Abstract
The genus Calonectria includes pathogens of various agricultural, horticultural, and forestry crops. Species of Calonectria are commonly collected from soils, fruits, leaves, stems, and roots. Some species of Calonectria isolated from soils are considered as important plant pathogens. Understanding the species diversity and distribution characteristics of Calonectria species in different soil layers will help us to clarify their long-term potential harm to plants and their patterns of dissemination. To our knowledge, no systematic research has been conducted concerning the species diversity and distribution characteristics of Calonectria in different soil layers. In this study, 1000 soil samples were collected from five soil layers (0–20, 20–40, 40–60, 60–80, and 80–100 cm) at 100 sampling points in one 15-year-old Eucalyptus urophylla hybrid plantation in southern China. A total of 1037 isolates of Calonectria present in all five soil layers were obtained from 93 of 100 sampling points. The 1037 isolates were identified based on DNA sequence comparisons of the translation elongation factor 1-alpha (tef1), β-tubulin (tub2), calmodulin (cmdA), and histone H3 (his3) gene regions, as well as the combination of morphological characteristics. These isolates were identified as C. hongkongensis (665 isolates; 64.1%), C. aconidialis (250 isolates; 24.1%), C. kyotensis (58 isolates; 5.6%), C. ilicicola (47 isolates; 4.5%), C. chinensis (2 isolates; 0.2%), and C. orientalis (15 isolates; 1.5%). With the exception of C. orientalis, which resides in the C. brassicae species complex, the other five species belonged to the C. kyotensis species complex. The results showed that the number of sampling points that yielded Calonectria and the number (and percentage) of Calonectria isolates obtained decreased with increasing depth of the soil. More than 84% of the isolates were obtained from the 0–20 and 20–40 cm soil layers. The deeper soil layers had comparatively lower numbers but still harbored a considerable number of Calonectria. The diversity of five species in the C. kyotensis species complex decreased with increasing soil depth. The genotypes of isolates in each Calonectria species were determined by tef1 and tub2 gene sequences. For each species in the C. kyotensis species complex, in most cases, the number of genotypes decreased with increasing soil depth. The 0–20 cm soil layer contained all of the genotypes of each species. To our knowledge, this study presents the first report of C. orientalis isolated in China. This species was isolated from the 40–60 and 60–80 cm soil layers at only one sampling point, and only one genotype was present. This study has enhanced our understanding of the species diversity and distribution characteristics of Calonectria in different soil layers.
1. Introduction
Species in the genus Calonectria (Hypocreales, Nectriaceae) are phytopathogenic fungi that cause serious losses to plant crops in tropical and subtropical regions of the world [1,2,3,4,5,6]. Many species of Calonectria are important pathogens of agricultural, horticultural, and forestry crops and these species occur in approximately 335 plant species in nearly 100 plant families [1]. Species of Calonectria have been isolated from soils, fruits, leaves, stems, and roots [1,4,7,8,9,10,11,12,13,14]. The fungi are best known as foliar, shoot, and root pathogens [1,2,4,5], and they are commonly associated with disease symptoms, including seedling damping-off, seedling rot, cutting rot, leaf spots, leaf blight, shoot blight, crown cankers, stem lesions, collar and root rots, and tuber rot [1,14,15,16,17,18,19,20,21,22,23].
Some species of Calonectria isolated from soils are important plant pathogens. Calonectria ilicicola is a soil-borne fungal pathogen of worldwide importance that causes black rot disease in peanut and red crown rot in soybean [21,24,25,26,27,28]. Recently, we isolated five Calonectria species, namely C. aconidialis, C. auriculiformis, C. hongkongensis, C. pseudoreteaudii, and C. reteaudii, from soils in a plantation of Eucalyptus trees [14]. Inoculation results showed that all five species caused leaf spot, leaf blight, and seedling rot to the tested Eucalyptus genotypes within three days [14].
Previous research results indicated a high level of species diversity of Calonectria in southern China, especially in soils [9,11,13,14,23]. Currently, a total of 125 Calonectria species have been described using DNA sequence-based phylogenetic analyses and morphological comparisons [5,13,29,30,31,32,33,34,35]. A total of 25 species of Calonectria have been identified and described in China based on DNA sequence data [5,9,11,13,14,36]. Of these species, 17 have been isolated from soils, with 11 from soils under plantation Eucalyptus trees [5,9,11,14].
Some Calonectria species can survive in soil for long periods, and microsclerotia are the primary survival structures [37]. Microsclerotia of some Calonectria species can survive in the absence of hosts for 15 years or more [38,39]. Calonectria microsclerotia have been recorded at depths of up to 66 cm below the soil surface [40]. Long-term survival and deep soil presence of microsclerotia are serious threats to the management of diseases caused by Calonectria species.
Understanding the diversity and distribution characteristics of Calonectria species in different soil layers will help us to clarify their potential long-term harm to plants and potential dissemination patterns. Very little research has been conducted concerning the distribution characteristics of microsclerotia in soils, and the few published studies have focused only on the surface soil [38,41]. In the past several years, studies have been conducted to understand Calonectria species diversity in forest soils [9,10,11,13,14,36], but all of the soil samples obtained for Calonectria isolation were collected from the 0–20 cm soil layer. In this study, a relatively large number of soil samples were collected from five different soil layers up to 100 cm depth in one 15-year-old Eucalyptus urophylla hybrid plantation. Isolates of Calonectria from this plantation were obtained and identified. The aims of this study were as follows: (1) to understand the species diversity of Calonectria in different soil layers; and (2) to understand the distribution characteristics of each Calonectria species in different soil layers.
2. Materials and Methods
2.1. Study Site, Soil Sampling, and Calonectria Isolation
This study was performed in a Eucalyptus urophylla hybrid plantation (21°15′31.74″ N, 110°06′35″ E; altitude 90 m) located in the South China Experimental Nursery, China Eucalypt Research Centre (CERC), ZhanJiang, GuangDong Province, China. The Eucalyptus plantation is located on the northern edge of the tropics, with a maritime monsoon climate [42]. The average annual precipitation is 1777 mm, and the period from May to October accounts for 84.1% of the annual precipitation. The annual average temperature is 23.4 °C (http://en.weather.com.cn; accessed date: 10 August 2021). The soil type is Rhodi-Udic Ferralosols, according to the Chinese Soil Taxonomy Classification [42,43]. The area of the Eucalyptus plantation is about 6 ha (400 × 150 m), and the planting density of Eucalyptus trees is 3 × 2 m. The Eucalyptus trees were 15 years old.
One hundred points in the Eucalyptus plantation were selected for soil sampling. The 100 points were randomly distributed in the plantation, and the distance between adjacent sampling points was 10 m. Soil samples were collected from five layers at each sampling point: 0–20, 20–40, 40–60, 60–80, and 80–100 cm. Two soil samples were collected in each soil layer for each sampling point. In total, 1000 soil samples were collected from the 100 sampling points. Each of the soil samples was placed in a resealable plastic bag and transferred to the laboratory for Calonectria isolation. The soil samples were collected from July to August 2020.
For Calonectria isolation, the collected soil was transferred into a plastic cylinder sampling cup (diameter = 4.5 cm, height = 5 cm, and volume = 80 mL) (Chengdu Rich Science Industry Co., Ltd., Chengdu, China); the soil sample occupied two-thirds of the volume of the whole sampling cup volume. The soil sample was moistened by spraying with sterile water and stirred evenly with a sterilized bamboo stick. Medicago sativa (alfalfa) seeds were scattered onto the soil surface after it was surface-disinfested (30 s in 75% ethanol and washed several times with sterile water) in the sampling cup. The sampling cup with soil and alfalfa seeds was incubated at 25 °C under 12 h of daylight and 12 h of darkness. After one week, sporulating conidiophores with typical morphological characteristics of Calonectria species [1] were produced on infected alfalfa tissue. Using a dissection microscope (AxioCam Stemi 2000C, Carl Zeiss, Germany), the single conidial mass was scattered onto 2% malt extract agar (MEA) (20 g malt extract powder and 20 g agar powder per liter of water: malt extract powder was obtained from Beijing Shuangxuan microbial culture medium products factory, Beijing, China; the agar powder was obtained from Beijing Solarbio Science & Technology Co., Ltd., Beijing, China) using a sterile needle. After incubation at 25 °C for three to four hours, the germinated conidia were individually transferred onto fresh MEA under the dissection microscope and incubated at 25 °C for one week to obtain single-conidium cultures. For each soil sample, the soil was transferred into two plastic sampling cups for Calonectria isolation.
2.2. DNA Extraction, PCR Amplification, and Sequencing
All isolates obtained in this study were used for DNA extraction and sequence comparisons. DNA was extracted from 10-day-old cultures. Mycelia were collected using a sterilized scalpel and transferred to 2-mL Eppendorf tubes. The total genomic DNA was extracted using the CTAB protocol described by van Burik and co-authors [44]. The extracted DNA was dissolved in 30 µL TE buffer (1 M Tris-HCl and 0.5 M EDTA, pH 8.0), and 2.5 µL RNase (10 mg/mL) was added at 37 °C for one hour to degrade RNA. Finally, the DNA concentration was measured using a NanoDrop 2000 spectrometer (Thermo Fisher Scientific, Waltham, MA, USA).
According to previous research results, sequences of partial gene regions of translation elongation factor 1-alpha (tef1) and β-tubulin (tub2), as well as calmodulin (cmdA) and histone H3 (his3), were used to successfully identify Calonectria species [5,14]. These four partial gene regions were amplified using the primer pairs EF1-728F/EF2, T1/CYLTUB1R, CAL-228F/CAL-2Rd, and CYLH3F/CYLH3R, respectively. The PCR procedure was conducted as described by Liu and Chen [36] and Wang and Chen [23].
To obtain accurate sequences for each of the sequenced isolates, all of the PCR products were sequenced in both forward and reverse directions using the same primers used for PCR amplification by the Beijing Genomics Institute, Guangzhou, China. All of the sequences obtained in this study were edited using MEGA v. 7.0 software [45] and were deposited in GenBank (https://www.ncbi.nlm.nih.gov; accessed date: 18 September 2021). The tef1 and tub2 gene regions were sequenced for all Calonectria isolates. The isolates were genotyped by the tef1 and tub2 sequences. Based on the genotypes generated by tef1 and tub2 sequences, up to eight isolates for each tef1-tub2 genotype were selected for sequencing the cmdA and his3 gene regions.
2.3. Multi-Gene Phylogenetic Analyses, Morphology, and Species Identification
A standard nucleotide BLAST search was conducted using the tef1, tub2, cmdA, and his3 sequences to preliminarily identify the species from which the isolates were obtained in this study. Sequences of tef1, tub2, cmdA, and his3 gene regions obtained in this study were compared with sequences of type specimen strains of published Calonectria species. Sequences of all of the published species in the relevant species complexes were used for sequence comparisons and phylogenetic analyses. The datasets of Liu and co-authors [5] were used as templates for analyses, while sequences of other recently described Calonectria species [13,32,33,34,35] were also used for sequence comparisons.
Sequences of each of the tef1, tub2, cmdA, and his3 gene regions, as well as the combination of these four gene regions, were aligned using the online version of MAFFT v. 7 (http://mafft.cbrc.jp/alignment/server; accessed date: 7 August 2021) with the alignment strategy FFT-NS-i (Slow; interactive refinement method). Sequence alignments were manually edited using MEGA v. 7.0 software [45] after initial alignments.
For Calonectria species, maximum parsimony (MP) and maximum likelihood (ML) are frequently used for phylogenetic analyses [5,9,12,14]. Both MP and ML were used for phylogenetic analyses of sequence datasets of each of the four genes and the combination of the four gene regions in order to test whether the analysis results between the two methods were consistent. The MP and ML analyses were conducted by the methods described by Liu and Chen [36]. Phylogenetic trees were viewed by MEGA v. 7.0 [45]. Sequence data of two isolates of Curvicladiella cignea (CBS 109167 and CBS 109168) were used as outgroups [5].
The isolates selected for sequencing tef1, tub2, cmdA, and his3 gene regions were used for morphological studies. Size of macroconidia and width of vesicles are the most typical asexual characteristics used for morphological comparisons for species of Calonectria [5,9,11,13,14,29,36]. In order to induce asexual structures, isolates were cultured on 2% MEA in Petri dishes at 25 °C for 10 days. Sterile water was then added to the Petri dishes, and a sterilized, soft-bristled paintbrush was used to dislodge the mycelium from the agar surface. The water was then removed, and the dishes were placed upside down and incubated at 25 °C for 2–3 days. This resulted in asexual structures being produced on the surface of the cultures for some Calonectria isolates, a pattern that has been noted for Calonectria pteridis by Graça and co-authors [46] and for Calonectria pentaseptata (synonymized as a synonym of C. pseudoreteaudii in Liu and co-authors [5]) by Wang and Chen [23]. Fifty measurements of macroconidia and vesicles were measured for the selected isolates that produced abundant macroconidia and vesicles.
2.4. Calonectria Species Diversity in Different Soil Layers
After all of the Calonectria isolates were identified, the number of isolates present in each identified species was counted. The species diversity associated with soil layers was computed. The distribution characteristics of each Calonectria species in each soil layer were recorded, including the number of sampling points from which each Calonectria species was obtained and the number of isolates of each Calonectria species in each of the five soil layers.
2.5. Genotyping of Isolates within Each Calonectria Species
After all of the Calonectria isolates were identified, we examined the genotype diversity of each identified Calonectria species in the five different soil layers. The genotypes of isolates within each species were determined based on tef1 and tub2 sequences, and the number of isolates belonging to each genotype was recorded.
2.6. Genotype Diversity of Calonectria Species in Different Soil Layers
Based on the results of genotype analysis of each isolate determined by the sequences of tef1 and tub2 gene regions, the numbers of genotypes of each Calonectria species in different soil layers were counted. To investigate possible evolutionary relationships among the observed tef1–tub2 genotypes for the Calonectria species identified in this study with the most dominant species, minimum spanning networks (MSN) were constructed using Bruvo’s distance with the R packages poppr and ape [47,48].
3. Results
3.1. Soil Sampling and Calonectria Isolation
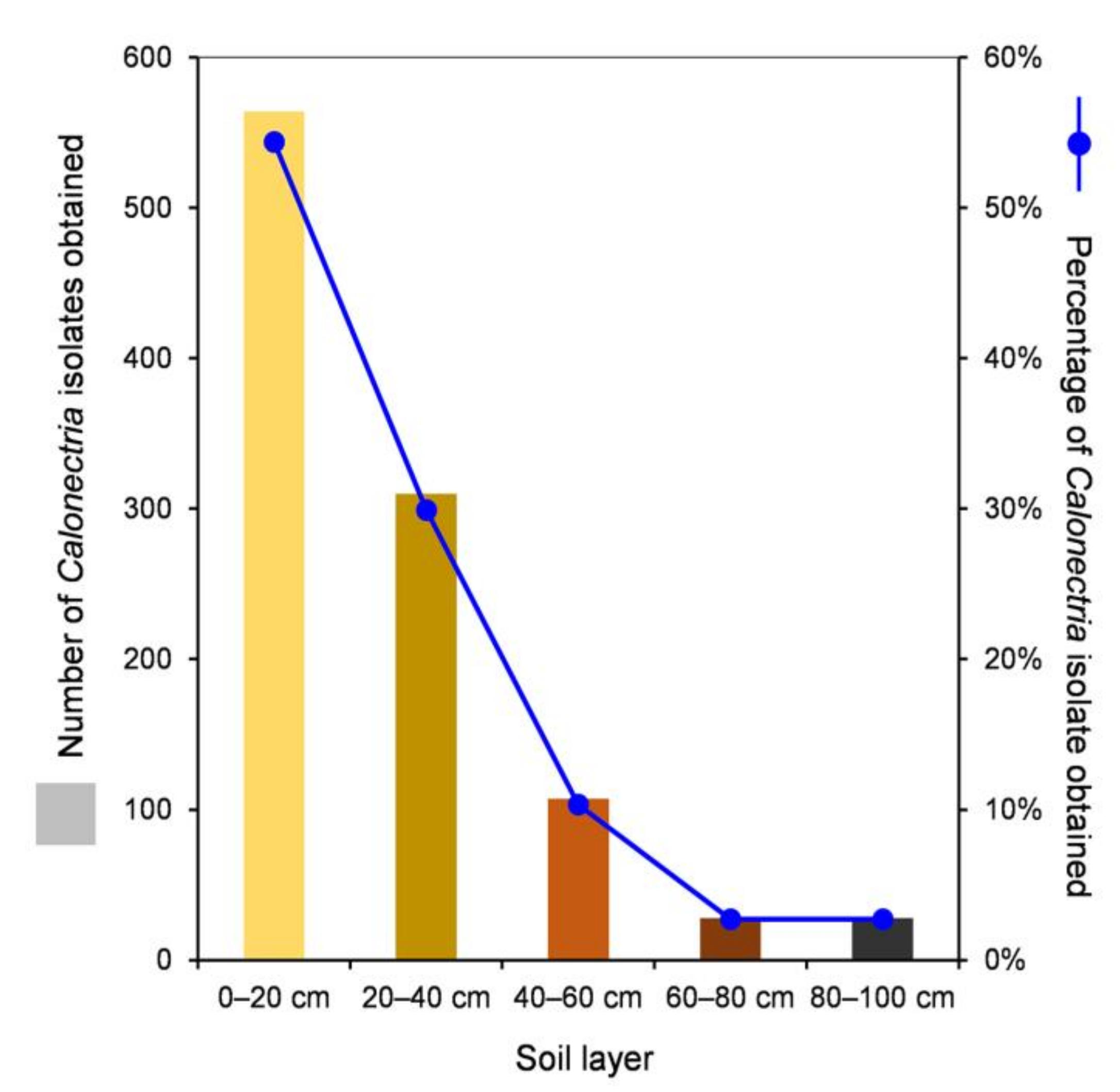
One thousand soil samples from 100 sample points were collected from the E. urophylla hybrid plantation, with 200 soil samples from each of the five soil layers. For each soil sample, two plastic sampling cups with soil and alfalfa seeds were used for the incubation of Calonectria. After the conidia were transferred onto fresh MEA and incubated at 25 °C, more than 90% of the conidia germinated within four hours. For each sampling cup, one to four single conidia were transferred onto fresh MEA to obtain one to four single-conidium cultures. In total, Calonectria fungi were isolated from 93 sampling points in the plantation; the totals were 92, 40, 20, 7, and 5 from the 0–20, 20–40, 40–60, 60–80, and 80–100 cm soil layers, respectively (Supplementary Table S1, Supplementary Figure S1). One thousand and thirty-seven isolates of Calonectria were obtained, with 564 (54.4%), 310 (29.9%), 107 (10.3%), 28 (2.7%), and 28 isolates (2.7%) from the 0–20, 20–40, 40–60, 60–80, and 80–100 cm soil layers, respectively, and 84.3% of the isolates were distributed in the 0–20 and 20–40 cm soil layers (Table 1, Supplementary Table S2, Figure 1). From the results, it was clear that the number of sampling points that yielded Calonectria and the number (and percentage) of Calonectria isolates obtained decreased with increasing soil depth (Supplementary Figure S1, Figure 1).

Table 1.
Number of isolates obtained for each Calonectria species from each soil layer.
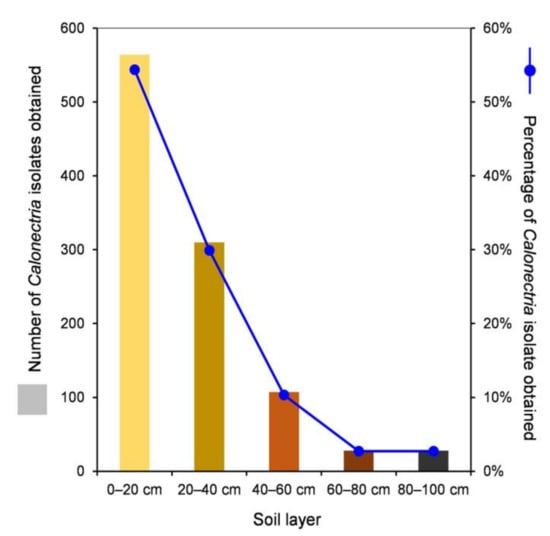
Figure 1.
Numbers and percentages of Calonectria isolates obtained in each of the five soil layers.
3.2. Sequencing
The tef1 and tub2 genes were amplified for all the 1037 isolates obtained in this study (Supplementary Table S2). Twenty-two genotypes were generated based on tef1 and tub2 gene sequences (Table 2). Depending on the isolate number of each tef1-tub2 genotype, one to eight isolates of each genotype were selected; finally, 85 isolates in total were selected to sequence the cmdA and his3 gene regions (Table 3). The sequence fragments were approximately 500, 565, 685, and 440 bp for the tef1, tub2, cmdA, and his3 gene regions, respectively.

Table 2.
Isolate numbers of each genotype from each Calonectria species.

Table 3.
Isolates sequenced and used for phylogenetic analyses and morphological studies in this study.
3.3. Multi-Gene Phylogenetic Analyses, Morphology, and Species Identification
The standard nucleotide BLAST search results conducted using the tef1, tub2, cmdA, and his3 sequences showed that the isolates obtained in the current study belonged to two species complexes of Calonectria, namely, the C. kyotensis species complex and the C. brassicae species complex. The 85 Calonectria isolates with four gene regions sequenced were used for phylogenetic analyses (Table 3). Based on the recently published results in Liu and co-authors [5] and Crous and co-authors [34], sequences of tef1, tub2, cmdA, and his3 of published species in the C. kyotensis species complex and C. brassicae species complex, respectively, were used for sequence comparisons and phylogenetic analyses (Table 4).

Table 4.
Isolates from other studies used in the phylogenetic analyses in this study.
The partition homogeneity test (PHT) comparing the tef1, tub2, cmdA, and his3 gene combination datasets generated a p-value of 0.001, indicating that the accuracy of the combined datasets did not suffer relative to the individual partitions [60]. Thus, sequences of the four loci were combined for analyses. Between the MP and ML trees, the overall topologies were similar for the phylogenetic trees based on tef1, tub2, cmdA, and his3 individually and the combination datasets, but the relative positions of some Calonectria species slightly differed. The five ML trees are presented in Figure 2 and Supplementary Figures S2–S5. The numbers of taxa and parsimony-informative characters, statistical values of the MP analyses, and parameters of the best-fit substitution models of ML analyses are provided in Table 5.
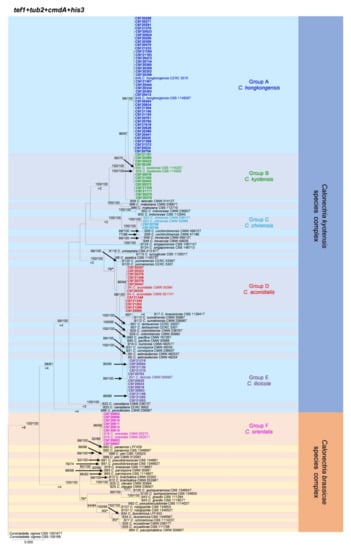
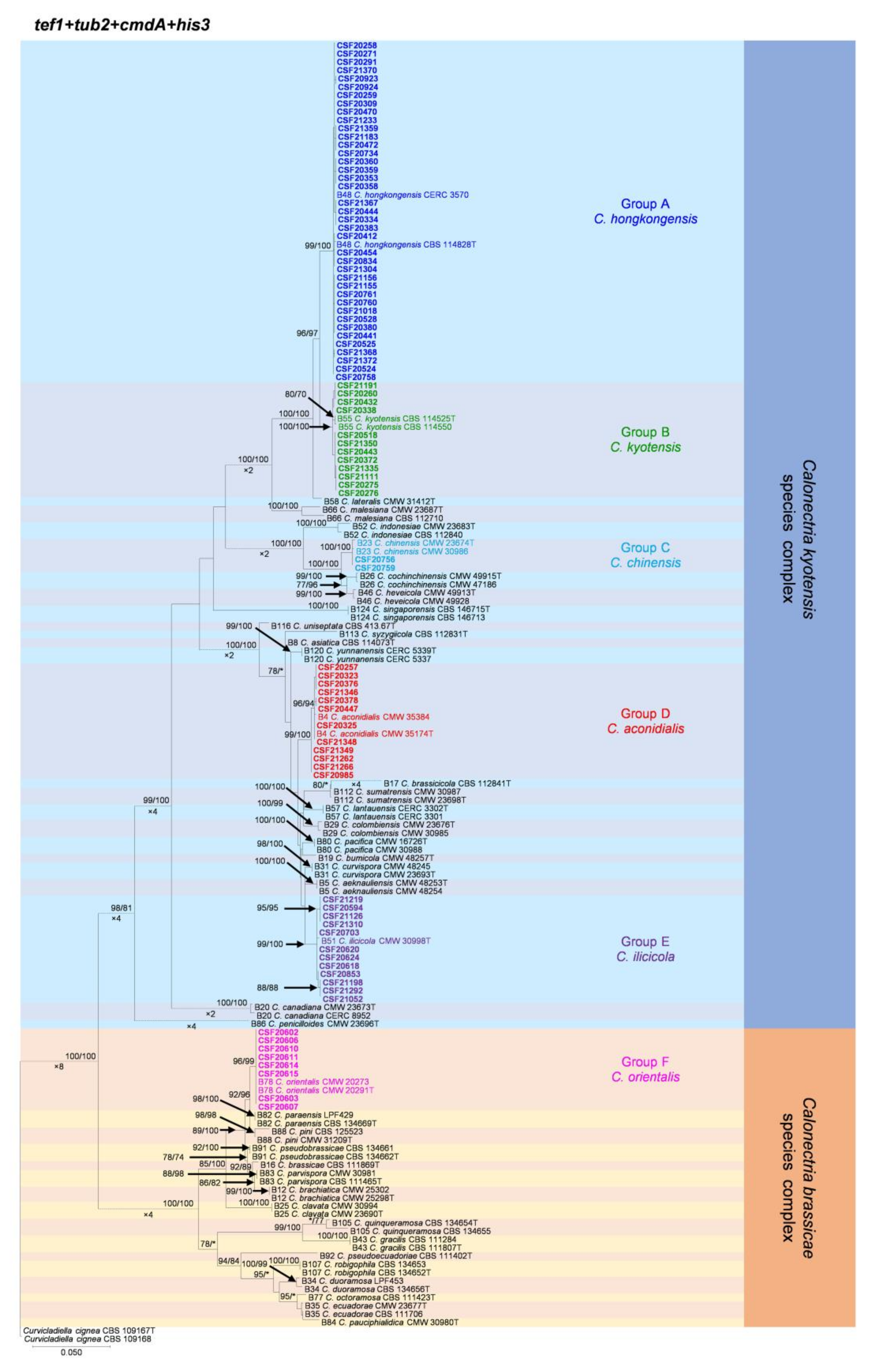
Figure 2.
Phylogenetic tree of Calonectria species based on maximum likelihood (ML) analyses of the dataset of combined tef1, tub2, cmdA, and his3 gene sequences in this study. Bootstrap support values ≥ 70% are presented above the branches as follows: ML/MP. Bootstrap values < 70% or absent are marked with “*”. Isolates highlighted in six different colors, and bold were obtained in this study. Ex-type isolates are marked with “T”. The “B” species codes are consistent with the recently published results in Liu and co-authors [5]. Curvicladiella cignea (CBS 109167 and CBS 109168) was used as the outgroup taxon.

Table 5.
Statistical values of datasets for maximum parsimony and maximum likelihood analyses in this study.
The phylogenetic analyses showed that the 85 Calonectria isolates were clustered in six groups (Group A, Group B, Group C, Group D, Group E, and Group F) based on tef1, tub2, cmdA, his3, and combined tef1/tub2/cmdA/his3 analyses (Figure 2, Supplementary Figures S2–S5). The analyses showed that isolates in Groups A, B, C, D, and E belonged to the C. kyotensis species complex. Isolates in Groups A, B, C, and E were clustered with or were closest to C. hongkongensis, C. kyotensis, C. chinensis, and C. ilicicola, respectively, based on the tef1, tub2, cmdA, his3, and combined tef1/tub2/cmdA/his3 trees (Figure 2, Supplementary Figures S2–S5). Therefore, isolates in Groups A, B, C, and E were identified as C. hongkongensis, C. kyotensis, C. chinensis, and C. ilicicola, respectively. Isolates in Group D were clustered in two sub-groups, sub-group D1 and sub-group D2, in the tub2 tree. Isolates in sub-group D1 were clustered with or were closest to C. aconidialis; isolates in sub-group D2 were clustered with C. asiatica (Supplementary Figure S3); isolates in Group D were clustered with or were closest to C. aconidialis based on the tef1, cmdA, his3, and combined tef1/tub2/cmdA/his3 trees (Figure 2, Supplementary Figures S2, S4, and S5). Isolates in Group D were identified as C. aconidialis. Isolates in Group F belonged to the C. brassicae species complex. These isolates were consistently only clustered with C. orientalis based on the tef1, tub2, his3, and combined tef1/tub2/cmdA/his3 trees and were clustered with both C. orientalis and C. brassicae in the cmdA tree (Figure 2, Supplementary Figures S2–S5). Isolates in Group F were identified as C. orientalis.
Based on the results of phylogenetic analyses and induction of asexual structures, 17 isolates representing six Calonectria species were selected for macroconidia and vesicle morphological comparisons (Table 3 and Table 6). These representative isolates could be classified into two groups based on the shape of the vesicles. Isolates of C. aconidialis, C. chinensis, C. hongkongensis, C. ilicicola, and C. kyotensis produce sphaeropedunculate vesicles, while the vesicles of C. orientalis are typically clavate. With the exception of C. ilicicola isolates, which produce 1(–3) septate macroconidia, isolates of the other five species all produced one septate macroconidium (Table 6). The shape of the vesicle and the number of macroconidia septations for each of the six Calonectria species found in this study were consistent with the described strains of relevant species in previous studies [1,9,29,49] (Table 6).

Table 6.
Morphological comparisons of Calonectria isolates and species obtained in the current study.
The morphological comparisons showed that significant variation existed in the size of macroconidia or width of vesicles among some isolates of each species of C. aconidialis, C. hongkongensis, and C. kyotensis identified in this study. For example, the macroconidia of C. aconidialis isolate CSF20985 were much longer than those of the other two tested C. aconidialis isolates CSF20323 and CSF20376. In C. hongkongensis, the macroconidia of isolate CSF20383 were longer than those of the other four isolates; the vesicles of isolate CSF20924 were wider than those of other isolates. In C. kyotensis, the macroconidia of isolate CSF20276 were much longer than those of isolate CSF21191 (Table 6).
The measurement results further showed that macroconidia size and vesicle width of isolates of some species obtained in this study were not always similar to the originally described strains of the same Calonectria species. For example, the macroconidia lengths of isolates of C. chinensis and C. orientalis obtained in this study were much shorter than the originally described strains of the relevant species [29,49] (Table 6).
3.4. Calonectria Species Diversity in Different Soil Layers
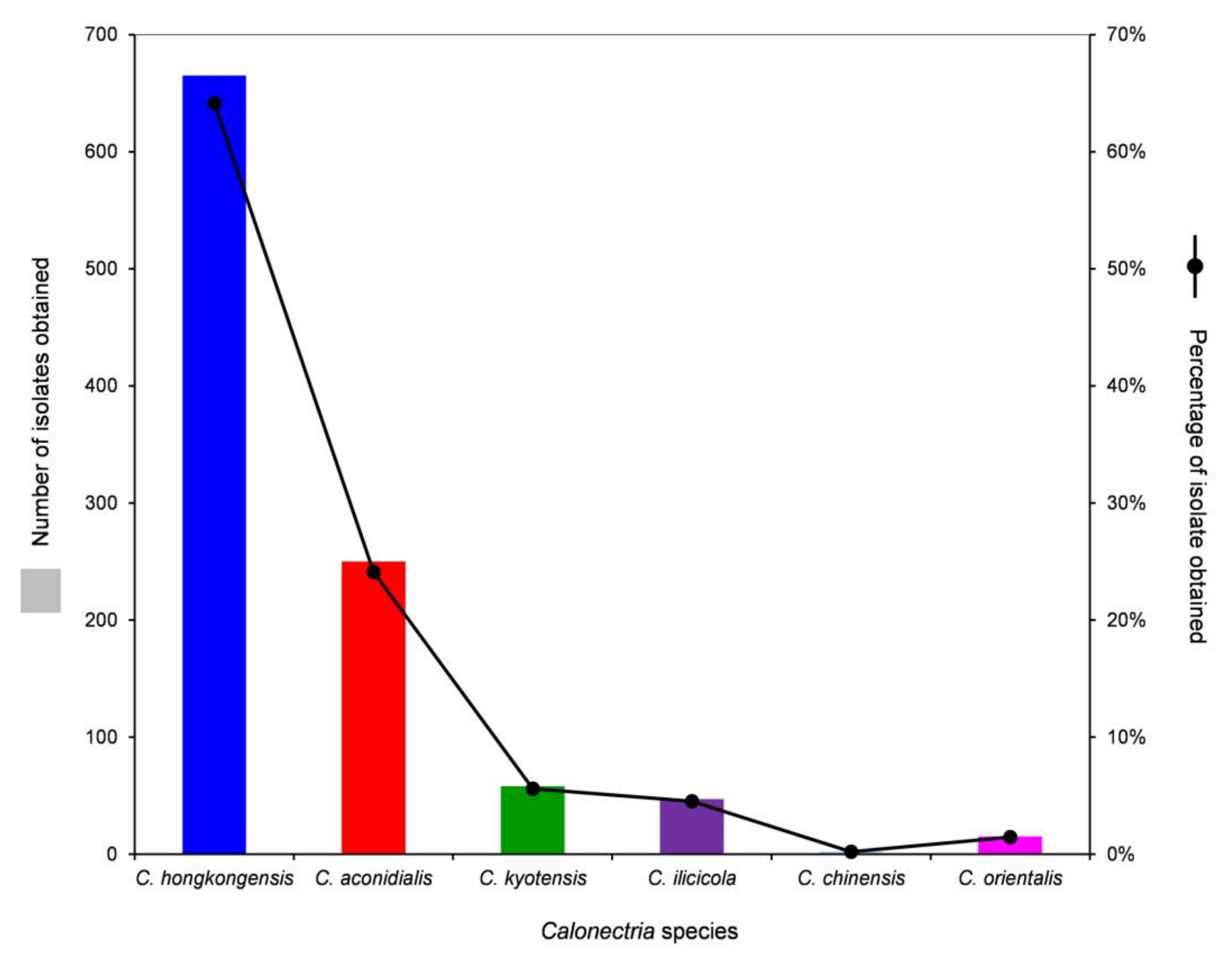
Based on the sequence comparisons of tef1, tub2, cmdA, and his3 sequences, the 1037 Calonectria isolates were identified as C. hongkongensis (665 isolates; 64.1%), C. aconidialis (250 isolates; 24.1%), C. kyotensis (58 isolates; 5.6%), C. ilicicola (47 isolates; 4.5%), C. chinensis (2 isolates; 0.2%), and C. orientalis (15 isolates; 1.5%) (Table 1). Calonectria hongkongensis was dominant, followed by C. aconidialis. Each of the two dominant species was isolated from more than or close to 50% of all of the sampling points, and the two species accounted for 88.2% of all of the Calonectria isolates obtained in this study (Table 1, Supplementary Table S1, Figure 3). Both C. chinensis and C. orientalis were only isolated from one sampling point; C. chinensis was only isolated from the 0–20 cm soil layer, and only two isolates were obtained; C. orientalis was isolated from the soil layers of 40–60 and 60–80 cm, and 11 and 4 isolates in the two soil layers were obtained, respectively (Table 1, Supplementary Table S1, Figure 3).
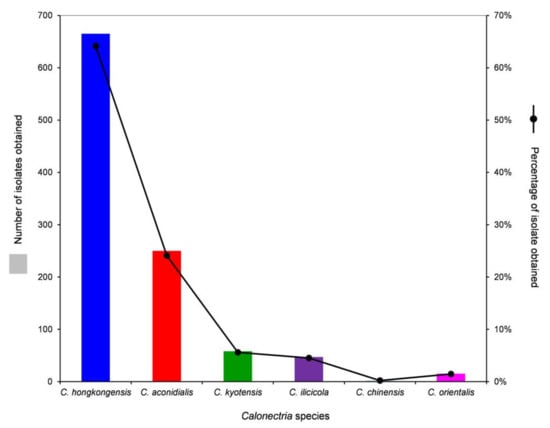
Figure 3.
Numbers and percentages of isolates obtained for each Calonectria species from all soil samples collected.
With the exception of C. orientalis in the C. brassicae species complex, the diversity of species in the C. kyotensis species complex decreased with increasing soil depth. Five, four, four, four, and two species were identified in the soil layers of 0–20, 20–40, 40–60, 60–80, and 80–100 cm, respectively (Table 1, Supplementary Table S1).
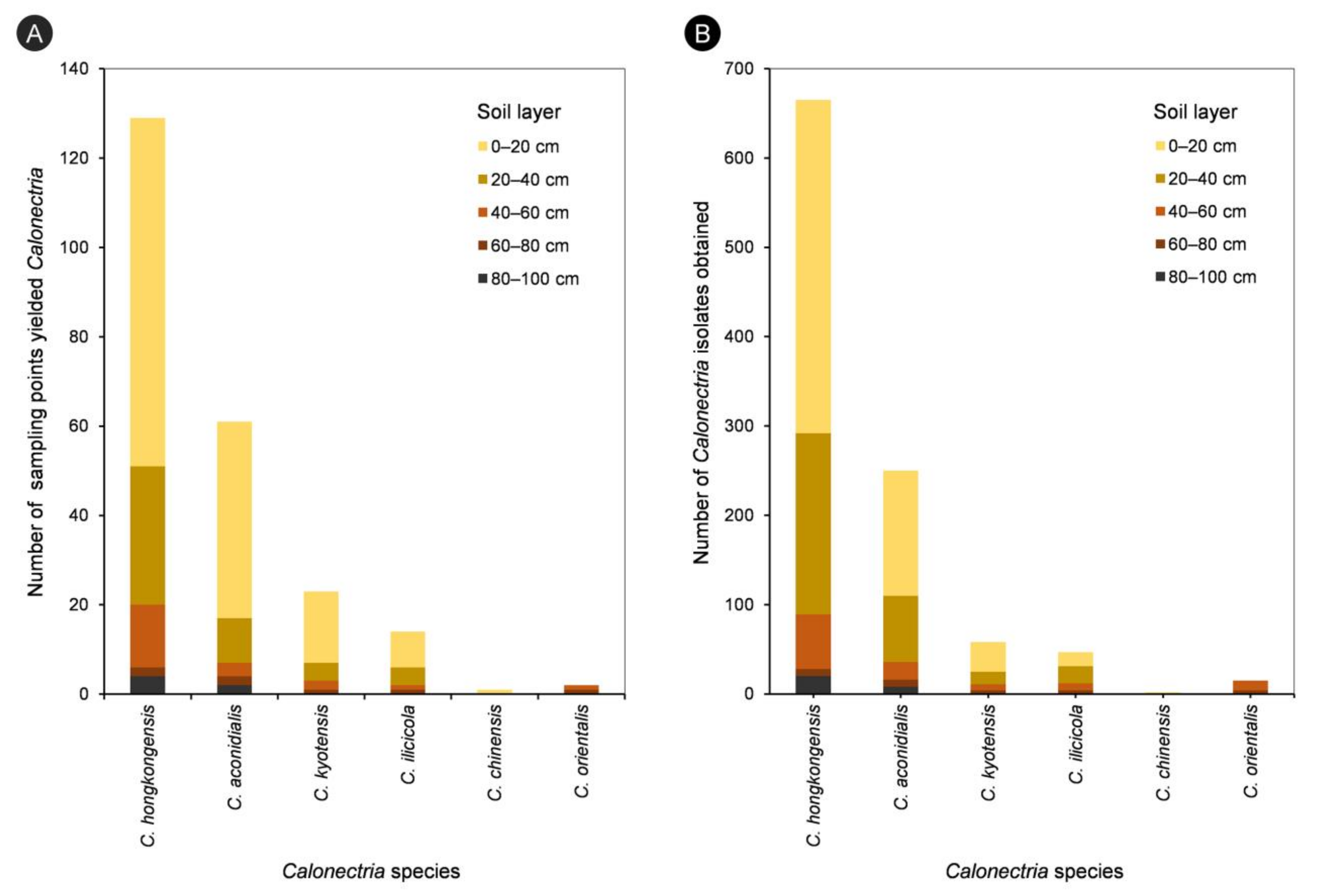
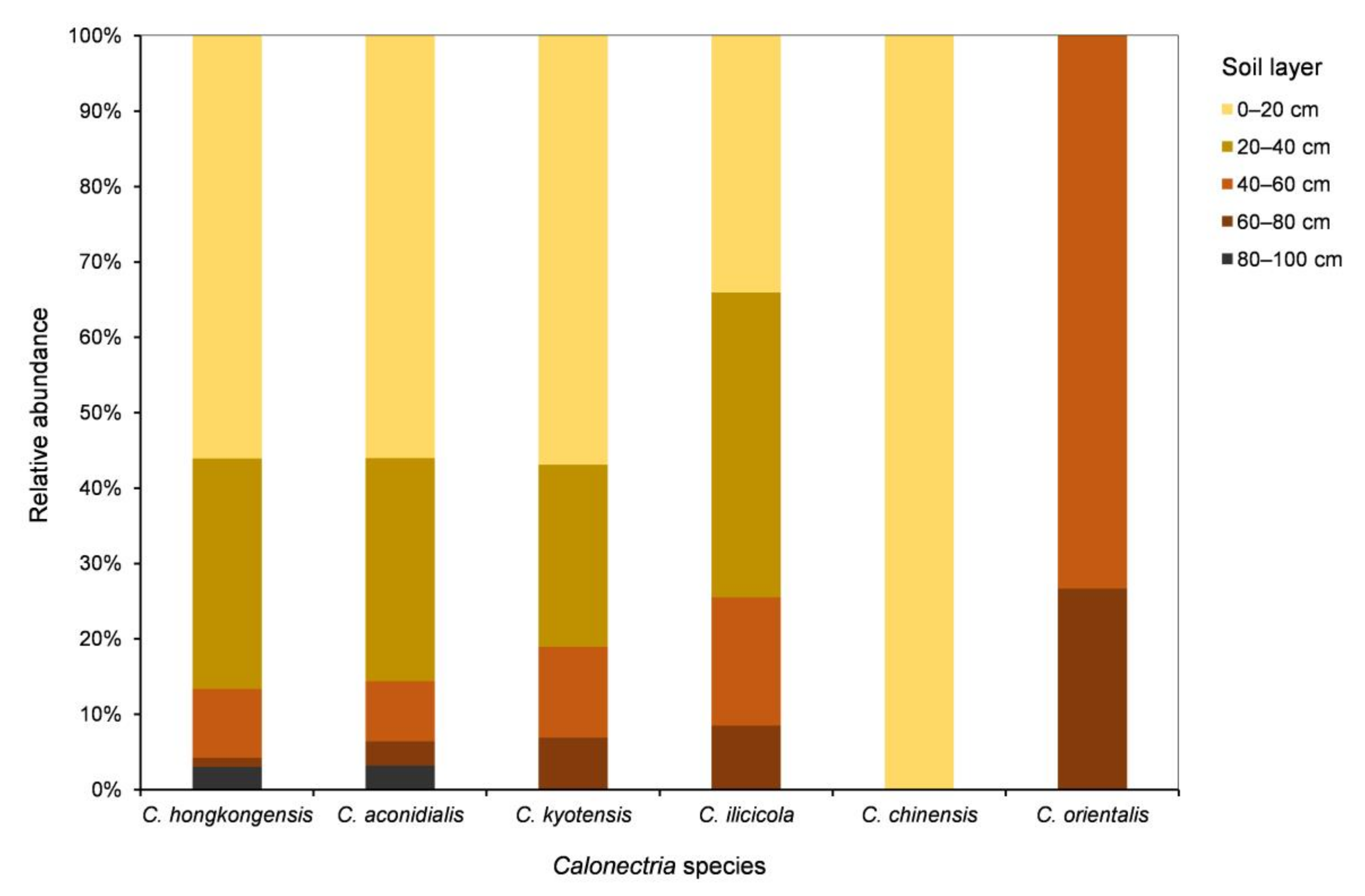
For each of the five species in the C. kyotensis species complex, the number of sampling points containing Calonectria decreased with increasing depth of the soil, with the exception of C. hongkongensis in soil layers of 60–80 cm (2 sampling points) and 80–100 cm (4 sampling points) (Supplementary Table S1, Figure 4A); the number of isolates obtained decreased with increasing soil depth, with the exception of C. hongkongensis in the 60–80 cm soil layer (8 isolates) and 80–100 cm (20 isolates) as well as C. ilicicola in the 0–20 cm (16 isolates) and 20–40 cm (19 isolates) soil layers (Table 1, Figure 4B). Most isolates were obtained from the soil layers 0–20 and 20–40 cm, accounting for 86.6%, 85.6%, 81%, 74.5%, and 100% of all of the obtained isolates within each species of C. hongkongensis, C. aconidialis, C. kyotensis, C. ilicicola, and C. chinensis, respectively (Figure 5).
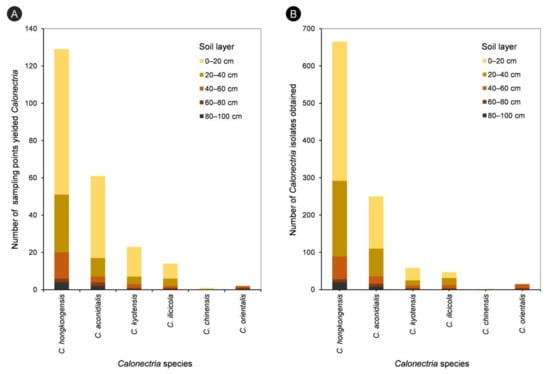
Figure 4.
Number of sampling points yielded different Calonectria species in each of the five soil layers (A), and numbers of isolates obtained for different Calonectria species in each of the five soil layers (B).
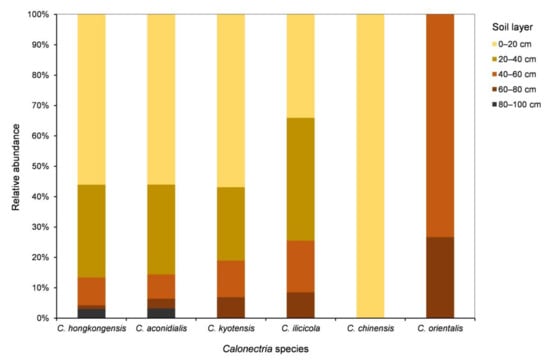
Figure 5.
Relative abundances of each Calonectria species in each of the five soil layers. Relative abundance was based on the proportional frequencies of isolates of each Calonectria species in each soil layer.
3.5. Genotyping of Isolates within Each Calonectria Species
For the 1037 Calonectria isolates obtained and identified in this study, the genotype results based on tef1 and tub2 sequences indicated that 11, 3, 3, 3, 1, and 1 genotype(s) existed in C. hongkongensis, C. aconidialis, C. kyotensis, C. ilicicola, C. chinensis, and C. orientalis, respectively (Table 2). The isolates presenting the dominant genotype (genotype AA) accounted for 84.4%, 62.4%, 56.9%, 55.3%, 100%, and 100% of all of the isolates obtained from C. hongkongensis, C. aconidialis, C. kyotensis, C. ilicicola, C. chinensis, and C. orientalis, respectively (Table 2).
3.6. Genotype Diversity of Calonectria Species in Different Soil Layers
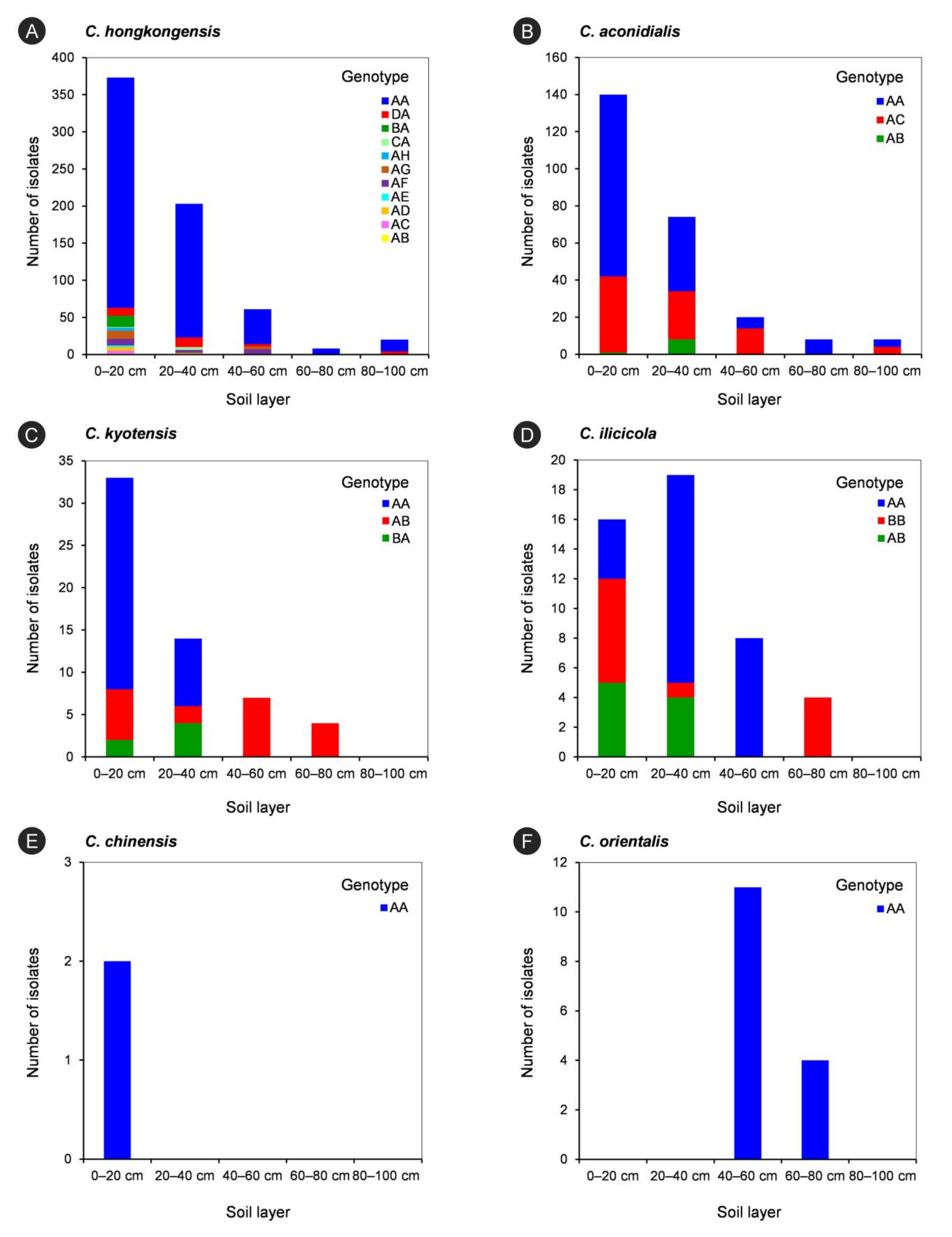
The tef1-tub2 genotypes of each Calonectria species in each soil layer are listed in Table 7 and are shown in Figure 6. For each species in the C. kyotensis species complex, the results showed that the number of genotypes decreased with increasing soil depth, with the exception of C. hongkongensis and C. aconidialis in the 60–80 cm (one genotype) and 80–100 cm (two genotypes) soil layers (Table 7, Figure 6A,B); the 0–20 cm soil layer contained all of the genotypes of each species in the C. kyotensis complex (Table 7, Figure 6A–E). For the genotype with the most isolates of each species in the C. kyotensis complex, the majority of isolates were obtained from 0–20 cm soil layer, with the exception of C. ilicicolla (Table 7, Figure 6A–E). Only one genotype of C. orientalis was present in the 40–60 and 60–80 cm soil layers (Table 7, Figure 6F).

Table 7.
Isolate numbers of each genotype in each soil layer for each Calonectria species.
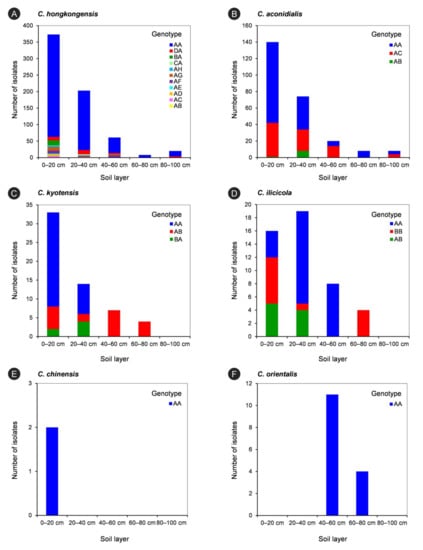
Figure 6.
The isolate numbers of each genotype of each Calonectria species in five soils layers. The genotypes were determined by DNA sequences of tef1 and tub2 gene regions. (A): C. hongkongensis; (B): C. aconidialis; (C): C. kyotensis; (D) C. ilicicola; (E): C. chinensis; (F): C. orientalis.
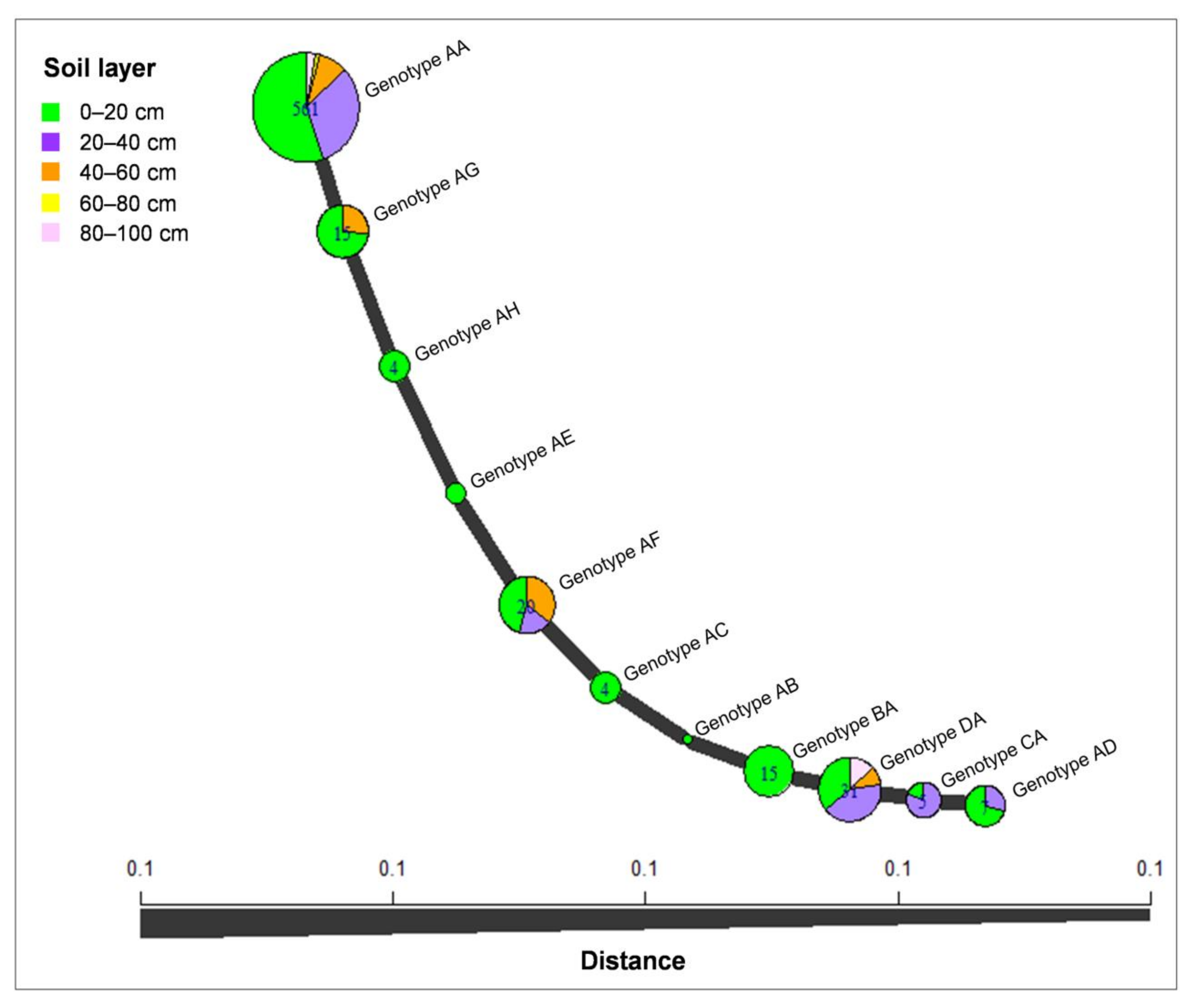
The minimum spanning network (MSN) analysis was conducted for C. hongkongensis, which was considered as the dominant species identified in this study. The analysis revealed that most isolates of C. hongkongensis were genotype AA (561 isolates), followed by genotypes DA (31 isolates) and AF (20 isolates); genotype AA was present in the isolates from all five soil layers; genotypes AB, AC, AE, AH, and BA were present only in the isolates from the 0–20 cm soil layer, and the other genotypes were present in isolates from two to four soil layers. Isolates from the 0–20 cm soil layer contained all of the genotypes (Figure 7).
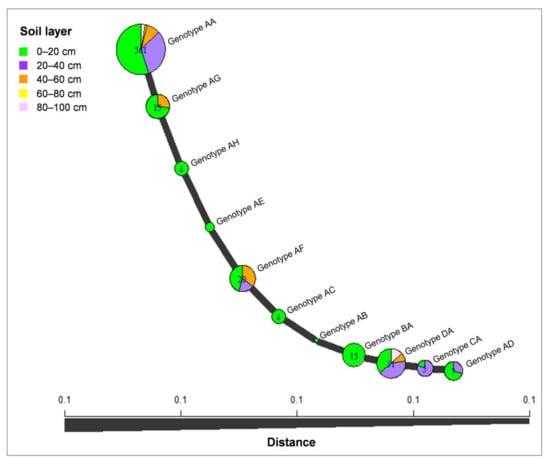
Figure 7.
Minimum spanning network constructed using Bruvo’s distances showing that the C. hongkongensis isolates were grouped into 11 genotypes based on tef1 and tub2 sequences. The size of a node is proportional to the number of represented tef1-tub2 genotypes.
4. Discussion
In this study, more than 1000 Calonectria isolates were obtained from five soil layers at 100 sampling points in one Eucalyptus plantation. All of the isolates were identified based on DNA sequence comparisons of multiple gene regions. Six Calonectria species were identified, namely, C. aconidialis, C. chinensis, C. hongkongensis, C. ilicicola, and C. kyotensis in the C. kyotensis species complex, and C. orientalis in the C. brassicae species complex. Calonectria hongkongensis (64.1% of all of the isolates) was the dominant species, followed by C. aconidialis (24.1% of all of the isolates). To our knowledge, this is the first report of C. orientalis in China. The species diversity and distribution characteristics of the six species in different soil layers were clarified. The results showed that the number of sampling points from which Calonectria was obtained, and the number of Calonectria isolates obtained decreased with increasing depth of the soil. The majority of isolates (84.3% of all the isolates) were obtained from soil layers of 0–20 and 20–40 cm. The diversity of the five species in the C. kyotensis species complex decreased with increasing soil depth. For each species in the C. kyotensis species complex, in most cases, the number of genotypes decreased with increasing soil depth, and the 0–20 cm soil layer contained all of the genotypes of each species.
Five species, namely, C. aconidialis, C. chinensis, C. hongkongensis, C. ilicicola, and C. kyotensis, in the C. kyotensis species complex were isolated from the soil of the Eucalyptus plantation in this study. These five species have been frequently isolated from soils in several other regions in southern China, especially from soils in Eucalyptus plantations [9,11,14,49]. Calonectria ilicicola is considered as a soil-borne fungal pathogen that has been isolated from a number of diseased plant species in China [21,61]. This study presents the first record of C. ilicicola isolated from soil in a Eucalyptus plantation. Results of this and previous studies suggest that all five of the species in the C. kyotensis species complex are potentially widely distributed in Eucalyptus plantation soils in other regions of southern China [9,11,14].
This study is the first report of C. orientalis in China, and this species is the first Calonectria species in the C. brassicae species complex found in China. Calonectria orientalis has been isolated from soil in Indonesia [29]. Some other species in the C. brassicae species complex have also been frequently isolated from soils. With the exception of C. orientalis, the other species in the C. brassicae species complex isolated from soils were all from Ecuador and Brazil in South America [5,10,29,30,31,56]. Most of the Calonectria species in the C. brassicae species complex have only been isolated from South America but not from Asia [5] and C. orientalis, in this study, was only isolated from one of the 100 sampling points. These results suggest that C. orientalis is not widely distributed in China.
For the five species in the C. kyotensis species complex, the results of this study indicate that the diversity of the five species decreased with increasing soil depth, and the number of sampling points containing Calonectria and the number of Calonectria isolates obtained also decreased with soil depth. Most isolates were obtained from the 0–20 and 20–40 cm soil layers. In most cases, the number of genotypes decreased with increasing soil depth for each species, and the 0–20 cm soil layer contained all of the genotypes of each species. These results suggest that 0–20 cm is the best soil depth for Calonectria isolation and for examining the species and genotype diversity of Calonectria in soils in Eucalyptus plantations in southern China. In several previous studies specialized in the research on Calonectria species diversity, soil samples were also exclusively collected from the surface layer, all from the 0–20 cm layer [9,10,11,13,14,36]. These studies have characterized the diversity of Calonectria species well. Results of a number of other studies indicated that microbial diversity and richness are typically affected by the soil depth [62,63,64,65,66,67], and shallower layers usually have a higher level of microbial diversity [62,63,66,67,68]. This pattern is consistent with the results of the present study. A possible reason for the vertical distribution of soil microbes is the harsher environment in deeper soil layers, where the soil density is higher, oxygen concentrations are lower, and carbon and nutrients are less available [69]. For Calonectria, which includes some important pathogens of various agricultural, horticultural, and forestry crops worldwide, as well as for other genera of fungi in forests, no systematic research has been conducted to examine the species diversity and distribution characteristics in different soil layers. This study showed that the deeper soil layers had comparatively fewer but still contained many Calonectria. It remains unknown whether the Calonectria were originally distributed in deeper soil layers or whether the fungi in deeper soil layers migrated from surface layers, perhaps through the infiltration of rainwater. Studies on the population diversity differences among different soil layers should be conducted to address this question. Furthermore, the Calonectria distributed in deeper soil layers increase the challenge of controlling the diseases caused by these fungi.
This study examined the species diversity and distribution characteristics of Calonectria in five soil layers in a Eucalyptus plantation in southern China. Six species were isolated from soils in a relatively small Eucalyptus plantation, indicating that the diversity of Calonectria species in these soils in southern China is relatively high. This study also revealed that the species diversity and number of genotypes of each Calonectria species decreased with increasing soil depth, a pattern that helps us to understand the distribution characteristics of Calonectria species in different layers of soil. For some Calonectria species, there were relatively large numbers of isolates obtained from different soil layers, especially for C. hongkongensis and C. aconidialis in the 0–20, 20–40, and 40–60 cm soil layers. The genetic structures and population biology of these species in the different soil layers are unknown, but additional studies may increase our understanding of the distribution characteristics and dissemination patterns of Calonectria species.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/jof7100857/s1, Table S1: Number of sampling points containing each Calonectria species in each soil layer, Table S2: All 1037 isolates obtained and sequenced in this study, Figure S1: Number of sampling points that yielded Calonectria in each of the five soil layers, Figure S2: Phylogenetic tree of Calonectria species based on maximum likelihood (ML) analyses of the tef1 gene sequences, Figure S3: Phylogenetic tree of Calonectria species based on ML analyses of the tub2 gene sequences, Figure S4: Phylogenetic tree of Calonectria species based on ML analyses of the cmdA gene sequences, Figure S5: Phylogenetic tree of Calonectria species based on ML analyses of the his3 gene sequences.
Author Contributions
S.C. conceived and designed the experiments. L.L. and S.C. collected the samples. L.L. performed the laboratory work. L.L., W.W. and S.C. analyzed the data. S.C. and L.L. wrote the paper. All authors have read and agreed to the published version of the manuscript.
Funding
This study was initiated through the bilateral agreement between the Governments of South Africa and China and supported by The National Key R&D Program of China (China-South Africa Forestry Joint Research Centre Project; project No. 2018YFE0120900), the National Ten-thousand Talents Program (Project No. W03070115) and the GuangDong Top Young Talents Program (Project No. 20171172).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data is contained within the article and Supplementary Materials.
Acknowledgments
We thank JiaLong Han, LanSen Sun, Ying Liu, and XueYing Liang for their assistance in collecting samples. We thank QianLi Liu for sequencing the tub2 gene region of some isolates in Table 4. We thank FeiFei Liu for analyzing the genotype data. We thank GuoQing Li, WenWen Li, and QuanChao Wang who provided assistance in laboratory work and checking the data. We thank LetPub (www.letpub.com; accessed date: 27 August 2021 and 6 October 2021) for providing linguistic assistance during the preparation of this manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Crous, P.W. Taxonomy and pathology of Cylindrocladium (Calonectria) and Allied Genera; APS Press: St. Paul, MN, USA, 2002. [Google Scholar]
- Lombard, L.; Crous, P.W.; Wingfield, B.D.; Wingfield, M.J. Species concepts in Calonectria (Cylindrocladium). Stud. Mycol. 2010, 66, 1–14. [Google Scholar] [CrossRef]
- Vitale, A.; Crous, P.W.; Lombard, L.; Polizzi, G. Calonectria diseases on ornamental plants in Europe and the Mediterranean basin: An overview. J. Plant Pathol. 2013, 95, 463–476. [Google Scholar]
- Daughtrey, M.L. Boxwood blight: Threat to ornamentals. Annu. Rev. Phytopathol. 2019, 57, 189–209. [Google Scholar] [CrossRef]
- Liu, Q.L.; Li, J.Q.; Wingfield, M.J.; Duong, T.A.; Wingfield, B.D.; Crous, P.W.; Chen, S.F. Reconsideration of species boundaries and proposed DNA barcodes for Calonectria. Stud. Mycol. 2020, 97, 100106. [Google Scholar] [CrossRef] [PubMed]
- Li, J.Q.; Barnes, I.; Liu, F.F.; Wingfield, M.J.; Chen, S.F. Global genetic diversity and mating type distribution of Calonectria pauciramosa: An important wide host-range plant pathogen. Plant Dis. 2021. [Google Scholar] [CrossRef]
- Lombard, L.; Rodas, C.A.; Crous, P.W.; Wingfield, B.D.; Wingfield, M.J. Calonectria (Cylindrocladium) species associated with dying Pinus cuttings. Persoonia 2009, 23, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Lombard, L.; Zhou, X.D.; Crous, P.W.; Wingfield, B.D.; Wingfield, M.J. Calonectria species associated with cutting rot of Eucalyptus. Persoonia 2010, 24, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Lombard, L.; Chen, S.F.; Mou, X.; Zhou, X.D.; Crous, P.W.; Wingfield, M.J. New species, hyper-diversity and potential importance of Calonectria spp. from Eucalyptus in South China. Stud. Mycol. 2015, 80, 151–188. [Google Scholar] [CrossRef]
- Alfenas, R.F.; Lombard, L.; Pereira, O.L.; Alfenas, A.C.; Crous, P.W. Diversity and potential impact of Calonectria species in Eucalyptus plantations in Brazil. Stud. Mycol. 2015, 80, 89–130. [Google Scholar] [CrossRef]
- Li, J.Q.; Wingfield, M.J.; Liu, Q.L.; Barnes, I.; Roux, J.; Lombard, L.; Crous, P.W.; Chen, S.F. Calonectria species isolated from Eucalyptus plantations and nurseries in South China. IMA Fungus 2017, 8, 259–286. [Google Scholar] [CrossRef]
- Pham, N.; Barnes, I.; Chen, S.F.; Liu, F.F.; Dang, Q.; Pham, T.; Lombard, L.; Crous, P.W.; Wingfield, M.J. Ten new species of Calonectria from Indonesia and Vietnam. Mycologia 2019, 111, 78–102. [Google Scholar] [CrossRef]
- Wang, Q.C.; Liu, Q.L.; Chen, S.F. Novel species of Calonectria isolated from soil near Eucalyptus plantations in southern China. Mycologia 2019, 111, 1028–1040. [Google Scholar] [CrossRef]
- Wu, W.X.; Chen, S.F. Species diversity, mating strategy, and pathogenicity of Calonectria species from diseased leaves and soils in the Eucalyptus plantation in southern China. J. Fungi 2021, 7, 73. [Google Scholar] [CrossRef]
- Old, K.M.; Wingfield, M.J.; Yuan, Z.Q. A Manual of Diseases of Eucalypts in South-East Asia; Center for International Forestry Research: Jakarta, Indonesia, 2003. [Google Scholar]
- Rodas, C.A.; Lombard, L.; Gryzenhout, M.; Slippers, B.; Wingfield, M.J. Cylindrocladium blight of Eucalyptus grandis in Colombia. Australas. Plant Pathol. 2005, 34, 143–149. [Google Scholar] [CrossRef]
- Lechat, C.; Crous, P.W.; Groenewald, J.Z. The enigma of Calonectria species occurring on leaves of Ilex aquifolium in Europe. IMA Fungus 2010, 1, 101–108. [Google Scholar] [CrossRef]
- Chen, S.F.; Lombard, L.; Roux, J.; Xie, Y.J.; Wingfield, M.J.; Zhou, X.D. Novel species of Calonectria associated with Eucalyptus leaf blight in Southeast China. Persoonia 2011, 26, 1–12. [Google Scholar] [CrossRef]
- Gehesquière, B.; Crouch, J.A.; Marra, R.E.; van Poucke, K.; Rys, F.; Maes, M.; Gobin, B.; Höfte, M.; Heungens, K. Characterization and taxonomic reassessment of the box blight pathogen Calonectria pseudonaviculata, introducing Calonectria henricotiae sp. nov. Plant Pathol. 2016, 65, 37–52. [Google Scholar] [CrossRef]
- Lopes, U.P.; Alfenas, R.F.; Zambolim, L.; Crous, P.W.; Costa, H.; Pereira, O.L. A new species of Calonectria causing rot on ripe strawberry fruit in Brazil. Australas. Plant Pathol. 2017, 47, 1–11. [Google Scholar] [CrossRef]
- Gai, Y.; Deng, Q.; Chen, X.; Guan, M.; Xiao, X.; Xu, D.; Deng, M.; Pan, R. Phylogenetic diversity of Calonectria ilicicola causing Cylindrocladium black rot of peanut and red crown rot of soybean in southern China. J. Gen. Plant Pathol. 2017, 83, 273–282. [Google Scholar] [CrossRef]
- Freitas, R.G.; Alfenas, R.F.; Guimarães, L.M.S.; Badel, J.L.; Alfenas, A.C. Genetic diversity and aggressiveness of Calonectria pteridis in Eucalyptus spp. Plant Pathol. 2019, 68, 869–877. [Google Scholar] [CrossRef]
- Wang, Q.C.; Chen, S.F. Calonectria pentaseptata causes severe leaf disease on cultivated Eucalyptus in Leizhou Peninsula of southern China. Plant Dis. 2020, 104, 493–509. [Google Scholar] [CrossRef] [PubMed]
- Bell, D.K.; Sobers, E.K. A peg, pod, and root necrosis of peanuts caused by a species of Calonectria. Phytopathology 1966, 56, 1361–1364. [Google Scholar]
- Johnston, S.A.; Beute, M.K. Histopathology of Cylindrocladium black rot of peanut. Phytopathology 1975, 64, 649–653. [Google Scholar] [CrossRef]
- Pataky, J.K.; Beute, M.K.; Wynne, J.C.; Carlson, G.A. Peanut yield, market quality and value reductions due to Cylindrocladium black rot. Peanut Sci. 1983, 10, 62–66. [Google Scholar] [CrossRef]
- Berner, D.K.; Berggren, G.T.; Snow, J.P.; White, E.P. Distribution and management of red crown rot of soybean in Louisiana. Appl. Agric. Res. 1988, 3, 160–166. [Google Scholar]
- Yamamoto, R.; Nakagawa, A.; Shimada, S.; Komatsu, S.; Kanematsu, S. Histopathology of red crown rot of soybean. J. Gen. Plant Pathol. 2016, 83, 23–32. [Google Scholar] [CrossRef]
- Lombard, L.; Crous, P.W.; Wingfield, B.D.; Wingfield, M.J. Phylogeny and systematics of the genus Calonectria. Stud. Mycol. 2010, 66, 31–69. [Google Scholar] [CrossRef] [PubMed]
- Lombard, L.; Wingfield, M.J.; Alfenas, A.C.; Crous, P.W. The forgotten Calonectria collection: Pouring old wine into new bags. Stud. Mycol. 2016, 85, 159–198. [Google Scholar] [CrossRef]
- Marin-Felix, Y.; Groenewald, J.Z.; Cai, L.; Chen, Q.; Marincowitz, S.; Barnes, I.; Bensch, K.; Braun, U.; Camporesi, E.; Damm, U.; et al. Genera of phytopathogenic fungi: GOPHY 1. Stud. Mycol. 2017, 86, 99–216. [Google Scholar] [CrossRef]
- Crous, P.W.; Luangsa-ard, J.J.; Wingfield, M.J.; Carnegie, A.J.; Hernández-Restrepo, M.; Lombard, L.; Roux, J.; Barreto, R.W.; Baseia, I.G.; Cano-Lira, J.F.; et al. Fungal Planet description sheets: 785–867. Persoonia 2018, 41, 238–417. [Google Scholar] [CrossRef]
- Crous, P.W.; Carnegie, A.J.; Wingfield, M.J.; Sharma, R.; Mughini, G.; Noordeloos, M.E.; Santini, A.; Shouche, Y.S.; Bezerra, J.D.P.; Dima, B.; et al. Fungal Planet description sheets: 868–950. Persoonia 2019, 42, 291–473. [Google Scholar] [CrossRef]
- Crous, P.W.; Hernández-Restrepo, M.; Schumacher, R.K.; Cowan, D.A.; Maggs-Kölling, G.; Marais, E.; Wingfield, M.J.; Yilmaz, N.; Adan, O.C.G.; Akulov, A.; et al. New and Interesting Fungi. 4. Fungal Syst. Evol. 2021, 7, 255–343. [Google Scholar] [CrossRef]
- Crous, P.W.; Cowan, D.A.; Maggs-Kölling, G.; Yilmaz, N.; Thangavel, R.; Wingfield, M.J.; Noordeloos, M.E.; Dima, B.; Brandrud, T.E.; Jansen, G.M.; et al. Fungal Planet description sheets: 1182–1283. Persoonia 2021, 46, 313–528. [Google Scholar]
- Liu, Q.L.; Chen, S.F. Two novel species of Calonectria isolated from soil in a natural forest in China. MycoKeys 2017, 26, 25–60. [Google Scholar] [CrossRef]
- Phipps, P.M.; Beute, M.K.; Barker, K.R. An elutriation method for quantitative isolation of Cylindrocladium crotalariae microsclerotia from peanut field soil. Phytopathology 1976, 66, 1255–1259. [Google Scholar] [CrossRef]
- Thies, W.F.; Patton, R.F. The biology of Cylindrocladium scoparium in Wisconsin forest tree nurseries. Phytopathology 1970, 60, 1662–1668. [Google Scholar] [CrossRef]
- Sobers, E.K.; Littrell, R.H. Pathogenicity of three species of Cylindrocladium to select hosts. Plant Dis. Report. 1974, 58, 1017–1019. [Google Scholar]
- Anderson, P.J. Rose Canker and Its Control; Massachusetts Agricultural Experiment Station Bulletin: Boston, MA, USA, 1918; Volume 183, pp. 11–46. [Google Scholar]
- Dumas, M.T.; Greifenhagen, S.; Halicki-Hayden, G.; Meyer, T.R. Effect of seedbed steaming on Cylindrocladium floridanum, soil microbes and the development of white pine seedlings. Phytoprotection 1998, 79, 35–43. [Google Scholar] [CrossRef][Green Version]
- Xu, Y.; Du, A.; Wang, Z.; Zhu, W.; Li, C.; Wu, L. Effects of different rotation periods of Eucalyptus plantations on soil physiochemical properties, enzyme activities, microbial biomass and microbial community structure and diversity. For. Ecol. Manag. 2020, 456, 117683. [Google Scholar] [CrossRef]
- Cooperative Research Group on Chinese Soil Taxonomy. Chinese Soil Taxonomy; Science Press: Beijing, China, 2001. [Google Scholar]
- van Burik, J.A.; Schreckhise, R.W.; White, T.C.; Bowden, R.A.; Myerson, D. Comparison of six extraction techniques for isolation of DNA from filamentous fungi. Med. Mycol. 1998, 36, 299–303. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Graça, R.N.; Alfenas, A.C.; Maffia, L.A.; Titon, M.; Alfenas, R.F.; Lau, D.; Rocabado, J.M.A. Factors influencing infection of eucalypts by Cylindrocladium pteridis. Plant Pathol. 2009, 58, 971–981. [Google Scholar] [CrossRef]
- Paradis, E.; Claude, J.; Strimmer, K. APE: Analyses of phylogenetics and evolution in R language. Bioinformatics 2004, 20, 289–290. [Google Scholar] [CrossRef] [PubMed]
- Kamvar, Z.N.; Tabima, J.F.; Grünwald, N.J. Poppr: An R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2014, 2, e281. [Google Scholar] [CrossRef] [PubMed]
- Crous, P.W.; Groenewald, J.Z.; Risede, J.M.; Simoneau, P.; Hywel-Jones, N.L. Calonectria species and their Cylindrocladium anamorphs: Species with sphaeropedunculate vesicles. Stud. Mycol. 2004, 50, 415–430. [Google Scholar]
- Kang, J.C.; Crous, P.W.; Schoch, C.L. Species concepts in the Cylindrocladium floridanum and Cy. spathiphylli complexes (Hypocreaceae) based on multi-allelic sequence data, sexual compatibility and morphology. Syst. Appl. Microbiol. 2001, 24, 206–217. [Google Scholar] [CrossRef] [PubMed]
- Victor, D.; Crous, P.W.; Janse, B.J.H.; Wingfield, M.J. Genetic variation in Cylindrocladium floridanum and other morphologically similar Cylindrocladium species. Syst. Appl. Microbiol. 1997, 20, 268–285. [Google Scholar] [CrossRef]
- Boedijn, K.B.; Reitsma, J. Notes on the genus Cylindrocladium (Fungi: Mucedineae). Reinwardtia 1950, 1, 51–60. [Google Scholar]
- Terashita, T. A new species of Calonectria and its conidial state. Trans. Mycol. Soc. Japan 1968, 8, 124–129. [Google Scholar]
- Tubaki, K. Studies on the Japanese Hyphomycetes. V. Leaf & stem group with a discussion of the classification of Hyphomycetes and their perfect stages. J. Hattori Bot. Lab. 1958, 20, 142–244. [Google Scholar]
- El-Gholl, N.E.; Alfieri, S.A., Jr.; Barnard, E.L. Description and pathogenicity of Calonectria clavata sp. nov. Mycotaxon 1993, 48, 201–216. [Google Scholar]
- Crous, P.W.; Groenewald, J.Z.; Risède, J.M.; Simoneau, P.; Hyde, K.D. Calonectria species and their Cylindrocladium anamorphs: Species with clavate vesicles. Stud. Mycol. 2006, 55, 213–226. [Google Scholar] [CrossRef]
- Crous, P.W.; Wingfield, M.J.; Alfenas, A.C. Additions to Calonectria. Mycotaxon 1993, 46, 217–234. [Google Scholar]
- Decock, C.; Crous, P.W. Curvicladium gen. nov., a new hyphomycete genus from French Guiana. Mycologia 1998, 90, 276–281. [Google Scholar] [CrossRef][Green Version]
- Lombard, L.; van der Merwe, N.A.; Groenewald, J.Z.; Crous, P.W. Generic concepts in Nectriaceae. Stud. Mycol. 2015, 80, 189–245. [Google Scholar] [CrossRef]
- Cunningham, C.W. Can three incongruence tests predict when data should be combined? Mol. Biol. Evol. 1997, 14, 733–740. [Google Scholar] [CrossRef] [PubMed]
- Pei, W.H.; Cao, J.F.; Yang, M.Y.; Zhao, Z.J.; Xue, S.M. First report of black rot of medicago sativa caused by Cylindrocladium parasiticum (teleomorph Calonectria ilicicola) in Yunnan province, China. Plant Dis. 2015, 99, 890. [Google Scholar] [CrossRef]
- Fierer, N.; Schimel, J.P.; Holden, P.A. Variations in microbial community composition through two soil depth profiles. Soil Biol. Biochem. 2003, 35, 167–176. [Google Scholar] [CrossRef]
- Jumpponen, A.; Jones, K.L.; Blair, J. Vertical distribution of fungal communities in tallgrass prairie soil. Mycologia 2010, 102, 1027–1041. [Google Scholar] [CrossRef] [PubMed]
- de Araujo Pereira, A.P.; Santana, M.C.; Bonfim, J.A.; de Lourdes Mescolotti, D.; Cardoso, E.J.B.N. Digging deeper to study the distribution of mycorrhizal arbuscular fungi along the soil profile in pure and mixed Eucalyptus grandis and Acacia mangium plantations. Appl. Soil Ecol. 2018, 128, 1–11. [Google Scholar] [CrossRef]
- Grishkan, I.; Lázaro, R.; Kidron, G.J. Vertical divergence of cultivable microfungal communities through biocrusted and bare soil profiles at the Tabernas Desert, Spain. Geomicrobiol. J. 2020, 37, 534–549. [Google Scholar] [CrossRef]
- Upton, R.N.; Sielaff, A.C.; Hofmockel, K.S.; Xu, X.; Polley, H.W.; Wilsey, B.J. Soil depth and grassland origin cooperatively shape microbial community co-occurrence and function. Ecosphere 2020, 11, e02973. [Google Scholar] [CrossRef]
- Frey, B.; Walthert, L.; Perez-Mon, C.; Stierli, B.; Köchli, R.; Dharmarajah, A.; Brunner, I. Deep soil layers of drought-exposed forests harbor poorly known bacterial and fungal communities. Front. Microbiol. 2021, 12, 674160. [Google Scholar] [CrossRef]
- Na, X.; Ma, S.; Ma, C.; Liu, Z.; Xu, P.; Zhu, H.; Liang, W.; Kardol, P. Lycium barbarum L. (goji berry) monocropping causes microbial diversity loss and induces Fusarium spp. enrichment at distinct soil layers. Appl. Soil Ecol. 2021, 168, 104107. [Google Scholar] [CrossRef]
- Lennon, J.T. Microbial life deep underfoot. mBio 2020, 11, e03201-19. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).