Feature Papers Related to Metabolomic Profiling Technology
Share This Topical Collection
Editor
Topical Collection Information
Dear Colleagues,
Metabolomics and lipidomics are emerging technologies which aim at the global characterization and quantification of small molecules and lipids within biological matrices including biofluids, cells, whole organs, and tissues. New emerging technology-based metabolite profiling has been used to track the change of levels of metabolites and lipids in biological systems due to many different situations. This Special Collection on metabolomic profiling technology aims to attract high-quality manuscripts in the metabolomics field. We encourage researchers from the metabolomics and lipidomics field to contribute manuscripts highlighting the latest development in their research field with new technology-based applications.
Dr. Thusitha Rupasinghe
Collection Editor
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 100 words) can be sent to the Editorial Office for announcement on this website.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Metabolites is an international peer-reviewed open access monthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript.
The Article Processing Charge (APC) for publication in this open access journal is 2700 CHF (Swiss Francs).
Submitted papers should be well formatted and use good English. Authors may use MDPI's
English editing service prior to publication or during author revisions.
Keywords
- Metabolomics
- Lipidomic
- High-resolution mass spectrometry
- Untargeted and targeted
- Data processing
- Pathway mapping
- Fluxomic
Published Papers (2 papers)
Open AccessArticle
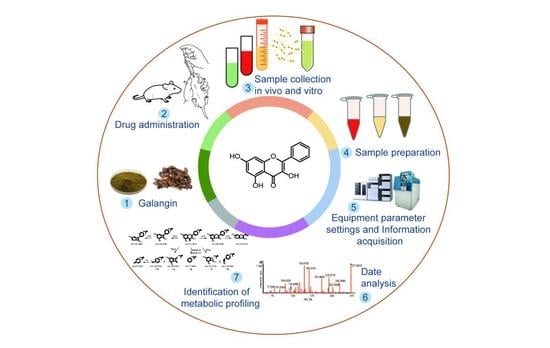
Analysis of Galangin and Its In Vitro/In Vivo Metabolites via Ultra-High-Performance Liquid Chromatography/Quadrupole Time-of-Flight Mass Spectrometry
by
Feng Zhao, Yinling Ma, Jintuo Yin, Ying Li, Yanli Cao and Lantong Zhang
Cited by 13 | Viewed by 2668
Abstract
Galangin, a naturally available flavonoid, induces a variety of pharmacological activities and biological effects via several mechanisms. However, in vivo metabolism of galangin has not been fully explored, which means knowledge of its pharmacodynamics and application potential is limited. The objective of this
[...] Read more.
Galangin, a naturally available flavonoid, induces a variety of pharmacological activities and biological effects via several mechanisms. However, in vivo metabolism of galangin has not been fully explored, which means knowledge of its pharmacodynamics and application potential is limited. The objective of this study was to establish an ultra-high-performance liquid chromatography–quadrupole time-of-flight mass spectrometry method for the rapid profiling and identification of galangin metabolites in vitro and in vivo using unique online information-dependent acquisition with multiple mass defect filtering combined with dynamic background subtraction in positive ion mode. A total of 27 metabolites were detected and characterized, among which eight metabolites in liver microsomes and four metabolites in intestinal microflora were characterized, and 27 metabolites from rat plasma, bile, urine, feces, and a number of different tissue samples were characterized. Thirteen major metabolic pathways including hydrogenation, hydroxylation, glycosylation, methylation, acetylation, glucuronidation, and sulfation were observed to be attributable to the biotransformation of the metabolites. This study provides evidence for the presence of in vitro and in vivo metabolites and the pharmacokinetic mechanism of galangin. Moreover, the study promotes the further development and utilization of galangin and the plant from which it is derived,
Alpinia officinarum Hance.
Full article
►▼
Show Figures
Open AccessArticle
Benchmarking Non-Targeted Metabolomics Using Yeast-Derived Libraries
by
Evelyn Rampler, Gerrit Hermann, Gerlinde Grabmann, Yasin El Abiead, Harald Schoeny, Christoph Baumgartinger, Thomas Köcher and Gunda Koellensperger
Cited by 7 | Viewed by 5206
Abstract
Non-targeted analysis by high-resolution mass spectrometry (HRMS) is an essential discovery tool in metabolomics. To date, standardization and validation remain a challenge. Community-wide accepted cost-effective benchmark materials are lacking. In this work, we propose yeast (
Pichia pastoris) extracts derived from fully
[...] Read more.
Non-targeted analysis by high-resolution mass spectrometry (HRMS) is an essential discovery tool in metabolomics. To date, standardization and validation remain a challenge. Community-wide accepted cost-effective benchmark materials are lacking. In this work, we propose yeast (
Pichia pastoris) extracts derived from fully controlled fermentations for this purpose. We established an open-source metabolite library of >200 identified metabolites based on compound identification by accurate mass, matching retention times, and MS/MS, as well as a comprehensive literature search. The library includes metabolites from the classes of (1) organic acids and derivatives (2) nucleosides, nucleotides, and analogs, (3) lipids and lipid-like molecules, (4) organic oxygen compounds, (5) organoheterocyclic compounds, (6) organic nitrogen compounds, and (7) benzoids at expected concentrations ranges of sub-nM to µM. As yeast is a eukaryotic organism, key regulatory elements are highly conserved between yeast and all annotated metabolites were also reported in the human metabolome database (HMDB). Orthogonal state-of-the-art reversed-phase (RP-) and hydrophilic interaction chromatography mass spectrometry (HILIC-MS) non-targeted analysis and authentic standards revealed that 104 out of the 206 confirmed metabolites were reproducibly recovered and stable over the course of three years when stored at −80 °C. Overall, 67 out of these 104 metabolites were identified with comparably stable areas over all three yeast fermentation and are the ideal starting point for benchmarking experiments. The provided yeast benchmark material enabled not only to test for the chemical space and coverage upon method implementation and developments but also allowed in-house routines for instrumental performance tests. Transferring the quality control strategy of proteomics workflows based on the number of protein identification in HeLa extracts, metabolite IDs in the yeast benchmarking material can be used as metabolomics quality control. Finally, the benchmark material opens new avenues for batch-to-batch corrections in large-scale non-targeted metabolomics studies.
Full article
►▼
Show Figures