the medicinal, agricultural product, and food handling industries [14,15]. The Malus fruit and related products, such as cider, vinegar or juice, are well received by consumers. Numerous studies have shown that compounds in Malus plants have a medicinal tonic function and therapeutic role [16,17]. Additionally, the plants of the Malus genus are used as materials that can potentially be used for the production of nutraceuticals and cosmeceuticals. The Malus species have an excellent horticultural trait that is used as an experimental research plant material, which is of great value to researchers. Previous studies have used microsatellite markers to assess a broad range of genetic diversity resources in Malus germplasm collections [18]. Additionally, in morphological and biochemical diversity analyses from the parts of Malus species, phylogenetic relationships have been conducted, however, the number of them is limited [19,20,21]. However, the taxonomy of the Malus genus is complex and unclear, and in light of new genomic resources, in need of revision [22]. Therefore, the Malus species complete chloroplast genome databases can make the contribution of a useful resource for researchers in identifying species, plant genetic improvements, biotic and abiotic resistance evaluations, and research on cell physiology and biochemistry.
Malus hupehensis, an indigenous wild crabapple cultivar of the Malus genus, grows naturally in the forests of slopes or valley thickets at an elevation of 50–2900 m and is widely distributed throughout China [12]. As an important traditional Chinese medicinal material, it is used to treat ailments related to the spleen stomach, and constipation [23,24]. The extracts of M. hupehensis possess abundant bioactive compounds, such as polyphenols, flavonoids and chalcon, which have the pharmacological action of potent anti-oxidative, anti-microbial, anti-inflammation and anti-fatigue properties [25,26,27]. Among these beneficial bioactive compounds from the M. hupehensis, polyphenols can significantly lower plasma glucose levels [28], flavonoids can protect doxorubicin-induced cell apoptosis and inhibit the occurrence of liver fibrosis [28,29]. Moreover, the young leaves of this plant are used for a tea drink in China due to being rich in a variety of essential trace elements of the human body, which have healthy activities and are very popular with people [30]. It has charming flowers in the spring, attractive foliage in the summer, beautiful fruit in the autumn, and is a steadfast component of the landscape industry that is widely cultivated. Furthermore, M. hupehensis is also a common apple rootstock, with apomixis traits, strong disease resistance, strong resistance to stress, strong grafting affinity with the main variety and a certain dwarfing effect [31].
Here, we sequenced the M. hupehensis cp genome applying Illumina sequencing technology and analyzed the genome features, and this was the first comprehensive complete cp genome analysis of M. hupehensis, combined with the whole cp genome sequences of eight other Malus species, previously published. Furthermore, we also used 26 complete cp genome sequences from GenBank to construct the phylogenetic relationships and infer the phylogenetic position of M. hupehensis. Our data will provide valuable information for further studies. Meanwhile, the data can contribute to the exploration and utilization of Malus plants.
2. Results and Discussions
2.1. Chloroplast Genome Features of M. hupehensis
We acquired approximately 7.3 Gb reads for M. hupehensis were through the Illumina HiSeq X Ten system (Illumina, San Diego, CA, USA). The complete cp genome sequence of M. hupehensis had been deposited into GenBank (No. MK020147). M. hupehensis cp genome has a quadripartite architecture, and has 160,065 nucleotides, which are geared to the size of a landplant cp genome [32], consisting of a pair of IRs (26,353 bp), a SSC region (19,193 bp) and a LSC region (88,166 bp), which is similar to other Malus complete chloroplast genomes (Table 1 and Figure 1). The GC content of the LSC (34.2%) and SSC regions (30.4%) was lower than that in IR regions (42.7%). The relatively high GC content of the IR regions was mostly attributable to the four rRNAs and tRNAs [33,34]. Additionally, the GC percentage in M. hupehensis complete chloroplast genome was 36.6%, which nearly the same as in the other eight Malus complete chloroplast genomes (Table 1).

Table 1.
Summary of complete chloroplast genomes for nine Malus species.
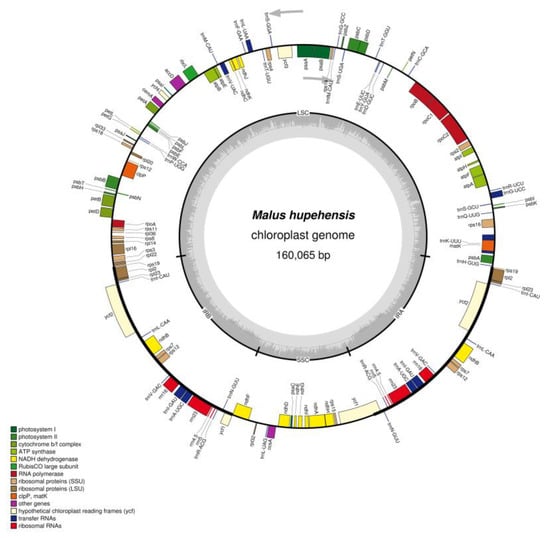
Figure 1.
Gene map of the M. hupehensis chloroplast genome. Genes shown outside the outer circle are transcribed clockwise and those inside are transcribed counterclockwise. The colored bars indicate different functional groups. The dark gray inner circle corresponds to the GC content, the light-gray to the AT content.
The studied chloroplast genomes of green plants usually comprise 110–130 genes, of which ~80 are PCGs, ~30 are tRNAs and four are rRNAs [35]. In the M. hupehensis chloroplast genome, 131 functional genes were identified, the positions of those genes are shown in Figure 1, which has 112 unique genes (Table 2), including 78 PCGs, 30 tRNAs, and four rRNAs. Among of those, six protein-coding genes (ndhB, rpl2, rpl23, rps7, rps12 and ycf2), seven tRNA genes (trnA-UGC, trnL-CAA, trnI-GAU, trnI-CAU, trnN-GUU, trnV-GAC, trnR-ACG), and four rRNA genes (4.5S, 5S, 16S, 23S) are located in IR regions, which were totally duplicated. Moreover, a total of 62 PCGs and 22 tRNA genes were located in the LSC region, also, there were 11 PCGs and one tRNA gene located in the SSC region.

Table 2.
Gene contents of the M. hupehensis chloroplast genome, based on genome annotation.
Among these annotated genes, a total of 15 genes (atpF, ndhA, ndhB, petB, petD, rpl16, rpl2, rpoC1, rps16, trnA-UGC, trnG-GCC, trnI-GAU, trnK-UUU, trnL-UAA, trnV-UAC) contained one intron, and three genes (clpP, rps12, and ycf3) contained two introns (Table 3). The clpP gene is essential for chloroplast development, which encodes ATP-dependent protease proteolytic subunit [36]. The past study have reported that the clpP splicing efficiency was increased under drought stress [37]. The clpP of M. hupehensis may be useful for further studies of this plant’s response to abiotic stresses in apple rootstock. A trans-spliced gene, with a 5′ exon situated in the LSC region and the duplicated 3′ end in the IR region, which is conserved in most other land plants [38], is found in rps12. The trnL-UAA was provided with the smallest intron (514 bp), whereas the intron of trnK-UUU possesses the largest intron (2497 bp), the matK gene is contained in it. Meanwhile, the matK gene is widely used as a molecular marker to research the phylogenetic relationships in other angiosperms [39,40,41,42,43]. Additionally, in previous studies, the matK region of Malus cp genome had been analyzed to contribute to the identification of potential germplasm donors for the cultivated Malus species [22].

Table 3.
Location and length of intron-containing genes within the M. hupehensis chloroplast genome.
Relative synonymous codon usage (RSCU) values as an availability source, which can make for the phylogenetic relationship studies [44]. The synonymous codons in angiosperms genomes possess usage frequencies differently, that is, a codon usage bias, which is a significant evolutionary character within genome that can provide essential information for studying organism evolution [45]. In the M. hupehensis chloroplast genome, the all PCGs included 78,564 bp that encoded codons numbers are 26,188. Among all these codons, there are up to 2747 (10.49%) codons encoded leucine. However, only a small amount of codons (300, 1.15%) encoded cysteine (Table S1, Figure 2). Of course, the used amino acids of leucine and cysteine were the most and the least frequently in the M. hupehensis cp genome, respectively. The use of the starting codon methionine AUG and tryptophan UGG had no bias (RSCU = 1). Moreover, 31 codons ending with A or U, which contained 29 preferred synonymous codons (RSCU > 1.0), the rest are trnL-UAG (RSCU = 0.78), trnI-CAU (RSCU = 0.95) and a stop codon (UAG) (RSCU = 0.54) (Table S1).
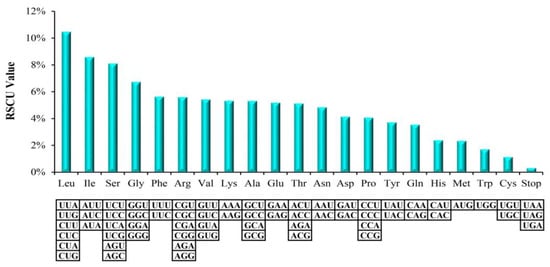
Figure 2.
Codon content of 20 amino acid and the stop codon of 84 coding genes of the M. hupehensis cp genome.
2.2. SSR and Long-Repeat Analysis
Simple sequence repeats, with high rate of mutation and diversity copy number, as shown by molecular markers for genetic diversity and evolutionary reseaches [46,47]. In a previous study, SSR markers were used to identify the germplasm and genetic relationship of M. hupehensis [31]. With MISA analysis, a total of 96 SSRs were identified, and there were 69, 19, 7, and 1, mononucleotide, dinucleotide, tetranucleotide, and pentanucleotide repeats, respectively (Figure 3A). These SSRs are very conducive to the Rosaceae complete chloroplast genomes A/T abundance [48,49,50]. In addition, the A/T mononucleotide repeats 69 (71.88%) were the most common. This result is in agreement with previous studies showing that the most abundant SSR pattern was generally composed of mononucleotides (A/T) [48]. Mononucleotides in all of the SSRs of nine Malus chloroplast genomes with the highest proportion reached 68.30%, followed by the dinucleotides (23.98%), tetranucleotides (6.43%), pentanucleotides (0.94%) and, finally, the hexanucleotide (0.35%) (Figure 3B). There were no trinucleotide repeats observed in all 9 Malus species. In all, 856 repeats were detected in the nine Malus species. The numbers of the SSR repeats were 96, 101, 91, 92, 97, 93, 97, 94, and 95 in M. hupehensis, M. trilobata, M. florentina, M. tschonoskii, M. baccata, M. micromalus, M. prunifolia, M. doumeri, and M. yunnanensis, respectively (Figure 3C). The results of these studies will allow chloroplast SSR markers to be used in the study of the genetic diversity in M. hupehensis, which can be valuable for comparing phylogenetic relationships and inferring the population genetic structure among related Malus species.
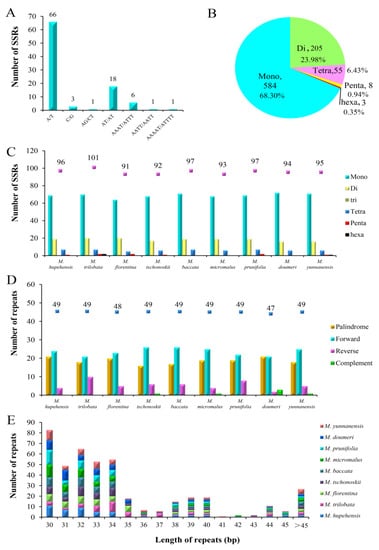
Figure 3.
Repeat analyses. (A) Repeat unit and amounts of SSR in the M. hupehensis cp genome. (B) Presence of different SSR types in all of the SSRs of nine Malus chloroplast genomes. (C) SSRs in the nine Malus cp genomes. (D) Repeated sequences in the nine Malus cp genomes. (E) Repeat frequency of four types by length in the nine Malus chloroplast genomes.
In total, 49 repeats were identified of chloroplast genome of M. hupehensis, including 24 forward repeats, 21 palindromic repeats, and four reverse repeats. This result agrees with the eight other Malus complete cp genomes, which vary in numbers, from 47 to 49. Of all nine Malus species, forward is the most abundant repeat type, palindrome and reverset are close behind; complements were detected in M. tschonoskii, M. micromalus, M. doumeri, and M. yunnanensis, and numbers of them were 1, 1, 3, and 1, respectively (Figure 3D). Most of these repeats were mainly fall within 30 bp to 40 bp. Furthermore, the maximum and minimum length are 69 and 30, respectively, and most of them are within this range for each species (Figure 3E). In M. hupehensis cp genome, we found that most repeats are situated in intergenic sequences (Table S2), which was in keeping with the other research results [51].
2.3. IR Contraction and Expansion
The IR boundary expansion and contraction is deemed to an evolutionary event and has been shown to be the primary mechanism of the size variation of chloroplast genomes in higher plants [52,53]. In this study, the junctions between the IR and LSC/SSC regions among the nine Malus chloroplast genomes were compared (Figure 4). The chloroplast genomes are highly conserved, although there are also slight length discrepancies between the nine chloroplast genomes. Some expansion and contraction was presented in M. hupehensis IR region lengths and other Malus species, with the IR regions ranging from 26,306 bp in M. yunnanensis to 26,392 bp in M. trilobata (Table 1). For the LSC/IR borders, the gene rps19 in the LSC of all complete chloroplast genomes extended from 69–120 bp into the IRb region. In M. hupehensis, M. trilobata, M. micromalus, and M. prunifolia complete chloroplast genomes, the ycf1 in the IRb regions was a long way from the IRb/SSC junction, 105 bp from the junction in M. trilobata and 0 bp from M. hupehensis, whereas it shifted by an identical distance (9 bp) from LSC to IRb at the LSC/IRb border in M. micromalus and M. prunifolia. Furthermore, the photosynthetic gene, ndhF, extended into the LSC region by 12 bp in all species. The position of ycf1 in the IRa regions varied from 1068 to 1080 bp. Similarly, the IRa/LSC border is located between the rpl2 and trnH genes, and the trnH gene is located in the LSC region, 72, 81, 183, 32, 38, 40, 38, 48, and 94 bp away from the IRa/LSC border in the nine Malus cp genomes (M. hupehensis, M. trilobata, M. florentina, M. tschonoskii, M. baccata, M. micromalus, M. prunifolia, M. doumeri and M. yunnanensis), respectively. The trnH gene in the LSC regions was 183 bp from the IRb/SSC border of M. florentina, which is much further than in other species. In general, among these nine Malus species cp genome, there is a slight difference in IR boundary regions.
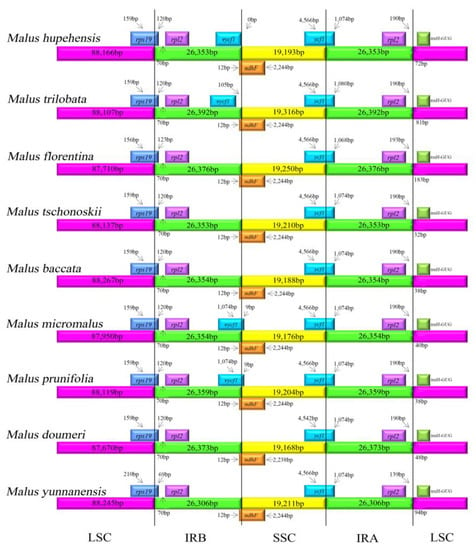
Figure 4.
Comparison of the border positions of LSC, SSC, and IR regions among the nine Malus chloroplast genomes.
2.4. Comparative Chloroplast Genomic Analysis
The comparative analysis of chloroplast genome can provide knowledge of complex evolutionary relationships [54]. In the present study, eight Malus chloroplast genomes, and M. hupehensis chloroplast genome were compared (Figure 5). The nine Malus cp genomes length between the confines of 159,584 to 160,207 bp. The chloroplast genome of M. trilobata has the largest size, whereas the chloroplast genome size of M. doumeri is the smallest. All nine Malus complete chloroplast genomes indicated that the length of IR regions ranges from 26,306–26,392 bp, that of the LSC regions ranges from 87,670–88,267 bp, and that of the SSC regions ranges from 19,168–19,316 bp, and all species showed a similar size in the LSC, SSC, and IR regions (Table 1). The complete chloroplast genome of M. hupehensis was compared with eight other genomes using the mVISTA program with a Shuffle-LAGAN model to investigate the level of sequence divergence, the alignment of which showed that the nine chloroplast genomes were conserved, with a high degree of synteny and gene order (Figure 4). However, some divergence was found within the intergenic spacers and introns of these nine chloroplast genomes, including trnH-psbA, trnK-rps16, rps16-trnQ, trnS-trnG, trnR-atpA, petN-psbM, trnE-trnT, trnT-psbD, trnS-psbZ, psbZ-trnG, psaA-ycf3, trnT-trnL, ndhC-trnV, rps8-rpl14, rpl16-rps3, ndhF-rpl32, rps32-trnL, ccsA-ndhD, as well as trnV, ndhA, and clpP introns. Additionally, the results of this study shown that the coding regions were more highly conserved than the non-coding regions, and IRs had a lower sequence divergence than the LSC and SSC regions, which is identical with other angiosperms [55]. The dissimilar coding regions of the nine cp genomes were matK, rpoA, ndhF, and ycf1, which are barcodes for land plants that have been indicated in past studies [56,57,58,59]. The possibility of further studying the trend of these regions used as molecular markers will allow for a deeper investigation of the phylogenetic development of the Malus.
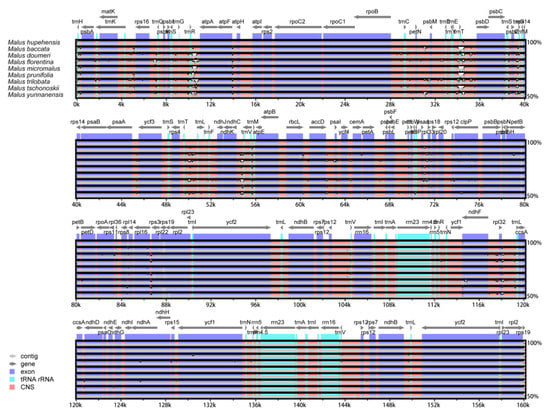
Figure 5.
Comparison of nine cp genomes using mVISTA. The chloroplast genome of M. hupehensis as a reference. The grey arrows and thick black lines above the alignment indicate the position and direction of each gene. The y-axis represents the percentage identity (shown: 50–100%).
2.5. Phylogenetic Analysis
Past research has shown that the chloroplast genome of terrestrial plants have been as a valuable source among related species, which is applied in phylogenetic studies [60,61]. In this paper, we completed an alignment of all chloroplast genomes of 26 species, which included nine Malus species, four Pyrus species, five Prunus species, three Fragaria species and three Rosa species, and two Moraceae species. As shown in the phylogenetic tree, Malus was closely related to Pyrus than with Prunus. Malus and Pyrus are included in the Maleae, and Prunoideae contain Prunus, which all were grouped within subfamily Amygdaloideae of morphological taxonomy. In addition, Fragaria (Potentilleae) and Rosa (Roseae) as sister, which revealed have a close relationship within subfamily Rosoideae. Among these relationships of genera are consistent with previous research [62,63,64]. Amygdaloideae and Rosoideae are two large subfamilies in Rosaceae, which including more than 1000 and 2000 species [65], respectively. Until recently, a lot of research has been focus on molecular phylogenetic studies in Rosaceae, to provide a theoretical basis of phylogenetic relationships [66]. However, Rosaceae includes about 100 genera and 3000 species [67], the relationships among them are still obscure, which makes phylogenetic analysis with difficulty. In this study, M. hupehensis is one of Malus species, phylogenetic tree showed that the chloroplast genome of it clustered most closely with M. baccata, M. micromalus, and M. prunifolia than with M. tschonoskii in Figure 6. The result here roughly agrees with previous studies [22] and, besides, this conclusion from in terms of genomics. Until now, little has been known about the chloroplast genome of the Malus, and a limited number of chloroplast genome sequences of the Malus species are recorded in GeneBank, which poses limitations for studying the phylogenetic relationships within the genus. Overall, M. hupehensis cp genome sequences are useful for genomic information studies, enhancing the understanding of the phylogenetic relationships of the Malus species.
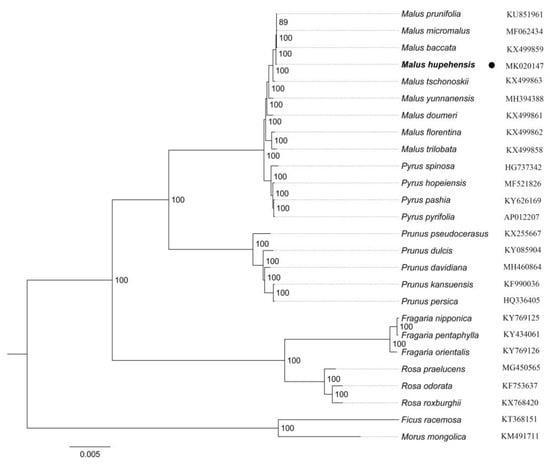
Figure 6.
A maximum likelihood (ML) phylogenetic tree based on 26 species chloroplast genomes was constructed. Ficus racemosa and Morus mongolica (Moraceae) were used as the outgroup.
3. Materials and Methods
3.1. Plant Materials and DNA Sequencing
Fresh leaves of a single individual of Malus hupehensis were collected from Yangling (34°30′49′′ N, 108°04′06′′ E), Shaanxi Province, China. A voucher specimen (AF-06-19) of M. hupehensis has been deposited in the Institute of College of Horticulture, Northwest A and F University, Yangling, China. The leaves were immediately preserved in liquid nitrogen before DNA extraction. The total genomic DNA was isolated with the DNeasy Plant Mini Kit (Qiagen, Valencia, CA, USA), following the manufacturer’s instructions. Subsequently, the concentration and quality of the extracted DNA were checked and inspected using spectrophotometry and agarose gel electrophoresis, respectively. Genome sequencing was carried out on the Illumina Hiseq X Ten platform, following the manufacturer’s protocol (Illumina, San Diego, CA, USA). Approximately 24,794,523 clean reads were obtained, with a quality value ≥Q30, accounting for 95.10%.
3.2. Genome Assembly and Genome Annotation
Before chloroplast genome assembly, adapters and low-quality sequences were removed. The MITObim v1.8 program (https://github.com/chrishah/MITObim) was used to genome assembly, based on the remaining clean data [68], and the reference sequence from the Malus baccata cp genome (Genebank accession number: KX499859). The complete Malus hupehensis chloroplast genome sequence was annotated using the online software, Dual Organellar GenoMe Annotator (DOGMA, http://dogma.ccbb.utexas.edu/) [35], and then manually corrected by comparing it with the complete cp genomes of the other published Malus species in Geneious R 11.0.4 (Biomatters Ltd., Auckland, New Zealand) [69]. Finally, the circular chloroplast genome map was completed using the online program, OGDRAW (http://ogdraw.mpimp-golm.mpg.de/) [70].
3.3. Sequence Analysis
Codon usage was determined for all protein-coding genes. To examine the deviations in the synonymous codon usage, the relative synonymous codon usage (RSCU) and GC content were determined using MEGA 6 software (Department of Biological Sciences, Tokyo, Japan) [71]. We used the online REPuter [72] software (University of Bielefeld, Bielefeld, Germany) to identify repeats (forward, palindrome, complement and reverse sequences). The minimal repeat size was set as 30 bp, and the identity of repeats was greater than 90% (hamming distance equal to 3). Perl script MISA (http://pgrc.ipk-gatersleben.de/misa/misa.html) [73] was used to detect microsatellites with minimal repeat numbers of 10, 5, 4, 3, 3, and 3 for mononucleotide, dinucleotide, trinucleotide, tetranucleotide, pentanucleotide, and hexanucleotide repeats, respectively.
3.4. Comparative Genome Analysis
The chloroplast genome size and organization were compared, and the differences of the IR border of nine Malus chloroplast genomes were analyzed. The M. hupehensis cp genome was used as a reference and was compared with other eight Malus species cp genomes using mVISTA software (Stanford University, Stanford, CA, USA) [74]. The whole-genome alignment for the cp genomes of eight species in the Malus genus, including M. hupehensis (MK020147), M. trilobata (KX499858), M. florentina (KX499862), M. tschonoskii (KX499863), M. baccata (KX499859), M. micromalus (MF062434), M. prunifolia (KU851961), M. doumeri (KX499861), and M. yunnanensis (MH394388) were analyzed.
3.5. Phylogenetic Analysis
The complete cp genome sequences of 26 species were downloaded from GenBank, using all genomes to ascertain the phylogenetic position of Malus hupehensis. Sequences were aligned using the MAFFT algorithm on the MAFFT version 7 alignment server (Osaka University, Suita, Japan) [75]. The maximum likelihood (ML) phylogenetic tree was generated using the MEGA 6 program (Department of Biological Sciences, Tokyo, Japan) [71], of which the bootstrap values of 1000 replicates to assess the branch support. In addition, Ficus racemosa and Morus mongolica (Moraceae) were set to the outgroup.
4. Conclusions
M. hupehensis is an economically important crabapple of the Malus genus in the Rosaceae family. In this study, we sequenced and annotated the whole chloroplast genome of Malus hupehensis, detected the arrangement of the genes, identified the SSRs and long repeats, and compared eight other complete chloroplast genomic characteristics of the Malus genus. M. hupehensis chloroplast genomes exhibited a typical quadripartite and circular structure in Malus, which is similar to those in other Malus species. The phylogenetic ML tree indicated that Malus was closely related to Pyrus, followed by Prunus, which indicated our data supports the position of Malus in the Amygdaloideae. Plus, the close relationships between Fragaria and Rosa were clustered into the clade as sister. The phylogenetic status of these genus is consistent with the previous report [48]. Because of the variety of Malus germplasm, the identification of evolutionary relationships is still vague, which has attracted a growing number of researchers that are trying to use biological, morphological, and molecular genetic classification analysis to classify Malus germplasm [21,76,77,78,79]. In this paper, M. hupehensis has a close relationship with M. baccata, M. micromalus and M. prunifolia than with M. tschonoskii. As recorded in book of Flora of China, M. hupehensis is similar to M. baccata. However, the leaf blade, calyx, and peduncle are slight purplish red, and the leaf edge is more acute, which are main distinguishing factors in two species. In the past, the AFLP marker system was used to analyze the genetic diversity of Malus, which indicated M. hupehensis and M. baccata within a group [80]. The matK sequence cluster analysis result indicated that M. hupehensis, M. baccata, and M. micromalus have a close relationship, M. doumeri and M. yunnanensis are within one clade, M. trilobata is closely related to M. florentina, and its sequence data also suggested M. hupehensis was close M. baccata [22]. Furthermore, our results are identical with the SRAP analysis, which indicated that M. hupehensis, M. doumeri, and M. yunnanensis are in different cluster groups [81]. China is an important primary area with rich Malus germplasm resources, with 17 wild species [82], including M. hupehensis, M. baccata, M. manshurica, M. kansuensis, M. rockii, M. sikkimensis, M. sieboldii, M. transitoria, M. sieverii, M. komarovii, M. melliana, M. xiaojinensis, M. toringoides, M. yunnanensis, M. ombrophila, M. honanensis, and M. prattii. It is necessary for more research of the complete cp genome within the Malus genus in the future. Obtaining the chloroplast genome of Malus hupehensis, which provided a possibility for further study to compare all wild Malus species in China, and other Malus species. In addition, our data also can provide a useful molecular basis, which can facilitate more extensive contributions to the exploration of the variation of Malus populations and further more studies.
Supplementary Materials
The following are available online. Table S1, Codon–anticodon recognition pattern and codon usage in the M. hupehensis chloroplast genome; Table S2, Long repeat sequences in the M. hupehensis chloroplast genome.
Author Contributions
X.Z. and M.Z. conceived and designed the research framework; L.F. and L.Q. prepared the sample and performed the experiments; J.Y., C.R., and C.M. analyzed the data; X.Z. and M.Z. wrote the paper. L.F. and M.Z. made revisions to the final manuscript. All authors have read and approved the final manuscript.
Funding
This research was funded by the earmarked fund for China Apple Research System (CARS-27).
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| cp | Chloroplast |
| PCGs | Protein-coding genes |
| tRNAs | Transfer RNA genes |
| rRNAs | Ribosomal RNA genes |
| IRs | Inverted repeat regions |
| LSC | Large single copy |
| SSC | Small single copy |
| SSR | Simple sequence repeat |
| ML | Maximum likelihood |
References
- Chaney, L.; Mangelson, R.; Ramaraj, T.; Jellen, E.N.; Maughan, P.J. The complete chloroplast genome sequences for four Amaranthus species (Amaranthaceae). Appl. Plant Sci. 2016, 4, 1600063. [Google Scholar] [CrossRef] [PubMed]
- Daniell, H.; Cohill, P.R.; Kumar, S.; Dufourmantel, N. Chloroplast Genetic Engineering. In Molecular Biology and Biotechnology of Plant Organelles; Springer: Dordrecht, The Netherlands, 2004; pp. 443–490. [Google Scholar] [CrossRef]
- Smith, D.R. Mutation rates in plastid genomes: They are lower than you might think. Genom. Biol. Evol. 2015, 7, 1227–1234. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, K.H.; Li, W.H.; Sharp, P.M. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc. Natl. Acad. Sci. USA 1987, 84, 9054–9058. [Google Scholar] [CrossRef] [PubMed]
- Nie, X.J.; Lv, S.Z.; Zhang, Y.X.; Du, X.H.; Wang, L.; Biradar, S.S.; Tan, X.F.; Wan, F.H.; Weining, S. Complete chloroplast genome sequence of a major invasive species, crofton weed (Ageratina adenophora). PLoS ONE 2012, 7, e36869. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Cui, L.; Feng, K.; Deng, P.; Du, X.; Wan, F.; Song, W.; Nie, X. Comparative analysis of asteraceae chloroplast genomes: Structural organization, RNA editing and evolution. Plant. Mol. Biol. Rep. 2015, 33, 1526–1538. [Google Scholar] [CrossRef]
- Wang, M.; Liu, H.; Ge, L.; Xing, G.; Wang, M.; Song, W.; Nie, X. Identification and analysis of RNA editing sites in the chloroplast transcripts of Aegilops tauschii L. Genes 2017, 8, 13. [Google Scholar] [CrossRef] [PubMed]
- Powell, W.; Morgante, M.; McDevitt, R.; Vendramin, G.G.; Rafalski, J.A. Polymorphic simple sequence repeat regions in chloroplast genomes-applications to the population genetics of pines. Proc. Natl. Acad. Sci. USA 1995, 92, 7759–7763. [Google Scholar] [CrossRef] [PubMed]
- Bock, R.; Khan, M.S. Taming plastids for a green future. Trends Biotechnol. 2004, 22, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Nie, X.; Zhao, X.; Wang, S.; Zhang, T.; Li, C.; Liu, H.; Tong, W.; Guo, Y. Complete chloroplast genome sequence of Broomcorn Millet (Panicum miliaceum L.) and comparative analysis with other Panicoideae species. Agronomy 2018, 8, 159. [Google Scholar] [CrossRef]
- Shaw, J.; Lickey, E.B.; Schilling, E.E.; Small, R.L. Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: The tortoise and the hare III. Am. J. Bot. 2007, 94, 275–288. [Google Scholar] [CrossRef] [PubMed]
- Ku, T.C.; Spongberg, S.A. Malus (Rosaceae). In Flora of China; Wu, Z.Y., Raven, P.H., Eds.; Science Press: Beijing, China; Missouri Botanical Garden Press: St. Louis, MO, USA, 2003; Volume 9, pp. 179–189. ISBN 9787030130440. [Google Scholar]
- Zhi-Qin, Z. The apple genetic resources in China: The wild species and their distributions, informative characteristics and utilisation. Genet. Res. Crop. Evol. 1999, 46, 599–609. [Google Scholar] [CrossRef]
- Cui, L.; Xing, M.; Xu, L.; Wang, J.; Zhang, X.; Ma, C.; Kang, W. Antithrombotic components of Malus halliana Koehne flowers. Food. Chem. Toxicol. 2018, 119, 326–333. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, F.A.N.; Rodrigues, S.; Carcel, J.A.; Garcia-Perez, J.V. Ultrasound-assisted air-drying of apple (Malus domestica L.) and its effects on the vitamin of the dried product. Food Bioprocess Technol. 2015, 8, 1503–1511. [Google Scholar] [CrossRef]
- Fang, L.; Meng, W.; Min, W. Phenolic compounds and antioxidant activities of flowers, leaves and fruits of five crabapple cultivars (Malus Mill. species). Sci. Hortic. 2018, 235, 460–467. [Google Scholar] [CrossRef]
- Huang, S.; Liu, H.F.; Meng, N.; Li, B.; Wang, J.L. Hypolipidemic and antioxidant effects of Malus toringoides (Rehd.) Hughes leaves in high-fat-diet-induced hyperlipidemic rats. J. Med. Food 2017, 20, 258–264. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Liu, F.Z.; Wang, K.; Wang, D.J.; Gong, X.; Liu, L.J.; Richards, C.M.; Henk, A.D.; Volk, G.M. Genetic diversity of Malus cultivars and wild relatives in the Chinese National Repository of Apple Germplasm Resources. Tree Genet. Genomes 2015, 11. [Google Scholar] [CrossRef]
- Kumar, C.; Singh, S.K.; Pramanick, K.K.; Verma, M.K.; Srivastav, M.; Singh, R.; Bharadwaj, C.; Naga, K.C. Morphological and biochemical diversity among the Malus species including indigenous Himalayan wild apples. Sci. Hortic. 2018, 233, 204–219. [Google Scholar] [CrossRef]
- Zhang, W.-X.; Zhao, M.-M.; Fan, J.-J.; Zhou, T.; Chen, Y.-X.; Cao, F.-L. Study on relationship between pollen exine ornamentation pattern and germplasm evolution in flowering crabapple. Sci Rep. 2017, 7, 39759. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, B.L.; Zhong, G.-Y.; Brown, S.K. Genetic diversity of dihydrochalcone content in Malus germplasm. Genet. Res. Crop. Evol. 2018, 65, 1485–1502. [Google Scholar] [CrossRef]
- Robinson, J.P.; Harris, S.A.; Juniper, B.E. Taxonomy of the genus Malus Mill. (Rosaceae) with emphasis on the cultivated apple, Malus domestica Borkh. Plant. Syst. Evol. 2001, 226, 35–58. [Google Scholar] [CrossRef]
- Wu, Z.Y.; Zhou, T.Y.; Xiao, P.G. Xinhua Materia Medica Outline. Jiangsu Institute of Botany; Shanghai Science and Technology Press: Shanghai, China, 1990; Volume 3, pp. 103–104. ISBN 7-5323-1421-9. [Google Scholar]
- State Administration of Traditional Chinese Medicine “Chinese Materia Medica Committee”. Zhong Hua Ben Cao (Chinese Materia Medica); Shanghai Science and Technology Press: Shanghai, China, 1999; Volume 4, pp. 158–159. ISBN 7-5323-5106-8. [Google Scholar]
- Chen, Y.L.; Tan, Z.X.; Peng, Y. Traditional uses and modern research of leaves of Malus hupehensis (Pamp.) rehd. Modern. Chin. Med. 2017, 10, 1505–1510. [Google Scholar] [CrossRef]
- Wen, C.; Wang, D.; Li, X.; Huang, T.; Huang, C.; Hu, K. Targeted isolation and identification of bioactive compounds lowering cholesterol in the crude extracts of crabapples using UPLC-DAD-MS-SPE/NMR based on pharmacology-guided PLS-DA. J. Pharm. Biomed. Anal. 2018, 150, 144–151. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Chen, Y.-Y.; Jiao, Q.-Y.; Khan, A.; Shan, J.; Cao, G.-D.; Li, F.; Zhang, C.; Lou, H.-X. Polyphenolic compounds from Malus hupehensis and their free radical scavenging effects. Nat. Prod. Res. 2018, 32, 2152–2158. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.-Q.; Zhu, X.-F.; Wang, X.-N.; Shen, T.; Xiang, F.; Lou, H.-X. Flavonoids from Malus hupehensis and their cardioprotective effects against doxorubicin-induced toxicity in H9c2 cells. Phytochemistry 2013, 87, 119–125. [Google Scholar] [CrossRef] [PubMed]
- You-Qin, D.; Tian-Yan, F.; Gai-Gai, D.; Ying, L.; Jun-Zhi, W. Inhibitory effect of total flavonoids of Malus hupehensis on hepatic fibrosis induced by Schistosoma japonicum in mice. Chin. J. Schi. Cont. 2011, 23, 551–554. [Google Scholar] [CrossRef]
- Chun-Nian, H.E.; Peng, Y.; Xiao, W.; Li-Jia, X.U.; Xiao, P.G. The prevention and treatment of cancer with non-camellia tea from China. Modern. Chin. Med. 2013, 16, 13–17. [Google Scholar] [CrossRef]
- Zhang, J.G.; Hu, H.J.; Xu, Y.H.; Luo, Z.R. Germplasm identification and genetic relationship of some Malus hupehensis (Pamp.) Rehd. accessions. J. Huazhong Agric. Univ. 2009, 6, 736–740. [Google Scholar] [CrossRef]
- Jansen, R.K.; Saski, C.; Lee, S.-B.; Hansen, A.K.; Daniell, H. Complete plastid genome sequences of three Rosids (Castanea, Prunus, Theobroma): Evidence for at least two independent transfers of rpl22 to the nucleus. Mol. Biol. Evol. 2011, 28, 835–847. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Wu, M.; Liao, B.; Liu, Z.; Bai, R.; Xiao, S.; Li, X.; Zhang, B.; Xu, J.; Chen, S. Complete chloroplast genome sequence and phylogenetic analysis of the medicinal plant Artemisia annua. Molecules 2017, 22, 1330. [Google Scholar] [CrossRef] [PubMed]
- Asaf, S.; Khan, A.L.; Khan, M.A.; Waqas, M.; Kang, S.-M.; Yun, B.-W.; Lee, I.-J. Chloroplast genomes of Arabidopsis halleri ssp. gemmifera and Arabidopsis lyrata ssp. petraea: Structures and comparative analysis. Sci. Rep. 2017, 7, 7556. [Google Scholar] [CrossRef] [PubMed]
- Wyman, S.K.; Jansen, R.K.; Boore, J.L. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 2004, 20, 3252–3255. [Google Scholar] [CrossRef] [PubMed]
- Shikanai, T.; Shimizu, K.; Ueda, K.; Nishimura, Y.; Kuroiwa, T.; Hashimoto, T. The chloroplast clpP gene, encoding a proteolytic subunit of ATP-dependent protease, is indispensable for chloroplast development in tobacco. Plant Cell Physiol. 2001, 42, 264–273. [Google Scholar] [CrossRef] [PubMed]
- Sy Nguyen, D.; Sai, T.Z.T.; Nawaz, G.; Lee, K.; Kang, H. Abiotic stresses affect differently the intron splicing and expression of chloroplast genes in coffee plants (Coffea arabica) and rice (Oryza sativa). Plant Physiol. 2016, 201, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Gu, C.; Dong, B.; Xu, L.; Tembrock, L.R.; Zheng, S.; Wu, Z. The complete chloroplast genome of Heimia myrtifolia and comparative analysis within Myrtales. Molecules 2018, 23, 846. [Google Scholar] [CrossRef] [PubMed]
- Jarvinen, P.; Palme, A.; Morales, L.O.; Lannenpaa, M.; Keinanen, M.; Sopanen, T.; Lascoux, M. Phylogenetic relationships of Betula species (Betulaceae) based on nuclear ADH and chloroplast matK sequences. Am. J. Bot. 2004, 91, 1834–1845. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.X.; Gadagkar, S.R.; Potter, D.; Xu, D.X.; Zhang, M.; Li, Z.Y. Phylogeny of Spiraea (Rosaceae) based on plastid and nuclear molecular data: Implications for morphological character evolution and systematics. Perspect. Plant Ecol. Evol. Syst. 2018, 34, 109–119. [Google Scholar] [CrossRef]
- Johnson, L.A.; Soltis, D.E. matK DNA sequences and phylogenetic reconstruction in Saxifragaceae s. str. Syst. Bot. 1994, 19, 143–156. [Google Scholar] [CrossRef]
- Steele, K.P.; Vilgalys, R. Phylogenetic analyses of polemoniaceae using nucleotide sequences of the plastid gene matK. Syst. Bot. 1994, 19, 126–142. [Google Scholar] [CrossRef]
- Plunkett, G.M.; Soltis, D.E.; Soltis, P.S. Clarification of the relationship between Apiaceae and Araliaceae based on matK and rbcL sequence data. Am. J. Bot. 1997, 84, 565–580. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.M.; He, R.; Zhang, H.Y.; Huang, Y.B.; Tian, M.L.; Zhang, J.J. Analysis of synonymous codon usage in Zea mays. Mol. Biol. Rep. 2010, 37, 677–684. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.Y.; Xing, H.X.; Yuan, Y.C.; Wang, X.L.; Saeed, M.; Tao, J.C.; Feng, W.; Zhang, G.H.; Song, X.L.; Sun, X.Z. Genome-wide analysis of codon usage bias in four sequenced cotton species. PLoS ONE 2018, 13, e0194372. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Shi, C.; Liu, Y.; Mao, S.Y.; Gao, L.Z. Thirteen Camellia chloroplast genome sequences determined by high-throughput sequencing: genome structure and phylogenetic relationships. BMC Evol. Biol. 2014, 14, 151. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.B.; Yin, J.L.; Guo, H.Y.; Zhang, Y.Y.; Xiao, W.; Sun, C.; Wu, J.Y.; Qu, X.B.; Yu, J.; Wang, X.M.; et al. The complete chloroplast genome provides insight into the evolution and polymorphism of Panax ginseng. Front. Plant Sci. 2015, 5, 696. [Google Scholar] [CrossRef] [PubMed]
- Gichira, A.W. The complete chloroplast genome sequence of an endemic monotypic genus Hagenia (Rosaceae): Structural comparative analysis, gene content and microsatellite detection. PeerJ 2017, 5, e2846. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Li, J.F.; Zhang, H.; Cai, B.H.; Gao, Z.H.; Qiao, Y.S.; Mi, L. The complete chloroplast genome sequence of strawberry (Fragaria X ananassa Duch.) and comparison with related species of Rosaceae. PeerJ 2017, 5, e3931. [Google Scholar] [CrossRef] [PubMed]
- Jian, H.Y.; Zhang, Y.H.; Yan, H.J.; Qiu, X.Q.; Wang, Q.G.; Li, S.B.; Zhang, S.D. The complete chloroplast genome of a key ancestor of modern roses, Rosa chinensis var. spontanea, and a comparison with congeneric species. Molecules 2018, 23, 389. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, Y.; Zang, M.; Li, M.; Fang, Y. Complete chloroplast genome sequence and phylogenetic analysis of quercus acutissima. Int. J. Mol. Sci. 2018, 19, 2443. [Google Scholar] [CrossRef] [PubMed]
- Chumley, T.W.; Palmer, J.D.; Mower, J.P.; Fourcade, H.M.; Calie, P.J.; Boore, J.L.; Jansen, R.K. The complete chloroplast genome sequence of Pelargonium x hortorum: Organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Mol. Biol. Evol. 2006, 23, 2175–2190. [Google Scholar] [CrossRef] [PubMed]
- Ogihara, Y.; Isono, K.; Kojima, T.; Endo, A.; Hanaoka, M.; Shiina, T.; Terachi, T.; Utsugi, S.; Murata, M.; Mori, N.; et al. Structural features of a wheat plastome as revealed by complete sequencing of chloroplast DNA. Mol. Genet. Genom. 2002, 266, 740–746. [Google Scholar] [CrossRef]
- Kim, K.J.; Lee, H.L. Complete chloroplast genome sequences from Korean ginseng (Panax schinseng Nees) and comparative analysis of sequence evolution among 17 vascular plants. DNA Res. 2004, 11, 247–261. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.P.; Xu, C.; Cheng, T.; Lin, K.; Zhou, S.L. Sequencing angiosperm plastid genomes made easy: A complete set of universal primers and a case study on the phylogeny of Saxifragales. Genome Biol. Evol. 2013, 5, 989–997. [Google Scholar] [CrossRef] [PubMed]
- Hollingsworth, P.M.; Graham, S.W.; Little, D.P. Choosing and using a plant DNA barcode. PLoS ONE 2011, 6. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.; Xu, C.; Li, C.; Sun, J.; Zuo, Y.; Shi, S.; Cheng, T.; Guo, J.; Zhou, S. Ycf1, the most promising plastid. DNA barcode of land plants. Sci. Rep. 2015, 5, 8348. [Google Scholar] [CrossRef] [PubMed]
- Krawczyk, K.; Szczecinska, M.; Sawicki, J. Evaluation of 11 single-locus and seven multilocus DNA barcodes in Lamium. L. (Lamiaceae). Mol. Ecol. Resour. 2014, 14, 272–285. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.A.; Gyulai, G.; Hidvegi, N.; Kerti, B.; Al Hemaid, F.M.A.; Pandey, A.K.; Lee, J.K. The changing epitome of species identification DNA barcoding. Saudi J. Biol. Sci. 2014, 21, 204–231. [Google Scholar] [CrossRef]
- Niu, Z.; Xue, Q.; Zhu, S.; Sun, J.; Liu, W.; Ding, X. The complete plastome sequences of four orchid species: Insights into the evolution of the Orchidaceae and the utility of plastomic mutational hotspots. Front. Plant. Sci. 2017, 8, 715. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.S.; Fu, P.C.; Zhou, X.J.; Cheng, Y.W.; Zhang, F.Q.; Chen, S.L.; Gao, Q.B. The complete plastome sequences of seven species in Gentianasect. Kudoa (Gentianaceae): Insights into plastid gene loss and molecular evolution. Front. Plant Sci. 2018, 9, 493. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.Z.; Huang, C.H.; Hu, Y.; Wen, J.; Li, S.S.; Yi, T.S.; Chen, H.Y.; Xiang, J.; Ma, H. Evolution of Rosaceae Fruit Types Based on Nuclear Phylogeny in the Context of Geological Times and Genome Duplication. Mol. Biol. Evol. 2017, 34, 262–281. [Google Scholar] [CrossRef] [PubMed]
- Cho, M.S.; Yang, J.Y.; Kim, S.C. Complete chloroplast genome of Ulleung Island endemic flowering cherry, Prunus takesimensis (Rosaceae), in Korea. Mitochondrial DNA Part B Resour. 2018, 3, 274–275. [Google Scholar] [CrossRef]
- Terakami, S.; Matsumura, Y.; Kurita, K.; Kanamori, H.; Katayose, Y.; Yamamoto, T.; Katayama, H. Complete sequence of the chloroplast genome from pear (Pyrus pyrifolia): genome structure and comparative analysis. Tree Genet. Genomes 2012, 8, 841–854. [Google Scholar] [CrossRef]
- Potter, D.; Eriksson, T.; Evans, R.C.; Oh, S.; Smedmark, J.E.E.; Morgan, D.R.; Kerr, M.; Robertson, K.R.; Arsenault, M.; Dickinson, T.A.; et al. Phylogeny and classification of Rosaceae. Plant Syst. Evol. 2007, 266, 5–43. [Google Scholar] [CrossRef]
- Zhang, S.D.; Jin, J.J.; Chen, S.Y.; Chase, M.W.; Soltis, D.E.; Li, H.T.; Yang, J.B.; Li, D.Z.; Yi, T.S. Diversification of Rosaceae since the late cretaceous based on plastid phylogenomics. New Phytol. 2017, 214, 1355–1367. [Google Scholar] [CrossRef] [PubMed]
- Lo, E.Y.Y.; Donoghue, M.J. Expanded phylogenetic and dating analyses of the apples and their relatives (Pyreae, Rosaceae). Mol. Phylogenet. Evol. 2012, 63, 230–243. [Google Scholar] [CrossRef] [PubMed]
- Hahn, C.; Bachmann, L.; Chevreux, B. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—A baiting and iterative mapping approach. Nucleic Acids Res. 2013, 41, e129. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Lohse, M.; Drechsel, O.; Kahlau, S.; Bock, R. OrganellarGenomeDRAW—A suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 2013, 41, W575–W581. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, S.; Choudhuri, J.V.; Ohlebusch, E.; Schleiermacher, C.; Stoye, J.; Giegerich, R. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001, 29, 4633–4642. [Google Scholar] [CrossRef] [PubMed]
- Thiel, T.; Michalek, W.; Varshney, R.; Graner, A. Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor. Appl. Genet. 2003, 106, 411–422. [Google Scholar] [CrossRef] [PubMed]
- Frazer, K.A.; Pachter, L.; Poliakov, A.; Rubin, E.M.; Dubchak, I. VISTA: Computational tools for comparative genomics. Nucl. Acids Res. 2004, 32, W273–W279. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Kaya, T.; Balta, F.; Sensoy, S. Fruit quality parameters and molecular analysis of apple germplasm resources from Van Lake Basin, Turkey. Turk. J. Agric. For. 2015, 39, 864–875. [Google Scholar] [CrossRef]
- Potts, S.M.; Han, Y.P.; Khan, M.A.; Kushad, M.M.; Rayburn, A.L.; Korban, S.S. Genetic diversity and characterization of a core collection of Malus germplasm using simple sequence repeats (SSRs). Plant Mol. Biol. Rep. 2012, 30, 827–837. [Google Scholar] [CrossRef]
- Potts, S.M.; Kushad, M.M.; Korban, S.S. Genetic diversity and classification of Malus germplasm using simple sequence repeats (SSRs). Hortscience 2009, 44, 1173. [Google Scholar]
- Linden, L.; Iwarsson, M. Identification of weeping crabapple cultivars by microsatellite DNA markers and morphological traits. Sci. Hortic. 2014, 179, 221–226. [Google Scholar] [CrossRef]
- Guo, Q.G.; Yu, Y.; He, Q.; Li, X.L.; Liang, G.L. AFLP analysis of four wild Malus Mill. Acta Hortic. 2007, 16, 131–135. [Google Scholar] [CrossRef]
- Xu, R.X.; Hu, D.C.; Chen, Z.Y.; Zhang, P.; Jiang, X.M.; Tang, G.G. SRAP analysis on genetic relationships of genotypes in the genus Malus Mill. Biotechnol. Biotechnol. Equip. 2014, 28, 602–607. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Liu, F.Z.; Gao, Y.; Wang, D.J.; Gong, X.; Liu, L.J. The natural distribution, diversity and utilization of wild in China. J. Plant Genet. Resour. 2013, 14, 1013–1019. [Google Scholar] [CrossRef]
Sample Availability: Samples of M. hupehensis are available from the authors. |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).